Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in East Asians (original) (raw)
. Author manuscript; available in PMC: 2011 Aug 19.
Published in final edited form as: Nat Genet. 2011 May 15;43(6):531–538. doi: 10.1038/ng.834
Abstract
We conducted a meta-analysis of genome-wide association studies of systolic (SBP) and diastolic (DBP) blood pressure in 19,608 subjects of East Asian ancestry from the AGEN-BP consortium followed by de novo genotypingin 2 stages of replication involving 10,518 and 20,247 East Asian samples. We identified novel genome-wide significant (P < 5 × 10−8) associations between SBP or DBP and variants at four novel loci: _ST7L_-CAPZA1, FIGN-GRB14, ENPEP, and NPR3, as well as a novel variant near TBX3. Except for NPR3, all novel findings were significantly replicated for SBP or DBP in independent samples. Sevenloci previously reported in populations of European descent were confirmed. On 12q24.13, we observed an ethnic specific association(implicating rs671 at the ALDH2 locus as the causal variant) that affected SBP, DBP and multiple traits related to coronary artery disease. These findings provide novel insights into blood pressure regulation and potential targets for intervention.
Hypertension is a leading risk factor of cardiovascular disease and premature deaths globally.1–4 It is especially common in Asian populations, contributing to a high incidence and mortality from stroke.5,6 Genetic and environmental factors and their interaction determine an individual’s risk for hypertension. Considerable efforts to elucidate the genetic determinants of hypertension, or elevated blood pressure (BP) levels,7,8 yielded little success until 2009 when two large-scale meta-analyses of genome-wide association studies (GWAS) from the Global BP Genetics (Global BPgen) and Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) consortia identified a total of 13 independent loci significantly associated with BP.9,10 The results from the 2 consortia represented an important advance in hypertension genomic research.11 However, these studies were conducted almost exclusively in populations of European descent. Studies in non-European populations will allow us to assess the relevance of these findings to other ethnic groups and potentially discover novel variants. The latter is important because some variants may be more common in specific ethnic groups, thereby providing greater power, or the effects of genetic variants on BP may be enlarged in specific ethnic groups.
The Asian Genetic Epidemiology Network BP (AGEN-BP) work group was established to facilitate the identification of genetic variants influencing BP among populations of East Asian ancestry. AGEN-BP includes 19,608 East Asian participants from 8 population-and family-based GWAS with standardized BP measurements. Here, we report the findings from our three-stage study which included stage 1, meta-analysis of BP GWAS from AGEN-BP (N_=19,608); stage 2, d_e novo genotyping of top loci in additional individuals (_N_=10,518); and stage 3, replication of findings in independent East Asian samples (_N_=20,247) (Supplementary Fig. 1).
RESULTS
Meta-analysis and follow-up of novel association signals
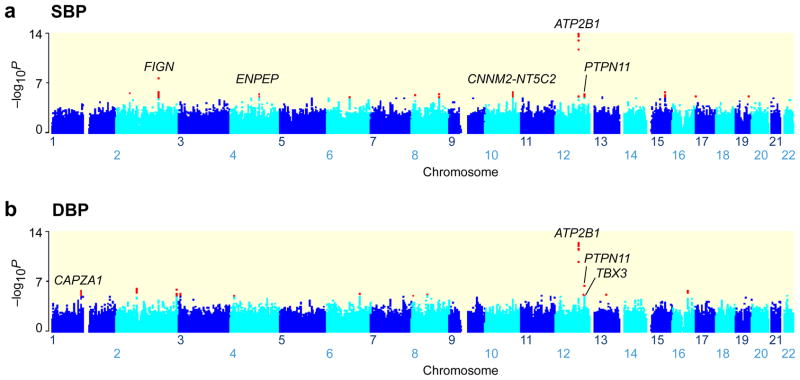
The stage-1 meta-analysis included 19,608 individuals from 8 GWAS in East Asian populations (Table 1, and Supplementary Table 1). The −log10 P_-values by chromosome location for SBP are showed in Fig. 1a and DBP in Fig. 1b. In addition, quantile-quantile plots for SBP and DBP are presented in Supplementary Fig. 2. The overall inflation post-meta-analysis was modest (λGC =1.03, SBP; λGC =1.02, DBP). The meta-analysis revealed two independent association signals reaching genome-wide significance (defined as P < 5 × 10−8). These included 1 locus known to harbor variants associated with BP—_ATP2B1_—and 1 novel locus—_FIGN-GRB14. Additional 11 variants that have not been previously implicated in the pathogenesis of hypertension were associated with SBP and/or DBP at a significance level of P < 1 × 10−5 (Supplementary Table 2). To increase the statistical power and strengthen support for the stage-1 findings, we undertook stage-2 follow-up de novo genotyping for 13 SNPs in an additional 10,518 Japanese individuals. These included 12 SNPs that showed associations with P < 1 × 10−5 and one additional SNP with P < 1 × 10−3, which was close to a biological candidate gene, NPR3.12 In the joint analysis of stage 1+2, one additional SNP (rs11066280 at the RPL6-PTPN11 locus) reached genome-wide significance. This SNP, and additional 5 SNPs attaining borderline significance (defined as P < 5 × 10−6) in our joint analysis of stage 1+2 were carried forward for stage-3 replication study involving 20,247 individuals. All 6 SNPs showed some evidence of replication in the stage-3 study, although the SBP association for rs1173766 (P = 0.016) did not reach a significance level after adjustment for multiple testing, i.e., P = 0.05/6 ≈ 0.008. All reached genome-wide significance in the joint analysis of stage 1+2+3 (Table 2 and Fig. 2). Moreover, 2 additional variants (rs880315 at the CASZ1 locus and rs155524 at the ITGA9 locus) had previously been genotyped in our stage 2 and stage 3 samples.13 When combined with data from stage 1, we found that rs880315 at CASZ1 showed an association with DBP that reached genome-wide significance as previously reported.13
Table 1.
Study design and sample characteristics.
| Study | N | Ancestry | BP Measurement (Device, Number of Measures) | Genotyping Platform | Women, % | Age (SD), y | SBP (SD), mm Hg | DBP (SD), mm Hg | BMI (SD), kg/m2 | HTN, %a | Antihyper tensive Therapy, % |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Stage 1: AGEN-BP GWAS meta-analysis (N=19,608) | |||||||||||
| CAGE | 1,547 | Japanese | Standard mercury sphygmomanometer, 2–3/Digital, 2–3 | Illumina 550/610K | 42.8 | 66.1 (8.0) | 134.1 (20.3) | 76.8 (11.9) | 23.5 (3.3) | 56.1 | 37.9 |
| GenSalt | 1,881 | Han Chinese | Random zero sphygmomanometer, 9 | Affymetrix 6.0 | 47.2 | 38.7 (9.5) | 116.9 (14.2) | 73.7 (10.3) | 23.3 (3.2) | 9.8 | 0.37 |
| KARE | 8,842 | Korean | Standard mercury sphygmomanometer, 3 | Affymetrix 5.0 | 52.7 | 52.2 (8.9) | 118.7 (19.4) | 75.6 (12.0) | 24.6 (3.1) | 22.3 | 10.9 |
| Shanghai-Ruijin | 455 | Han Chinese | Standard mercury sphygmomanometer, 2–3 | Illumina 610K | 49.5 | 54.2 (7.0) | 109.1 (8,6) | 72.7 (8.6) | 21.9 (1.9) | 0 | 0 |
| SiMES | 2,519 | Malay | Digital, 2–3 | Illumina 610K | 50.5 | 59.0 (11.0) | 147.9 (23.9) | 80.1 (11.3) | 26.4 (5.1) | 66.6 | 22.9 |
| SP2 | 2,431 | Han Chinese | Digital, 2–3 | Illumina 550K/610K/1M | 46.5 | 48.1 (11.2) | 129.4 (19.8) | 76.9 (10.8) | 22.9 (3.7) | 34.6 | 14.3 |
| Suita (1) | 933 | Japanese | Random zero sphygmomanometer, 3 | Illumina 550K | 56.3 | 59.0 (7.0) | 119.7 (17.5) | 75.0 (10.6) | 22.7 (2.9) | 14.8 | 0 |
| Taiwan | 1,000 | Han Chinese | Digital, 2–3 | Illumina 550K | 49.8 | 51.2 (17.8) | 121.9 (18.5) | 76.2 (10.8) | 23.8 (3.5) | 10.9 | 6.8 |
| Stage 2: De novo genotyping follow-up study (N=10,518) | |||||||||||
| CAGE-Amagasaki | 5,331 | Japanese | Digital, 2–3 | TaqMan | 39.8 | 47.8 (12.3) | 124.3 (17.3) | 75.9 (11.0) | 23.0 (3.2) | 21.5 | 9.0 |
| Ehime | 2,895 | Japanese | Digital, 3 | TaqMan | 56.6 | 61.1 (14.0) | 137.7 (22.2) | 81.0 (11.8) | 23.4 (3.2) | 53.4 | 25.7 |
| Suita (2) | 2,292 | Japanese | Random zero sphygmomanometer, 3 | TaqMan | 53.8 | 67.2 (11.0) | 127.4 (19.0) | 76.5 (10.3) | 22.9 (3.2) | 48.1 | 36.6 |
| Stage 3: Replication study (N=20,247) | |||||||||||
| CAGE-Fukuoka | 12,569 | Japanese | Digital, 2 | TaqMan | 54.9 | 62.6 (6.8) | 138.9 (21.2) | 83.9 (11.7) | 23.1 (3.0) | 57.9 | 23.9 |
| CAGE-KING | 3,975 | Japanese | Digital, 2 | TaqMan | 56.9 | 63.6 (6.6) | 132.3 (19.8) | 76.9 (11.2) | 22.9 (3.0) | 48.4 | 24.3 |
| HEXA-shared control | 3,703 | Korean | Standard mercury sphygmomanometer, 3 | Affymetrix 6.0 | 55.4 | 53.2 (8.3) | 121.7 (14.4) | 77.1 (9.9) | 24.0 (2.9) | 18 | 0 |
Fig. 1.
Genome-wide association results for the AGEN-BP meta-analysis for blood pressure. Manhattan plots show the significance of association between all SNPs and SBP (a) and DBP (b) in the stage 1 meta-analysis, highlighting signals with suggestive levels of significance (P < 10−5) in red. Seven named loci showed genome-wide significance (P < 5 × 10−8) in the joint analysis (stage 1 + 2 + 3; 2 reported loci—CNNM2-NT5C2 and _ATP2B1_—were followed up in part of stage 2 and stage 3). The genes used to name signals have been chosen on the basis of proximity to the lead SNP and should not be presumed to indicate causality.
Table 2.
Top genome-wide association results for SBP and DBP.
| Chr | SNP ID (pos Build 36.3) | Coded/Other allele | Nearby Gene(s) | Stage | N | Coded allele freq. | SBP | DBP | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Beta (SE), mm Hg | P | Beta (SE), mm Hg | P | |||||||
| Loci newly identified in or unique to Asians | ||||||||||
| 1 | rs17030613 (112,971,190) | C/A | ST7L CAPZA1 | 1 | 19,251 | 0.47 | 0.52 (0.17) | 0.002 | 0.50 (0.11) | 3.2E-06 |
| 2 | 10,465 | 0.50 | 0.43 (0.24) | 0.073 | 0.25 (0.14) | 0.078 | ||||
| 1+2 | 29,716 | 0.48 | 0.49 (0.14) | 4.0E-04 | 0.41 (0.09) | 1.8E-06 | ||||
| 3 | 20,236 | 0.50 | 0.49 (0.18) | 0.007 | 0.34 (0.11) | 0.002 | ||||
| Joint Analysis | 49,952 | 0.49 | 0.49 (0.11) | 8.4E-06 | 0.38 (0.07) | 1.2E-08 | ||||
| 2 | rs16849225 (164,615,066) | C/T | FIGN GRB14 | 1 | 18,867 | 0.60 | 0.97 (0.17) | 2.2E-08 | 0.40 (0.11) | 3.4E-04 |
| 2 | 10,465 | 0.62 | 0.56 (0.25) | 0.022 | 0.26 (0.15) | 0.068 | ||||
| 1+2 | 29,332 | 0.61 | 0.84 (0.14) | 3.8E-09 | 0.35 (0.09) | 7.7E-05 | ||||
| 3 | 20,179 | 0.62 | 0.60 (0.19) | 0.001 | 0.20 (0.11) | 0.076 | ||||
| Joint Analysis | 49,511 | 0.61 | 0.75 (0.11) | 3.5E-11 | 0.29 (0.07) | 2.7E-05 | ||||
| 4 | rs6825911 (111,601,087) | C/T | ENPEP | 1 | 19,033 | 0.48 | 0.79 (0.17) | 3.3E-06 | 0.40 (0.11) | 1.7E-04 |
| 2 | 10,463 | 0.54 | 0.66 (0.24) | 0.007 | 0.40 (0.14) | 0.005 | ||||
| 1+2 | 29,496 | 0.50 | 0.75 (0.14) | 7.8E-08 | 0.40 (0.09) | 2.5E-06 | ||||
| 3 | 20,019 | 0.52 | 0.33 (0.18) | 0.070 | 0.36 (0.11) | 9.1E-04 | ||||
| Joint Analysis | 49,515 | 0.51 | 0.60 (0.11) | 7.3E-08 | 0.39 (0.07) | 9.0E-09 | ||||
| 5 | rs1173766 (32,840,285) | C/T | NPR3 | 1 | 19,414 | 0.62 | 0.64 (0.17) | 2.2E-04 | 0.38 (0.11) | 6.1E-04 |
| 2 | 10,461 | 0.58 | 0.94 (0.24) | 1.3E-04 | 0.54 (0.14) | 1.8E-04 | ||||
| 1+2 | 29,875 | 0.61 | 0.74 (0.14) | 1.7E-07 | 0.44 (0.09) | 5.9E-07 | ||||
| 3 | 20,095 | 0.58 | 0.45 (0.19) | 0.016 | 0.25 (0.11) | 0.026 | ||||
| Joint Analysis | 49,970 | 0.60 | 0.63 (0.11) | 1.9E-08 | 0.36 (0.07) | 1.2E-07 | ||||
| 12 | rs11066280 (111,302,166) | T/A | RPL6 PTPN11 ALDH2 | 1 | 16,268 | 0.78 | 1.03 (0.22) | 3.8E-06 | 0.73 (0.14) | 3.1E-07 |
| 2 | 10,453 | 0.75 | 1.41 (0.28) | 3.2E-07 | 0.83 (0.16) | 3.5E-07 | ||||
| 1+2 | 26,721 | 0.77 | 1.18 (0.17) | 1.0E-11 | 0.77 (0.11) | 5.9E-13 | ||||
| 3 | 20,236 | 0.74 | 2.13 (0.21) | 2.6E-23 | 1.34 (0.13) | 6.5E-27 | ||||
| Joint Analysis | 46,957 | 0.75 | 1.56 (0.13) | 7.9E-31 | 1.01 (0.08) | 1.3E-35 | ||||
| 12 | rs35444 (114,036,820) | A/G | TBX3 | 1 | 19,286 | 0.75 | 0.69 (0.19) | 3.7E-04 | 0.54 (0.12) | 8.1E-06 |
| 2 | 10,460 | 0.75 | 0.49 (0.28) | 0.077 | 0.48 (0.16) | 0.003 | ||||
| 1+2 | 29,746 | 0.75 | 0.63 (0.16) | 8.6E-05 | 0.52 (0.10) | 9.6E-08 | ||||
| 3 | 20,238 | 0.75 | 0.64 (0.21) | 0.003 | 0.46 (0.13) | 3.0E-04 | ||||
| Joint Analysis | 49,984 | 0.75 | 0.63 (0.13) | 7.5E-07 | 0.50 (0.08) | 1.3E-10 | ||||
| Loci previously identified in Europeans | ||||||||||
| 1 | rs880315 (10,719,453) | C/T | CASZ1 | 1 | 10,765 | 0.61 | 0.29 (0.24) | 0.226 | 0.26 (0.14) | 0.073 |
| Follow-up a | 21,846 | 0.67 | 1.03 (0.19) | 8.1E-08 | 0.72 (0.11) | 5.9E-11 | ||||
| Joint Analysis | 32,611 | 0.65 | 0.74 (0.15) | 7.3E-07 | 0.56 (0.09) | 3.1E-10 | ||||
| 4 | rs16998073 (81,541,520) | T/A | FGF5 | 1 | N/A | N/A | N/A | |||
| Follow-up a | 21,864 | 0.30 | 1.43 (0.20) | 3.9E-13 | 0.76 (0.11) | 2.0E-11 | ||||
| 10 | rs11191548 (104,836,168) | T/C | CNNM2 NT5C2 CYP17A1 | 1 | 19,457 | 0.74 | 0.91 (0.19) | 2.1E-06 | 0.49 (0.12) | 5.6E-05 |
| Follow-up a | 21,858 | 0.73 | 1.47 (0.20) | 4.9E-13 | 0.66 (0.12) | 1.6E-08 | ||||
| Joint Analysis | 41,315 | 0.74 | 1.18 (0.14) | 3.9E-17 | 0.58 (0.08) | 6.6E-12 | ||||
| 12 | rs17249754 (88,584,717) | G/A | ATP2B1 | 1 | 18,856 | 0.65 | 1.38 (0.18) | 7.6E-15 | 0.83 (0.11) | 3.2E-13 |
| Follow-up a | 21,863 | 0.63 | 0.94 (0.19) | 4.2E-07 | 0.35 (0.11) | 1.2E-03 | ||||
| Joint Analysis | 40,719 | 0.64 | 1.17 (0.13) | 7.7E-20 | 0.58 (0.08) | 1.9E-13 |
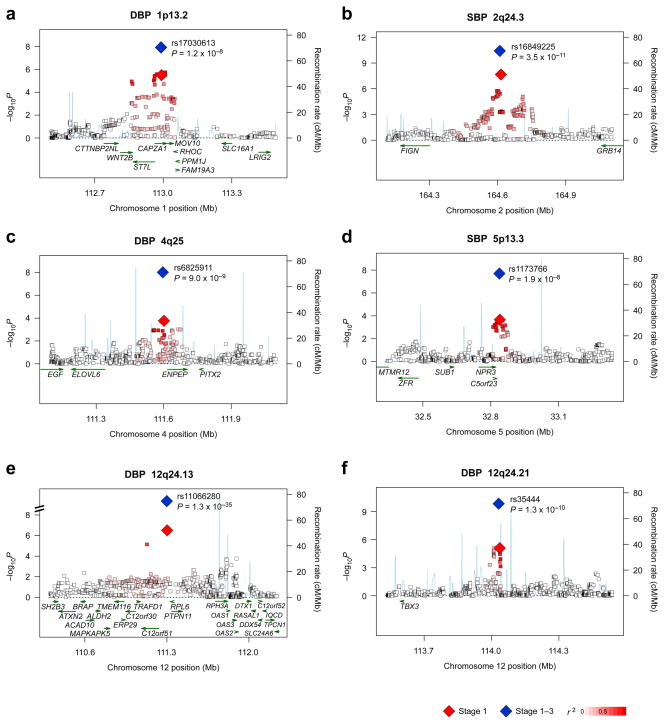
Fig. 2.
Regional association plots of six blood pressure loci. Genotyped and imputed SNPs passing quality control measures in stage 1 are plotted with their meta-analysis P values (as −log10 values) as a function of genomic position (build 36). In each panel, the lead SNP is represented by a diamond, with stage 1 meta-analysis results denoted by a red diamond and the joint analysis (combined P) results denoted by a blue diamond. The correlation of the lead SNP to other SNPs at the locus is shown on a scale from minimal (white) to maximal (red). Superimposed on the plot are gene locations (green) and recombination rates (blue). The regional plots were drawn using the SNAP software.35 On 12q24.13, rs671 was not included in the construction of regional association plots (Fig. 2e) because the genotype data for this SNP were unavailable from two GWAS in stage 1, KARE and Suita (1), which resulted in an effective sample size of 9,828.
Finally, on 12q24.21 near TBX3, we detected an association between BP and rs35444, which is approximately 200-kb apart from the reported lead SNP (rs2384550) identified in populations of European descent and its proxies. Since there is no linkage disequilibrium (LD) between the two clusters of SNPs on 12q24.21 in both ethnic groups (r2 = 0.000, _D_′ = 0.018 in HapMap JPT/CHB; r2 = 0.003, _D_′ = 0.063 in HapMap CEU between rs35444 and rs2384550), we consider the BP association at rs35444 independent and novel, presumably indicating the presence of allelic heterogeneity between the ethnic groups.
East Asian-specific associations on 12q24.13
One of the most prominent BP associations was detected on 12q24.13 [rs11066280, effect allele frequency = 0.75, per-allele effect = 1.56 mm Hg for SBP (_P_ = 7.9 × 10−31) and 1.01 mm Hg for DBP (_P_ = 1.3 × 10−35) in the joint analysis of stage 1+2+3, Table 2]. This association is likely to derive from a known functional variant rs671 at ALDH2, which is common in East Asians and has been reported to associate with hypertension principally through modification of alcohol consumption.14–17 Among the proxy SNPs in the surrounding region, a strong LD (r2 = 0.87) was found between rs11066280 and rs671 (Supplementary Table 3). In addition, both SNPs showed an association with BP in the CAGE-Amagasaki, Fukuoka, and KING Studies (_N_=21,875; Supplementary Table 4). When both SNPs were simultaneously included in the regression model, statistical significance remained at rs671 (P = 0.018 for SBP and P = 8.9 × 10−4 for DBP) but not at rs11066280 (P > 0.05). However, conclusive evidence for the primary source of association on 12q24.13 remains to be provided. The two SNPs are close to SH2B3 (Fig. 3a), one of the BP loci identified in populations of European-descent.9,10
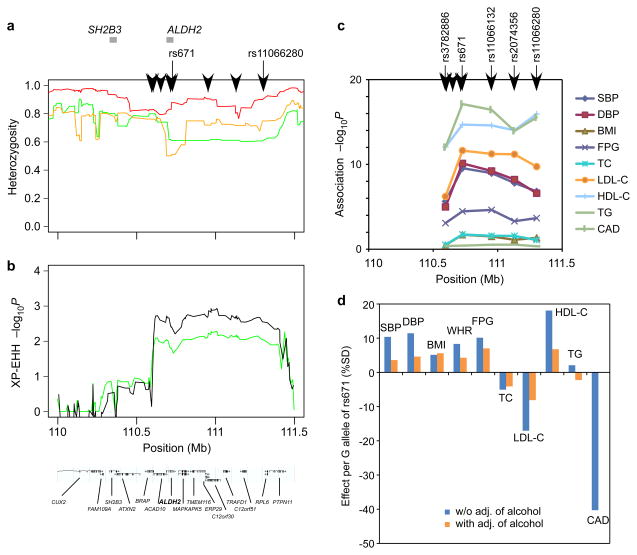
Fig. 3.
Evidence for positive selection and pleiotropic effects on 12q24.23. a. Lines show heterozygosity calculated in a sliding window of 20 SNPs in the East Asian (green), European (orange), and African (red) populations of the Human Genome Diversity Panel.36 Heterozygosity in East Asians dropped to 0.61 in a range from 110.7 to 111.4 Mb. Black arrowheads at the top of the plot represent the positions of SNPs forming an East Asian-specific haplotype with ALDH2 rs671. The positions of the SH2B3 and ALDH2 genes are highlighted by gray bars at the top of the plot. Signals of selection detected using a haplotype-based test, XP-EHH (b)19, in the East Asian population. Since there are multiple SNP clusters showing high LD within a cluster but relatively modest LD between clusters and no ancestral haplotype in East Asians (Supplementary Fig. 4), XP-EHH is assumed to provide more appropriate signals of selection than iHS18 on 12q24.23. Vertical axes represent empirical P values, where a suggestive level for positive selection is set at P < 0.01; i.e., −log10(_P_) >2. The results are shown for all East Asians combined (green line) and for Japanese (black line). c. The −log10(P) for associations with cardio-metabolic traits at 5 of the 8 SNPs forming a common East Asian-specific haplotype. The position of each SNP is denoted at the top of the plot. d. Per-allele effects of rs671 (G vs. A) on cardio-metabolic traits with and without adjustment for alcohol intake.
The SNP associated with BP in populations of European descent (rs3184504 at the SH2B3 locus) is not polymorphic in East Asians and the SNP associated with BP in our study (rs671 at the ALDH2 locus) is not polymorphic in Europeans. Also, we found modest signatures of recent selection, which was supported by several types of population genetic evidence, e.g., reduction of haplotype diversity in East Asians, and suggestive evidence of a selective sweep for East Asians at 110.7–111.4 Mb (empirical p-values < 0.01) by two haplotype-based tests—iHS scores18 and XP-EHH19 (Fig. 3a and 3b and Supplementary Fig. 3). These could be further supported by data from the HapMap Project (release 22) [http://www.hapmap.org] and 1000 Genomes Project (pilot 1) [http://www.1000genomes.org/]; i.e., 8 common SNPs (minor allele frequency [MAF] = 0.23–0.24 in the Japanese) appeared to identify a common haplotype (H5), which arose on a haplotype (H4) more common in East Asians but absent in Europeans and rare in Africans (Supplementary Fig. 4), spanning almost the same length of interval (0.7 Mb) as that of a suggestive selective sweep. Moreover, in the haplotype analysis involving 5 (of 8) SNPs genotyped on 12q24.13, rs671 and rs11066132 (r2 = 0.99 between the 2 SNPs) clearly differentiated a group of BP-increasing haplotypes—H1 to H3—from that of BP-decreasing haplotypes H4 to H6 (Supplementary Fig. 5).
These SNPs showed substantial pleiotropic effects on risk factors of cardiovascular disease17 as well as on susceptibility to coronary artery disease (CAD) in the Japanese (a paper submitted, F.T., et al.) with the strongest association being detected at rs671 for several traits besides SBP/DBP in the Japanese population studied (Fig. 3c and Supplementary Table 4). Two additional observations are of note. Firstly, the alleles associated with increased BP were associated with relatively small elevations in fasting glucose and body mass index (BMI), which would be expected to result in elevated risk of CAD [OR = 1.69 for A-allele vs. G-allele (95% CI 1.50–1.91), _P_ = 7.7 × 10−18; Supplementary Table 4]. However, the effects of rs671 on LDL cholesterol and HDL cholesterol were larger than those on BP, glucose and BMI, and in a direction that would be expected to decrease CAD risk. Indeed, overall, rs671 was associated with reduced risk of CAD. Here, a previous European GWAS claimed significant evidence supporting CAD association of a haplotype involving rs3184504 (at SH2B3) identical to that for BP association and signatures of natural selection in an overlapping region on 12q24.13.20 By inspecting the phylogeny of haplotypes in the relevant region, we confirmed that these haplotypes arose independently and each of the haplotypes was responsible for independent association signals in the 2 populations (Supplementary Fig. 4). Secondly, given the biological role of ALDH2 in the propensity for alcohol consumption, we further examined the following issue: to what extent alcohol intake could mediate the effects of rs671 on various cardiovascular risk factors. We showed that most of the associations between rs671 and each of the cardiovascular risk factors were substantially attenuated after adjustment for alcohol intake (Fig. 3d and Supplementary Table 4).
Associations at BP loci identified in populations of European descent
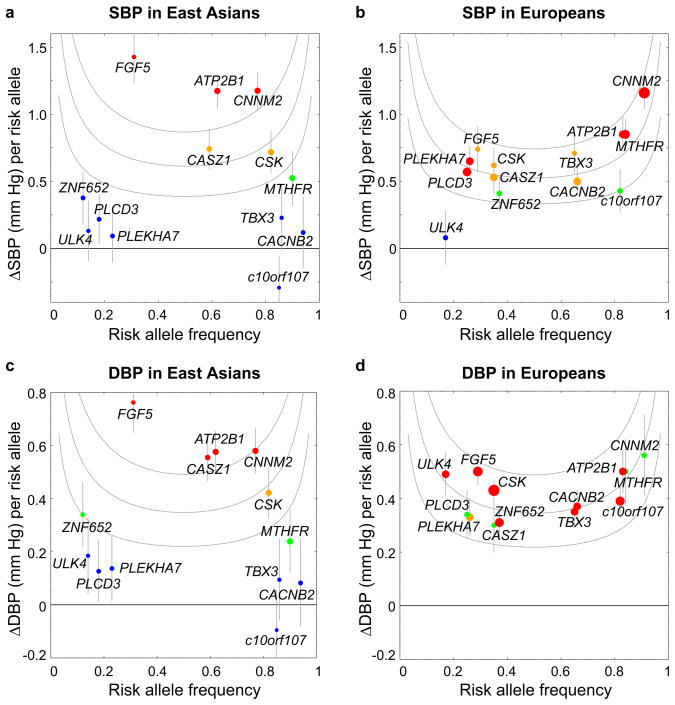
We performed replication study of the genome-wide significant and suggestive associations previously identified in populations of European descent9,10 in the AGEN-BP samples (Fig. 4 and Supplementary Table 5). We had genotyped variants at 7 loci—CASZ1, MTHFR, ITGA9, FGF5, CNNM2-NT5C2, ATP2B1 and CSK-ULK3_—in the CAGE-Amagasaki, CAGE-Fukuoka and CAGE-KING samples and 2 loci—_CACNB2 and _PLEKHA7_—in the CAGE-Amagasaki samples as part of the previous replication study.13 In the present study, we additionally genotyped in the CAGE Amagasaki samples 3 (in c10orf107, PLCD3, and ZNF652) of 7 variants that showed directionally consistent and nominally significant BP associations in the stage 1 study; the remaining 4 (of 7) variants were overlapped with those previously genotyped in the CAGE study samples (Supplementary Table 2). When these data were combined with our stage 1 data, 6 of the 7 previously replicated loci,13 and 1 additional locus (ZNF652) still showed directionally consistent associations with BP with p-values < 0.05.
Fig. 4.
Plots of effect size (β) versus risk-allele frequency of 13 loci previously identified in European GWAS meta-analysis. The list of 13 loci includes 1 locus (CASZ1) that did not reach genome-wide significance in Europeans but is found to show a significant association with BP in the present AGEN-BP meta-analysis. Plots for SBP [East Asians(a); Europeans(b)] and DBP [East Asians(c); Europeans(d)] are depicted separately. Each point refers to a single BP association signal, with colors denoting the strength of the BP association (red, P < 5 × 10−8; orange, 5 × 10−8 ≤ P < 10−5; green, 10−5 ≤ P < 0.05; blue, P ≥ 0.05) and with sizes being proportional to the tested sample size. Whiskers are ± standard errors. The gene names associated with each signal have been chosen on the basis of proximity to the lead SNP and should not be presumed to indicate causality. The gray curves represent the coefficient of determination (R2), i.e., BP variance explained by an SNP (%); those from the top to the bottom correspond to R2 = 0.1, 0.05, and 0.02. For estimating R2, the standard deviations of BP were assumed to be 19.4 mm Hg (SBP) and 11.0 mm Hg (DBP) in East Asians, and 16.6 mm Hg (SBP) and 10.9 mm Hg (DBP) in Europeans.9 The risk alleles were designated as previously reported in European GWAS meta-analysis. See the details in Supplementary Table 5.
In the 6 variants that we did not detect BP associations in the East Asian populations (Supplementary Fig. 6), we compared the effects in our study to those in the follow-up panels in Europeans (to exclude the possible winner’s curse effect21). Cross-population heterogeneity in effect size was detected for c10orf107 and PLEKHA7 (Supplementary Table 5). One possibility for our failure to detect an association between these 6 variants and BP in our population is the lack of power. To claim replication of BP associations previously identified in populations of European descent with p-values < 0.05 in East Asians, sample size was not sufficient for 3 (of the 6) variants—CACNB2, c10orf107, and TBX3-TBX5 (Supplementary Table 6). Another possibility could relate to differences in LD patterns between the ethnic groups. We examined LD differences between Europeans (HapMap CEU) and East Asians (HapMap JPT+CHB) at these loci by using the varLD program22 (Supplementary Fig. 7). For the 7 reported variants that showed an association with BP in East Asians, there was limited evidence of LD differences between European and East Asian populations at 5 (of 7) loci that harbor these variants. On the other hand, for the 6 reported variants for which we did not detect an association with BP in East Asians, we found significant differences in LD between the ethnic groups at 4 (of 6) loci harbouring these variants—CACNB2, c10orf107, PLEKHA7, and TBX3-TBX5.
Cumulative impact of risk alleles and genetic association with dichotomous hypertension
In total, 10 variants showed associations that reached genome-wide significance after all stages of our study. These 10 variants had a cumulative impact on SBP and DBP (Supplementary Note and Supplementary Fig. 8) and were also associated with odds of hypertension (as a dichotomous trait) in directions consistent with the continuous trait effect (Supplementary Note, Supplementary Fig. 9 and Supplementary Tables 7 and 8).
eQTL analysis
In search of putative functional variation at the newly identified loci, we found that among the 6 genes on 1p13.2, expression of ST7L was associated with expression-associated SNP (eSNP) rs17030613 (P = 9.4 × 10−15 and 1.4 × 10−8, reported in 2 independent studies)23,24 in lymphoblastoid cell lines (LCLs) derived from samples of European and East Asian ancestry (Supplementary Fig. 10). In the current study, the rs17030613 A-allele associated with lower ST7L transcript levels is also associated with lower BP. No other eSNPs or non-synonymous SNPs, which were in LD with BP-associated variants, were identified for the 5 novel association signals in the public databases currently available.
DISCUSSION
By using a three-stage GWAS meta-analysis of up to 50,373 individuals of East Asian ancestry, we identified a total of 5 novel BP association signals with genome-wide significance. These include: 1p13.2 in _ST7L_-CAPZA1 (P = 1.2 × 10−8 for DBP), 2q24.3 in FIGN-GRB14 (P = 3.5 × 10−11 for SBP), 4q25 in ENPEP (P = 9.0 × 10−9 for DBP), and 5p13.3 near NPR3 (P = 1.9 × 10−8 for SBP). On 12q24.21, near TBX3 (a known BP-associated locus in populations of European descent), we also identified a novel BP-associated variant, illustrating the possible presence of allelic heterogeneity at this locus in relation to BP regulation.
The glutamyl aminopeptidase encoded by ENPEP plays a pivotal role in BP regulation by facilitating conversion of angiotensin II, the main effector protein of the renin-angiotensin-aldosterone system, to angiotensin III and the knock out mouse develops hypertension.25 The natriuretic peptide receptor C/guanylase cyclase C encoded by NPR3 is one of three receptor subtypes that mediate specific binding of the natriuretic peptides, which are important in the maintenance of BP and extracellular fluid volume, to the cell surface.12 On 2q24.3, the lead SNP (rs16849225) is located in between the FIGN and GRB14 genes. FIGN encodes fidgetin, a member of ATPases associated with diverse cellular activities.26 GRB14 encodes the growth factor receptor-bound protein 14 that interacts with insulin receptors and insulin-like growth factor receptors, suggesting the role of GRB14 in signaling pathways that regulate growth and metabolism.27 A variant close to the GRB14 locus (rs10195252) has been identified in association with a waist–to-hip ratio (WHR) in populations of European descent.28 However, the variant we identified for the BP association in this study (rs16849225) is distant (>600 kb) from that identified for the WHR association with the two SNPs being in linkage equilibrium. While the novel variant on 1p13.2 is in close proximity to 6 annotated genes, it is an eSNP for ST7L, with the consistent results supporting an association between rs17030613 and ST7L transcript levels in the LCLs being demonstrated in two independent studies23,24 in three ethnic groups (HapMap CEU, YRI, and JPT+CHB, P > 0.05 for testing inter-population per-allele effect difference).23 ST7L (suppression of tumorigenicity 7 like) has been identified by its similarity to the ST7 tumor suppressor gene found in the 7q31 region known to be deleted and rearranged in a variety of cancers,29 although the function of this gene remains to be determined.
Our study provides important information on the genetic architecture of BP and CAD in relation with known loci. In this study, we provided evidence suggesting that the association between variants on 12q24.13 and BP is likely to relate to a non-synonymous SNP (rs671, E504K) in the ALDH2 gene. Furthermore, the associations seem to be largely mediated by alcohol intake, a finding that is supported by pre-existing studies showing that this variant determines an individual’s tolerance of alcohol intake by altering ALDH2 enzymatic activity.30,31 Variantsat this locus also appear to have undergone natural selection, while phylogenetic analysis suggests that rs671 is not responsible for the BP association identified in the overlapping chromosomal region (near SH2B3) in populations of European descent.9,10 Thus, the associations described in our study appear to be East Asian specific. Moreover, our study highlights the importance of extending studies examining associations with surrogate markers of cardiovascular risk factors (such as BP) to include clinically relevant events (such as CAD) when trying to identify therapeutic targets, especially if the variant of interest could have pleiotropic effects. In this instance, the allele (of rs671) associated with elevated BP was associated with reduced risk of CAD, a finding that is counter-intuitive based on what is known about the relationship between BP and CAD. Further analysis suggests that the deleterious effect of the variant on BP was balanced by protective effects on HDL cholesterol and LDL cholesterol, resulting in a net reduction in CAD risk. This is reminiscent of the effects of the CETP inhibitor, torcetrapib, which increased HDL cholesterol and decreased LDL cholesterol. Despite these cardioprotective effects, its use was associated with increased risk of CAD and mortality, which may relate to an off-target effect causing BP elevation.32 Secondly, on examining BP associations for 13 variants identified in European GWAS meta-analyses (Supplementary Table 5), we found 7 of the 13 loci (54%) to show nominally significant associations of the reported lead SNPs in East Asians. This is consistent with previous studies showing that findings from populations of European ancestry may be relevant to other ethnic groups.13,33,34 We also found another locus (_TBX3_-TBX5) to show some evidence of allelic heterogeneity in relation to BP. These data suggest that although some inter-population differences may exist in the pathways involved in the pathogenesis of BP elevation (or hypertension) between Europeans and East Asians, the majority of pathways are common.
A number of factors are considered to influence our ability to detect associations at the remaining 6 loci in the East Asian populations. Firstly, the sample size for genome-wide exploration (stage 1) was smaller in AGEN-BP (N = 19,608) than two previous consortia of European-descent populations, the Global BPgen (N = 34,433) and CHARGE (N = 29,136),9,10 and this provided limited power to detect several of these loci, even using a p-value threshold of <0.05 (Supplementary Table 6). This limited power also means that additional novel loci could be identified by enlarging the sample size in the discovery stage, providing motivation for larger meta-analyses in the future. Secondly, we found some evidence of LD differences between European and East Asian populations at 2 of 7 loci (29%) for which we did detect an association in our study, versus at 4 of 6 loci (67%) for which we did not detect an association (Supplementary Fig. 7). This could also contribute to our failure to detect associations for some of the known variants in the present study.
In conclusion, even though variants identified in European populations are often relevant to other ethnic groups, by conducting a large-scale GWAS meta-analysis in East Asians, we have identified previously unreported association signals for BP—at _ST7L_-CAPZA1, FIGN-GRB14, ENPEP, and _NPR3_—that reach genome-wide significance. Near TBX3, we have identified some evidence for allelic heterogeneity in East Asians compared to Europeans in relation to BP associations. Further, our data have provided evidence of East Asian-specific BP association at ALDH2 which has pleiotropic effects on other metabolic traits and CAD, highlighting the importance of fine mapping efforts to pinpoint causal variants and causal genes, thereby providing new insights into the physiology of complex diseases.
METHODS
Stage 1 samples
The Asian Genetic Epidemiology Network (AGEN) is a consortium of genetic epidemiology studies of cardiovascular disease related phenotypes, such as BP (or hypertension), diabetes, and obesity, conducted among Asian populations. AGEN-BP consists of 19,608 East Asian participants who underwent standardized collection of BP measurements in eight population-and family-based GWAS, including: the Cardio-metabolic Genome Epidemiology (CAGE) Network, Genetic Epidemiology Network of Salt-Sensitivity (GenSalt), Korean Association Resource (KARE) Project, Shanghai Hypertension Study, Singapore Malay Eye Survey (SiMES), Singapore Prospective Study (SP2) Program, Suita Study, and Taiwan Super Control Study. Each study established a consensus on phenotype harmonization and analytical plan for within-study GWAS and meta-analysis of results across studies. Each study received an approval from the institutional review board and all participants in each study provided written informed consent for participation in the study. A detailed description of the study design and phenotype measurement for each study (or cohort) is provided in the Supplementary Note and Supplementary Table 1 online. Overall design of the three-staged GWAS meta-analysis is depicted in Supplementary Fig. 1. Stage 1 was a meta-analysis of directly genotyped and imputed SNPs from individuals of East Asian descent, drawn from population-based or control samples in case-control studies in AGEN-BP.
Genome-wide genotyping and quality control
Genotyping arrays and quality control filters applied to the individual studies are provided in Supplementary Table 1.
Genotype imputation
Imputation of genotypes to the HapMap Phase 2 (JPT+CHB except for SiMES, which used HapMap JPT+CHB+CEU+YRI) set was carried out using MACH,37 IMPUTE,38 or BEAGLE39 with preimputation filters as specified in Supplementary Table 1. Imputation results are summarized as an ‘allele dosage’ defined as the expected number of copies of the coded allele at that SNP (a fractional value between 0 to 2) for each genotype. In total, up to 2.4 million genotyped or imputed autosomal SNPs were analyzed.
Phenotype modeling and SNP association analysis
For participants taking antihypertensive therapies, BP was imputed by adding 10 mm Hg and 5 mm Hg to SBP and DBP values, respectively.40 Within each study, continuous SBP and DBP were adjusted for age, age2, sex, BMI, and any study-specific covariates in linear regression models.
In secondary and confirmatory analyses of hypertension, hypertensive cases were defined as follows:(i) SBP ≥ 160 mm Hg and/or DBP ≥ 100 mm Hg for untreated subjects; (ii) patients receiving chronic antihypertensive treatments; and (iii) age of onset ≤ 65 years. Normotensive controls were defined as follows: (i) SBP < 130 mm Hg and DBP < 85 mm Hg without antihypertensive treatments and (ii) age ≥ 50 years. Within each study, a dichotomous trait of hypertension status was adjusted for age, age2, sex, and BMI in logistic regression models, except for the CAGE samples used for follow-up and replication (3,294 cases and 6,831 controls; Supplementary Table 7), in which age was not adjusted for due to its relation to case-control status; i.e., the mean age was younger in cases than in controls.
Stage 1 meta-analysis
All study-specific effect estimates and coded alleles were oriented to the forward strand of the NCBI36 reference sequence of the human genome. If an SNP from a study did not meet quality standards, it was reported as missing from that study for the purpose of meta-analysis. Results for this SNP were pooled among the other contributing studies. SNPs were excluded if they had study-specific imputation quality _R_2 < 0.5. Genomic control41 was carried out on study-specific test statistics: λGC estimates are given in Supplementary Table 1. We used inverse variance-weighted meta-analysis to combine association results for stage 1 with METAL software (http://www.sph.umich.edu/csg/abecasis/Metal). Evidence for heterogeneity of effect sizes was investigated using Cochran’s _Q_ statistic.42 We retained 1.7 million SNPs with minor allele frequency (MAF) of >0.05 and an effective sample size of >10,000; in Fig. 1, we show the Manhattan plots drawn using the WGAViewer software43 and the quantile-quantile plots.
SNP prioritization for stage 2
We selected 13 SNPs for follow-up in stage 2 using two methods in parallel. Using the first method, the most strongly associated SNP was chosen from each of 12 distinct regions containing ≥1 SNPs with P < 1 × 10−5 for SBP and/or DBP based on the stage-1 data, in which samples from ≥7 (of 8) GWAS were available for the meta-analysis, in principle, to decrease the potential noise of false positive associations due to the small sample size. We excluded SNPs within 50 kb of the lead SNPs for BP associations previously identified in Europeans at _CNNM2-NT5C2_, _ATP2B1_, and _CSK-ULK3_.9,10 Although attaining a significance level of _P_ < 1 × 10−5 for SBP, we did not subject 2 SNPs (rs4671977 and rs12547784) to follow-up in stage 2, because their association with the DBP trait was not significant (_P_ > 0.05) in stage 1. Using the second method, we generated a list of the next most significant SNPs (21 unique loci showed P ≤ 1 × 10−3 for both SBP and DBP and consistent association signals across the studies). We selected a SNP at 5p13.3 ad hoc, due to physiological candidacy of the nearby gene, NPR3.12
Stage 2 samples and genotyping
We genotyped 13 SNPs in 10,518 individuals of Japanese descent from 3 studies—CAGE-Amagasaki Study, Suita (2) Study, and Ehime Study. Summary characteristics are shown in Table 1. Study information and genotyping methods are provided in the Supplementary Note.
Stage 3 samples, genotyping and joint analysis
For SNP selectionin the stage-3 study, we set a threshold of P < 5 × 10−6 for joint analysis of stage 1+2. Six of 13 SNPs exceeded this significance level and were followed up in 20,247 individuals of East Asian descent from 3 studies by de novo genotyping (for Fukuoka Study and Kita-Nagoya Genomic Epidemiology [KING]Study of the CAGE Network) and in silico replication (for HEXA-shared control).
We carried out meta-analysis of stage 1+2+3 results and considered associations genome-wide significant if they attained P < 5 × 10−8. Association results for multiple stages (1+2 or 1+2+3) were combined using inverse variance weighting with the rmeta package of the R software (http://www.r-project.org).
Replication of previously reported loci
Along with genome-wide exploration of novel loci, we examined the BP associations at 27 loci reported by the European GWAS meta-analyses: 9,10 13 genome-wide significant loci and 14 loci with suggestive association (Supplementary Table 5). A one-tailed P < 0.05 (two-tailed P < 0.1) was considered statistically significant for the 13 loci previously shown to have genome-wide significant (P < 5 × 10−8) associations in Europeans; for an association to be considered significant, it had to involve the same risk allele as that reported in Europeans, and was accordingly assessed with the one-tailed test. However, two-tailed P values are presented throughout the text for readability. For the 14 loci with suggestive association in the original European studies,9,10 a one-tailed P < 0.05/14 = 0.0036 was considered statistically significant after Bonferroni correction.
Haplotype phylogeny and positive selection on 12q24.13
Previous studies in population of European descent claimed significant evidence supporting BP associations9,10 and signatures of natural selection20 in a 1.6 Mb interval on 12q24.13, near which the present East Asian study also identified strong BP associations. To clarify phylogenic differences in susceptibility variants between the 2 populations, we inferred the haplotypes and constructed their phylogeny across 3 HapMap panels—JPT, CEU, and YRI—in the 12q24.13 region of interest. Further, using haplotype-based tests, iHS18 and XP-EHH,19 we tested the hypothesis that a long-range, evolutionarily derived haplotype, upon which BP-decreasing alleles could lie, arose from a positive selection in East Asians independently of Europeans.
Pleiotropic effects on cardio-metabolic traits
We tested SNP—trait associations of 5 SNPs located in the target interval on 12q24.13 for risk factors of cardiovascular disease and CAD. The associations with SBP/DBP, BMI, fasting plasma glucose, and lipids were analyzed in CAGE-Amagasaki Study (for 5 SNPs) and CAGE-Fukuoka Study (for rs11066280 and rs671 alone) samples with and without adjustment for alcohol intake; where the amount of alcohol consumed was denoted in terms of servings of sake (1 gou [180 ml] of Japanese rice wine is considered equal to 22 g of ethanol).17 The association with CAD was analyzed in the CAD case-control panel (1,347 CAD cases and 1,337 controls) derived from the CAGE Network (a paper submitted, F.T., et al.).
eQTL analysis
Using publicly available data [GTEx (Genotype-Tissue Expression) eQTL Browser (http://www.ncbi.nlm.nih.gov/gtex/test/GTEX2/gtex.cgi); http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/; http://www.sph.umich.edu/csg/liang/imputation/; SNPExpress (http://people.genome.duke.edu/~dg48/SNPExpress/)], we examined the _cis_-associations (defined as genes within 1 Mb) between each of the 5 novel BP SNPs and expression of nearby genes in a variety of cells/tissues; e.g., LCLs, monocytes, fibroblasts, liver and brain tissues. After initial screening, we narrowed the targets of eQTLs down to LCLs in two databases.23,24 Briefly, LCLs, derived from peripheral blood lymphocytes, were available for 209 HapMap samples (comprising 60 CEU, 60 YRI, and 89 JPT+CHB individuals)23 and 378 individuals of white European descent24. The expression of 47,294 (for 209 HapMap samples) and 54,675 (for 378 Europeans) transcripts was assessed for each individual, whose genotypes were also imputed using the corresponding HapMap dataset. SNPs were tested for cis associations, assuming an additive genetic model, adjusting for non-genetic effects in the gene expression value.
Supplementary Material
Supl_figures
Supl_tables
Supl_text
Acknowledgments
The authors acknowledge the essential role of the Asian Genetic Epidemiology Network (AGEN)in development and support of this manuscript. AGEN members include the Cardio-metabolic Genome Epidemiology (CAGE) Network, Genetic Epidemiology Network of Salt-Sensitivity (GenSalt), Korean Association Resource (KARE) Project, Shanghai Hypertension Study, Singapore Malay Eye Survey (SiMES), Singapore Prospective Study (SP2) Program, Suita Study, and Taiwan Super Control Study.
CAGE: The CAGE Network Studies were supported by grants for the Core Research for Evolutional Science and Technology (CREST) from the Japan Science Technology Agency; the Program for Promotion of Fundamental Studies in Health Sciences, National Institute of Biomedical Innovation Organization (NIBIO); KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and the Grant of National Center for Global Health and Medicine (NCGM).
GenSalt: The Genetic Epidemiology Network of Salt Sensitivity is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA. Dr. Kelly is supported partially by Award Number K12HD043451 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, USA.
KARE: KARE and HEXA-shared control studies were supported by grants from Korea Centers for Disease Control & Prevention, Republic of Korea (4845-301, 4851-302, 4851-307).
Shanghai: This work was supported by the Chinese National Key Program for Basic Research (Grant 973:2004CB518603) and Chinese National High Tech Program (Grant 863:2009AA022703).
SiMES: The Singapore Malay Eye Study (SiMES) was funded by the National Medical Research Council (NMRC 0796/2003 and NMRC/STaR/0003/2008) and Biomedical Research Council (BMRC, 09/1/35/19/616).
SP2: The Singapore Prospective Study Program (SP2) was funded through grants from the Biomedical Research Council of Singapore (BMRC 05/1/36/19/413 and 03/1/27/18/216) and the National Medical Research Council of Singapore (NMRC/1174/2008).
Y.Y.T. acknowledges support from the Singapore National Research Foundation (NRF-RF-2010-05).
E.S.T. also receives additional support from the National Medical Research Council through a clinician scientist award (NMRC/CSA/008/2009).
Suita/Ehime Study: The Ehime study was supported by Grants for Scientific Research (Priority Areas “Medical Genome Science (Millennium Genome Project)” and “Applied Genomics”) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; a Grants-in-Aid (H15-Longevity-005, H17-longevity-003, H16-kenko-001, H18-longevity (kokusai)) from the Ministry of Health, Labor and Welfare, Health and Labor Sciences Research Grants, Japan; a Science and Technology Incubation Program in Advanced Regions, Japan Science and Technology Agency; and the Japan Atherosclerosis Prevention Fund.
Taiwan Super Control Study: This study was supported by Academia Sinica Genomic Medicine Multicenter Study; National Research Program for Genomic Medicine, National Science Council, Taiwan (National Clinical Core, NSC97-3112-B-001-014; and National Genotyping Center, NSC97-3112-B-001-015).
Footnotes
Author contributions
Principal investigators: N.K., J.H.
Project coordination leaders: N.K., J.H., Y.T.
Manuscript writing group: N.K., F.T., T.N.K., J.H., Y.Y.T., Y.S.C., E.S.T.
Project data management: T.N.K.
Genotyping and QC: F.T., M.I., K.Y., Y.T., N.I., Y.K., X.S., W.T.T., Y.Y.T.
Phenotype collection, data management: CAGE: N.K., K.Y., T.K., T.N., M.Y., K.O., Y.Y., E.N., T.S., R.T., T.O.; Gensalt: T.N.K., D.G., J.H.; KARE: J.-P.J., S.S.K., Y.S.C.; Shanghai: Y.Z., X.G.Z., X.Z., D.L.Z.; SiMES/SP2: T.A., T.Y.W., E.S.T.; Suita: N.I., Y.K., Y.T., T.M.; Taiwan: C.-H.C. L.-C.C., Y.-T.C., J.-Y.W.
Genome-wide genotyping: CAGE: N.K., M.I.; Gensalt: J.E.H., Y.J.S.; KARE: J.-Y.L., B.G.H., Y.S.C.; Shanghai: W.H.; SiMES/SP2: M.S., J.J.L.; Suita: N.I.; Taiwan: Y.-T.C., J.-Y.W.
Data analysis and data interpretation: N.K., F.T., T.N.K., Y.T., Y.Y.T., E.S.T., M.J.G., Y.J.K., X.S., W.T.T., R.T.H.O., C.-H.C., L.-C.C., C.E.J., J.H.
References
- 1.Ezzati M, et al. Selected major risk factors and global and regional burden of disease. Lancet. 2002;360:1347–1360. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- 2.Lopez AD, et al. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 3.Lawes CMM, Vander Hoorn S, Rodgers A. International Society of Hypertension. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371:1513–1518. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 4.Kearney PM, et al. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 5.He J, et al. Premature deaths attributable to blood pressure in China: a prospective cohort study. Lancet. 2009;374:1765–1772. doi: 10.1016/S0140-6736(09)61199-5. [DOI] [PubMed] [Google Scholar]
- 6.Eastern Stroke and Coronary Heart Disease Collaborative Research Group. Blood pressure, cholesterol, and stroke in eastern Asia. Lancet. 1998;352:1801–1807. [PubMed] [Google Scholar]
- 7.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet. 2006;7:829–840. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 8.Kurtz TW. Genome-Wide Association Studies Will Unlock the Genetic Basis of Hypertension. Con Side of the Argument. Hypertension. 2010 Nov 8; doi: 10.1161/HYPERTENSIONAHA.110.156190. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominiczak AF, Munroe PB. Genome-Wide Association Studies Will Unlock the Genetic Basis of Hypertension. Pro Side of the Argument. Hypertension. 2010 Nov 8; doi: 10.1161/HYPERTENSIONAHA.110.156208. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Anand-Srivastava MB. Natriuretic peptide receptor-C signaling and regulation. Peptides. 2005;26:1044–1459. doi: 10.1016/j.peptides.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi F, et al. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation. 2010;121:2302–2309. doi: 10.1161/CIRCULATIONAHA.109.904664. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Davey Smith G, Harbord RM, Lewis SJ. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med. 2008;5:e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchihashi-Makaya M, et al. Gene-environmental interaction regarding alcohol-metabolizing enzymes in the Japanese general population. Hypertens Res. 2009;32:207–213. doi: 10.1038/hr.2009.3. [DOI] [PubMed] [Google Scholar]
- 16.Li H, et al. Refined geographic distribution of the oriental ALDH2*504Lys (nee 487Lys) variant. Ann Hum Genet. 2009;73:335–345. doi: 10.1111/j.1469-1809.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi F, et al. Confirmation of ALDH2 asa major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in Japanese. Circ J. doi: 10.1253/circj.cj-10-0774. In press. [DOI] [PubMed] [Google Scholar]
- 18.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zollner S, Pritchard JK. Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong RT, Teo YY. varLD: a program for quantifying variation in linkage disequilibrium patterns between populations. Bioinformatics. 2010;26:1269–1270. doi: 10.1093/bioinformatics/btq125. [DOI] [PubMed] [Google Scholar]
- 23.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 25.Mizutani S, et al. New insights into the importance of aminopeptidase A in hypertension. Heart Fail Rev. 2008;13:273–284. doi: 10.1007/s10741-007-9065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox GA, Mahaffey CL, Nystuen A, Letts VA, Frankel WN. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat Genet. 2000;26:198–202. doi: 10.1038/79923. [DOI] [PubMed] [Google Scholar]
- 27.Goenaga D, et al. Molecular determinants of Grb14-mediated inhibition of insulin signaling. Mol Endocrinol. 2009;23:1043–1051. doi: 10.1210/me.2008-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh M. Molecular cloning and characterization of ST7R (ST7-like, ST7L) on human chromosome 1p13, a novel gene homologous to tumor suppressor gene ST7 on human chromosome 7q31. Int J Oncol. 2002;20:1247–1253. [PubMed] [Google Scholar]
- 30.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6:e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada S, Misawa S, Agarwal DP, Goedde HW. Liver alcohol dehydrogenase and aldehyde dehydrogenase in the Japanese: isozyme variation and its possible role in alcohol intoxication. Am J Hum Genet. 1980;32:8–15. [PMC free article] [PubMed] [Google Scholar]
- 32.Barter PJ, et al. Effects of Torcetrapib in Patients at High Risk for Coronary Events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 33.Tabara Y, et al. Common variants in the ATP2B1 gene are associated with susceptibility to hypertension: the Japanese Millennium Genome Project. Hypertension. 2010;56:973–980. doi: 10.1161/HYPERTENSIONAHA.110.153429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong KW, et al. Recapitulation of two genomewide association studies on blood pressure and essential hypertension in the Korean population. J Hum Genet. 2010;55:336–341. doi: 10.1038/jhg.2010.31. [DOI] [PubMed] [Google Scholar]
- 35.Johnson AD, et al. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 39.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui JS, Hopper JL, Harrap SB. Antihypertensive treatments obscure familial contributions to blood pressure variation. Hypertension. 2003;41:207–210. doi: 10.1161/01.hyp.0000044938.94050.e3. [DOI] [PubMed] [Google Scholar]
- 41.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 42.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge D, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supl_figures
Supl_tables
Supl_text