RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries (original) (raw)
Abstract
Sequencing of small RNA cDNA libraries is an important tool for the discovery of new RNAs and the analysis of their mutational status as well as expression changes across samples. It requires multiple enzyme-catalyzed steps, including sequential oligonucleotide adapter ligations to the 3′ and 5′ ends of the small RNAs, reverse transcription (RT), and PCR. We assessed biases in representation of miRNAs relative to their input concentration, using a pool of 770 synthetic miRNAs and 45 calibrator oligoribonucleotides, and tested the influence of Rnl1 and two variants of Rnl2, Rnl2(1–249) and Rnl2(1–249)K227Q, for 3′-adapter ligation. The use of the Rnl2 variants for adapter ligations yielded substantially fewer side products compared with Rnl1; however, the benefits of using Rnl2 remained largely obscured by additional biases in the 5′-adapter ligation step; RT and PCR steps did not have a significant impact on read frequencies. Intramolecular secondary structures of miRNA and/or miRNA/3′-adapter products contributed to these biases, which were highly reproducible under defined experimental conditions. We used the synthetic miRNA cocktail to derive correction factors for approximation of the absolute levels of individual miRNAs in biological samples. Finally, we evaluated the influence of 5′-terminal 5-nt barcode extensions for a set of 20 barcoded 3′ adapters and observed similar biases in miRNA read distribution, thereby enabling cost-saving multiplex analysis for large-scale miRNA profiling.
Keywords: small RNAs, high-throughput sequencing, next-generation sequencing, sequence-specific bias, adapter ligation, oligonucleotide linker ligation
INTRODUCTION
MicroRNAs (miRNAs) are 20–23-nt RNAs that destabilize target mRNAs (Bhattacharyya and Filipowicz 2007; Bartel 2009). These molecules are typically processed by RNase III enzymes from longer precursor transcripts, first in the nucleus by Drosha and then in the cytoplasm by Dicer, resulting in mature miRNAs with characteristic 5′-phosphate (p) and 3′-hydroxyl (OH) termini. Distinct tissue-, cell-type-, and developmental-stage-specific miRNA expression patterns have been identified in plants and animals (Stefani and Slack 2008; Voinnet 2009), and dysregulation or mutation of miRNAs was found to associate with or contribute to human disease (Hebert and de Strooper 2007; Croce 2009; Latronico and Condorelli 2009; Inui et al. 2010). Several methodologies have been developed for capturing miRNA expression changes (Aravin and Tuschl 2005; Hunt et al. 2009); the most widely used ones include hybridization-based miRNA microarrays (Wang et al. 2007; Bissels et al. 2009), quantitative reverse transcription and PCR (Schmittgen et al. 2008), and small RNA cDNA library sequencing (Berninger et al. 2008; Hafner et al. 2008).
Sequencing of cDNA libraries allows for the discovery of new miRNAs or mutations within known miRNAs in addition to capturing miRNA expression changes; however, the correlation of read counts or read frequencies to original RNA abundance in the sample may be variable (Linsen et al. 2009). Biases in the read distributions are expected because several steps during the small RNA cDNA library preparation are enzymatic reactions that are sensitive to varying degrees to sequence and structure of their nucleic acid substrates. Understanding the absolute abundance of miRNA molecules in samples, and not only their relative changes between samples, allows for prioritization of follow-up studies by selecting miRNAs abundant enough to warrant biological effects (Krutzfeldt et al. 2005; Linsley et al. 2007; Landthaler et al. 2008; Hafner et al. 2010).
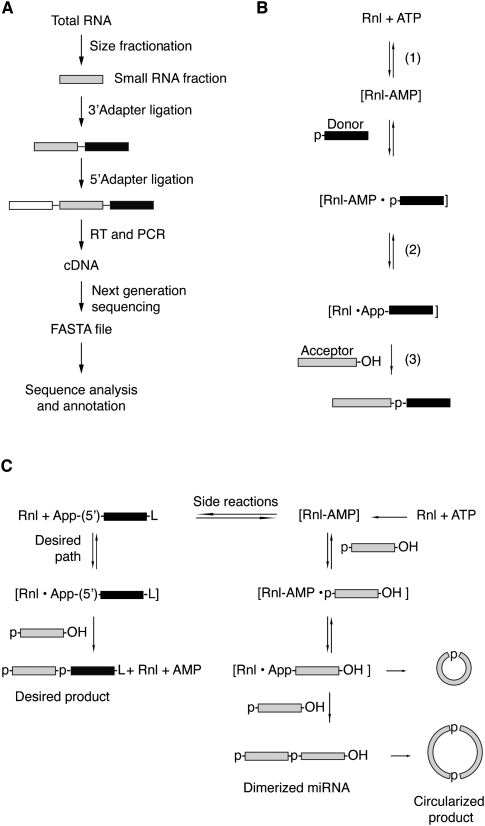
Small RNA cDNA library preparation involves sequential ligation of adapter oligonucleotides using RNA ligases that introduce primer-binding sites for subsequent reverse transcription (RT) and PCR amplification prior to deep sequencing (Fig. 1A). AGO/PIWI-protein-family-associated RNAs are typically characterized by 5′-p and 3′-OH groups, although some may be additionally 2′-_O_-methylated at their 3′ ends, such as piRNAs and Drosophila melanogaster Ago2-bound siRNAs (Farazi et al. 2008), or have 5′-triphosphate termini, such as secondary siRNAs in Caenorhabditis elegans (Pak and Fire 2007). Various approaches have been developed to generate cDNA libraries that are enriched for small RNAs with specific types of 5′ and/or 3′ ends (Pak and Fire 2007; Hafner et al. 2008; Sharma et al. 2010).
FIGURE 1.
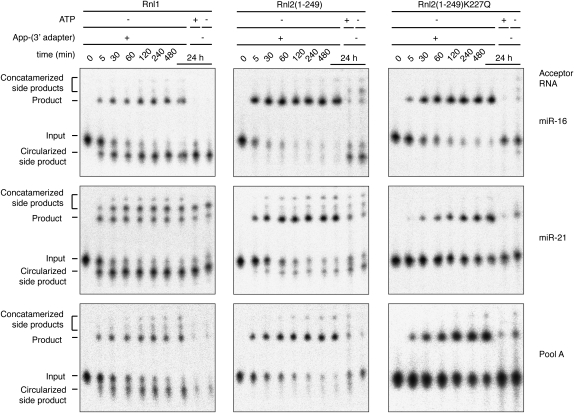
Scheme of the reactions for the small RNA cDNA library preparation. (A) Overview of the workflow for the generation of small RNA profiles by cDNA library sequencing. (B) Ligation reaction catalyzed by RNA ligases involving three nucleotidyl transfer steps: (1) the ligase forms a covalent Rnl-(lysyl-N)-AMP intermediate from ATP; (2) the AMP is subsequently transferred to the 5′ P of the donor RNA (black box) to form an RNA-adenylate (AppRNA); and (3) the 3′-OH of the acceptor RNA (light gray box) attacks the AppRNA, releasing AMP and forming a molecule where the donor RNA is ligated to the 3′ end of the acceptor RNA. (C) Reactions taking place in the 3′-adapter ligation step using RNA ligases 1 and 2 and 5′ pre-adenylylated adapters in the absence of ATP leading to the desired adapter ligated small RNA (left panel) or the undesired circularized or concatamerized small RNA products by adenylate transfer.
Typically, the first step of the small RNA cDNA library preparation involves ligation of an oligonucleotide adapter to the 3′ end of the sample RNA using either T4 RNA ligase 1 or 2 (Rnl1 or Rnl2) to allow RT priming and subsequent PCR (Lau et al. 2001; Pfeffer et al. 2005). Alternatively, addition of a nucleotide homopolymer sequence by poly(A) polymerase (Sharma et al. 2010) or terminal deoxyribonucleotidyl transferase (Deng and Wu 1983) has been used, but prevents the unambiguous determination of the termini of the input RNAs.
Rnl1 was identified as a T4 phage enzyme that, together with a polynucleotide kinase, repairs bacterial host tRNAs nicked at the anticodon positions during phage infection (Silber et al. 1972). T4 phage encodes for a second Rnl enzyme, Rnl2, which is also able to repair nicked tRNA substrates (Ho and Shuman 2002), but contains a structurally distinct nucleotidyl transferase domain (Pascal 2008).
RNA ligases join the 5′-p terminus of the so-called donor RNA to the 3′-hydroxyl (OH) terminus of the acceptor RNA requiring ATP (pppA). The reaction proceeds in three nucleotidyl transfer steps (Fig. 1B), in which (1) the RNA ligase reacts with pppA to form a covalent Rnl-(lysyl-N)-pA intermediate and pyrophosphate (pp); (2) the pA is subsequently transferred to the 5′-p of the donor RNA yielding an RNA-adenylylate (AppRNA); and (3) the 3′-OH of the acceptor RNA attacks the AppRNA forming the new phosphodiester bond and releasing pA (Walker et al. 1975).
A major concern using RNA ligases for joining of 3′-adapter molecules to 5′-p small RNAs is adenylylation of the small RNA 5′-p, leading to undesired circularization and concatamerization of the input RNA as well as the 3′ adapter (Fig. 1C). The circularization of the 3′ adapter is prevented by using chemically synthesized oligonucleotides carrying a 3′ aminolinker or inverted nucleotide. Side reactions of the input RNA side reactions are prevented by dephosphorylation prior to the 3′-adapter ligation and rephosphorylation of the ligation product before joining the 5′ adapter. However, dephosphorylation of the input RNA leads to a loss of enrichment for RNAs carrying 5′-p and 3′-OH due to the undesired recovery of abundant rRNA and tRNA degradation and turnover products with 5′-OH and 2′-p, 3′-p, or 2′,3′-cyclic p termini (Hafner et al. 2008).
Adenylylation of input RNAs during 3′-adapter ligation can be reduced by providing chemically pre-adenylylated 3′-adapter oligonucleotides and Rnl1-catalyzed ligation in the absence of ATP (Lau et al. 2001), but because ligase-catalyzed adenylylate transfer from 3′ adapter via the adenylylated ligase intermediate to 5′-pRNA is rapid, circularization and/or concatenation of input 5′-pRNA circulation still compete with 3′-adapter ligation. These side reactions are attenuated by the use of the truncated form of Rnl2, Rnl2(1–249), which has reduced affinity to 5′-pRNA donors compared with Rnl1, resulting in less efficient adenylyl transfer from the adenylated enzyme to the donor 5′-pRNA (Ho et al. 2004). Conservative mutation of the lysine residue K227 to glutamine in Rnl2(1–249)K227Q further compromised the adenylyltransferase activity while retaining ligation activity (Yin et al. 2003).
Some species of AGO/PIWI-interacting small RNAs, such as piRNAs and D. melanogaster endogenous siRNAs, are 2′-_O_-methylated in addition to having a 5′-p. The T4 ligases accept 2′-_O_-methylated substrates, albeit ∼1.5-fold less efficient compared to 2′-OH substrates (Munafo and Robb 2010). A way to specifically enrich for 2′-_O_-methylated 5′-p small RNAs in small RNA cDNA libraries is the pretreatment of total RNA by diol-specific oxidation, e.g., by NaIO4, thereby cleaving and eliminating vicinal 2′-OH and 3′-OH without affecting an isolated 3′-OH adjacent to _O_-methylated or phosphorylated 2′-OH (Alefelder et al. 1998; Vagin et al. 2006).
The next step of small RNA cDNA library preparation consists of joining the 3′-OH of the 5′-adapter oligonucleotide to the 5′ end of the small RNA/3′-adapter ligation product. Side reactions as they can occur during 3′-adapter ligation are of no concern because the 3′-OH of the small RNA/3′-adapter ligation product has been modified, and the 5′ adapter does not have a reactive 5′-p. Therefore, ordinary ligation in the presence of ATP and Rnl1 can be used; chemical adenylylation of the RNA/3′-adapter product and subsequent use of Rnl2(1–249)K227Q would be possible, but would be less convenient given the additional steps required.
Following 5′- and 3′-adapter ligation, cDNA is generated by reverse transcription using a primer complementary to the 3′-adapter sequence. Secondary structures such as stem–loops formed by small RNAs potentially impede uniform reverse transcription of all members of the small RNA pool. To minimize these effects, RNase H–deficient and/or thermostable RT enzymes, such as the SuperScript family, are preferably used. A final PCR amplification step is required to prepare cDNA for deep-sequencing platforms such as Solexa, 454, and SOLiD.
RNA sequence and structure influence the activity of nucleic acid–processing enzymes, and it is anticipated that the series of enzymatic steps required for small RNA cDNA library preparation and deep sequencing leads to a bias in the representation of input RNA abundance based on sequence read frequencies (Landgraf et al. 2007; Linsen et al. 2009; Munafo and Robb 2010). These enzyme- and RNA-sequence-dependent biases are not unique to RNA sequencing approaches but also affect other small RNA detection methods, such as ligase- and polymerase-based fluorescent labeling of RNA followed by array hybridization. Unless certain RNAs were inert to enzymatic modification and therefore undetectable by the approaches described above, these biases were treated as systematic during the quantification of relative changes in miRNA expression between samples (Bissels et al. 2009; Linsen et al. 2009; Schulte et al. 2010). Absolute RNA expression values may be derived from calibration experiments using pools of concentration-defined input RNAs and spiking of samples by external oligonucleotide standards (Bissels et al. 2009).
To evaluate possible biases and their sources in miRNA cDNA library preparation and sequencing approaches, we generated concentration-defined pools of 770 synthetic, 5′-phosphorylated synthetic oligoribonucleotides representing human, mouse, rat, and viral miRNA sequences identified in previous small RNA sequencing studies (Landgraf et al. 2007) and 45 5′-phosphorylated 22-nt calibrator oligoribonucleotides with no match to the human or rodent genomes. These pools were carried through various small RNA cDNA library generation protocols and were then sequenced using Solexa or 454 technologies. We tested three different RNA ligases in the 3′-adapter joining step: Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q and different temperature 5′-adapter ligation, as well as the impact of RT and PCR steps on small RNA cDNA library generation. Finally, we evaluated the effect of short barcodes placed within the 3′ adapter and demonstrate the feasibility of this approach for parallel processing of 20 samples.
RESULTS
We obtained a set of 770 5′-phosphorylated synthetic miRNAs and 45 oligoribonucleotides noncognate to human or rodent genomes (Supplemental Table 1) and prepared two pools of oligoribonucleotides. Pool A contained all 815 oligoribonucleotides in equimolar concentrations, whereas pool B contained all 770 miRNAs in four subpools combined in a 10-fold serial dilution so that the concentration of the last subpool was 1000-fold less than that of the first subpool (Supplemental Table 2).
Comparison of RNA ligase activities on the efficiency of 3′-adapter ligation
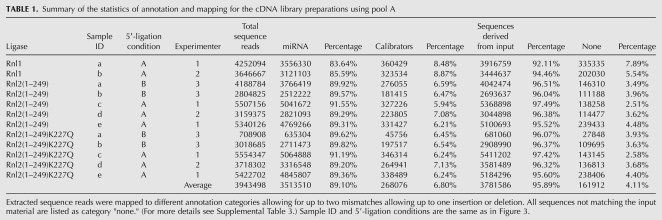
We first tested miR-16, miR-21, and pool A as acceptors in individual 3′-adapter ligation reactions. The 3′-adapter donor oligodeoxynucleotide was chemically pre-adenylylated and blocked at its 3′ end by an aminolinker residue to protect it from participation in inter- and intramolecular ligation. Although RNA ligases are generally specific to ribonucleotides, pre-adenylylation of the phosphorylated donor moiety enables the use of oligodeoxynucleotides as donors (England et al. 1977). Ligation reactions were performed in the presence of an excess of RNA ligase and pre-adenylylated 3′ adapter over 5′ 32P-labeled acceptor RNAs in the absence of ATP and on ice; Rnl2(1–249) and Rnl2(1–249)K227Q were inactive at 37°C (data not shown). The time courses of 3′-adapter ligation are shown in Figure 2. The reaction rates are RNA-sequence- and ligase-dependent. Rnl1-mediated ligation yielded the largest fraction of byproducts due to acceptor RNA circularization and multimerization. Although >90% of the input sequences had reacted after 2 h, the yield of desired 3′-adapter ligation product for Rnl1 was only 45%, 20%, and 25%, for miR-16, miR-21, and pool A, respectively. Side-reactions were significantly reduced using Rnl2(1–249), yielding 70%, 50%, and 60% of desired product for miR-16, miR-21, and pool A, respectively. The use of Rnl2(1–249)K227Q eliminated all side-product formation, yielding 85%, 55%, and 50%, for miR-16, miR-21, and pool A, respectively, after 24 h of reaction time. Compared with unmutated Rnl2(1–249), however, the ligation rate was somewhat reduced. To test if unreacted miRNA was trapped in a stable nonreactive secondary structure or whether the ligase had gradually lost activity during the 24-h incubation period, we added fresh Rnl2(1–249)K227Q after 24 h with or without a 30-sec, 95°C heat-denaturing step. Addition of ligase after the heat shock further increased the yield of ligation product, whereas the addition of new enzyme without heat shock had little effect (data not shown). Together, these experiments documented the influence of RNA sequence, structure, and ligase on the yield of ligation product. We were therefore interested in evaluating the sequence and structural biases contributing to miRNA profiles from deep sequencing.
FIGURE 2.
Comparison of 3′-adapter ligation by three RNA ligases. 5′-32P-radiolabeled oligoribonucleotides (miR-16, miR-21, and a pool containing 770 different miRNAs) were reacted in the absence of ATP with a pre-adenylylated adapter oligodeoxyribonucleotide on ice for the indicated time using either Rnl1 (left panels), the truncated Rnl2(1–249) (middle panels), and the truncated and mutated Rnl2(1–249)K227Q (right panels). To monitor the progress of the reaction, samples for each time point were fractionated by denaturing gel electrophoresis on a 15% denaturing polyacrylamide gel and visualized by phosphorimaging. The different reaction products and the input are marked. Control reactions in the presence of ATP and in the absence of the adapter were included. The experiments were performed in triplicate, and a representative phosphorimage picture is shown.
Sequence read distribution and biases in small RNA cDNA libraries
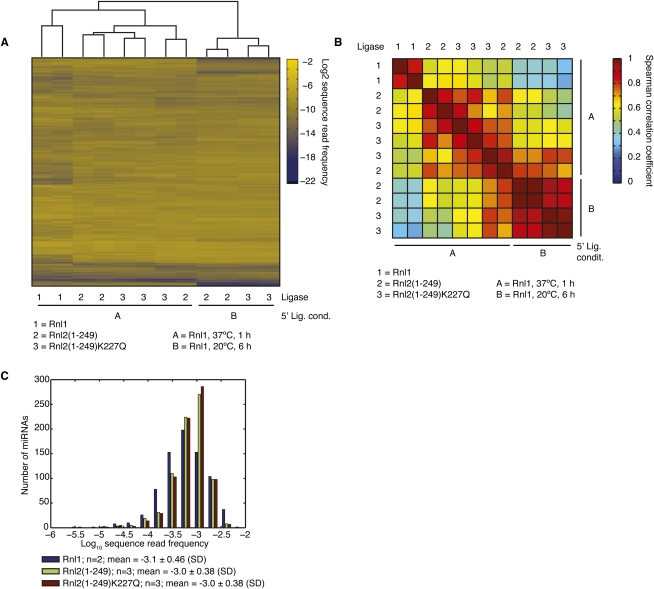
We prepared small RNA cDNA libraries of pools A and B using Rnl1, Rnl2(1–249), or Rnl2(1–249)K227Q (Supplemental Table 3) for 3′-adapter ligation, followed by 5′-adapter ligation using Rnl1 in the presence of ATP for either 1 h at 37°C or for 6 h at 20°C, and RT/PCR (Table 1; Hafner et al. 2008). Replicates of pool A libraries were prepared by three separate individuals at two separate locations to assess experimental reproducibility. cDNA libraries were sequenced using either Solexa or 454 sequencing at an average depth of 4,100,000 or 17,500 sequence reads, respectively. On average, 96% of all reads matched sequences present in the input material allowing for up to two mismatches, including one insertion or deletion (see Supplemental Table 3 for statistics of annotation and mapping and Supplemental Table 4 for sequence read abundance of miRNAs present in pool A for 454 and Solexa sequencing). The remaining nonmatching sequences were repetitive sequences, which arise as artifacts of the sequencing instrument data acquisition and processing.
TABLE 1.
Summary of the statistics of annotation and mapping for the cDNA library preparations using pool A
miRNA profiles report miRNA read frequencies and thereby normalize for experimental variation in sequence read numbers between libraries. If all miRNAs were represented equally without biases in pool A samples, the expected read frequency for each miRNA would be 130 ± 1.3 × 10−5 (corresponding to ∼5000 ± 50 reads for Solexa or 21 ± 0.2 reads for 454-based sequencing). Solexa sequencing detected every sequence present in the miRNA input pool in at least one of the biological replicates; however, read frequencies were spread across three orders of magnitude, ranging between 500 and 700 × 10−5 for miR-567 and between 0.2 and 4.0 × 10−5 for miR-31. For 10 of the most under-represented miRNAs, we performed additional quality controls determining 5′-phosphorylation status, sequence, and integrity and found no irregularities, suggesting that RNA structure was the likely reason affecting ligation efficiency. Although we had fewer reads by 454 sequencing, similar variations in read frequencies were noted. Rank correlations between Solexa- and 454-sequenced small RNA cDNA libraries were 0.7 and 0.8, the same as for biological replicates sequenced by the same method (see below), indicating that the sequencing methods were not a major source of sequence-specific biases (data not shown).
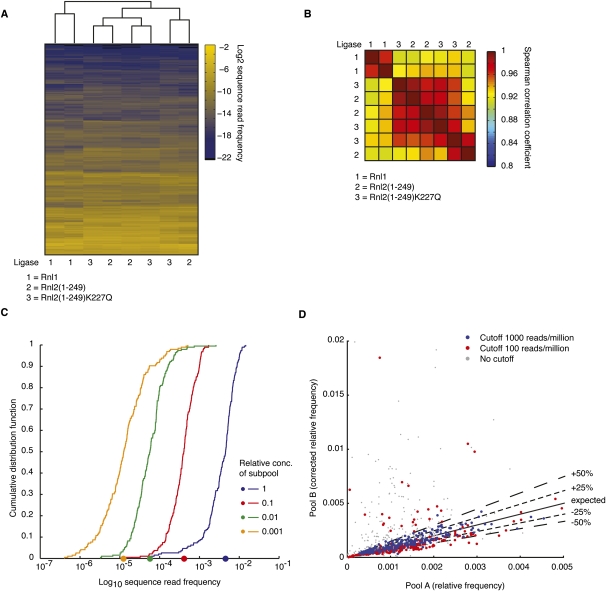
Sequence read frequency profiles of pool A libraries were subjected to unsupervised hierarchical clustering (Fig. 3A). The samples clustered according to the two 5′-adapter ligation protocols. Within the 37°C 5′-adapter ligation samples, the replicates of the cDNA libraries using Rnl1 for 3′-adapter ligation clustered together and were well separated from the libraries prepared by Rnl2 3′-adapter ligation. The differences between samples using different Rnl2 variants were small, and in some instances they clustered with a similar correlation as their biological replicates (Spearman correlation > 0.8) (Fig. 3B). Together, these results indicated that incubation temperatures in the ligation reactions have a stronger effect on miRNA representation than the choice of ligases, and that Rnl2 variants shared the same sequence and/or structural specificity.
FIGURE 3.
miRNA representation by sequencing varies by three orders of magnitude and is dependent on the structure of the mature miRNA and miRNA-adapter product. (A) Unsupervised hierarchical clustering of miRNA profiles derived from cDNA libraries generated from the pool of 815 oligoribonucleotides present in equimolar concentrations (pool A, Supplemental Table 1) using Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q for the 3′-adapter ligation step and sequenced by Solexa next-generation sequencing platform. (B) Pairwise comparison of Spearman rank correlation coefficients of the miRNA profiles from A. (C) Distribution of average sequence read frequencies of the 770 miRNAs present in equimolar concentrations in pool A in cDNA libraries generated using Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q in the 3′-adapter ligation step. The number of biological replicates for each distribution is indicated. miRNA relative frequencies vary by 1000-fold.
To best represent the non-Gaussian distribution of miRNA read frequencies, we log-transformed the average miRNA sequence read frequencies (Fig. 3C). The log10 of the mean values were −3.1 ± 0.46, −3.0 ± 0.39, and −3.0 ± 0.39 for Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q, respectively. Overall, we noted that more miRNAs were under- than over-represented based on a cutoff defined by a two–standard deviation interval around the average log-transformed read frequencies. For Rnl1-, Rnl2(1–249)-, and Rnl2(1–249)K227Q-based pool A cDNA libraries, we identified 22, 35, and 36 miRNAs, respectively, that were sequenced less frequently, and five, two, and one miRNAs that were sequenced more frequently, respectively (Supplemental Table 5). As 97% of all miRNAs fell within two standard deviations from the mean and the mean values were very similar, we concluded that the magnitude of the biases was similar for the different 3′-adapter ligation reactions.
The yield of the ligation of the 3′ adapter to miR-16 versus miR-21 was 2.3-fold, 1.4-fold, and 1.6-fold higher for Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q, respectively (Fig. 2; Supplemental Table 4). However, the sequence read frequency for miR-16 versus 21 was 2.7-fold, 4.6-fold, and 2.1-fold lower when the 3′-adapter ligation was performed using Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q, respectively. These findings indicate that steps subsequent to 3′-adapter ligation, comprising 5′-adapter ligation and RT/PCR, introduced further biases.
Dissection of biases for 5′- and 3′-adapter ligation
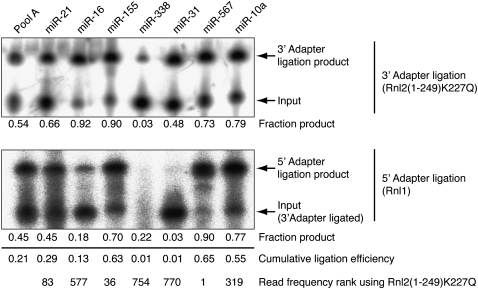
To define the molecular steps responsible for biases in the miRNA cDNA library preparation, we selected a subset of miRNAs and determined the cumulative yield of the ligation product formation. We examined miR-567, miR-155, and miR-21, which were over-represented; miR-10a and miR-16, which were average; and miR-31 and miR-338, which were under-represented in the cDNA libraries prepared with the Rnl2 variants in the 3′-adapter ligation step. The radiolabeled synthetic miRNAs were first incubated with 3′ adapter and Rnl2(1–249)K227Q under standard conditions. Consistent with the under-representation by library sequencing, miR-338 yielded only 10% ligation product, whereas the average and over-represented miRNAs yielded between 34% and 92% (Fig. 4, upper panel). We noted that miR-31, which was among the five least sequenced miRNAs, yielded 63% 3′-adapter ligation product, indicating that steps other than 3′-adapter ligation are critically contributing to sequence read distribution.
FIGURE 4.
5′-Adapter ligation introduces sequence-specific biases. Autoradiographs of the 3′-adapter ligation step using Rnl2(1–249)K227Q with 5′-32P-radiolabeled oligoribonucleotide sequences shown in Supplemental Table 1 (upper panel), the products of which were purified and used as input into the 5′-adapter ligation step using Rnl1 (lower panel). The fraction of adapter-ligated material was calculated from the ratio of intensity of the product band and the sum of the intensities of input and product band. The cumulative adapter ligation efficiencies are indicated.
Following the isolation of miRNA/3′-adapter product, the yield of joining the 5′ adapter was determined, using Rnl1 and ATP for 1 h at 37°C (Fig. 4, lower panel). The ligation efficiency for the under-represented miR-31 was <1% and was also very low for miR-338, although it could not be determined reliably because of the poor yield of 3′-adapter ligation. In contrast, the efficiency for the over-represented miRNAs varied between 43% and 87%, and the average distributed miR-16 and miR-10a products ligated with 14% and 80% efficiency, respectively.
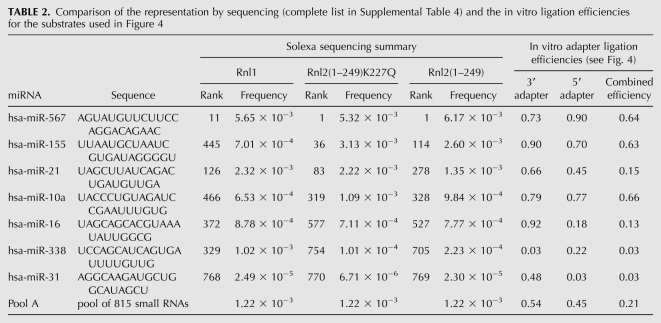
The cumulative ligation efficiency was calculated by multiplying the yields of the 3′- and 5′-adapter ligations. It ranged between <1% for miR-31 and miR-338 (among the least efficiently sequenced miRNAs) and 64% for miR-567 (the most frequently sequenced miRNA). This efficiency range explained well the observed broad sequence read frequency distribution as well as the rank of miRNAs. Furthermore, the up to threefold over-representation of some miRNAs relative to the mean read frequency is consistent with the 21% cumulative ligation yield of pool A RNA (Table 2).
TABLE 2.
Comparison of the representation by sequencing (complete list in Supplemental Table 4) and the in vitro ligation efficiencies for the substrates used in Figure 4
We further isolated the products from the 5′-adapter ligation for pool A, miR-567, miR-155, miR-10a, miR-16, and miR-21 and performed reverse transcription reactions with equimolar amounts of adapter-ligated material, using a 25-fold excess of radioactively labeled reverse transcription primer followed by hydrolysis of the RNA template. The yields of primer extension products were comparable (data not shown), indicating that reverse transcription was not a significant source of sequence-specific biases.
Lastly, we examined the influence of excessive PCR on small RNA read frequency distribution. A small RNA cDNA library generated from pool A using Rnl2(1–249)K227Q for the 3′-ligation step followed by Solexa sequencing. This library was subjected to five rounds of 1:1000 sample dilution followed by 10 PCR cycles, corresponding to a total of 50 additional cycles of PCR amplification. Re-sequencing revealed no appreciable distortion in miRNA representation according to this protocol (Supplemental Table 6).
In summary, we have defined the 5′- and 3′-adapter ligation reactions as the critical steps in introducing biases in miRNA cDNA sequence read representation. Next, we wanted to identify miRNA sequence or structural features predictive of adapter ligation efficiencies. However, within the small set of miRNAs studied above, there was no obvious sequence, secondary structure, or predicted free energy change correlating with the ligation efficiency (Supplemental Table 4).
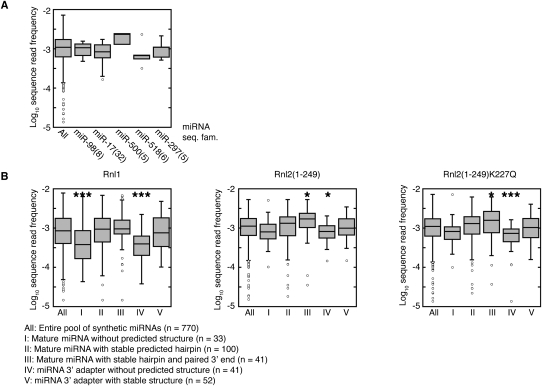
To detect more subtle correlations between sequence features and miRNA representation, we grouped all miRNAs into sequence families (Supplemental Table 7). We found that sequence-related miRNAs were often sequenced with comparable efficiency, and that large miRNA sequence families, e.g., the 32-member miR-17 family, displayed a narrower distribution of sequence read frequencies than the entire pool (Fig. 5A). This suggested that RNA sequence and structure indeed influenced read distribution. We therefore calculated their minimal free-energy structures by RNAfold (Supplemental Table 4; Hofacker 2003) and compared the read frequency distribution for several structurally distinct groups of miRNAs in Figure 5B. The structural groups were defined as follows: (I) miRNAs without predicted stable structure, (II) miRNAs folding into a hairpin with more than 14 paired bases, (III) miRNAs with more than 14 paired bases involving the 3′ end, (IV) miRNA/3′-adapter products without any or with a weak structure comprising less than eight paired bases, and (V) miRNA-adapter predicted to fold into strong secondary structures with more than 28 paired bases (Supplemental Table 7). Groups I and IV, characterized by instable secondary structures, underperformed in 3′- and 5′-adapter ligation, especially when using Rnl1, whereas Rnl2 variants performed better on substrates with strong secondary structure and paired 3′ ends (group III).
FIGURE 5.
miRNA representation is influenced by folding. (A) Members of the same miRNA sequence families are represented with similar sequence read frequencies. The log-transformed sequence-read frequencies for the indicated sequence families (see Supplemental Table 7 for miRNA sequence family classification) are compared with the distribution of sequence reads for the entire pool. Sequence families are named after the member with the lowest number; the number of members in the sequence family are in brackets. Results are shown for the experiments using Rnl2(1–249)K227Q in the 3′-adapter ligation; the experiments using the other ligases yielded similar results (data not shown). (B) miRNA representation depends on structure. miRNAs present in the pool were classified according to their predicted structure: (I) mature miRNA without predicted structure; (II) mature miRNA folds into stable structure with at least 14 paired bases; (III) same as II, in addition, the 3′ end is paired; (IV) miRNA-3′-adapter-ligation product has weak predicted structure (less than nine paired bases); and (V) miRNA-3′-adapter ligation product has strong predicted structure with more than 28 paired bases. The log-transformed sequence-read frequencies of the individual categories were compared with the distribution of sequence reads for the entire pool, and the statistical significance of the difference between the populations was calculated; two-tailed _t_-test, (*) p < 0.05; (**) p < 0.01; (***) p < 0.001.
We conclude that biases in miRNA read distribution are reflective of different RNA structures with different reactivity toward 5′- and 3′-adapter ligation. Although transitions between RNA structures may occur at different rates, they represent intramolecular rearrangements that are independent of the input RNA concentration. It is therefore expected that for a given adapter ligation protocol, the biases in read frequencies should be miRNA concentration-independent, and that differences in miRNA read frequencies between samples correspond to the differences in relative miRNA concentration between samples. Therefore, as long as miRNA adapter ligations are performed with an excess of adapter and the reaction conditions including time and temperature are held constant, differences in miRNA read frequencies between samples should be directly proportional to the differences in RNA input concentration.
Range of miRNA detection using deep sequencing
To evaluate the influence of miRNA concentration on miRNA read representation, we prepared pool B from four subpools of synthetic oligoribonucleotides at 1×, 10×, 100×, and 1000× dilution, respectively (Supplemental Table 2). cDNA libraries were again prepared using the three different ligases—Rnl1, Rnl2(1–249), or Rnl2(1–249)K227Q—for the 3′-adapter ligation step and the same reaction conditions as above. The resulting cDNA libraries were sequenced at an average depth of four Mio reads by Solexa or 12,000 reads by 454 sequencing (see Supplemental Table 3 for statistics of annotation and mapping and Supplemental Table 9 for miRNA read counts). On average, 97% of the Solexa reads and 98% of the 454 reads matched sequences represented in pool B, and only three to five miRNAs were not recovered from the subset of oligoribonucleotides present at the lowest concentration. In 454-sequenced cDNA libraries, an average of 230 miRNAs were not detected, compared to four in libraries from pool A sequenced at similar depth (see above). The missing miRNAs mostly corresponded to those in the 1000× dilution and 100× dilution subsets.
The miRNA profiles from the Solexa-sequenced libraries were subjected to unsupervised hierarchical clustering (Fig. 6A). Given the large spread of miRNA input concentration and the comparatively modest ligase-based read frequency biases seen for the majority of miRNAs, pairwise correlation coefficients between individual libraries were much higher than seen for pool A samples composed of equimolar concentrated miRNAs (Fig. 6B). Analogous to the results described for pool A (Fig. 3), the pool B libraries generated using Rnl1 were similar to (average Spearman correlation 0.92), but clustered separately from those generated using either of the two Rnl2 variants, which were more similar (average Spearman correlation 0.95).
FIGURE 6.
miRNA profiles generated by sequencing are able to reflect relative differences in miRNAs of approximately three orders of magnitude. (A) miRNA profiles derived from individual cDNA libraries generated from the pool of 770 oligoribonucleotides divided into four subpools present in concentrations spanning three orders of magnitude (pool B, Supplemental Table 2) using Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q for the 3′-adapter ligation step and sequenced either on a 454 or Solexa next-generation sequencing platform were subjected to unsupervised hierarchical clustering. (B) Pairwise comparison of Spearman rank correlation coefficients of the miRNA profiles clustered in A. (C) Increased sequencing depth makes relative comparison between samples of miRNAs present in very different concentrations more reliable. Sequence read frequencies of miRNAs from pool B and Rnl2(1–249)K227Q in the 3′-adapter ligation step were sorted according to the four subpools (Supplemental Table 2) making up pool B and their cumulative distribution function plotted for the Solexa-sequenced libraries. The same plot for 454-sequenced cDNA libraries is appended to Supplemental Table 9. The median sequence read frequency of each subpool is indicated on the _x_-axis. (D) Sequence read frequencies for the Solexa-sequenced cDNA libraries from C were corrected by the known input concentration from pool B and plotted against the sequence read frequencies from pool A. Blue dots are miRNAs sequenced with at least 1000 reads per million in pool B; red dots with at least 100 reads per million; and gray dots without cutoff. The full line denotes the expected corrected values, the dashed lines an error margin of 25% and 50%, respectively.
To assess whether sequence read frequencies from pool B cDNA libraries accurately reflected the 10-fold subpool dilution steps, miRNA sequences were grouped according to their relative concentration in pool B, and we plotted the cumulative distribution function (CDF) for the miRNAs from each subpool against their relative sequence read frequency for the Solexa-sequenced cDNA libraries (Fig. 6C). The curves for the subpools at the onefold, 10-fold, and 100-fold dilution were parallel to each other and separated by steps of 10. The slope of the CDF at the lowest concentration of the subpool present at 1000-fold dilution was less steep due to the influence of the limited read coverage of these miRNAs (Supplemental Table 9). The CDF curve separation for the less deep 454-sequenced library was similar, except that the slope of the 100-fold dilution already showed distortion (Supplemental Table 9). In summary, average miRNA abundance was accurately captured over a 100- to 1000-fold range depending on the depth of sequencing.
We next explored whether the sequence-specific biases in cDNA preparation for pool B were, as expected, independent of miRNA concentration. We first corrected the relative sequence read frequencies for pool B for their input concentration and plotted the values against the relative frequency observed in pool A (Fig. 6D). The miRNAs were expected to scatter along the diagonal. We found this, indeed, to be the case, except for miRNAs with low sequence coverage, where statistical scattering leads to misrepresentation. At a cutoff of 100 sequence reads per million, the concentration of ∼20% of miRNAs was misestimated by >50%, while at a cutoff of 1000 sequence reads per million, <1% of the miRNAs were misestimated by >50% error (Supplemental Table 10). We conclude that sequencing of pool A or similar reagents can be used to infer the absolute concentration of abundantly sequenced miRNAs.
Detection of synthetic oligoribonucleotides by barcoded small RNA sequencing
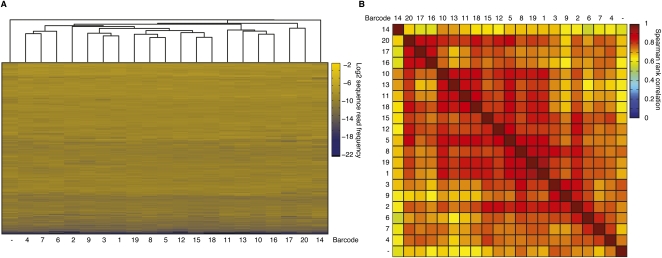
To reduce costs and increase sample throughput for clinical studies monitoring miRNA expression, we wanted to develop a barcoding strategy. We generated a set of 20 different pre-adenylylated 3′ adapters with inserted 5-nt sequence tags at their 5′ ends. The pentamer barcode sequences were designed to have a difference of at least 2 nt between them, to ensure that a single sequencing error cannot lead to a misclassification of the sequence read. Placement at the 5′ end of the 3′ adapter was critical in order to capture the barcode when sequencing small RNAs in a conventional 36-nt Solexa read. To quantify any additional biases introduced by the barcode segment into the miRNA cDNA library, we joined the barcoded 3′ adapters to pool A using Rnl2(1–249)K227Q followed by pooling of the ligation products and standard 5′-adapter ligation, RT and PCR. Solexa sequencing of this library yielded more than 6.4 Mio reads of which more than four Mio (67%) contained recognizable pentameric barcode sequences without mismatch (Supplemental Table 11). The sequence reads were grouped into 20 sublibraries, each corresponding to an individual sample. Total sequence reads averaged 203,000 (range: 107,000–292,000), indicating comparable efficiencies for adapter ligation reactions and subsequent RT and PCR amplification steps (Supplemental Table 11). Following annotation, 97% of the extracted sequences matched either input miRNA (91%) or calibration marker (6%) sequences; few (3%) sequences were classified as either nonmatching or repeat sequences, similar to the nonmultiplexed libraries (see Supplemental Table 3). Next, we generated miRNA expression profiles (Supplemental Table 12) for each sublibrary and compared by pairwise correlation these profiles as well as the averaged profile for the nonbarcoded library (Figure 3; Supplemental Table 4) prepared under the equivalent conditions. The nonbarcoded profile clustered separately and with Spearman correlations of 0.6–0.75 to the barcoded libraries indicating that either the changed adapter length or the additional pentamer sequence subtly changes the adapter ligation biases. Within the barcoded samples, average Spearman correlation coefficients ranged from 0.7 to 0.95 (Fig. 7), in the same range as the correlations of biological replicates of individual cDNA libraries (Fig. 3B), making multiplexing a viable alternative to the sequencing of individual cDNA libraries.
FIGURE 7.
Barcoded 3′-adapters allow multiplexed miRNA profiling. (A) Unsupervised hierarchical clustering of miRNA profiles derived from cDNA libraries generated from pool A in Supplemental Table 1 using Rnl2(1–249)K227Q for the 3′-adapter ligation step with a panel of 20 chemically adenylylated 3′-adapter oligonucleotides bearing a pentameric barcode sequence at their 5′ end. Samples were pooled after 3′-adapter ligation and subjected to standard 5′-adapter ligation, RT and PCR. (B) Pairwise comparison of Spearman rank correlation coefficients of the miRNA profiles from A.
DISCUSSION
Deep sequencing of small RNA cDNA libraries not only allows for derivation of small RNA expression profiles but also provides RNA length and sequence information, which, in turn, provides insights into small RNA biogenesis, RNA editing, and mutational events. With sequencing depths exceeding hundreds of millions of reads, the sensitivity range of this technology has increased tremendously, and sequencing of RNA is becoming increasingly important for quantification of gene expression.
Here, we evaluated the biases in small RNA cDNA library preparation approaches. We detected moderate biases for the majority of input RNA (≥95%) that amount to fourfold over- and 10-fold under-representation. Two percent of small RNAs were more than 50-fold under-represented in the cDNA libraries. Given that all of the miRNA sequences in our pool were discovered by small RNA cDNA library sequencing across various cell types and tissues by less than 200,000 reads total (Landgraf et al. 2007), it was expected to rediscover these RNAs by deep sequencing. Although represented by a much smaller sample size, our set of artificial calibrator sequences showed similar biases, suggesting that we sampled enough sequences to provide a representative view for biases encountered in small RNA cDNA library sequencing.
The influence of RNA and adapter secondary structure on RNA ligation
We observed that the biases were mostly inherent to the sequences of small RNAs, and their secondary and tertiary self-structures predominantly affected the efficiency of 3′-adapter and 5′-adapter ligation during the first steps of library preparation; whereas subsequent steps, such as RT, PCR, or sequencing technology, showed little effect. Although statistical analysis involving many RNA sequences documented a general influence of RNA secondary structure on the efficiency of RNA ligation, several of the best and worst represented RNAs were not readily distinguished in terms of computable sequence or structural parameters, indicating that experimental approaches using complex and concentration-defined pools of RNAs remain important to determine biases.
RNA secondary self-structure, in contrast to intermolecular paired structures, are RNA concentration-independent, yet dependent on temperature, as well as the concentration of mono- and polyvalent cations (Mathews and Turner 2006), and are influenced by the addition of destabilizing organic solvents, such as DMSO (von Ahsen et al. 2001). Variation of these physical parameters was only possible within a window compatible with RNA ligase function. Our ligation reaction uses 15% DMSO, but otherwise optimal salt and buffer conditions established for these ligases (England and Uhlenbeck 1978; Yin et al. 2003). A 90°C heat denaturation step prior to the addition of ligase disrupted secondary structures formed during storage or freeze–thawing. Following the addition of ligase, ligation products accumulated rapidly within the first 30 min and then approached a sequence-dependent threshold value asymptotically, suggesting that the nonreactive fraction of RNA was trapped in a kinetically stable secondary structure, which only upon additional heat denaturing and addition of new ligase could be refolded and thereby made amenable for further ligation. To ensure reproducibility in cDNA library preparation, it was critical to allow the ligation reaction to proceed close to its endpoint for capturing reactive secondary structure conformation and maintaining constant heat-shock procedures and incubation temperatures for comparison of samples.
The thermodynamic stability of secondary structure also depends on nucleic acid backbone modification, DNA being less stable than RNA. In an attempt to minimize the influence of secondary structure during sequential adapter ligation, we used DNA rather than RNA as 3′-adapter oligonucleotide. Chemical pre-adenylation of 5′-phosphorylated donor molecules has been shown to extend dramatically the range of substrates amenable to RNA ligation (England et al. 1977). The 5′ adapter was composed of RNA, although formally only the 3′-terminal nucleotides were required to be RNA for optimal RNA ligation. Given the process we have established for analysis of biases in library preparation, it is possible to further test and optimize the influence of backbone-modified adapters, e.g., using chimeric DNA/RNA oligonucleotides.
The pair of adapter oligonucleotides used in our study was based on Solexa sequencing primer pairs, and their sequential performance in adapter ligations was comparable to our previously used adapter pairs (Hafner et al. 2008). Our previously used pair was obtained from a small screen of a collection of 5′- and 3′-adapter oligonucleotides (data not shown). Testing of various 3′-adapter sequences on model substrates indicated that the ligation yields were rather insensitive to the adapter sequence, and similar findings were obtained when various 5′-adapter sequences were tested for ligation to model substrates that had not yet joined a 3′ adapter. However, when we tested the same set of 5′ adapters after joining of a 3′ adapter to the model substrate, the sequential ligation yield was often much lower than predicted from isolated adapter ligation, indicating that 5′- and 3′-adapter sequences can interact and needed to be optimized in a sequential manner. Initially we aimed at eliminating 5′- and 3′-adapter pairing interactions; however, secondary structure prediction suggest that 5′ and 3′ adapters in both cases were able to form base pairs over 6–8 nucleotides, depending on the miRNA insert. Given that substrates with stronger secondary structure appeared to ligate more efficiently, further stabilization of the 5′- and 3′-adapter interaction may lead to increased 5′-adapter ligation yield.
In future studies, it will also be interesting to test thermostable RNA ligases (Blondal et al. 2003, 2005) as well as the influence of additives such as PEG-8000 (Miyoshi and Sugimoto 2008) or compounds such as cationic comb-type copolymers (CCC), both of which enhance the dynamic of RNA folding (Kim et al. 2003; Choi et al. 2007). It is possible that these conditions lead to an increased rate of interconversion between secondary structure and thereby allow for more product accumulation, given that adapters are always used in excess over input RNAs.
Guide to reagents and critical experimental steps
Although our adenylated and regular oligonucleotides and RNA ligases used for small RNA cDNA library preparation were prepared in our laboratory, pre-adenylylated oligonucleotides can be purchased from several oligonucleotide synthesis companies. The various RNA ligases, including Rnl2(1–249)K227Q, can be obtained from New England Biolabs. Pools of synthetic miRNAs (miRXplore Universal Reference) can be obtained commercially from Miltenyi Biotec.
For the obvious reasons of reproducibility, it is important to keep the reaction conditions constant throughout series of experiments. The most critical parameters influencing biases in miRNA representation are the 5′- and 3′-adapter ligation reaction temperatures and incubation times, preceded by heat denaturation steps prior to addition of enzymes. The concentration of input total RNA also has to be maintained constant and should not be altered. Finally, it is critical to limit the cycles of PCR amplification after RT to avoid nonexponential amplification in late PCR cycles.
Barcoding in small RNA cDNA library sequencing
The increasing read depth for sequencing prompted the development of cost-saving multiplexing of samples by barcoding approaches (Parameswaran et al. 2007; Vigneault et al. 2008; Xu et al. 2009; Buermans et al. 2010; Schulte et al. 2010; Farazi et al. 2011). It is preferable to place the barcoding step as early as possible in the RNA processing protocol to minimize parallel handling of samples rather than at the end stage of cDNA library preparation as in current commercial kits (Illumina TruSeq Small RNA Sample Prep kit). We therefore placed the adapter within the 3′-adapter sequence at its 5′ end, keeping in mind that when sequencing is initiated from the 5′ primer, the barcode immediately follows the small RNA sequence even before the constant 3′-primer region is sequenced, thereby allowing the use of the less expensive and more rapid 36-nt read length sequencing reagent sets.
A further advantage of barcoding by the 3′-adapter ligation step was the opportunity to reduce the amount of sample RNA to 2 μg of unfractionated total RNA for routine applications or even less should the sample be limiting. Following 3′-adapter ligation, we pooled up to 20 samples corresponding to an overall input of 40 μg of total RNA. Processing larger-quantity pooled samples minimizes subsequent losses due to adsorption or presence of traces of nuclease contaminations otherwise faced when working with little starting material. Furthermore, multiple pooled samples can be readily processed in parallel, thereby generating more than a hundred small RNA profiles per batch of Solexa sequencing.
Mutation of general miRNA biogenesis factors has been implicated in certain cancers (Kumar et al. 2007; Mudhasani et al. 2008), and it is therefore not only important to capture miRNA abundance rank and the differences between samples but also to determine the absolute abundance of miRNAs and its change between samples. This is feasible by spiking samples with a known amount of 5′-phosphorylated calibrator RNAs noncognate to the genome. We have included in our characterization of biases a set of 45 such calibrator sequences, which displayed a similar range of biases compared to miRNAs. The read frequency and its distribution for calibrators and miRNAs can then be used to derive the absolute amount of small RNAs present in the respective samples. We have recently tested this approach studying normal and breast cancer tissue samples and found between 9 and 15 fmol of miRNA per microgram of total RNA without any significant differences between normal or disease samples (Farazi et al. 2011). We were also able to use the synthetic pool to derive correction factors that allow the assessment of absolute amounts of abundant miRNAs analogous to reported approaches using an array-based expression profiling platform and similar synthetic pools and calibrator sequences (Bissels et al. 2009).
The depth of sequencing required for derivation of miRNA profiles remains a matter of debate. Typically, 90% of the sequence reads captured from a biological sample originate from its top expressed about 50 miRNA genes (Hafner et al. 2010), and only the top-expressed miRNAs were so far shown to yield measurable target mRNA stability in antagomirs (Krutzfeldt et al. 2005; Esau et al. 2006; Landthaler et al. 2008) or miRNA-overexpression assays (Lim et al. 2005; Linsley et al. 2007). Therefore, miRNA profiles derived from sequencing of several thousand reads are already likely to capture the biologically important changes in abundance of regulatory important miRNAs and provide critical starting points for further follow-up experiments. Nevertheless, accurately capturing changes in low-abundant miRNAs may be useful in diagnostic or prognostic studies as they are considered reporters of altered gene expression.
MATERIALS AND METHODS
Pools
Seven hundred seventy 5′-phosphorylated oligoribonucleotide sequences corresponding to human, viral, mouse, and rat miRNAs from miRbase V12 and 45 5′-phosphorylated oligoribonucleotides with no match in the human, mouse, and rat genomes (Bissels et al. 2009) that can be used as spike-in controls were purchased from Sigma. The integrity of these RNAs was confirmed by denaturing polyacrylamide electrophoresis (PAGE) followed by UV shadowing. Furthermore, the presence of 5′ phosphate and the accuracy of the sequence were confirmed for a random subset and the 10 least sequenced miRNAs by MALDI-TOF mass spectrometry. The concentration of the individual miRNA aliquots was calculated based on their absorbance at 260 nm using the extinction coefficients listed in Supplemental Table 1. The miRNA sequences were pooled into four subpools of 188 to 195 oligoribonucleotides each (Supplemental Table 2).
RNA ligases
cDNAs of Rnl1, Rnl2(1–249), and Rnl2(1–249)K227Q were cloned into pET16b and recombinantly expressed as N-terminal hexahistidine-tagged proteins in Escherichia coli BL21DE3. The plasmid pET16b-Rnl2(1–249)K227Q is deposited at http://www.addgene.org. Recombinant proteins were purified on Ni-NTA agarose (QIAGEN) and polished by anion-exchange chromatography. The protein concentration was determined by Coomassie staining of a dilution series of the proteins on SDS-PAGE compared with a dilution series of BSA.
In vitro ligation assays
Fifty nanomolar (50 nM) radiolabeled synthetic oligoribonucleotide sequences corresponding to miR-16, miR-21, miR-155, miR-338, miR-31, miR-567, and miR-10a, or of 815 pooled synthetic oligoribonucleotides (see Supplemental Table 4 for sequences) were reacted with 5 μM adenylated 3′ adapter (App-TCGTATGCCGTCTTCTGCTTGT) in 50 mM Tris-HCl (pH 7.6); 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mg/mL acetylated BSA (Sigma, B-8894), and 15% DMSO at 4°C with 0.05 μg/μL the respective RNA ligase at 4°C. As a control for circularization, two samples without the adenylated adapter, with and without 0.2 mM ATP, were incubated for 24 h at 4°C. At the indicated time points, aliquots of 5 μL were taken, and the reaction was stopped by addition of 5 μL of formamide stop mix (50 mM EDTA, 0.05% [w/v] bromophenol blue in formamide). The RNA was fractionated on a 15% denaturing polyacrylamide gel and visualized by autoradiography and quantified.
To generate substrate for in vitro 5′-adapter ligation assays, bands corresponding to miR-3′-adapter product after 3′-adapter ligation overnight using Rnl2(1–249)K227Q were cut from the gel, passively eluted overnight at 4°C using 3 volumes 0.3 M NaCl, and precipitated with 3 volumes of EtOH and collected by centrifugation. The 5′-adapter ligation was performed using ∼50 nM RNA-3′-adapter product (calculated from the yield of 3′-adapter ligation) and ligated to 100 pmol of 5′-oligoribonucleotide adapter (GUUCAGAGUUCUACAGUCCGACGAUC) using 1 μg of Rnl1 in 50 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mg/mL acetylated BSA (Sigma, B-8894), 0.2 mM ATP, and 15% DMSO for 1 h at 37°C. Ligated small RNAs were purified on a 12% polyacrylamide gel and visualized by autoradiography.
cDNA preparation from synthetic pools
cDNA libraries representing pools A and B were prepared for massive parallel (454 or Solexa) sequencing as described in Hafner et al. (2008). Briefly, in a total reaction volume of 20 μL, 2 pmol of RNA was ligated to 100 pmol of adenylated 3′ adapter (App-TCGTATGCCGTCTTCTGCTTGT) using 1 μg of Rnl1, Rnl2(1–249), or Rnl2(1–249)K227Q, in 50 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mg/mL acetylated BSA (Sigma, B-8894), and 15% DMSO for 16 h at 4°C. Products were purified on a 15% denaturing polyacrylamide gel and ligated to 100 pmol of 5′-oligoribonucleotide adapter (GUUCAGAGUUCUACAGUCCGACGAUC) using 1 μg of Rnl1 in 50 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mg/mL acetylated BSA (Sigma, B-8894), 0.2 mM ATP, and 15% DMSO for 1 h at 37°C. Ligated small RNAs were purified on a 12% polyacrylamide gel, reverse-transcribed using SuperScript III (Invitrogen), and amplified by PCR using appropriate primers compatible with either Solexa-sequencing (forward primer: AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA; RT and reverse primer: CAAGCAGAAGACGGCATACGA) or 454-sequencing (RT primer: CAAGCAGAAGACGGCATACGA; forward primer: GCCTCCCTCGCGCCATCAGAATGATACGGCGACCAC, reverse primer: GCCTTGCCAGCCCGCTCAGCAAGCAGAAGACGGCAT). The appearance of the PCR product during the exponential amplification phase was monitored in a pilot PCR by running aliquots of the PCR reaction after 10, 12, 14, 16, or more cycles on an agarose gel stained with ethidium bromide (Hafner et al. 2008). The large-scale PCR was then performed using the cycle number determined prior to the loss of exponential amplification. The PCR product of the expected length was purified on an agarose gel from the insert-less shorter byproduct and submitted for sequencing. Sequencing was carried out using Solexa sequencing (Illumina, Rockefeller University Genomics Facility) or 454 sequencing (Roche, Memorial Sloan Kettering Cancer Center). Bioinformatics analysis was carried out as described by Berninger et al. (2008).
Barcoded cDNA library preparation
Barcoded cDNA library preparation from synthetic pools was carried out analogous to the standard cDNA library preparation. 3′-Adapter ligations were carried out separately for each barcoded adapter sequence (Supplemental Table 12) with the same reaction volumes and conditions as described above. The reaction was stopped, and the individual samples were pooled by adding all ligation reactions into a reaction tube containing 1200 μL of ethanol. RNA was collected by centrifugation and purified on a 15% denaturing polyacrylamide gel. 5′-Adapter ligation, reverse transcription, and PCR were performed for the pooled sample as described above. Sequencing was carried out using Solexa sequencing at the Rockefeller University Genomics Facility.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
T.T. is cofounder and scientific advisor to Alnylam Pharmaceuticals and an advisor to Regulus Therapeutics.
ACKNOWLEDGMENTS
We thank S. Juranek for helpful comments on the manuscript. We are grateful to A. Viale (Genomics core facility, MSKCC) for 454 sequencing and S. Dewell (Genomics Resource Center, Rockefeller University) for Solexa sequencing. M.H. is supported by a fellowship of the Charles Revson, Jr. Foundation. N.R. is supported by an NIH K08 award (K08NS072235-01). T.T. is an HHMI investigator, and work in his laboratory was supported by NIH grants GM073047, MH08442, NIH Challenge Grant RC1CA145442, and the Starr Foundation.
Footnotes
REFERENCES
- Alefelder S, Patel BK, Eckstein F 1998. Incorporation of terminal phosphorothioates into oligonucleotides. Nucleic Acids Res 26: 4983–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Tuschl T 2005. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett 579: 5830–5840 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M 2008. Computational analysis of small RNA cloning data. Methods 44: 13–21 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Filipowicz W 2007. Argonautes and company: Sailing against the wind. Cell 128: 1027–1028 [DOI] [PubMed] [Google Scholar]
- Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A 2009. Absolute quantification of microRNAs by using a universal reference. RNA 15: 2375–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T, Hjorleifsdottir SH, Fridjonsson OF, Üvarsson A, Skirnisdottir S, Hermannsdottir AG, Hreggvidsson GO, Smith AV, Kristjansson JK 2003. Discovery and characterization of a thermostable bacteriophage RNA ligase homologous to T4 RNA ligase 1. Nucleic Acids Res 31: 7247–7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T, Thorisdottir A, Unnsteinsdottir U, Hjorleifsdottir S, Aevarsson A, Ernstsson S, Fridjonsson OH, Skirnisdottir S, Wheat JO, Hermannsdottir AG, et al. 2005. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res 33: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermans HPJ, Ariyurek Y, van Ommen G, den Dunnen JT, 't Hoen PAC 2010. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics 11: 716 doi: 10.1186/1471-2164-11-716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Kano A, Maruyama A 2007. Activation of DNA strand exchange by cationic comb-type copolymers: effect of cationic moieties of the copolymers. Nucleic Acids Res 36: 342–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM 2009. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G, Wu R 1983. Terminal transferase: Use of the tailing of DNA and for in vitro mutagenesis. Methods Enzymol 100: 96–116 [DOI] [PubMed] [Google Scholar]
- England TE, Uhlenbeck OC 1978. 3′-terminal labelling of RNA with T4 RNA ligase. Nature 275: 560–561 [DOI] [PubMed] [Google Scholar]
- England TE, Gumport RI, Uhlenbeck OC 1977. Dinucleoside pyrophosphate are substrates for T4-induced RNA ligase. Proc Natl Acad Sci 74: 4839–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87–98 [DOI] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T 2008. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135: 1201–1214 [DOI] [PubMed] [Google Scholar]
- Farazi TA, Horlings HM, Ten Hoeve J, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, et al. 2011. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res doi: 10.1158/0008-5472.CAN-11-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, de Strooper B 2007. Molecular biology. miRNAs in neurodegeneration. Science 317: 1179–1180 [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S 2002. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc Natl Acad Sci 99: 12709–12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Wang LK, Lima CD, Shuman S 2004. Structure and mechanism of RNA ligase. Structure 12: 327–339 [DOI] [PubMed] [Google Scholar]
- Hofacker IL 2003. Vienna RNA secondary structure server. Nucleic Acids Res 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt EA, Goulding AM, Deo SK 2009. Direct detection and quantification of microRNAs. Anal Biochem 387: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S 2010. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Kim WJ, Sato Y, Akaike T, Maruyama A 2003. Cationic comb-type copolymers for DNA analysis. Nat Mater 2: 815–820 [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M 2005. Silencing of microRNAs in vivo with “antagomirs.” Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39: 673–677 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latronico MV, Condorelli G 2009. MicroRNAs and cardiac pathology. Nat Rev Cardiol 6: 419–429 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Turner DH 2006. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol 16: 270–278 [DOI] [PubMed] [Google Scholar]
- Miyoshi D, Sugimoto N 2008. Molecular crowding effects on structure and stability of DNA. Biochimie 90: 1040–1051 [DOI] [PubMed] [Google Scholar]
- Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN 2008. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol 181: 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo DB, Robb GB 2010. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 16: 2537–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire AZ 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, Fire AZ 2007. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res 35: e130 doi: 10.1093/nar/gkm760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM 2008. DNA and RNA ligases: structural variations and shared mechanisms. Curr Opin Struct Biol 18: 96–105 [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C 2008. Real-time PCR quantification of precursor and mature microRNA. Methods 44: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, Thor T, Vandesompele J, Eggert A, Schreiber S, et al. 2010. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res 38: 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber R, Malathi VG, Hurwitz J 1972. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci 69: 3009–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ 2008. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev VA, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vigneault F, Sismour AM, Church GM 2008. Efficient microRNA capture and bar-coding via enzymatic oligonucleotide adenylation. Nat Methods 5: 777–779 [DOI] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- von Ahsen N, Wittwer C, Schütz E 2001. Oligonucleotide melting temperatures under PCR conditions: Nearest-neighbor corrections for Mg2+, deoxynucleotide triphosphate, and dimethyl sulfoxide concentrations with comparison to alternative empirical formulas. Clin Chem 47: 1956–1961 [PubMed] [Google Scholar]
- Walker GC, Uhlenbeck OC, Bedows E, Gumport RI 1975. T4-induced RNA ligase joins single-stranded oligoribonucleotides. Proc Natl Acad Sci 72: 122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ach RA, Curry B 2007. Direct and sensitive miRNA profiling from low-input total RNA. RNA 13: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Schlabach MR, Hannon GJ, Elledge SJ 2009. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc Natl Acad Sci 106: 2289–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Ho CK, Shuman S 2003. Structure–function analysis of T4 RNA ligase 2. J Biol Chem 278: 17601–17608 [DOI] [PubMed] [Google Scholar]