Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1 (original) (raw)
. Author manuscript; available in PMC: 2011 Aug 28.
Published in final edited form as: Science. 2011 Jul 28;333(6046):1154–1157. doi: 10.1126/science.1206923
Abstract
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide. To explore the genetic origins of this cancer, we used whole exome sequencing and gene copy number analyses to study 32 primary tumors. Tumors from patients with a history of tobacco use had more mutations than did tumors from patients who did not use tobacco, and tumors that were negative for human papilloma virus (HPV) had more mutations than did HPV-positive tumors. Six of the genes that were mutated in multiple tumors were assessed in up to 88 additional HNSCCs. In addition to previously described mutations in TP53, CDKN2A, PIK3CA and HRAS, we identified mutations in FBXW7 and NOTCH1. Interestingly, nearly 40% of the 28 mutations identified in NOTCH1 were predicted to truncate the gene product, suggesting that NOTCH1 may function as a tumor suppressor gene rather than an oncogene in this tumor type.
More than half a million new cases of head and neck squamous cell carcinoma (HNSCC) will occur in 2011, including 50,000 cases in the United States, making it the sixth most common cancer in the world (1–3). HNSCC and its treatment can result in cosmetic deformity and functional impairment of vital functions, including breathing, swallowing, speech, phonation, taste, hearing and smell. These cancers are frequently lethal, with a five-year survival of only ~50% (4). HNSCCs, like all solid tumors, are thought to be initiated and to progress through a series of genetic alterations. Indeed several cellular signaling pathways are dysregulated in this tumor type through genetic and epigenetic alterations, such as those involving TP53 and CDK2NA (4). HNSCCs also exhibit many chromosomal abnormalities, including amplifications of region 11q13 containing the cyclin D1 gene and region 7p11 encoding EGFR (5). Tobacco use and excessive alcohol consumption are major risk factors for HNSCC in the United States (6). More recently, human papilloma virus (HPV) has emerged as an additional risk factor for the development of cancers of the oropharynx (7). Patients with HPV-associated cancers have an improved overall and disease-specific survival, suggesting that these tumors have distinct biological features (8).
To gain a comprehensive view of the genetic alterations underlying HNSCC, we sequenced ~18,000 protein-encoding genes in tumors from 32 patients. Thirty of the thirty-two patients had not been treated with chemotherapy or radiation prior to their tumor biopsy, so the spectrum of changes we observed largely reflects those of tumors in their naturally occurring state. Tumor samples were carefully selected or microdissected to achieve a neoplastic cellularity of >60%. DNA was purified from these tumors as well as matched non-neoplastic tissue and used to generate libraries suitable for massively parallel sequencing. After capture of the coding sequences with a SureSelect (Agilent) or CCDS (Nimblegen) Enrichment System, the DNA was sequenced using an Illumina GAIIx/HiSeq (17 tumors) or SOLiD V3/V4 (15 tumors) instruments. The average coverage of each base in the targeted regions was 77-fold and 44-fold for the Illumina and SOLiD instruments, and 92.6% and 90% of targeted bases were represented by at least 10 reads in these platforms, respectively (table S1).
Using stringent criteria for analysis of these data (9) we identified 911 candidate somatic mutations in 725 genes among the 32 tumors. To ensure that our algorithms for identifying mutations were reliable, we evaluated the candidate mutations by Sanger sequencing or by 454 sequencing and confirmed 609 of them (67%)(table S2). One hundred and fifty two (17%) mutations did not confirm and 150 (16%) mutations could not be tested because of an unusually high GC content, difficulty in the design of unique primers, or other unknown factors preventing specific amplification and sequencing of the locus. The range of confirmed mutations per tumor was 2 to 78, with a mean and standard deviation of 19 ± 16.5 mutations per tumor (table S1).
There were obvious differences in the genetic landscapes of HPV-associated and HPV-negative HNSCCs. First, far fewer genes were mutated per tumor in the HPV-associated tumors compared to those tumors not epidemiologically related to HPV (table S3A, 4.8 ± 3 versus 20.6 ± 16.7, p < 0.05, Welch two sample t-test). The difference in the number of mutations between HPV-associated and HPV-negative tumors was independent of smoking status. Second, TP53 mutations were not identified in any of the HPV-associated tumors but were found in 78% of the HPV-negative tumors. These data are consistent with previous results on HNSCC as well as HPV-associated cervical cancers (10–12).
As expected, more mutations were identified in tumors from patients with a history of tobacco use compared with those from patients who did not use tobacco (table S3B, 21.6 ± 17.8 versus 9.5 ± 6.5, p<0.05, Welch two sample t-test). Surprisingly, and in contrast to data from lung cancer, the mutational spectrum was not enriched for G:C>T:A transversions in those tumors associated with smoking (table S4). Nearly all of the HNSCC tumors analyzed had a mutation spectrum similar to that of non-smoking related lung cancers and other non-smoking related tumors. These data suggest that the effects of tobacco on the mutational spectra vary among different tumor types.
We selected genes for further analysis if they or closely related genes were altered in at least two of the 32 tumors sequenced. The genes included were PIK3AP1, RIMBP2, SI, NRXN2, NRXN3, EPHA7, RASA1, RXFP3, PIK3CA, HRAS, TP53, CDKN2A, NOTCH1, and FBXW7 (table S2). We then analyzed the sequences of these genes in additional HNSCC and their corresponding normal tissues (9). In total, somatic mutations in TP53, NOTCH1, CDKN2A, PIK3CA, FBXW7, and HRAS were identified in 47%, 15%, 9%, 6%, 5% and 4% of patients, respectively (table S5). The remaining genes were not observed to be mutated in more than one of the additional samples analyzed. Of the 63 TP53 mutations in the 120 samples analyzed (32 in the Discovery Set and 88 in the Prevalence set), 25 were predicted to be inactivating mutations [ten nonsense, ten insertions or deletions (“indels”), five splice-site mutations], and 38 were missense. Two of these were homozygous, that is, there was no remaining normal allele, and seven tumors demonstrated two mutations in TP53, likely representing inactivation of both alleles. In the other samples, the presence of contaminating DNA from non-neoplastic cells may have made it difficult to reliably distinguish heterozygous from homozygous changes. There were 11 CDKN2A mutations observed among the 120 tumors analyzed, of which nine were definitely inactivating (3 nonsense, 4 indels, and 2 splice site) and 2 were missense. The frequency and types of mutations we observed in TP53 and CDKN2A, as well as in HRAS and PIK3CA, were consistent with previous studies of HNSCC (4).
A total of 28 NOTCH1 mutations were identified. Seven of 21 patients with NOTCH1 mutations had two independent mutations, presumably on different alleles. Eleven of the NOTCH1 mutations were predicted to truncate the protein product (7 nonsense, 4 indels), whereas 17 were missense (Tables 1 and 2). Next to TP53, NOTCH1 was the most frequently mutated gene found in the combined Discovery and Prevalence set, with alterations present in 15% of patients. To date, NOTCH1 has not been reported to be mutated at a significant frequency in other solid tumor types (13).
Table 1.
Truncating mutations in NOTCH1. Samples in bold were used for the initial (Discovery Set) screen. Underlined samples were from Human Genome Sequencing Center and the remaining samples were from Johns Hopkins University. OC, oral cavity; OP, oropharynx; L, larynx; N, no; Y, yes; WT, wild-type; unk, unknown; na, not applicable; +, positive.
| Sample | Site | Tobacco | TP53 status | HPVstatus | Amino acid (protein) | Mutation type |
|---|---|---|---|---|---|---|
| HN12PT | OC | Y | WT | na | p.W1843X | Nonsense |
| 385 | OC | N | Mutant | na | p.R1984X | Nonsense |
| 600 | OC | Y | WT | unk | Q290X | Nonsense |
| HN 105 PT | OC | Y | WT | na | fs | Indel |
| HN 107 PT | OC | Y | Mutant | na | p.S402X | Nonsense |
| HN 115 PT | OC | Y | Mutant | na | fs | Indel |
| HN 130 PT | L | Y | WT | na | p.E949X | Nonsense |
| HN 139 PT | OP | Y | Mutant | unk | p.C554X | Nonsense |
| HN 183 PT | OC | unk | Mutant | na | p.Q1958X | Nonsense |
| HN 194 PT | OC | N | Mutant | na | fs | Indel |
| HN 245 PT | OP | Y | WT | + | fs | Indel |
Table 2.
Missense mutations in NOTCH1. Samples in bold were used for the initial (Discovery Set) screen. Underlined samples were from Human Genome Sequencing Center and the remaining samples were from Johns Hopkins University. OC, oral cavity; OP, oropharynx; HP, hypopharynx; N, no; Y, yes; WT, wild-type; unk, unknown; na, not applicable; +, positive.
| Sample | Site | Tobacco | TP53 status | HPVstatus | Amino acid (protein) |
|---|---|---|---|---|---|
| HN14PT | OC | N | Mutant | na | p.P391S |
| 478 | OC | N | WT | na | p.G484V |
| HN 102 PT | OC | N | WT | na | p.E450K |
| HN 105 PT | OC | Y | WT | na | p.G812W |
| HN 115 PT | OC | Y | Mutant | na | p.G1340A |
| HN 117 PT | OC | N | WT | na | p.G310R |
| HN 117 PT | OC | N | WT | na | p.C456R |
| HN 142 PT | OP | Y | WT | + | p.R353H |
| HN 142 PT | OP | Y | WT | + | p.M2011R |
| HN 148 PT | OP | Y | Mutant | unk | p.G1638V |
| HN 148 PT | OP | Y | Mutant | unk | p.P2272S |
| HN 208 PT | OC | Y | Mutant | na | p.R365C |
| HN 208 PT | OC | Y | Mutant | na | p.R1280C |
| HN 227 PT | HP | Y | Mutant | na | p.R365C |
| HN 251 PT | OP | N | WT | + | p.F1292L |
| HN 255 PT | OP | Y | WT | unk | p.V2039L |
| HN 255 PT | OP | Y | WT | unk | p.V2039E |
FBXW7 mutations have not been previously observed in HNSCC, though they are frequent in other tumor types. Of the 6 FBXW7 mutations we identified, two were indels and the other four were missense; none were homozygous. Although both alleles of most tumor suppressor genes are mutated in tumors in which they play a role, FBXW7 is an exception. FBXW7 is a member of the F-box protein family and constitutes a component of the ubiquitin protein ligase complex. It acts as a tumor suppressor in several tumors and one of its major targets is NOTCH1, which it targets for degradation. The FBXW7 mutations we observed were in a hotspot known to block the degradation of active NOTCH1 (14). It is attractive to hypothesize that FBXW7 mutations are modulating the Notch pathway, although FBXW7 also targets other cancer related proteins for degradation, including cyclin E and c-myc. Further studies will be necessary to elucidate the function of these FBXW7 mutations in HNSCC.
To complement the sequencing data, we performed copy number analysis with Affymetrix SNP6.0 microarrays on 42 tumor and normal sample pairs, including 25 of the samples used for massively parallel sequencing. We found that the most frequently mutated genes often were affected by copy number changes. For example, loss of heterozygosity (LOH) was observed in the tumor samples with mutations in TP53, and CDKN2A was frequently deleted (table S6). In addition, LOH at the NOTCH1 locus was detected in two of the three tumors with NOTCH1 mutations that were analyzed for copy number. Given that 7 other tumors in our cohort had two NOTCH1 mutations, inactivation of both alleles likely occurred in at least 9 of the 21 patients with NOTCH1 mutations. These data support the idea that NOTCH1 acts as a tumor suppressor in HNSCC. Additionally, recurrent gains and losses were observed in several specific regions of the genome (table S7 and figs. S1 to S6). Recurrent focal changes were identified by filtering for segments with more than 3 copies or losses of 1 or more copies in at least 3 samples. The focal changes included deletions of 9p21.3 (containing CDKN2A) and amplifications of small segments on 11q, 3q and 7p. These regions have previously been implicated in HNSCC and contain known oncogenes (CCND1, PIK3CA and EGFR, respectively).
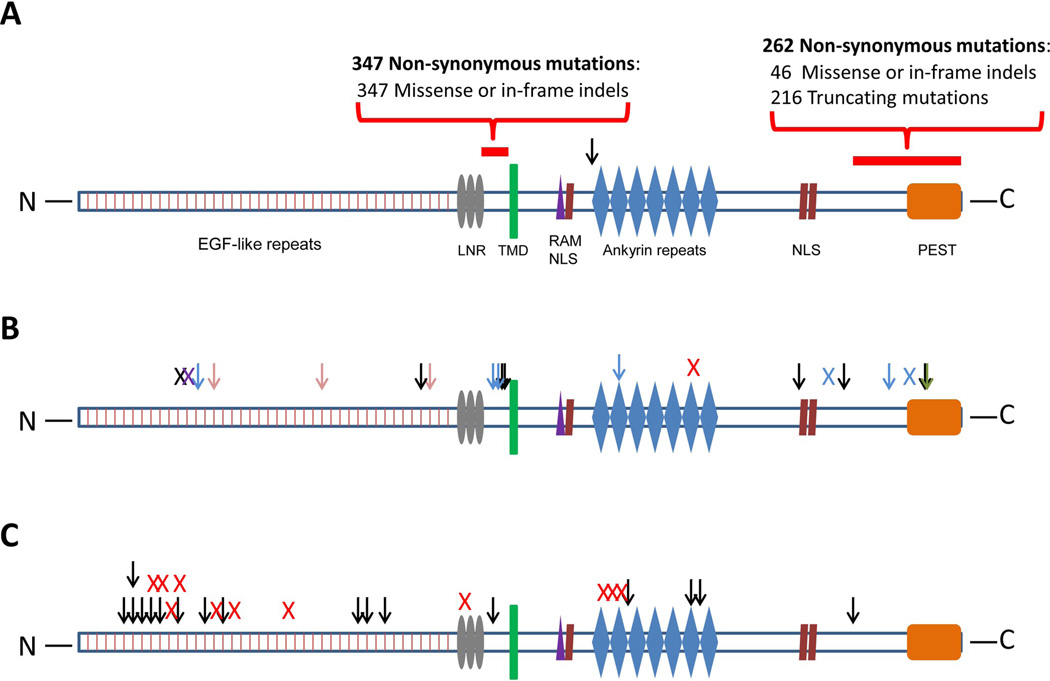
Our data raise intriguing questions about the role of NOTCH1 in tumorigenesis. An involvement of NOTCH1 in human cancers was first demonstrated through the discovery of translocations in T-cell leukemias (15). Subsequently, more subtle mutations of NOTCH1 were identified in a variety of hematopoietic tumors. Most of the mutations in hematopoietic tumors are clustered in two hot spots in the heterodimerization (HD) and C-terminal polypeptide enriched proline, glutamate, serine and threonine (PEST) domain (Fig. 1A)(14). Exogenous expression of these mutants, as well as of the translocated NOTCH1, lead to neoplastic transformation in vitro and in vivo. In contrast, while this manuscript was under review, Klinakis et.al. showed that reduced activity of the Notch signaling pathway is associated with the development of chronic myelomonocytic leukemia (CMML), suggesting a tumor suppressor role for Notch (16). Additionally, a small number of mutations, some truncating, have been previously observed in solid tumors (Fig. 1B) (17). This pattern is consistent with a suppressor gene rather than an oncogene function, but the mutations were present in only a small fraction of any individual tumor type and were difficult to distinguish from passenger mutations. In contrast, the relatively large number of mutations we observed in HNSCC strongly implicates them as drivers (P < 10−8)(8). Moreover, the spectrum of NOTCH1 mutations we observed in HNSCC was fundamentally different from those identified in hematopoietic tumors, as a high fraction were in the N-terminal EGF-like ligand binding domain and the majority of the mutations were predicted to alter the protein N-terminal to the transmembrane region (Fig. 1C). The location and nature of these alterations, together with the observation that two NOTCH1 mutations were found in each of seven patients, provides strong genetic evidence that NOTCH1 often acts as a tumor suppressor gene in HNSCC. This interpretation is consistent with functional studies of the role of NOTCH1 in squamous epithelial cells, as _NOTCH1_−/− mice develop epithelial tumors (18). The function of NOTCH1 in cancer - oncogene in some leukemias and tumor suppressor gene in CMML, HNSCC, and perhaps other cancers - may recapitulate its dual role in normal biology, where activation can lead to stem cell maintenance in some tissues but terminal differentiation in others (19). These results also emphasize the importance of assessing the functional attributes of cancer-associated mutations in a cell-type specific fashion, either in tissue culture or in model organisms.
Fig. 1.
Schematic depiction of mutations in NOTCH1. (A) Previously observed NOTCH1 mutations in hematopoietic malignancies. EGF = Epidermal growth factor, LNR = Lin12-Notch repeats, TMD = trans-membrane domain, RAM = Recombination signal-binding protein 1 for J-Kappa (_RBP_jk) association module, NLS = nuclear localization signal, PEST = proline, glutamic acid, serine/threonine-rich motifs. Red bars represent previously described mutation hotspots (amino acids 1575–1630 and 2250–2550). (B) Previously observed NOTCH1 mutations in solid tumors. Colored arrow (missense mutation) and “X” (truncating mutation) depict mutations found in different tumor types: pink = breast cancer, black = glioma, blue = lung cancer, green = pancreatic adenocarcinoma, red = esophageal squamous cell carcinoma, purple = tongue squamous cell carcinoma. (C) Mutations in NOTCH1 in HNSCC observed in this study. Black arrow = missense mutation, red “X” = truncating mutation (13, 22–24).
The results of this study provide evidence that HNSCCs, although morphologically similar, are comprised of distinct diseases at the molecular level. The different genetic landscapes associated with HPV and tobacco exposure are consistent with clinical and epidemiologic data that has suggested the importance of these environmental factors in prognosis and response to therapeutic interventions. Another important observation is that only 18% of the 28 tumors not associated with HPV from the Discovery set had activating mutations in a bona fide oncogene. In contrast, 89% of these tumors harbored inactivating mutations in at least one bona fide tumor suppressor gene. This distinction is critical because the new generation of molecularly-targeted therapies is directed toward activated oncogenes but such drugs cannot directly target mutated tumor suppressor genes because they are already inactivated. Tumor suppressor gene predominance is not limited to HNSCC as exomic analysis of other tumor types, such as renal cell carcinomas and pancreatic endocrine neoplasms, have revealed similar patterns (20, 21). Our finding that HNSCCs have few directly targetable mutations has implications for controlling this disease in the future. In particular, it suggests that prevention, careful surveillance of patients at risk, and early detection are the optimal approaches for reducing morbidity and mortality from this disease.
Supplementary Material
Supplementary Data
Acknowledgements
We thank our patients for their courage and generosity. We thank N. Silliman, J. Ptak, M. Whalen, L. Dobbyn, J. Schaeffer, X. Li, O. Folawiyo and Z. Khan for expert technical assistance. This work was supported by the National Institutes of Health/National Institute of Dental and Craniofacial Research grants RC2DE020957, RC2DE020958 and P50DE019032, as well as NIH grants CA121113, CA43460 and CN43302, NIH Specialized Program of Research Excellence Grant P50CA097007, Cancer Prevention Research Institute of Texas Grant RP100233, AACR Stand Up To Cancer-Dream Team Translational Cancer Research Grant, and the Virginia and D.K. Ludwig Fund for Cancer Research. Under agreements between the Johns Hopkins University, Genzyme, Exact Sciences, Inostics, Qiagen, Invitrogen and Personal Genome Diagnostics, N.P., B.V., K.W.K. and V.E.V are entitled to a share of the royalties received by the University on sales of products related to genes and technologies described in this manuscript. N.P., B.V., K.W.K. and V.E.V are co-founders of Inostics and Personal Genome Diagnostics are members of their Scientific Advisory Boards, and own Inostics and Personal Genome Diagnostics stock, which is subject to certain restrictions under Johns Hopkins University policy. J.C. is the Director of Research of the Milton J. Dance Head and Neck Endowment. C. B. is a recipient of T32 CA009574 NIH/NCI Inst. National Research Service Award. C. R. P. is a TRIUMPH Fellow and supported by the GSK Translational Research Fellowship. This paper is based on a web database application provided by Research Information Technology Systems (RITS) https://www.rits.onc.jhmi.edu/.
Footnotes
The terms of these arrangements are managed by the Johns Hopkins University in accordance with its conflict-of-interest policies.
References and Notes
- 1.Haddad RI, Shin DM. N Engl J Med. 2008 Sep 11;359:1143. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. CA Cancer J Clin. 2010 Sep–Oct;60:277. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. CA Cancer J Clin. 2005 Mar–Apr;55:74. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Leemans CR, Braakhuis BJ, Brakenhoff RH. Nat Rev Cancer. 2011 Jan;11(9) doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 5.Berenson JR, Yang J, Mickel RA. Oncogene. 1989 Sep;4:1111. [PubMed] [Google Scholar]
- 6.Brennan JA, et al. N Engl J Med. 1995 Mar 16;332:712. doi: 10.1056/NEJM199503163321104. [DOI] [PubMed] [Google Scholar]
- 7.D'Souza G, et al. N Engl J Med. 2007 May 10;356:1944. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 8.Ang KK, et al. N Engl J Med. 2010 Jul 1;363:24. [Google Scholar]
- 9.Materials and methods are available as supporting material on Science Online
- 10.Klussmann JP, et al. Clin Cancer Res. 2009 Mar 1;15:1779. doi: 10.1158/1078-0432.CCR-08-1463. [DOI] [PubMed] [Google Scholar]
- 11.Hafkamp HC, et al. Int J Cancer. 2003 Nov 10;107:394. doi: 10.1002/ijc.11389. [DOI] [PubMed] [Google Scholar]
- 12.Balz V, et al. Cancer Res. 2003 Mar 15;63:1188. [PubMed] [Google Scholar]
- 14.Baldus CD, et al. Haematologica. 2009 Oct;94:1383. doi: 10.3324/haematol.2008.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellisen LW, et al. Cell. 1991 Aug 23;66:649. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 16.Klinakis A, et al. Nature. May 12;473:230. [Google Scholar]
- 17.Westhoff B, et al. Proc Natl Acad Sci U S A. 2009 Dec 29;106:22293. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas M, et al. Nat Genet. 2003 Mar;33:416. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 19.Roy M, Pear WS, Aster JC. Curr Opin Genet Dev. 2007 Feb;17:52. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Varela I, et al. Nature. 2011 Jan 27;469:539. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao Y, et al. Science. 2011 Mar 4;331:1199. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aster JC, et al. J Biol Chem. 1997 Apr 25;272:11336. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 23.Kopan R, Ilagan MX. Cell. 2009 Apr 17;137:216. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen LY, et al. Cell Death Differ. 2005 Mar;12:243. doi: 10.1038/sj.cdd.4401568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data