Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease (original) (raw)
. Author manuscript; available in PMC: 2012 Oct 1.
Abstract
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder that threatens to reach epidemic proportions as our population ages. Although much research has examined molecular pathways associated with AD, relatively few such studies have focused on the disease’s critical early stages. In a prior microarray study we correlated gene expression in hippocampus with degree of Alzheimer’s disease and found close associations between upregulation of apparent glial transcription factor/epigenetic/tumor suppressor genes and incipient AD. The results suggested a new model in which AD pathology spreads along myelinated axons (Blalock et al., 2004). However, the microarray analyses were performed on RNA extracted from frozen hand-dissected hippocampal CA1 tissue blocks containing both gray and white matter, limiting the confidence with which transcriptional changes in gray matter could be distinguished from those in white matter. Here, we used laser capture microdissection (LCM) to exclude major white matter tracts while selectively collecting CA1 hippocampal gray matter from formalin-fixed, paraffin-embedded (FFPE) hippocampal sections of the same subjects assessed in our prior study. Microarray analyses of this gray matter-enriched tissue revealed many transcriptional changes similar to those seen in our past study and in studies by others, particularly for downregulated neuron-related genes. Additionally, the present analyses identified several previously undetected pathway alterations, including downregulation of molecules that stabilize ryanodine receptor Ca2+ release and upregulation of vasculature development. Conversely, we found a striking paucity of the upregulated changes in the putative glial and growth-related genes that had been strongly overrepresented in the prior mixed-tissue study. We conclude that FFPE tissue can be a reliable resource for microarray studies of brain tissue, that upregulation of growth-related epigenetic/transcription factors during incipient AD is predominantly localized in and around white matter (supporting our prior findings and model), and that novel alterations in vascular and ryanodine receptor-related pathways in gray matter are closely associated with incipient AD.
Keywords: Cognitive impairment, ageing, neurodegeneration, neurofibrillary tangles, myelin, glia
1. Introduction
The major pathological hallmarks of Alzheimer’s disease (AD) include beta amyloid accumulation (Hardy and Selkoe, 2002; Klein et al., 2001; Morgan, 2003; Mucke et al., 2000; Mullan and Crawford, 1994; Price and Sisodia, 1998; Tanzi and Bertram, 2001), neurofibrillary tangles (NFTs) (Davies and Koppel, 2009; Johnson and Bailey, 2002; Morris et al., 2011; Noble et al., 2003) and synaptic dysfunction or loss (Masliah et al., 1994; Scheff and Price, 2001; Sze et al., 1997; Yao et al., 2003). In addition, AD is associated with other pathological processes, including failing mitochondrial function and oxidative stress (Bickford et al., 2000; Butterfield and Sultana, 2007; Perry et al., 2003; Wang et al., 2010), increased inflammatory response (Finch et al., 2002; Gemma et al., 2002; Ginsberg et al., 2006; Mrak and Griffin, 2001; Mucke et al., 2000; Rogers et al., 1996), protein misfolding (Forman et al., 2003; Stefani and Dobson, 2003), altered growth factor signaling (Mufson et al., 2007; Tuszynski and Gage, 1990; Williams et al., 2006), aberrant reentry of neurons into the cell cycle (Arendt et al., 2000; Bowser and Smith, 2002), lysosomal activation (Ginsberg et al., 2010; Nixon et al., 2001), endocrine alteration (Brinton, 2008; Landfield et al., 2007; Simpkins et al., 2005; Sohrabji and Lewis, 2006), insulin resistance (Craft, 2007; Gustafson, 2006; Whitmer et al., 2007; Yaffe et al., 2004), cholesterol dyshomeostasis (Petanceska et al., 2002; Puglielli et al., 2001), and calcium dysregulation. The latter plays an important role in normal brain aging as well as in some models of AD (Bezprozvanny and Mattson, 2008; Disterhoft et al., 1994; Foster and Norris, 1997; Gibson and Peterson, 1987; Khachaturian, 1989; Landfield, 1987; Michaelis et al., 1996; Nixon et al., 1994; Norris et al., 1998; Stutzmann, 2005; Stutzmann et al., 2007; Thibault et al., 2007; Wang et al., 2010), and may arise in part from a decrease in immunophilin-mediated stabilization of ryanodine receptors (Gant et al., 2011).
The complexity and number of changes associated with AD has impeded attempts to disentangle the processes important for pathogenesis or to define the roles of specific cell types in disease progression. Microarray analysis of the simultaneous expression of thousands of genes is well-suited to address complex processes and has been used effectively to provide overviews of the gene networks disturbed in AD (Colangelo et al., 2002; Ginsberg et al., 2006; Ginsberg et al., 2000; Liang et al., 2008; Loring et al., 2001; Pasinetti, 2001; Wang et al., 2010; Yao et al., 2003). Nevertheless, expression profiles vary substantially across cell types and regions (Bishop et al., 2010; Blalock et al., 2010; Burger, 2010; Ginsberg et al., 2006; Zahn et al., 2007; Zhao et al., 2001), and it is not yet clear how the expression signatures of specific cell/tissue types are related to early-stage AD.
In a prior study, we combined microarray technology with statistical correlation analyses to identify hippocampal gene expression changes associated with cognitive dysfunction and neurofibrillary tangles across a spectrum from incipient to severe AD (Blalock et al., 2004). Results of that study led us to propose that incipient AD was in part driven by aberrant activation of growth factors in oligodendrocytes and myelinated fiber tracts and, in turn, suppressive responses in other cell types (Blalock et al, 2004). However, because our study was conducted on hand-dissected frozen hippocampal CA1 blocks containing both white and gray matter, it was not possible to clearly distinguish their transcriptional changes. Further, when heterogeneous cell types/regions are mixed together, alterations in one component may oppose or dilute those in another, potentially obscuring important tissue-specific changes.
To overcome tissue heterogeneity problems, laser capture microdissection (LCM) technology and other techniques to isolate specific subregions and individual cell types have been employed in several AD studies (Ginsberg et al., 2010; Ginsberg et al., 2006; Liang et al., 2008). Here, we used a similar LCM approach to address some of the tissue-specific questions that were not clearly resolved in our prior study. In the present LCM study, we laser dissected formalin-fixed, paraffin-embedded (FFPE) sections from the same subjects (N = 30) analyzed in our earlier study (Blalock et al., 2004) and captured hippocampal CA1 gray matter (neuropil/neuronal somata) regions while largely excluding prominent white matter tracts (the perforant path, fimbria and alveus). Thus, this re-sampling of the same subjects provided a unique opportunity to directly compare results from similar samples containing or lacking significant white matter components, as well as to compare data from two disparate tissue fixation/collection approaches. If RNA integrity were generally comparable across these methods, we predicted many findings would correspond between the two studies, as both methods collected neuropil. However, we also hypothesized that, compared to our past analysis of white-matter-enriched hand-dissected CA1 tissue, the present analysis of white-matter-sparse tissue should detect fewer upregulated AD-associated genes, while also uncovering some previously obscured neuronal-specific genes/processes. The data reported here support these hypotheses, strengthening our model of white matter pathogenesis and revealing novel gray matter alterations with potential mechanistic and/or therapeutic implications.
2. Materials and Methods
2.1 Subjects, specimen preparation
Descriptions and categorization of the subjects were reported previously (Blalock et al, 2004). Brain sections from a total of 30 of the original 31 subjects were analyzed here. As in the prior study, we correlated gene expression with quantitative values on two AD marker scales (MiniMental Status Exam (MMSE) and neurofibrillary tangle (NFT) density), irrespective of clinical diagnosis. This strategy relied on quantitative markers to assess extent of disease progression rather than on categorization by clinico-pathologic criteria, which can be uncertain in borderline cases or when diagnostic markers disagree. However, for comparison and validation, the 30 subjects were also categorized into four categories of varying AD severity. Briefly, subjects (11 male, 19 female; average age 86.3 ± 1.4 years) were separated into four categories (control n = 8; incipient n = 7; moderate n = 8; severe n = 7) largely based on MMSE results (control- 27.6 ± 0.6; incipient- 24.3 ± 1.1; moderate- 16.5 ± 0.6; severe- 6.0 ± 1.4). For borderline cases, NFT count (control- 3.0 ± 1.1; incipient- 17.5 ± 8.2; moderate- 25.6 ± 3.5; severe- 32.7 ± 3.2) and Braak staging (control- 2.3 ± 0.4; incipient- 5.0 ± 0.5; moderate- 5.5 ± 0.2; severe- 5.8 ± 0.2) also informed categorization decisions.
For this study, formalin fixed, paraffin embedded (FFPE) specimens were used. Post mortem interval (PMI: 3.7 ± 0.6 hours); duration of direct formalin exposure (4.0 ± 1.2 days); and time in paraffin block (7.3 ± 0.2 years) did not differ significantly among groups (p > 0.05 in all tests; 1-ANOVA). Eight-μm sections from FFPE hippocampal blocks were mounted on PALM 1mm polyethylene naphthalate (PEN) membrane slides (Zeiss, Germany). Slides were de-waxed (2 × 10 min xylene, 2 × 10 min 100% EtOH, 1 × 10 min 90% EtOH, 1 × 10 min 70% EtOH, 1 × dH20 5 min) and dehydrated (70% EtOH 10 min, 90 % EtOH 2 min, 2 × 100% EtOH 3 & 5 min, 2 × 100% xylene 5 & 10 min) to facilitate dissection and laser capture. Neuropathologists defined the CA1 region for each specimen using photomicrographs.
2.2 Laser Capture Microdissection
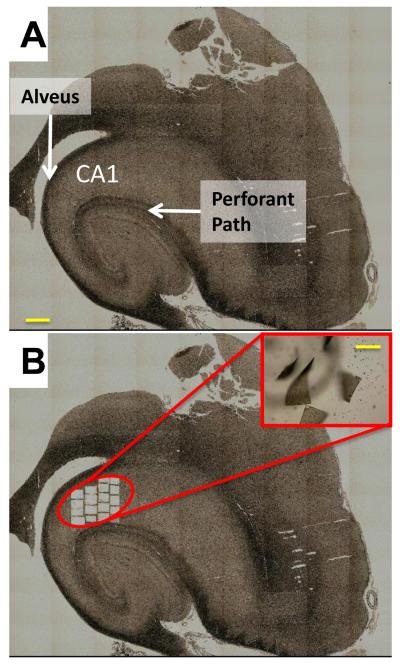
A Zeiss AxioObserver PALM Microbeam with RoboMover cap system was used to image, cut and capture specimens (Woods Hole, MA; courtesy of Zeiss- Fig. 1). The pathologist-identified CA1 region for each specimen (Fig. 1A) was used as a guide during laser capture. Specimen sections were targeted using a 5x cutting objective and were collected from regions of grossly defined gray matter (as defined in The Human Brain, Nolte, 2002) in hippocampal sub-field CA1, comprising the pyramidal cell layer and surrounding neuropil, largely excluding regions containing major white matter tracts (e.g., fimbria, alveus and perforant path) (Fig. 1B). Note that gray matter still contains astrocytes, capillaries, and other non-neuronal cells, as well as myelinated fibers (Vercellino et al., 2009), albeit considerably fewer of the latter than found in whole tissue dissections. The laser was set to cut through the PEN membrane to which the specimens were adhered, and then defocused and activated to capture (using the light pressure catapult) the cut regions of each sample. After each catapult, the capture area (siliconized eppendorf-style 0.5 ml centrifuge tube cap- one per slide) was visualized (Fig. 1B inset) to validate collection. On average, ~20 individual capture attempts were performed on each specimen and 1-2 capture attempts failed per sample. Captured material was stored dry at room temperature prior to RNA extraction and analysis.
Figure 1. Representative images of laser capture process.
A. Reconstructed image of human hippocampal section of CA1 region before laser cut and capture process. Associated white matter tracts, which appear as dark bundles, are labeled (alveus, perforant path). Calibration bar (yellow) = 1 mm. B. Same section after cut and capture process. Note that sampling region largely excludes white matter tracts. Inset: Laser pressure micro-catapulted sections in the capture region. Calibration bar (yellow) = 300 μm.
2.3 RNA isolation and amplification
RNA was extracted using RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Ambion) according to manufacturer’s instructions (3h incubation at 55° C followed by glass fiber filtration). This system has recently been shown to outperform other FFPE methods/ kits regarding yield of amplifiable RNA (Okello et al., 2010). Quality assessment of extracted material was performed with the Paradise Reagent Quality Assessment Kit (Molecular Devices), as well as via NanoDrop (Thermoscientific). All samples yielded sufficient genetic material (>50 ng) for subsequent reactions. 50 ng of extracted purified nucleic acid underwent RNA amplification using WT-Ovation FFPE System (NuGen) followed by FL-Ovation cDNA Biotin Module V2 (NuGen) for labeling and microarray (Affymetrix HGU133 v2) hybridization. All 30 microarrays (one per each subject sample) performed within acceptable limits (Scaling factor: 32.6 +/− 3.7; RawQ: 1.28 +/− 0.01; GapDH 3′:5′: 1.48 +/− 0.08; % present 35.4 +/− 1.5) and were not significantly different across condition (p < 0.5 for all measures, 1-ANOVA). In comparison to prior work on frozen samples from the same subjects (Scaling factor: 5.9 +/− 0.6; RawQ 2.7 +/− 0.04; % present: 44.6 +/− 1.1), the increased scaling factor, decreased RawQ, and reduced % present all indicate reduced signal intensity, consistent with other reports of the dynamics of small FFPE sample extraction (Turner et al., 2011). Finally, the % present call, while lower than found in frozen tissue, is much greater than would be expected by chance (5%),suggesting that the extracted material contains substantial amounts of valid mRNA.
2.4 Microarray analysis and statistics
Probe sets were annotated, and transcriptional profiles were generated, using the MAS5 algorithm and annotation data sets (Affymetrix GCOS v. 1.1; HGU133 annotation October, 2003) in order to facilitate comparison with our prior work (Blalock et al., 2004). Raw data are available from the Gene Expression Omnibus (Barrett and Edgar, 2006) under accession ID GSE28146. Results were filtered for presence, redundancy, and annotation status and analyzed by Pearson’s test for correlation with each subject’s Mini-Mental Status Exam score and Neurofibrillary Tangle count. The false discovery rate (FDR) (Hochberg and Benjamini, 1990) was used to estimate the error of multiple testing’s contribution to False Positives (see FDR, Fig 2). The DAVID suite of bioinformatic tools (Huang da et al., 2009), which identifies functional categories and biological processes/pathways that are statistically overrepresented by genes of interest, was used to identify processes/pathways from the Gene Ontology (Ashburner et al., 2000) associated with the various lists of AD-correlated genes, using the ‘table cluster’ option. To reduce redundancy, only one representative pathway (p ≤ 0.05; between 3 and 100 genes) from within each cluster is reported.
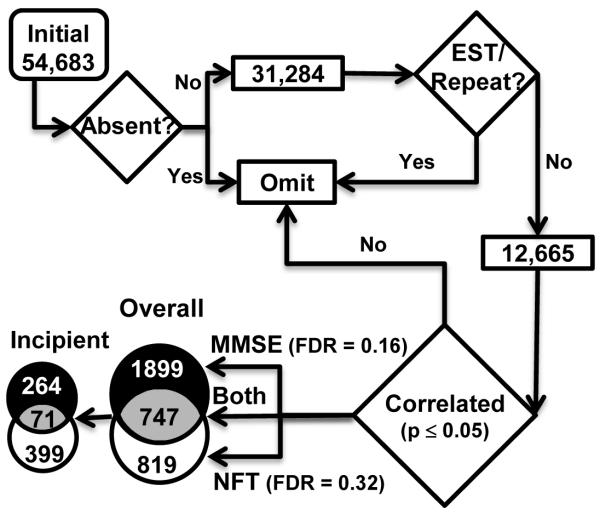
Figure 2. Flowchart of microarray correlation analysis procedure.
Pre-statistical filtering omitted probe sets with poor annotation and/or low quality signal. Each of the remaining genes was correlated (Pearson’s test) with MMSE scores and NFT counts across all 30 subjects (Overall correlation). If genes were found to correlate significantly with either marker overall, then a post-hoc correlation test across the subset of 15 subjects in the control and incipient groups (incipient correlation) was also performed. Numbers of significant genes were separated based on the AD marker (MMSE or NFT, or both) with which they were correlated. All genes significant in MMSE and/or NFT were used for subsequent functional overrepresentation analysis (Table 1).
3. Results
3.1 Microarray analyses in laser captured samples
The comprehensive laser dissection data set was acquired from formalin-fixed, paraffin-embedded microscope slide specimens (from the same subjects as the original frozen tissue block study). Quality control results (Methods 2.3) showed that the laser dissection samples averaged 35% presence calls, ~7x the presence call rate that would be expected by chance (5%). Further, concordance at the presence call level among the 30 laser dissection arrays was also quite high. Genes rated present/absent on a single microarray tended to agree across all arrays (78.4% agreement).
Microarray signal intensities and presence calls were transferred to Excel and analyzed as in prior work (Blalock et al., 2004) (Fig. 2). Briefly, results were filtered to remove probe sets that were rated absent (< 4 presence calls across 30 chips) or poorly annotated (lacking or redundant gene symbol). The remaining 12,665 genes were correlated (Pearson’s test) with MMSE scores as well as NFT counts across all subjects (Overall correlation). Genes with significant (p ≤ 0.05) overall correlations were also sub-correlated with MMSE and NFT across only the control and incipient subjects (Incipient correlation).
3.1.1 AD-related genes
Results for all significantly correlated genes are provided as supplementary information (Supplemental Table 1). As seen in Fig. 2, correlation with MMSE appeared to explain a greater proportion of transcriptional profile variability than correlation with NFT, although there was a strong, statistically significant overlap between the two (747 genes; p < 0.0001, binomial test). However, 470/ 1566 (30%) of the NFT-correlated genes were also correlated across the incipient and control groups, whereas only 335/ 2646 (12.6%) of MMSE-correlated genes showed a similar incipient AD correlation. Hence, a greater proportion of NFT-correlated genes were associated with early disease progression, while cognitive measures appeared more strongly tied to later disease stages.
3.1.2 AD-related functional categories and pathways
Table 1 shows the functional processes found by DAVID analysis to be overrepresented by either downregulated or upregulated overall AD-correlated genes. Downregulated processes were strongly associated with neuronal function (synaptic components, ion channel activity, neurotransmitter receptors, axon, Ca2+ signaling) and, to a lesser extent, with energy production (glycolysis, mitochondrial components) and cell development (differentiation, transport, and filament structure). Conversely, upregulated categories included the now-stereotypical inflammatory (inflammatory response and complement activation) and iron homeostasis responses often associated with glial activation, as well as strong apoptotic signals. These patterns of downregulated neuronal and mitochondrial processes in conjunction with upregulated inflammatory and apoptotic pathways are highly similar to results reported in our prior work analyzing AD-associated microarray signatures from excised blocks of frozen hippocampal CA1 tissue (Blalock et al, 2004), as well to results from a number of studies by others (Wang et al., 2010). All significantly correlated genes in the present study are listed in Supplemental Table 1.
Table 1. Functional categories/ processes overrepresented by AD-correlated genes identified in laser-captured hippocampal gray matter.
Categories/ processes shown are those significantly overrepresented (p ≤ 0.05) by genes correlated with AD markers in the overall AD and the incipient AD analyses. Categories are separated by direction of change and sorted by p-value. To reduce redundancy, only one representative functional category from each identified cluster of functions was selected. Processes not identified in the previous tissue block study include regulation of ryanodine-sensitive Ca2+ release and vasculature development. Column headings: Overall: results from genes significant in the overall correlation analysis; Incipient: results from genes also significant in the incipient correlation analysis; #: Number of genes significant; P- val: probability of overrepresentation; modified Fisher’s exact test p-value provided by DAVID bioinformatic analysis software. Supplemental Table 1 lists all significant genes.
| Overall | Incipient | |||
|---|---|---|---|---|
| Downregulated with AD | # | P- val | # | P- val |
| synapse | 73 | 1.25×10−10 | 17 | 0.0005 |
| synaptic transmission | 59 | 7.23×10−09 | 12 | 0.0171 |
| extracellular ligand-gated ion channel activity | 15 | 2.77×10−05 | 04 | 0.0469 |
| axon | 33 | 2.97×10−05 | 09 | 0.0070 |
| synaptic vesicle | 19 | 9.55×10−05 | 06 | 0.0090 |
| ion transmembrane transporter activity | 84 | 1.22×10−04 | 18 | 0.0348 |
| regulation of neuron differentiation | 26 | 3.35×10−04 | 09 | 0.0026 |
| cation-transporting ATPase activity | 12 | 4.09×10−04 | 04 | 0.0298 |
| glycolysis | 12 | 6.89×10−04 | 04 | 0.0372 |
| clathrin-coated vesicle | 24 | 0.0030 | 08 | 0.0084 |
| regulation of transport | 47 | 0.0038 | 12 | 0.0395 |
| mitochondrial part | 76 | 0.0113 | 18 | 0.0364 |
| GABA-A receptor activity | 06 | 0.0263 | 03 | 0.0443 |
| *regulation of ryanodine-sensitive calcium-release channel activity | 04 | 0.0414 | 03 | 0.0106 |
| intermediate filament organization | 04 | 0.0414 | 03 | 0.0106 |
| mitochondrial membrane part | 20 | 0.0419 | 08 | 0.0079 |
| Upregulated with AD | ||||
| *vasculature development | 48 | 2.62×10−06 | 14 | 0.0165 |
| negative regulation of developmental process | 46 | 8.26×10−06 | 14 | 0.0140 |
| positive regulation of programmed cell death | 68 | 2.11×10−04 | 22 | 0.0103 |
| serine-type endopeptidase inhibitor activity | 14 | 0.0049 | 06 | 0.0232 |
| inflammatory response | 37 | 0.0060 | 14 | 0.0140 |
| iron ion homeostasis | 09 | 0.0091 | 05 | 0.0105 |
| complement activation | 08 | 0.0114 | 04 | 0.0363 |
| apoptosis | 80 | 0.0140 | 29 | 0.0071 |
| induction of apoptosis by extracellular signals | 22 | 0.0142 | 10 | 0.0088 |
| actin cytoskeleton organization | 37 | 0.0148 | 13 | 0.0448 |
| response to vitamin | 13 | 0.0207 | 06 | 0.0310 |
Nonetheless, some notable differences were found between processes identified in this study vs. those identified in our prior study. Among the most prominent was the present study’s paucity, relative to the prior study, of upregulated genes related to growth, tumor suppression, transcription, chromatin remodeling, lipid metabolism, extracellular matrix, cytoskeletal organization, oxidative stress, and some immune processes. For downregulated processes, the absence here of ubiquitin-dependent protein catabolic categories was particularly evident. Conversely, potentially important processes seen in laser dissected gray matter samples, including decreased regulation of Ca2+ release from ryanodine receptors, and increased vascular development, were not found in our prior work.
3.2 Direct comparisons with our prior study (Blalock et al, 2004)
The comprehensive analysis (section 3.1) of laser-dissected gray matter from the same subjects generally confirmed and extended many of our prior observations from frozen tissue blocks of CA1 material (Blalock et al., 2004). However, to more systematically test the degree of similarity between the two data sets (as well as to identify unique or missing signatures), we statistically compared the two studies as follows: The same probe level algorithm (MAS5) and annotation files were used for both the “laser dissection” and “tissue block” data. Further, testing for significant correlations was restricted to 10,475 probe sets that were perfect matches between the laser microdissection (Affymetrix HG U133 v.2) and tissue block (Affymetrix HG U133A) microarrays. At the presence call level, 59.4% (6,227/10,475) of probe sets were present in laser dissection, and 60.1% (6,338/ 10,475) were present in tissue block, with 5,065 present in both (~81% agreement). These results are highly unlikely by chance (p < 0.0001; binomial test), supporting the idea that detection sensitivities for the laser dissection and tissue block samples, at least at the presence call level prior to evaluation of AD-related gene signatures, were highly similar. Because presence in either study indicates a potential for correlation in at least one dataset, the list of common annotated genes available for testing correlations was expanded from 5,065 present in both studies to include genes present in at least one of the two studies, for a total of 7,537 testable genes.
All genes found to correlate overall with at least one AD marker (NFT or MMSE) were partitioned according to the data set in which they correlated (“tissue block”, “laser dissection”, or “both”; Fig. 3-Venn diagram) and the complete list of correlated genes is provided (Supplemental Table 2). Functional overrepresentation analysis (see Methods) was used to identify biological processes overrepresented by significant genes. The primary processes identified for each of the three Venn component gene lists are shown below (Tables 2-4).
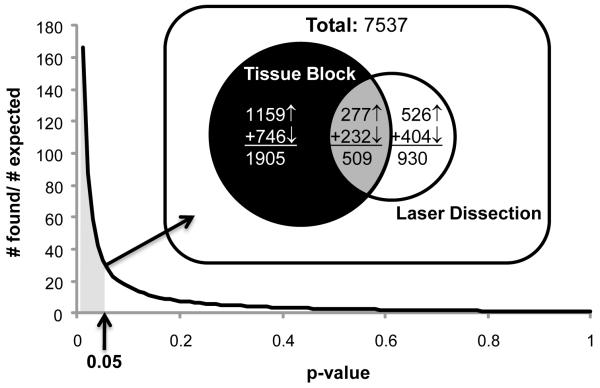
Figure 3. Comparing tissue block and laser capture dissection gene signatures.
The two studies were conducted six years apart using different microarray platforms. For the 7537 gene probes that were present and annotated in both studies, the number of genes that showed correlations with an AD marker in both the tissue block and laser dissection samples (overlap) was determined. At extremely high (relaxed) p-value criteria for correlation significance (right), all genes are expected by chance to correlate with AD in both studies and all genes were found in the overlap (found:expected = 1.0). As p-value criterion stringency increases toward the left, the number of genes expected to correlate significantly in both studies by chance alone falls precipitously, as would the number actually found if there were little true overlap (maintaining a ratio of ~ 1.0). However, with increasing p-value stringency the found:expected ratio rises sharply, particularly at p-values below 0.05, indicating that far more genes were found to agree between studies than would be expected by chance. _Inset_- Venn Diagram: Using the 0.05 cutoff (gray shaded area, arrow), ~34 times as many overlapping genes as expected by chance was identified. Functional overrepresentation analysis was performed for genes encompassed within each region of the Venn diagram (section 3.2).
Table 2. Functional categories overrepresented by genes correlated with AD markers in both studies (Venn diagram overlap-Supplemental Table 2A).
Functional categories/ processes significantly (p ≤ 0.05) downregulated (left) or upregulated (right) and correlated with overall AD in both laser-dissected and tissue block samples. The # genes significant as well as probability of overrepresentation (P-val) are given.
| Downregulated | # | P -val | Upregulated | # | P -val |
|---|---|---|---|---|---|
| neuron projection | 24 | 5.55×10−7 | kinase activity | 31 | 0.004 |
| precursor metabolites and energy | 23 | 1.00×10−6 | transcription repressor | 16 | 0.006 |
| transmission of nerve impulse | 23 | 1.36×10−6 | negative reg. differentiation | 12 | 0.006 |
| transporter activity | 39 | 5.67×10−6 | complement activation | 05 | 0.007 |
| synapse part | 18 | 6.60×10−6 | regulation of proliferation | 30 | 0.009 |
| neuron differentiation | 21 | 5.54×10−5 | extracellular matrix | 13 | 0.009 |
| glycolysis | 07 | 1.20×10−4 | inflammatory response | 14 | 0.014 |
| cellular ion homeostasis | 17 | 0.001 | transcr. RNA pol II promoter | 26 | 0.017 |
| intermediate filament assembly | 03 | 0.003 | mitotic interphase | 08 | 0.027 |
| cytoplasmic vesicle | 21 | 0.006 | microtub. organizing center | 10 | 0.027 |
| cognition | 17 | 0.006 | calmodulin binding | 08 | 0.049 |
| cellular macromolecule localization | 18 | 0.008 | |||
| regulation of hormone levels | 08 | 0.008 | |||
| microtubule associated complex | 07 | 0.009 | |||
| apoptosis cell structure disassembly | 04 | 0.009 | |||
| regulation of neuron differentiation | 08 | 0.009 | |||
| cellular respiration | 08 | 0.009 | |||
| unfolded protein binding | 08 | 0.013 | |||
| voltage-gated channel activity | 08 | 0.016 | |||
| calcium ion binding | 22 | 0.019 | |||
| protein oligomerization | 09 | 0.029 | |||
| extracell.glutamate-gated ion channel | 03 | 0.037 |
Table 4. Functional categories overrepresented by genes correlated with overall AD uniquely in laser-dissected samples (Venn diagram, left-Supplemental Table 2C).
Significant downregulated (left) and upregulated (right) biological categories/ processes overrepresented (p ≤ 0.05) by AD-correlated genes found exclusively in laser-dissected samples. Note novel blood vessel development category (right). # genes significant as well as probability (P-val) are given.
| Downregulated | # | P -val | Upregulated | # | P -val |
|---|---|---|---|---|---|
| protein transport | 40 | 0.0050 | blood vessel development | 22 | 0.0011 |
| ion channel activity | 19 | 0.0062 | tube morphogenesis | 13 | 0.0013 |
| transporter activity | 46 | 0.0069 | release of cyt. c from mitochondria | 06 | 0.0029 |
| N-methyltransferase activity | 06 | 0.0083 | regulation of protein kinase cascade | 22 | 0.0037 |
| chloride channel activity | 07 | 0.0105 | secretory granule | 15 | 0.0066 |
| heat shock protein binding | 08 | 0.0113 | extracellular structure organization | 14 | 0.0136 |
| glucose metabolic process | 12 | 0.0157 | chromatin | 16 | 0.0152 |
| cytoskeletal protein binding | 25 | 0.0281 | pteridine metabolic process | 05 | 0.0182 |
| learning | 07 | 0.0304 | protein pptase 1 reg. activity | 03 | 0.0275 |
| cellular carbohydrate biosynthesis | 07 | 0.0304 | chromatin binding | 13 | 0.0312 |
| ribonucleoprotein biogenesis | 12 | 0.0340 | apoptosis | 37 | 0.0337 |
| regulation of lipase activity | 07 | 0.0369 | phosphoprotein pptase activity | 13 | 0.0361 |
| F-actin capping protein complex | 03 | 0.0375 | zinc ion binding | 79 | 0.0386 |
| neurotransmitter secretion | 05 | 0.0385 | positive regulation of transcription | 28 | 0.0396 |
| ubiquitin-protein ligase activity | 10 | 0.0403 | axonemal dynein complex | 03 | 0.0433 |
| translational initiation | 06 | 0.0435 | zinc transmembrane transporter | 03 | 0.0437 |
| negative reg. developmental process | 17 | 0.0456 | |||
| stress-activated protein kinase pathway | 07 | 0.0492 |
3.2.1 Correlated with overall AD in both tissue block and laser captured samples
(Overlap in Venn diagram, Fig. 3 inset center; Table 2; Supplemental Table 2A). If the results from laser dissection samples are generally valid, then the laser dissection and tissue block data should have many significantly correlated genes in common. That is, the number of AD-correlated overlapping genes should be much higher than expected by chance. Further, that overlap should be relatively enriched in neuron-specific genes and relatively impoverished in white matter genes, compared with results that are unique to the tissue block analysis. As in prior work examining similarities among gene signatures (Blalock et al., 2010), we plotted the found:expected ratio for number of overlapping genes as a function of the p-value cutoff (α level) used to test for correlation in both datasets (Fig. 3). There was very strong agreement between the two data sets and at a p-value cutoff of 0.05, 509 genes, or more than 34x as many genes as would be expected by chance, were found in the overlap (Fig. 3 Venn diagram). This highly significant result (p < 1.0×10−16; binomial test) shows that the overlapping genomic signature is very unlikely to have been identified by chance. Of the 509 genes that exhibited correlation with overall AD in both studies (Supplemental Table 2A), nearly twice as many also exhibited correlation with incipient AD in the laser capture study (202 genes, ~40%) as in the tissue block study (110 genes, ~22%) (p = 1.9×10−14, binomial test), suggesting that laser captured gray matter provided higher resolution and better signal-to-noise than mixed samples of gray and white matter.
Table 2 shows the functional processes found by DAVID analysis to be overrepresented by the 509 genes correlated with overall AD in both studies. It can be seen that these functional processes are generally similar to the processes identified in the comprehensive analysis of all genes in the laser capture study (Table 1).
3.2.2 Correlated with overall AD exclusively in tissue block samples
(Fig. 3 Venn diagram left; Table 3; Supplemental table 2B). The largest number of significantly correlated genes (1905, 1159 of which were upregulated), was found among those correlated exclusively within tissue block samples (Supplemental Table 2B). Finding more significant gene expression changes in tissue block specimens is perhaps not surprising, given that the blocks contained a more varied cell type population. In addition, the functional categories identified by DAVID analysis for these 1905 tissue block genes tended to have the strongest statistical significance (lowest p-values). The tissue block study’s relative abundance of upregulated AD-associated genes is reflected statistically by a highly significant reduction (p = 8.7×10−6, binomial test) in the ratio of upregulated to downregulated AD-correlated genes in the laser dissection (1.26) vs. tissue block (1.46) studies (Fig. 3), indicating that many upregulated correlated genes were related to the white matter tracts included exclusively in those tissue block samples. Among downregulated categories, the 5 most significant (Table 3, left) appeared closely related to mitochondrial bioenergetics, while other downregulated categories were associated with axonal and synaptic activity. The upregulated AD gene signature in tissue block appeared heavily enriched with epigenetic and transcription factor-related processes (Table 3 right- chromosome organization, chromatin assembly, promoter binding, etc.).
Table 3. Functional categories overrepresented by genes uniquely correlated with AD in tissue block samples (Venn diagram, left-Supplemental Table 2B).
Significant downregulated (left) and upregulated (right) biological processes (p ≤ 0.05) overrepresented by correlated genes found exclusively in tissue block. Note the high proportion of processes associated with transcription, chromatin modification, immune functions, oxidative stress and lipid handling among upregulated categories (right). # genes significant as well as probability of overrepresentation (P-val) are given.
| Downregulated | # | P -val | Upregulated | # | P -val |
|---|---|---|---|---|---|
| precursor metabolites and energy | 62 | 1.38×10−14 | chromosome organization | 67 | 2.63×10−05 |
| cellular respiration | 32 | 1.05×10−12 | chromatin assembly or disassembly | 21 | 9.23×10−04 |
| H+transmembrane transporter activity | 28 | 1.84×10−12 | promoter binding | 15 | 0.0025 |
| mitochondrial matrix | 40 | 6.00×10−08 | transcription activator activity | 57 | 0.0031 |
| aerobic respiration | 13 | 2.91×10−05 | extracellular matrix | 37 | 0.0038 |
| glutamate catabolic process | 05 | 0.0012 | T cell activation | 22 | 0.0047 |
| transporter activity | 81 | 0.0014 | histone acetyltransferase complex | 12 | 0.0061 |
| cation-transporting ATPase activity | 10 | 0.0017 | positive regulation of lipid storage | 05 | 0.0066 |
| phosphorylation | 70 | 0.0021 | immune system development | 36 | 0.0088 |
| transmission of nerve impulse | 36 | 0.0037 | intracellular induction of apoptosis | 13 | 0.0099 |
| glutamate metabolic process | 06 | 0.0042 | regulation of transcription factor activity | 19 | 0.0115 |
| +reg. ubiquitin-protein ligase activity | 14 | 0.0044 | regulation of immune response | 28 | 0.0173 |
| ubiquitin-dependent catabolic process | 27 | 0.0047 | regulation of ARF GTPase activity | 08 | 0.0193 |
| nervous system development | 83 | 0.0053 | nuclear chromatin | 12 | 0.0288 |
| synaptic vesicle membrane | 07 | 0.0069 | histone deacetylase activity | 05 | 0.0378 |
| ubiquitin thiolesterase activity | 10 | 0.0082 | interphase of mitotic cell cycle | 19 | 0.0397 |
| nucleoside diphosphate metabolic process | 05 | 0.0124 | transcription repressor activity | 41 | 0.0405 |
| mitochondrial ribosome | 08 | 0.0158 | cellular response to oxidative stress | 10 | 0.0408 |
| response to glucose stimulus | 08 | 0.0207 | regulation of lipid transport | 07 | 0.0481 |
| regulation of translational initiation | 08 | 0.0207 | |||
| microtubule associated complex | 12 | 0.0234 | |||
| peptidase activity | 35 | 0.0268 | |||
| exocytosis | 13 | 0.0330 | |||
| translation | 31 | 0.0380 |
3.2.3 Correlated with overall AD exclusively in laser dissection samples
(Fig. 3 Venn diagram right; Table 4; Supplemental table 2C). Downregulated genes that correlated with AD exclusively in laser dissected material were similar to those seen in both the comprehensive analysis (Table 1) as well as the overlapping region of the Venn diagram, suggesting that different genes from the same processes may be identified in the different analyses (see Discussion). Nonetheless, proportionately more downregulated genes identified exclusively in LCM gray matter samples represented processes associated with declining protein transport and processing, compared to those in the prior tissue block study. Among upregulated genes, the more than 500 that were associated with secretory, epigenetic, stress and transcription processes were correlated with AD at much weaker average significance levels than in the tissue block samples. However, LCM sample genes uniquely identified previously undetected alterations correlated with incipient AD, including upregulated vasculature development Table 4), as well as downregulation of genes important for stabilization of ryanodine receptor-related intracellular Ca2+ release (the latter in the comprehensive laser capture analysis, Table 1).
4. Discussion
The laser dissection protocol’s gray matter-enriched collection technique appears to have facilitated identification of novel AD-associated processes, including downregulated stabilization of ryanodine-sensitive Ca2+ release and upregulated vasculature development. Although the laser microdissected region of CA1 is heavily vascularized (Marinkovic et al., 1992) and contains high densities of ryanodine receptors (Gant et al., 2011), these signals may have been diluted or masked by white matter expression patterns in the prior tissue block study. Accordingly, the appearance of novel processes in the present study confirms and extends results of others (Ginsberg et al., 2010; Ginsberg et al., 2006; Liang et al., 2008) showing that laser dissection techniques can provide a higher resolution assessment of region-specific transcriptional profiles. Moreover, when contrasted with our prior data on hand-dissected CA1 that included white matter tracts, these selective gray matter analyses provide strong support for the view that the previously reported upregulated transcriptional, epigenetic, lipid transport, glial-immune and tumor suppressor responses in AD (Blalock et al., 2004), which were largely absent in the present study, are predominantly localized in and around the excluded white matter.
Comparison of the two studies also highlighted important technical issues. While there have been numerous investigations demonstrating that RNA extracted from FFPE tissue is suitable for quantitative assessment (Abdueva et al., 2010; Coudry et al., 2007; Farragher et al., 2008; Scicchitano et al., 2006; Turner et al., 2011), to our knowledge the present study is the first to make head-to-head comparisons of brain microarray data between FFPE tissue and frozen tissue blocks from the same human subjects. Apart from the apparent white-matter-related differences, there was substantial concordance between the two microarray studies, given that they were performed several years apart on samples collected and preserved very differently (laser dissected, formalin-fixed vs. hand dissected, frozen). Multiple gene networks altered in incipient AD and correlated with MMSE cognitive scores agreed closely across these studies, at both the gene and pathway levels (Overlap, Tables 2-4), particularly for downregulated neuronal processes.
Thus, this general concordance strongly indicates that the FFPE data are valid, and that the expression differences found between tissue block and laser dissection reflect, at least in part, differences in the cell types and sub-regions collected, rather than technical issues related to RNA integrity or extraction efficiency. Nonetheless, some non-specific procedural effects cannot be ruled out.
4.1 Similarities to other array studies
The conclusion that the FFPE material provides valid data is supported further by general agreement between the present LCM study and other gene expression analyses. One of the first studies of transcriptional profiles in AD selectively isolated tangle-bearing neurons (Ginsberg et al., 2000). Results showed significant reductions in genes associated with canonical AD neuropathology, including phosphatases/ kinases, glutamate and dopamine receptors, and cytoskeletal proteins, as well as upregulation of Cathepsin D. A subsequent study that surveyed multiple brain regions (Loring et al., 2001) found now-canonical upregulated inflammation, cell adhesion, cell proliferation and protein synthesis pathways in AD, as well as downregulated signal transduction, energy metabolism, stress response, synaptic vesicle synthesis/ function, calcium binding, and cytoskeletal pathways. An analysis of temporal gyrus found that downregulated synaptophysin directly correlated with, while downregulated alpha synapsin expression appeared to precede, AD-related changes (Pasinetti, 2001). Microarray analysis in the CA1 hippocampal subregion (Colangelo et al., 2002) found decreased transcription factor, neurotrophic factor, signaling, and synaptic activity along with increased inflammation in AD. A focused study of synapse-related gene expression changes in superior frontal gyrus (Yao et al., 2003) found downregulated dynamin 1 and syntaxin 1A expression. Studies of individually isolated brain neurons in AD have shown widespread downregulation of synaptic and cytoskeletal genes, as well as mitochondrial genes associated with reduced metabolism and altered expression of pathological hallmarks (Ginsberg et al., 2006; Liang et al., 2008). Moreover, a recent study in laser-dissected tangle-bearing neurons (Ginsberg et al., 2010) reported that Rab 5 and 7 upregulation correlated with cognitive decline during AD progression, supporting the hypothesis that increased endosomal activity, as reflected by Rab component upregulation, may enhance TrkB degradation and lead to neuronal dysfunction. Thus, among microarray studies of AD, there appears to be relatively good agreement across analyses, at least for downregulated changes in neurons and gray matter and for upregulated inflammatory responses. However, the expression signatures of different regions/tissues/ cell types, and their selective vulnerability in AD remain relatively unexplored (reviewed in Wang et al., 2010).
4.2 Differences from previous studies in the functional processes identified here
As noted, some of the most prominent incipient AD-related changes in our earlier study (increases of growth-related and oncogenic transcription factors, chromatin modifiers, lipid regulators and tumor suppressors (Blalock et al., 2004) were largely absent in the laser dissection material (Tables 1, 2, and 3; Supplemental Table 2). This result is consistent with our model of incipient AD (Blalock et al., 2004) in which aberrant transcriptional processes in oligodendrocytes are proposed to trigger cascades that affect other cell types, providing a potential mechanistic explanation for the apparent progression of AD pathology along myelinated tracts. Of course, additional studies will be needed to further elucidate the role of white matter transcriptional changes in incipient AD.
Another notable difference from our previous study and studies of others is the detection here of novel changes in potentially important functional processes, including upregulation of vasculature development and downregulation of ryanodine receptor stabilization, both of which correlated strongly with incipient AD. These processes may well have important mechanistic implications. For example, Ca2+ dyshomeostasis has long been suspected of a role in brain aging and AD (see Introduction) and it was recently found that downregulation or disruption of the immunophilin FKBP1b (and possibly, FKBP1a) can destabilize and increase ryanodine receptor Ca2+ release in hippocampal neurons, creating an aging-like phenotype (Gant et al., 2011). Here, FKBP1a, two junctophilins and CALM 3, which also participate in ryanodine receptor stabilization, were found to be significantly downregulated and correlated with incipient AD (Supplemental Table 1), as was FKBP1b in the previous study (Blalock et al, 2004; their Table 3). Thus, further studies investigating the significance of ryanodine receptor destabilization in the progression of AD appear warranted by the present results.
Similarly, the upregulation of genes related to vasculature development also may have mechanistic implications. Although many, or most, of the 48 correlated genes populating this category (see Supplemental Table 3) have roles in development and growth of other tissue types, all have also been linked to vasculature development. While the functional significance of this is difficult to discern at present, it may be relevant that angiogenesis is a well-established co-factor in tumor growth, and plays a key role in retinal macular degeneration (Ambati, 2011).
A major challenge for future research on molecular pathogenesis of AD will be to determine how, or if, these novel gray matter changes interact with the putative white matter alterations or, for that matter, with any of the multiple other processes previously identified in our studies and studies of others. Clearly, some of the alterations found may be secondary or incidental correlates of AD. Nonetheless, multiple processes identified in our studies were correlated with incipient as well as with late-stage AD, and consequently appear at least to be candidates for primary processes.
4.3 Statistical issues
The overlap analysis (Fig. 3) reduces false positives, but does so at the cost of a likely increase in false negatives, in that a gene must meet the p ≤ 0.05 criterion in two studies (Blalock et al., 2005). Therefore, the ‘true’ agreement between these two studies may be substantially greater than reported here. Nonetheless, the more than 34x increase in genes agreeing between the two studies, compared to what should be expected by chance, illustrates that genes in the overlap analysis likely contain a very high proportion of true positive findings. It also should be noted that, because this approach assigns sets of genes to each exclusive Venn region based on a single p-value cutoff, genes attributed to one Venn region (Fig. 3, inset) may be borderline significant in another, effectively splitting a functional category across Venn regions. Upregulated immune activity, which appears in both the “overlap” and “tissue block” analyses may reflect this borderline effect.
5. Conclusions
Our results provide strong reciprocal validation for two technically dissimilar microarray analyses of Alzheimer’s disease. Gene expression values derived from laser dissected gray matter in formalin fixed, paraffin embedded specimens yielded a signature highly similar in many aspects to that measured in frozen tissue block specimens, including calcium regulation, synaptic, neuronal, inflammatory, and endosomal pathways- most of which have been identified in multiple studies (see section 4.1). Therefore, the discrepancies between the tissue block and laser dissection signatures did not appear to be due to random error, but to differences in the expression profiles of gray and white matter (the latter perhaps including the synaptic fields and glial processes immediately adjacent to myelinated tracts) . Although technical differences to some extent probably contribute to discordance between the two data sets, such effects appear minor and, further, it is highly unlikely that technical influences would exert themselves at the functional grouping level. That is, there is no a priori reason to expect that a particular functional category of genes would be more sensitive to formalin, laser dissection, or amplification protocols- these effects should seemingly distribute randomly, or at least, not partition discreetly within biological pathways. Thus, we conclude that the targeted difference in tissue components is the most likely and most parsimonious explanation for many of those aspects of the transcriptional profile that differed between our two studies.
Supplementary Material
01
Supplemental Table 1. Complete list of 3,465 genes in the laser-captured tissue that were significant in at least one correlation analysis (Fig. 2). Genes are listed in alphabetical order by gene symbol. Column: (A) _Probe set_- Affymetrix probe set ID; (B) _Symbol_- gene symbol; (C) _Description_- gene description; (D- G) _Overall_- correlation results across all 30 subjects; (H- K) _Incipient_- correlation across the subset of control and incipient subjects only; (D, H) _MMSE r_- Pearson’s r value for MiniMental Status Exam correlation; (E, I) _MMSE p_- p-value for adjacent r value; (F, J) _NFT r_- Pearson’s r value for neurofibrillary tangle correlation; (G, K) _NFT p_- p-value for adjacent r value. Note: p-value results are color-coded if significant (p ≤ 0.05), red for increased, and blue for decreased expression with worsening AD.
02
Supplemental Table 2. Complete list of all 3,344 genes significantly correlated with overall AD from among the 7537 total genes used in the Venn diagram overlap analysis between tissue block and laser dissection results (Fig. 3). Genes are listed alphabetically by symbol and are separated by Venn region (overlap, unique tissue block, unique laser dissection) and direction in the following rows: 4-235 overlapping downregulated; 236-512 overlapping upregulated; 513-1258 tissue downregulated; 1259-2417 tissue upregulated; 2418-2821 laser downregulated; 2822-3347 laser upregulated. Columns (A) Venn and (B) Dir give Venn region and direction of change, and columns C-E give probe set ID, gene symbol and d_escription_ information. All correlation results are provided. (F-M) Tissue Block Results; (N-U) Laser Dissection Results; (F-I; N-Q)- Overall correlation results; (J-M; R-U) Incipient correlation results); _MMSE_- delineates MiniMental Status Exam related results, NFT describes neurofibrillary tangle results (the suffix ‘r’ refers to Pearson’s r value, the suffix ‘p’ refers to p-value calculated based on adjacent r value). Note: p-values are color coded if significant (p ≤ 0.05), red for increased and blue for decreased expression with AD severity.
03
Supplemental Table 3. Complete list of all 48 genes comprising the vasculature response category. Genes are listed alphabetically by gene symbol, along with descriptions, and overall MMSE and NFT correlation R values and p-values. Significant results are color coded red.
Research Highlights.
- Subjects (N=30) spanned control, incipient, moderate and severe Alzheimer’s disease
- Focused on microarray analyses of laser-dissected hippocampal gray matter
- Array profiles largely agreed with a prior study of combined white and gray matter
- Unique gray-matter AD changes included ryanodine and vascular pathways
- Unique white-matter AD changes included epigenetic and transcriptional pathways
Acknowledgements
We thank Sonya Anderson and Ela Patel from the Sanders-Brown Center on Aging for sectioning and mounting hippocampal specimens, Jeff Gilbert and Zeiss Microscopy, as well as Woods Hole Research Center for generously allowing us to use their Zeiss PALM Microbeam system, Anjali Chhabra for performing RNA isolation and amplification protocols, and Dr. Kuey Chu Chen and Donna Wall of the University of Kentucky Microarray Core Facility for microarray hybridization and scanning. This work was supported in part by grants from the University of Kentucky Alzheimer’s Disease Center (AG028383) and the NIH Research Resources Center (S10RR024704) (to E.M.B.), and by grants AG034605, AG004542 and P01AG010836 from the National Institute on Aging (to P.W.L.).
Footnotes
1
Abbreviations: AD- Alzheimer’s disease; CA- Cornu Ammonis; FFPE: Formalin-fixed, paraffin-embedded; LCM- laser capture microdissection; MMSE- mini-mental status exam; NFT- neurofibrillary tangle
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdueva D, Wing M, Schaub B, Triche T, Davicioni E. Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J Mol Diagn. 2010;12:409–417. doi: 10.2353/jmoldx.2010.090155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati J. Age-related macular degeneration and the other double helix. The Cogan Lecture. Investigative ophthalmology & visual science. 2011;52:2165–2169. doi: 10.1167/iovs.11-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt T, Holzer M, Stobe A, Gartner U, Luth HJ, Bruckner MK, Ueberham U. Activated mitogenic signaling induces a process of dedifferentiation in Alzheimer’s disease that eventually results in cell death. Ann N Y Acad Sci. 2000;920:249–255. doi: 10.1111/j.1749-6632.2000.tb06931.x. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I, Mattson MP. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci. 2008;31:454–463. doi: 10.1016/j.tins.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford PC, Gould T, Briederick L, Chadman K, Pollock A, Young D, Shukitt-Hale B, Joseph J. Antioxidant-rich diets improve cerebellar physiology and motor learning in aged rats. Brain Res. 2000;866:211–217. doi: 10.1016/s0006-8993(00)02280-0. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Chen KC, Stromberg AJ, Norris CM, Kadish I, Kraner SD, Porter NM, Landfield PW. Harnessing the power of gene microarrays for the study of brain aging and Alzheimer’s disease: statistical reliability and functional correlation. Ageing Res Rev. 2005;4:481–512. doi: 10.1016/j.arr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Grondin R, Chen KC, Thibault O, Thibault V, Pandya JD, Dowling A, Zhang Z, Sullivan P, Porter NM, Landfield PW. Aging-related gene expression in hippocampus proper compared with dentate gyrus is selectively associated with metabolic syndrome variables in rhesus monkeys. J Neurosci. 2010;30:6058–6071. doi: 10.1523/JNEUROSCI.3956-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser R, Smith MA. Cell cycle proteins in Alzheimer’s disease: plenty of wheels but no cycle. J Alzheimers Dis. 2002;4:249–254. doi: 10.3233/jad-2002-4316. [DOI] [PubMed] [Google Scholar]
- Brinton RD. Estrogen regulation of glucose metabolism and mitochondrial function: therapeutic implications for prevention of Alzheimer’s disease. Adv Drug Deliv Rev. 2008;60:1504–1511. doi: 10.1016/j.addr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger C. Region-specific genetic alterations in the aging hippocampus: implications for cognitive aging. Front Aging Neurosci. 2010;2:140. doi: 10.3389/fnagi.2010.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Sultana R. Redox proteomics identification of oxidatively modified brain proteins in Alzheimer’s disease and mild cognitive impairment: insights into the progression of this dementing disorder. Journal of Alzheimer’s disease : JAD. 2007;12:61–72. doi: 10.3233/jad-2007-12107. [DOI] [PubMed] [Google Scholar]
- Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- Coudry RA, Meireles SI, Stoyanova R, Cooper HS, Carpino A, Wang X, Engstrom PF, Clapper ML. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2007;9:70–79. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft S. Insulin resistance and Alzheimer’s disease pathogenesis: potential mechanisms and implications for treatment. Current Alzheimer research. 2007;4:147–152. doi: 10.2174/156720507780362137. [DOI] [PubMed] [Google Scholar]
- Davies P, Koppel J. Mechanism-based treatments for Alzheimer’s disease. Dialogues Clin Neurosci. 2009;11:159–169. doi: 10.31887/DCNS.2009.11.2/pdavies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disterhoft JF, Moyer JR, Jr., Thompson LT. The calcium rationale in aging and Alzheimer’s disease. Evidence from an animal model of normal aging. Ann N Y Acad Sci. 1994;747:382–406. doi: 10.1111/j.1749-6632.1994.tb44424.x. [DOI] [PubMed] [Google Scholar]
- Farragher SM, Tanney A, Kennedy RD, Harkin D. Paul. RNA expression analysis from formalin fixed paraffin embedded tissues. Histochem Cell Biol. 2008;130:435–445. doi: 10.1007/s00418-008-0479-7. [DOI] [PubMed] [Google Scholar]
- Finch CE, Morgan T, Rozovsky I, Xie Z, Weindruch R, Prolla T. Microglia and aging in the brain. In: WJ S, editor. Microglia in the Degenerating and Regenerating CNS. Springer-Verlag; New York: 2002. [Google Scholar]
- Forman MS, Lee VM, Trojanowski JQ. ‘Unfolding’ pathways in neurodegenerative disease. Trends Neurosci. 2003;26:407–410. doi: 10.1016/S0166-2236(03)00197-8. [DOI] [PubMed] [Google Scholar]
- Foster TC, Norris CM. Age-associated changes in Ca(2+)-dependent processes: relation to hippocampal synaptic plasticity. Hippocampus. 1997;7:602–612. doi: 10.1002/(SICI)1098-1063(1997)7:6<602::AID-HIPO3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Gant JC, Chen KC, Norris CM, Kadish I, Thibault O, Blalock EM, Porter NM, Landfield PW. Disrupting function of FK506-binding protein 1b/12.6 induces the Ca(2)+-dysregulation aging phenotype in hippocampal neurons. J Neurosci. 2011;31:1693–1703. doi: 10.1523/JNEUROSCI.4805-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemma C, Mesches MH, Sepesi B, Choo K, Holmes DB, Bickford PC. Diets enriched in foods with high antioxidant activity reverse age-induced decreases in cerebellar beta-adrenergic function and increases in proinflammatory cytokines. J Neurosci. 2002;22:6114–6120. doi: 10.1523/JNEUROSCI.22-14-06114.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GE, Peterson C. Calcium and the aging nervous system. Neurobiol Aging. 1987;8:329–343. doi: 10.1016/0197-4580(87)90072-8. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Counts SE, Cataldo AM, Neve RL, Jiang Y, Wuu J, Chao MV, Mufson EJ, Nixon RA, Che S. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol Psychiatry. 2010;68:885–893. doi: 10.1016/j.biopsych.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Che S, Counts SE, Mufson EJ. Single cell gene expression profiling in Alzheimer’s disease. NeuroRx. 2006;3:302–318. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- Gustafson D. Adiposity indices and dementia. Lancet Neurol. 2006;5:713–720. doi: 10.1016/S1474-4422(06)70526-9. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Johnson GV, Bailey CD. Tau, where are we now? J Alzheimers Dis. 2002;4:375–398. doi: 10.3233/jad-2002-4505. [DOI] [PubMed] [Google Scholar]
- Khachaturian ZS. The role of calcium regulation in brain aging: reexamination of a hypothesis. Aging (Milano) 1989;1:17–34. doi: 10.1007/BF03323872. [DOI] [PubMed] [Google Scholar]
- Klein WL, Krafft GA, Finch CE. Targeting small Abeta oligomers: the solution to an Alzheimer’s disease conundrum? Trends Neurosci. 2001;24:219–224. doi: 10.1016/s0166-2236(00)01749-5. [DOI] [PubMed] [Google Scholar]
- Landfield PW. ‘Increased calcium-current’ hypothesis of brain aging. Neurobiol Aging. 1987;8:346–347. doi: 10.1016/0197-4580(87)90074-1. [DOI] [PubMed] [Google Scholar]
- Landfield PW, Blalock EM, Chen KC, Porter NM. A new glucocorticoid hypothesis of brain aging: implications for Alzheimer’s disease. Curr Alzheimer Res. 2007;4:205–212. doi: 10.2174/156720507780362083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Ramsey K, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette CM, Schmechel D, Reiman EM, Rogers J, Stephan DA. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring JF, Wen X, Lee JM, Seilhamer J, Somogyi R. A gene expression profile of Alzheimer’s disease. DNA Cell Biol. 2001;20:683–695. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- Marinkovic S, Milisavljevic M, Puskas L. Microvascular anatomy of the hippocampal formation. Surg Neurol. 1992;37:339–349. doi: 10.1016/0090-3019(92)90001-4. [DOI] [PubMed] [Google Scholar]
- Masliah E, Mallory M, Hansen L, DeTeresa R, Alford M, Terry R. Synaptic and neuritic alterations during the progression of Alzheimer’s disease. Neurosci Lett. 1994;174:67–72. doi: 10.1016/0304-3940(94)90121-x. [DOI] [PubMed] [Google Scholar]
- Michaelis ML, Bigelow DJ, Schoneich C, Williams TD, Ramonda L, Yin D, Huhmer AF, Yao Y, Gao J, Squier TC. Decreased plasma membrane calcium transport activity in aging brain. Life sciences. 1996;59:405–412. doi: 10.1016/0024-3205(96)00319-0. [DOI] [PubMed] [Google Scholar]
- Morgan D. Learning and memory deficits in APP transgenic mouse models of amyloid deposition. Neurochem Res. 2003;28:1029–1034. doi: 10.1023/a:1023255106106. [DOI] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak RE, Griffin WS. Interleukin-1, neuroinflammation, and Alzheimer’s disease. Neurobiol Aging. 2001;22:903–908. doi: 10.1016/s0197-4580(01)00287-1. [DOI] [PubMed] [Google Scholar]
- Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, Counts SE, Fahnestock M, Ginsberg SD. Cholinotrophic molecular substrates of mild cognitive impairment in the elderly. Current Alzheimer research. 2007;4:340–350. doi: 10.2174/156720507781788855. [DOI] [PubMed] [Google Scholar]
- Mullan M, Crawford F. The molecular genetics of Alzheimer’s disease. Mol Neurobiol. 1994;9:15–22. doi: 10.1007/BF02816100. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Mathews PM, Cataldo AM. The neuronal endosomal-lysosomal system in Alzheimer’s disease. J Alzheimers Dis. 2001;3:97–107. doi: 10.3233/jad-2001-3114. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Saito KI, Grynspan F, Griffin WR, Katayama S, Honda T, Mohan PS, Shea TB, Beermann M. Calcium-activated neutral proteinase (calpain) system in aging and Alzheimer’s disease. Ann N Y Acad Sci. 1994;747:77–91. doi: 10.1111/j.1749-6632.1994.tb44402.x. [DOI] [PubMed] [Google Scholar]
- Noble W, Olm V, Takata K, Casey E, Mary O, Meyerson J, Gaynor K, LaFrancois J, Wang L, Kondo T, Davies P, Burns M, Veeranna, Nixon R, Dickson D, Matsuoka Y, Ahlijanian M, Lau LF, Duff K. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron. 2003;38:555–565. doi: 10.1016/s0896-6273(03)00259-9. [DOI] [PubMed] [Google Scholar]
- Norris CM, Halpain S, Foster TC. Reversal of age-related alterations in synaptic plasticity by blockade of L-type Ca2+ channels. J Neurosci. 1998;18:3171–3179. doi: 10.1523/JNEUROSCI.18-09-03171.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okello JB, Zurek J, Devault AM, Kuch M, Okwi AL, Sewankambo NK, Bimenya GS, Poinar D, Poinar HN. Comparison of methods in the recovery of nucleic acids from archival formalin-fixed paraffin-embedded autopsy tissues. Anal Biochem. 2010;400:110–117. doi: 10.1016/j.ab.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Pasinetti GM. Use of cDNA microarray in the search for molecular markers involved in the onset of Alzheimer’s disease dementia. J Neurosci Res. 2001;65:471–476. doi: 10.1002/jnr.1176. [DOI] [PubMed] [Google Scholar]
- Perry G, Nunomura A, Raina AK, Aliev G, Siedlak SL, Harris PL, Casadesus G, Petersen RB, Bligh-Glover W, Balraj E, Petot GJ, Smith MA. A metabolic basis for Alzheimer disease. Neurochemical research. 2003;28:1549–1552. doi: 10.1023/a:1025678510480. [DOI] [PubMed] [Google Scholar]
- Petanceska SS, DeRosa S, Olm V, Diaz N, Sharma A, Thomas-Bryant T, Duff K, Pappolla M, Refolo LM. Statin therapy for Alzheimer’s disease: will it work? J Mol Neurosci. 2002;19:155–161. doi: 10.1007/s12031-002-0026-2. [DOI] [PubMed] [Google Scholar]
- Price DL, Sisodia SS. Mutant genes in familial Alzheimer’s disease and transgenic models. Annu Rev Neurosci. 1998;21:479–505. doi: 10.1146/annurev.neuro.21.1.479. [DOI] [PubMed] [Google Scholar]
- Puglielli L, Konopka G, Pack-Chung E, Ingano LA, Berezovska O, Hyman BT, Chang TY, Tanzi RE, Kovacs DM. Acyl-coenzyme A: cholesterol acyltransferase modulates the generation of the amyloid beta-peptide. Nat Cell Biol. 2001;3:905–912. doi: 10.1038/ncb1001-905. [DOI] [PubMed] [Google Scholar]
- Rogers J, Webster S, Lue LF, Brachova L, Civin WH, Emmerling M, Shivers B, Walker D, McGeer P. Inflammation and Alzheimer’s disease pathogenesis. Neurobiol Aging. 1996;17:681–686. doi: 10.1016/0197-4580(96)00115-7. [DOI] [PubMed] [Google Scholar]
- Scheff SW, Price DA. Alzheimer’s disease-related synapse loss in the cingulate cortex. J Alzheimers Dis. 2001;3:495–505. doi: 10.3233/jad-2001-3509. [DOI] [PubMed] [Google Scholar]
- Scicchitano MS, Dalmas DA, Bertiaux MA, Anderson SM, Turner LR, Thomas RA, Mirable R, Boyce RW. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J Histochem Cytochem. 2006;54:1229–1237. doi: 10.1369/jhc.6A6999.2006. [DOI] [PubMed] [Google Scholar]
- Simpkins JW, Wen Y, Perez E, Yang S, Wang X. Role of nonfeminizing estrogens in brain protection from cerebral ischemia: an animal model of Alzheimer’s disease neuropathology. Annals of the New York Academy of Sciences. 2005;1052:233–242. doi: 10.1196/annals.1347.019. [DOI] [PubMed] [Google Scholar]
- Sohrabji F, Lewis DK. Estrogen-BDNF interactions: implications for neurodegenerative diseases. Front Neuroendocrinol. 2006;27:404–414. doi: 10.1016/j.yfrne.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- Stutzmann GE. Calcium dysregulation, IP3 signaling, and Alzheimer’s disease. Neuroscientist. 2005;11:110–115. doi: 10.1177/1073858404270899. [DOI] [PubMed] [Google Scholar]
- Stutzmann GE, Smith I, Caccamo A, Oddo S, Parker I, Laferla F. Enhanced ryanodine-mediated calcium release in mutant PS1-expressing Alzheimer’s mouse models. Ann N Y Acad Sci. 2007;1097:265–277. doi: 10.1196/annals.1379.025. [DOI] [PubMed] [Google Scholar]
- Sze CI, Troncoso JC, Kawas C, Mouton P, Price DL, Martin LJ. Loss of the presynaptic vesicle protein synaptophysin in hippocampus correlates with cognitive decline in Alzheimer disease. J Neuropathol Exp Neurol. 1997;56:933–944. doi: 10.1097/00005072-199708000-00011. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L. New frontiers in Alzheimer’s disease genetics. Neuron. 2001;32:181–184. doi: 10.1016/s0896-6273(01)00476-7. [DOI] [PubMed] [Google Scholar]
- Thibault O, Gant JC, Landfield PW. Expansion of the calcium hypothesis of brain aging and Alzheimer’s disease: minding the store. Aging Cell. 2007;6:307–317. doi: 10.1111/j.1474-9726.2007.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L, Heath JD, Kurn N. Gene expression profiling of RNA extracted from FFPE tissues: NuGEN technologies’ whole-transcriptome amplification system. Methods Mol Biol. 2011;724:269–280. doi: 10.1007/978-1-61779-055-3_17. [DOI] [PubMed] [Google Scholar]
- Tuszynski MH, Gage FH. Potential use of neurotrophic agents in the treatment of neurodegenerative disorders. Acta Neurobiol Exp (Wars) 1990;50:311–322. [PubMed] [Google Scholar]
- Vercellino M, Masera S, Lorenzatti M, Condello C, Merola A, Mattioda A, Tribolo A, Capello E, Mancardi GL, Mutani R, Giordana MT, Cavalla P. Demyelination, inflammation, and neurodegeneration in multiple sclerosis deep gray matter. Journal of neuropathology and experimental neurology. 2009;68:489–502. doi: 10.1097/NEN.0b013e3181a19a5a. [DOI] [PubMed] [Google Scholar]
- Wang X, Michaelis ML, Michaelis EK. Functional genomics of brain aging and Alzheimer’s disease: focus on selective neuronal vulnerability. Curr Genomics. 2010;11:618–633. doi: 10.2174/138920210793360943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmer RA, Gunderson EP, Quesenberry CP, Jr., Zhou J, Yaffe K. Body mass index in midlife and risk of Alzheimer disease and vascular dementia. Current Alzheimer research. 2007;4:103–109. doi: 10.2174/156720507780362047. [DOI] [PubMed] [Google Scholar]
- Williams BJ, Eriksdotter-Jonhagen M, Granholm AC. Nerve growth factor in treatment and pathogenesis of Alzheimer’s disease. Prog Neurobiol. 2006;80:114–128. doi: 10.1016/j.pneurobio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Yaffe K, Blackwell T, Kanaya AM, Davidowitz N, Barrett-Connor E, Krueger K. Diabetes, impaired fasting glucose, and development of cognitive impairment in older women. Neurology. 2004;63:658–663. doi: 10.1212/01.wnl.0000134666.64593.ba. [DOI] [PubMed] [Google Scholar]
- Yao PJ, Zhu M, Pyun EI, Brooks AI, Therianos S, Meyers VE, Coleman PD. Defects in expression of genes related to synaptic vesicle trafficking in frontal cortex of Alzheimer’s disease. Neurobiol Dis. 2003;12:97–109. doi: 10.1016/s0969-9961(02)00009-8. [DOI] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, Lakatta EG, Boheler KR, Xu X, Mattson MP, Falco G, Ko MS, Schlessinger D, Firman J, Kummerfeld SK, Wood WH, 3rd, Zonderman AB, Kim SK, Becker KG. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Lein ES, He A, Smith SC, Aston C, Gage FH. Transcriptional profiling reveals strict boundaries between hippocampal subregions. J Comp Neurol. 2001;441:187–196. doi: 10.1002/cne.1406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
Supplemental Table 1. Complete list of 3,465 genes in the laser-captured tissue that were significant in at least one correlation analysis (Fig. 2). Genes are listed in alphabetical order by gene symbol. Column: (A) _Probe set_- Affymetrix probe set ID; (B) _Symbol_- gene symbol; (C) _Description_- gene description; (D- G) _Overall_- correlation results across all 30 subjects; (H- K) _Incipient_- correlation across the subset of control and incipient subjects only; (D, H) _MMSE r_- Pearson’s r value for MiniMental Status Exam correlation; (E, I) _MMSE p_- p-value for adjacent r value; (F, J) _NFT r_- Pearson’s r value for neurofibrillary tangle correlation; (G, K) _NFT p_- p-value for adjacent r value. Note: p-value results are color-coded if significant (p ≤ 0.05), red for increased, and blue for decreased expression with worsening AD.
02
Supplemental Table 2. Complete list of all 3,344 genes significantly correlated with overall AD from among the 7537 total genes used in the Venn diagram overlap analysis between tissue block and laser dissection results (Fig. 3). Genes are listed alphabetically by symbol and are separated by Venn region (overlap, unique tissue block, unique laser dissection) and direction in the following rows: 4-235 overlapping downregulated; 236-512 overlapping upregulated; 513-1258 tissue downregulated; 1259-2417 tissue upregulated; 2418-2821 laser downregulated; 2822-3347 laser upregulated. Columns (A) Venn and (B) Dir give Venn region and direction of change, and columns C-E give probe set ID, gene symbol and d_escription_ information. All correlation results are provided. (F-M) Tissue Block Results; (N-U) Laser Dissection Results; (F-I; N-Q)- Overall correlation results; (J-M; R-U) Incipient correlation results); _MMSE_- delineates MiniMental Status Exam related results, NFT describes neurofibrillary tangle results (the suffix ‘r’ refers to Pearson’s r value, the suffix ‘p’ refers to p-value calculated based on adjacent r value). Note: p-values are color coded if significant (p ≤ 0.05), red for increased and blue for decreased expression with AD severity.
03
Supplemental Table 3. Complete list of all 48 genes comprising the vasculature response category. Genes are listed alphabetically by gene symbol, along with descriptions, and overall MMSE and NFT correlation R values and p-values. Significant results are color coded red.