Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p (original) (raw)
. 2000 Feb 15;14(4):493–503.
Abstract
Spo11p is a key mediator of interhomolog interactions during meiosis. Deletion of the SPO11 gene decreases the length of S phase by ∼25%. Rec8p is a key coordinator of meiotic interhomolog and intersister interactions. Deletion of the REC8 gene increases S-phase length, by ∼10% in wild-type and ∼30% in a spo11Δ background. Thus, the progression of DNA replication is modulated by interchromosomal interaction proteins. The spo11–Y135F DSB (double strand break) catalysis-defective mutant is normal for S-phase modulation and DSB-independent homolog pairing but is defective for later events, formation of DSBs, and synaptonemal complexes. Thus, earlier and later functions of Spo11 are defined. We propose that meiotic S-phase progression is linked directly to development of specific chromosomal features required for meiotic interhomolog interactions and that this feedback process is built upon a more fundamental mechanism, common to all cell types, by which S-phase progression is coupled to development of nascent intersister connections and/or related aspects of chromosome morphogenesis. Roles for Rec8 and/or Spo11 in progression through other stages of meiosis are also revealed.
Keywords: SPO11, REC8, DNA replication, meiosis
During meiosis, a single round of DNA replication is followed by two successive rounds of chromosome segregation. A central unique feature of meiosis is homolog juxtaposition, which occurs primarily during meiotic prophase via the combined effects of three component processes: DSB-independent homolog pairing, programmed interhomolog recombination, and synaptonemal complex (SC) formation (for reviews, see Kleckner 1996; Roeder 1997; Zickler and Kleckner 1999).
It has been suggested that preparation of chromosomes for these events occurs as early as the time of DNA replication. Although the meiotic replication program is generally quite similar to that of cycling cells, meiotic S phase is universally several times longer than that in the same organism (Discussion; Bennett and Smith 1972; Callan 1973; Holm 1977). That difference has been proposed to be related to the need for the laying down of specialized chromosome features utilized at later stages for interhomolog interactions (Holm 1977). This proposal raised a further possibility: that progression of S-phase might be directly coupled to the laying down of such specialized features, presumably on a region-by-region basis. Such coupling could help to ensure that the necessary modifications occur correctly, in concert with other chromosomal features that develop during S phase (below). Moreover, the longer duration of meiotic S phase, as compared to its mitotic counterpart, could then be explained as a direct consequence of the extra time required for completion of the necessary events.
Such a model could also make sense from another point of view. It appears that several aspects of meiotic interhomolog interactions may be built on the foundation of intersister interactions (Kleckner 1996; see also Klein et al. 1999; Parisi et al. 1999). And intersister interactions are initially formed during S phase, in close coordination with DNA replication (Weiner and Kleckner 1994; Skibbens et al. 1999; Toth et al. 1999). Thus, progression of S phase from region to region could, fundamentally, be coupled to establishment of sister chromatid connections, or to some closely correlated event, as a universal feature of replication in all cell types (a possibility also suggested by Guacci and colleagues 1997). Then, during meiosis, coupling of replication progression to events required for interhomolog interactions could be achieved by linking meiosis-, interhomolog-specific components to a more basic intersister interaction feedback process. Such a mechanism would permit proper spatial and temporal coordination between intersister and interhomolog interactions (for further discussion, see Kleckner 1996; Burgess et al. 1999).
Links between chromosomal morphogenesis and S phase have been established with respect to gene silencing (Fox et al. 1997) and certain epigenetic states mediated by chromatin assembly factor 1 (Shibahara and Stillman 1999). Also, the temporal program of replication during S phase is determined, at least in part, by basic chromosome structure (e.g., Stevenson and Gottschling 1999).
According to the scenario outlined above, deletion or alteration of genes encoding critical interchromosomal interaction proteins should result in a change in the length of S phase. In contrast, if the length of S phase is determined by other features, with chromosome morphogenesis ensuing passively, such genetic alterations should have no effect. We have therefore examined the length of meiotic S phase in Saccharomyces cerevisiae in wild-type and selected isogenic mutant strains.
We began by analyzing the effects of mutations in the SPO11 gene. Spo11 protein is the catalytic subunit of the meiotic double-strand break (DSB) transesterase and is thus directly responsible for initiation of meiotic recombination (Bergerat et al. 1997; Keeney et al. 1997). In addition, Spo11 is absolutely required for DSB-independent juxtaposition of homologs at interstitial positions during early prophase (Weiner and Kleckner 1994). We examined the effects of both a complete gene deletion (spo11Δ) and an allele that is specifically defective in DSB catalysis due to surgical elimination of a specific catalytic −OH group (spo11–Y135F). The latter mutant was also characterized with respect to DSB-independent homolog pairing and formation of SC.
We also examined the effects of deleting the REC8 gene. REC8 encodes a meiosis-specific member of the Rad21 cohesin family (Mcd1/Scc1 in budding yeast; Guacci et al. 1997; Michaelis et al. 1997). Rec8p is required for both meiotic intersister cohesion and meiotic recombination (DeVeaux and Smith 1994; Klein et al. 1999; Krawchuck et al. 1999; Parisi et al. 1999; Watanabe and Nurse 1999) and, thus, is a strong candidate to mediate interplay between these two aspects. Moreover, a non-null mcd1 mutation has been shown to confer longer mitotic S phase (Guacci et al. 1997) in accord with the ideas described above. The effect of a complete deletion mutation, rec8Δ has been examined in both wild-type and spo11Δ backgrounds.
Twelve other genes besides SPO11 are known to be involved in meiotic DSB formation (Roeder 1997). We have examined meiotic S phase in strains carrying null mutations in three of these genes, RED1, HOP1, and REC102, which represent three different types of DSB-promoting activities: REC102 is required very strongly (Bhargava et al. 1992), HOP1 is required strongly but not absolutely (Mao-Draayer et al. 1996; L. Xu and N. Klecker, unpubl.), and RED1 is required differentially for DSBs along the interhomolog-only pathway of meiotic recombination (Schwacha and Kleckner 1997). Furthermore, although Red1p and Hop1p are both abundant axis-associated structural components of meiotic chromosomes, their kinetics of localization differ significantly (Smith and Roeder 1997). These three genes also differ significantly from SPO11 in that none seem to be involved in DSB-independent homolog pairing (Loidl et al. 1994; Weiner and Kleckner 1994; Nag et al. 1995). Moreover, although a spo11Δ mutant appears to be normal with respect to sister chromatid cohesion (Klein et al. 1999; B.M. Weiner, unpubl.), a red1Δ mutant exhibits abnormalities in sister chromatid cohesion (Bailis and Roeder 1998).
The results presented below demonstrate that meiotic S-phase progression is modulated by key interchromosomal interaction proteins and imply key roles for Spo11 and Rec8 in integrating diverse aspects of chromosome morphogenesis and regulatory progression throughout meiosis.
Results
Meiotic S phase is longer than mitotic S phase in a diploid SK1 strain
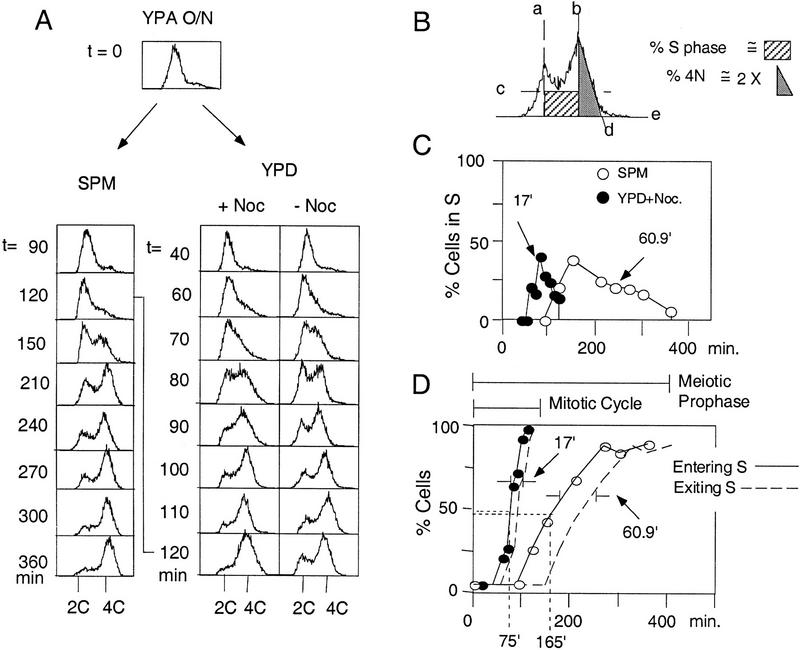
Previous studies in S. cerevisiae have reported mitotic S-phase lengths of 20–30 min (e.g., Slater et al. 1977; Brewer et al. 1984) and meiotic S-phase lengths of 65–80 min (e.g., Williamson et al. 1983; Padmore et al. 1991). As a baseline for the current study we compared mitotic and meiotic S-phase lengths directly in a diploid SK1 strain (Fig. 1). A single culture of NKY611 was grown to stationary phase in YPacetate (YPA) medium, in which condition cells accumulate at G1/G0. Cells were then washed and resuspended, in parallel, in medium that supported either sporulation (SPM) or vegetative growth (YPD without or with nocodazole, which limits cells to a single mitotic cycle). Cells in the three cultures then proceeded through either synchronous meiosis or a synchronous mitotic cell cycle. The occurrence of DNA replication was monitored by FACS analysis (Fig. 1A).
Figure 1.
Measurement of meiotic and mitotic S-phase lengths. (A) Progression of DNA replication was monitored by FACS analysis of samples collected at specified time points during meiotic (SPM) and mitotic (YPD) time courses. Cells progress from an unreplicated state indicated by their presence in a single peak of fluorescence (2C), to a state in which bulk DNA replication is complete, as indicated by movement into a second peak of twice the fluorescence intensity of the first peak (4C). (B) The fraction of cells undergoing DNA replication at each time point was evaluated from the corresponding FACS profile. The fractional area under the indicated rectangle was taken to represent the fraction of cells in S phase; the fractional area under the indicated triangle, doubled, was taken to represent the fraction of cells in G2/M. The appropriateness of these approximations is discussed in Materials and Methods. (C) Fraction of cells undergoing DNA replication at each time point in A (SPM and YPD + Noc cultures) is estimated as in Materials and Methods and plotted as a function of time. The lengths of mitotic (17.0 min) and meiotic (60.9 min) S phases were calculated as described in the text (also see Materials and Methods). (D) Cumulative curves for mitotic and meiotic DNA replication. The fraction of cells that have entered (solid line) and exited (broken line) S phase at each time point is calculated (Materials and Methods) as a function of time. The times at which 50% of active cells have entered mitotic (75 min) and meiotic (165 min) S phases are indicated. The lengths of mitotic (17 min) and meiotic (60.9 min) S phases is the time distance between the Entering S and Exiting S lines as depicted.
To determine the length of S phase in a given culture, the fraction of cells in S phase at each time point was estimated (Fig. 1B; Materials and Methods) and plotted as a function of time (Fig. 1C). The length of S phase is given by the area under each noncumulative curve in Figure 1C divided by the percentage of cells that progress through S phase in a given culture. (This “active S-phase fraction” is 100% for mitotic cultures and slightly less for meiosis; see below). This method provides a population average value for S-phase length; notably, this determination is independent of the degree of synchrony within the culture. In the meiotic and mitotic (+ nocodazole) cultures, S-phase lengths were 60.9 and 17.0 min, respectively (Fig. 1C); analogous kinetics were observed in the mitotic culture incubated in the absence of nocodazole (Fig. 1A, cf. two right panels). These values are comparable to those reported previously (above).
The kinetics with which cells in a culture enter S phase can also be evaluated, by converting noncumulative curves to cumulative curves (Materials and Methods; Padmore et al. 1991; Burgess et al. 1999). In the meiotic culture, 50% of cells had entered S phase at t = 165 min (Fig. 1D), highly consistent with previous reports of Williamson et al. (1983) and Padmore et al. (1991), who observed that 50% of cells entered S phase at t = 180 and t = 160 min, respectively. In the mitotic culture, cells enter S phase sooner, 50% at t = 75 min (Fig. 1D). The kinetics with which cells exit S phase in each case is given by a curve parallel to the entry curve but displaced along the time axis by the corresponding S-phase duration (Fig. 1D).
Comparison of S-phase lengths in wild-type and mutant strains
For quantitatively assessing the effects of mutation(s) on the length of meiotic S phase, we designed an experimental system that minimizes strain background differences and, at the same time, provides an internal wild-type control for every experiment (Fig. 2A). NKY3000 is a homothallic diploid SK1 strain that arose by self-diploidization of a single spore clone of NKY730. The maternal and paternal genome complements of NKY3000 should be essentially identical. Specific mutations of interest were introduced into NKY3000 by integrative transformation to give corresponding diploid derivatives heterozygous for the introduced mutation. Sporulation of such a diploid yields asci containing four spores that are essentially perfectly isogenic except that two contain the mutation of interest and two are wild type. Germination of separated spores from such an ascus, with concomitant efficient self-diploidization, yields four very closely related homozygous diploid strains, two wild type and two mutant, which can then be analyzed in parallel for the duration of meiotic S phase.
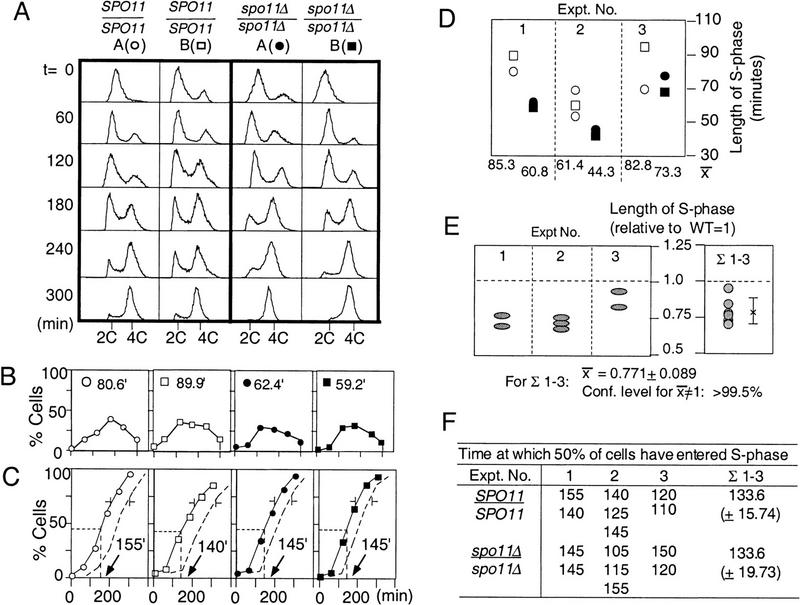
Figure 2.
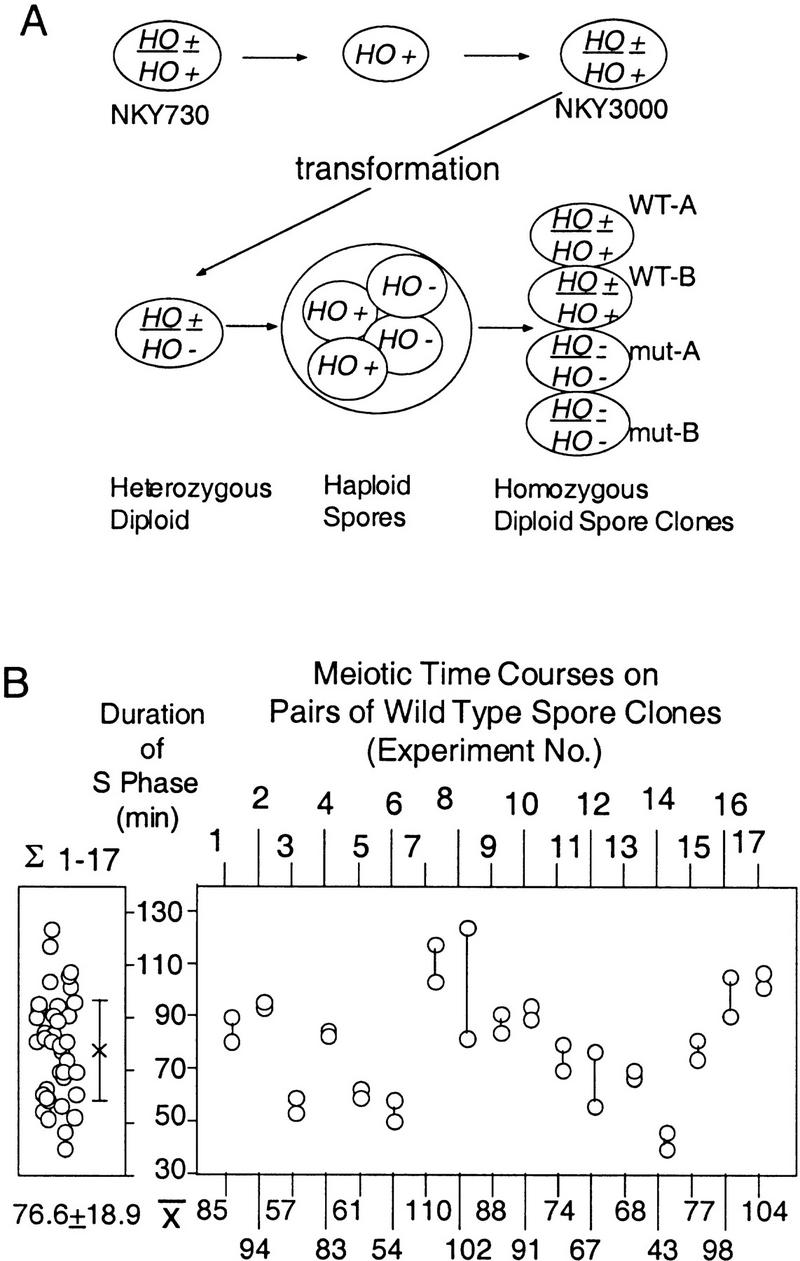
Experimental system. (A) NKY3000 is a self-diploidized spore clone of NKY730. A panel of homothallic heterozygous strains were generated by standard yeast transformation procedures. Each heterozygotic derivative of NKY3000 was sporulated and the four haploid spore clones from the same ascus were allowed to undergo self-diploidization. The resulting four diploid strains, two wild type and two mutant, were analyzed simultaneously during parallel synchronous meiotic time courses. (B) Distribution of wild-type meiotic S-phase lengths. (Left) The distribution of S-phase lengths observed for pairs of wild-type spore clones derived as described in A. The mean and s.d., 76.7 ± 18.9 min, is of 34 measurements from 17 different experiments. (Right) The variation between the two sibling cultures examined in parallel in a single experiment. Average length of S phase for each pair of wild-type cultures from a single experiment is summarized below.
The distributions of S-phase lengths observed for the wild-type strains from such heterozygotes are shown in Figure 2B. Data are shown for 17 different experiments, each involving parallel analysis of the two wild-type spore clones of a single ascal quartet. The mean S-phase length among the 34 cultures was 77 min, again consistent with previously reported values (above). Interestingly, among the entire set of 34 cultures, meiotic S-phase lengths varied by >80 min, from 40 to 123.8 min (Fig. 2B, left). In contrast, S phase varied relatively little between the two sibling cultures examined in parallel in any single experiment: Among the 17 such pairs examined, 9 had S-phase lengths that differed by 10 min or less and only four had S-phase lengths that differed by >15 min (Fig. 2B, right). Genetic heterogeneity among these strains, all derived directly from NKY3000, should be minimal. Thus, variation from experiment to experiment likely reflects day-to-day variability. Consistent with this interpretation, the variability observed for a given pair of strains analyzed on two different days was comparable to that observed among the various different pairs of strains in different experiments (e.g., Fig. 2B).
As mentioned above, the fraction of cells that progress through S phase in each culture, defined as active S-phase fraction, was taken into account in calculating the life span of S phase. This fraction was estimated by measuring the percentage of cells in the 4C peak (Fig. 1; Materials and Methods) at the time the last FACS sample was collected for a given culture, typically 5–7 hr after transfer of cells to SPM. For wild-type strains, the fraction of cells that have progressed through S phase was uniformly high, 86.3 ± 7.2%; n = 37) and corresponds well to final sporulation efficiencies (87.3 ± 4.0%; n = 14), as found in previous studies (Padmore et al. 1991). In no case was a significant fraction of cells present between the 2C and 4C peaks at the latest time point, implying that by the latest time point, the cell had either started and completed DNA replication or never entered S phase at all.
Deletion of SPO11 decreases the length of meiotic S phase
The lengths of meiotic S phase in wild-type and spo11Δ strains were measured and compared by the approach outlined above. In one such experiment, shown in detail, the four strains derived from a single ascus exhibited S-phase lengths of 80.6 and 89.9 min for the two wild-type cultures and of 62.4 and 59.2 min for the two spo11Δ cultures (Fig. 3A,B). Comparable results were obtained in two other independent experiments (Fig. 3D, cf. open symbols to closed symbols; strain details in legend). The effect of the spo11Δ mutation can be compared among the three experiments by normalizing the length observed in each mutant culture to the average of the lengths observed for the two wild-type cultures in the same experiment (Fig. 3E, left). By this criterion, the spo11Δ mutation decreases the length of S phase by ∼25% (Fig. 3E, right). This effect corresponds to a predicted reduction of ∼18 min of the average S-phase length of 77 min observed for wild type (Table 1). Cumulative curve analysis shows that the spo11Δ mutation has no effect on the time at which cells enter S phase, which occurs, on average, 133 min after transfer to sporulation medium in both wild-type and mutant cultures (Fig. 3F). Parallel analysis of a pair of isogenic heterothallic wild-type (NKY611) and spo11Δ (NKY648) strains also yielded similar results (data not shown), indicating that the effect of spo11Δ is not specific to the homothallic nature of the strains analyzed.
Figure 3.
Meiotic S-phase is shorter in spo11Δ. (A) Progression of meiotic DNA replication in two wild-type (○,□) and two spo11Δ (●,█) strains derived from NKY3002. (B) Lengths of S phases in wild type and spo11Δ. (see Fig. 1 and text for details). (C) The fraction of cells that have entered (solid line) and exited (broken line) S phase is calculated for the four cultures in A (Materials and Methods). Arrows indicate the time at which 50% of the active cells have entered S phase in each culture. (D) Lengths of meiotic S phase in wild-type and spo11Δ cultures from three different experiments. Open and solid symbols represent the calculated lengths of meiotic S phase in wild type and spo11Δ, respectively. Results from A are summarized in experiment 1. NKY3002 was analyzed for experiments 1 and 2, and NKY3196 for experiment 3. For experiment 2, one of the two wild-type and spo11Δ spore clones was analyzed in duplicate giving rise to a total of six cultures analyzed. Also shown are the average lengths of meiotic S phase for wild-type and mutant spore clones in each experiment. (E) The relative lengths of meiotic S phase in spo11Δ cultures (see text for detail). (Left) Each shaded oval corresponds to a specific ● and █ in D. (Right) Σ1–3 is the distribution of normalized S-phase length in seven spo11Δ cultures. The mean and standard deviation are used in a one-sample _t_-test; the confidence level that the mean value for spo11Δ (0.771) is different from that of the wild type (1.0) is >99.5%. (F) Kinetics of entry into meiotic S phase is comparable in SPO11 and spo11Δ strains. The time at which 50% of active cells have entered S-phase in each SPO11 and spo11Δ culture of the above-mentioned three independent experiments are summarized. Mean values of seven SPO11 and spo11Δ cultures are shown (right) (Σ1–3).
Table 1.
Statistical evaluation of relative lengths of meiotic S phases
| | spo11Δ | spo11–Y135F | rec102Δ | hop1Δ | red1Δ | rec8Δ | spo11Δ rec8Δ | | | ---------------------------------------- | ------------- | ------------- | ------------- | ------------- | ------------- | -------------- | ------------- | | Relative lengh of S phase (mean; WT = 1) | 0.771 ± 0.089 | 0.962 ± 0.121 | 0.933 ± 0.121 | 1.013 ± 0.102 | 0.941 ± 0.131 | 1.083 ± 0.100 | 1.052 ± 0.075 | | n = | 7 | 6 | 6 | 4 | 4 | 6 | 4 | | Confidence level (%) for mean ≠′ 1 | >99.5 | 75–90 | 75–90 | <60 | 75–90 | >95 | 75–90 | | Length of S (WT = 76.6 min)a | 59.1 | 73.7 | 71.5 | 77.6 | 72.1 | 83.0 | 80.6 | | Δ from WT (min)b | −17.5 | −2.9 | −5.1 | +1.0 | −4.5 | +6.4 | +4.0 |
The fraction of cells entering S phase for spo11Δ (91.9 ± 5.9%; n = 7) was comparable to that of wild type (89.8 ± 3.0%; n = 7). The percentage of spo11Δ cells that successfully completed meiosis I, meiosis II, and spore formation were also not statistically different from that of the wild type or from the fraction of spo11Δ cells that have progressed through S phase (data not shown). Thus, just as in wild type, nearly all the spo11Δ cells that complete meiotic S phase go on to complete the other basic steps of meiosis and spore formation.
Because SPO11 is not transcribed in mitotic yeast cells (Atcheson et al. 1987), the effect of the spo11Δ mutation on S-phase length is expected to be limited to meiosis. This expectation is supported by parallel analysis of isogenic wild-type (NKY611) and spo11Δ (NKY648) cultures, which exhibited mitotic S-phase lengths of 18.9 min (±2.7; n = 2) and 19.8 min (±3.0; n = 2), respectively.
The spo11–Y135F DSB catalysis-defective mutant and other DSB-defective mutants exhibit normal meiotic S-phase length
Residue 135 of Spo11p encodes a conserved tyrosine residue whose −OH residue is likely the chemical group that directly catalyzes DSB formation (Bergerat et al. 1997; Keeney et al. 1997). Correspondingly, surgical elimination of this critical −OH group, by a mutation that changes the corresponding residue to phenylalanine (spo11–Y135F), completely eliminates both DSB formation and essentially all meiotic recombination (Bergerat et al. 1997; R.S. Cha, data not shown).
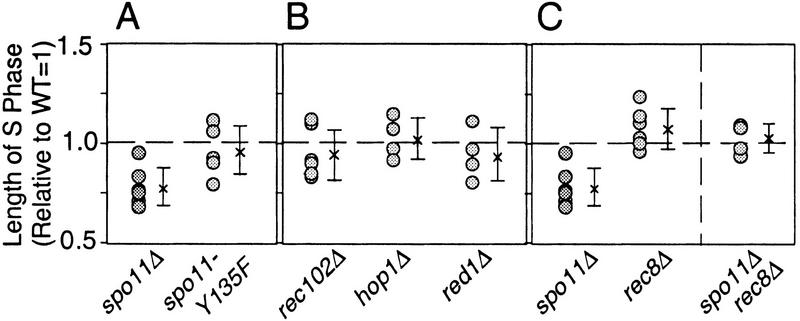
The duration of meiotic S phase was examined in the spo11–Y135F mutant and shown to be indistinguishable from that of wild type (Fig. 4A; Table 1). The kinetics of S-phase entry as well as the active S-phase fraction in spo11–Y135F cultures (89.2 ± 5.8%; n = 6) were also comparable to those observed for wild-type cultures (88.3 ± 2.7%; n = 6). Thus, the functions of Spo11p for S-phase regulation and DSB catalysis can be separated; more specifically, DSB catalysis can be eliminated without effect on S-phase regulation. The length of S phase was also the same as for wild type in rec102Δ, red1Δ, and hop1Δ mutants (Fig. 4B; Table 1). The active S-phase fraction and the kinetics of entry into S phase in the three mutants were also comparable to that observed in isogenic wild-type controls (data not shown).
Figure 4.
Relative lengths of meiotic S-phase in various mutants. (A–D) Lengths of meiotic S-phase in various mutants relative to wild type (defined as 1.0) are calculated as described in Fig. 3E. The number of circles in each strain represents the number of cultures analyzed. (A) (spo11Δ) NKY3002 and NKY3196; (spo11–Y135F) NKY3001. Analysis of NKY3197 indicated that HA-epitope tagging of Spo11p did not affect the timely occurrence of major events during meiosis, including meiotic S phase (data not shown). (B) (rec102Δ) NKY3135; (red1Δ) NKY3190; (hop1Δ) NKY3191. (C) (rec8Δ) NKY3157; (rec8Δ spo11Δ) NKY3194.
spo11–Y135F exhibits normal DSB-independent meiotic homolog pairing
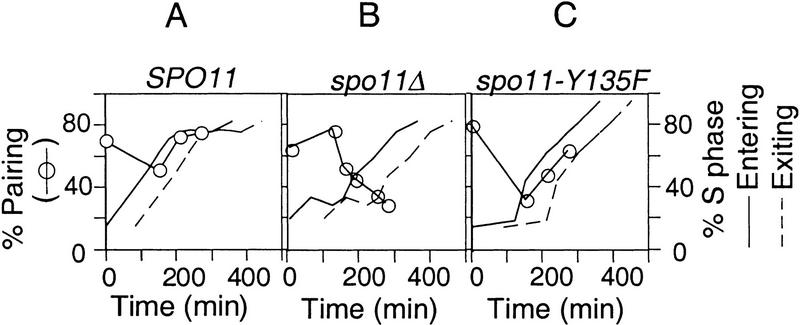
The existence and basic features of homolog pairing in budding yeast have been shown by fluorescence in situ hybridization (FISH) analysis of well-spread chromatin with locus-specific probes (Weiner and Kleckner 1994; Burgess et al. 1999). As illustrated in Figure 5A, pairing is present at high levels in wild-type premeiotic G1 cells, is disrupted during S-phase, and is restored at or immediately prior to early prophase. In contrast, essentially no restoration of pairing is observable in a spo11Δ mutant (Loidl et al. 1994; Weiner and Kleckner 1994; Rockmill et al. 1995; also see Fig. 5B).
Figure 5.
DSB-independent homolog pairing in SPO11, spo11Δ, and spo11–Y135F. Synchronous meiotic cultures for NKY1992 (SPO11; A), BWY168A (spo11Δ; B), and BWY119 (spo11–Y135F; C) were set up as described in Materials and Methods. Samples were harvested at specified time points and the extent of homolog pairing is measured by FISH analysis (Weiner and Kleckner 1994). Two homologs are defined as paired if the distance between them, visualized by a fluorescence labeled probe, is ≤0.7 μm (Weiner and Kleckner 1994). The kinetics of loss and re-establishment of pairing, (○) as well as the entry into (solid line) and exit from meiotic (broken line) S-phase are summarized.
The spo11Δ mutant is also defective with respect to both S-phase progression (above) and DSB formation. Earlier work has shown, however, that homolog pairing is independent of DSBs; for example, by the approach illustrated here, high levels of homolog pairing is restored at early prophase in a hop1Δ mutant in which the level of DSBs is extremely low (Weiner and Kleckner 1994). Thus, the role of Spo11 in homolog pairing should be independent of its role in DSB catalysis. It was therefore of interest to know whether the spo11–Y135F mutant retains pairing proficiency, in correlation with its proficiency for modulation of S-phase progression. FISH analysis demonstrates that a homozygous spo11–Y135F diploid strain is indistinguishable from an isogenic wild-type strain with respect to premeiotic pairing and the pattern of pairing disruption and restoration during early meiosis (Fig. 4A,C); minor differences between the two cases are within the range observed for day-to-day differences among different wild-type cultures. Control experiments show that sister chromatid cohesion is normal in both spo11Δ and spo11–Y135F, as judged by analogous FISH analysis of a diploid strain heterozygous for a probed region. Furthermore, no SC forms in either mutant, as judged by immunostaining of the SC component Zip1 (B.M. Weiner, unpubl.). These results provide the most clear-cut evidence to date for DSB-independent (and SC-independent) early meiotic homolog pairing and show that the functions of Spo11 for pairing and meiotic S-phase regulation can be separated cleanly from its function for DSB catalysis.
SPO11 has a role in progression of meiotic prophase independent of DSB and SC formation
Among meiotic mutants defective in DSB formation, some undergo the first meiotic division (meiosis I) with normal timing after transfer of cells to sporulation conditions, whereas others undergo meiosis I significantly earlier than normal (Jiao et al. 1999). It was proposed that this effect reflects accelerated progression through the prophase stage of meiosis and that the functions defined by the latter group of mutants are involved in generating a signal that delays progression through that stage. Interestingly, however, spo11 mutants were among those found to exhibit accelerated occurrence of meiosis I (Klapholz et al. 1985). Thus, the results presented above raised the possibility that early occurrence of meiotic divisions in certain mutants might be due, at least in part, to a reduction in the length of S phase rather than to a reduction in the length of prophase.
To address this issue, we compared wild-type and spo11 mutant strains with respect to the length of time that elapses between the time at which 50% of (active) cells complete S phase and the time at which 50% of (active) cells complete meiosis I, that is, the length of prophase. Such comparisons reveal that SPO11 is involved in delaying progression through both stages of meiosis, prophase as well as during S phase. In wild-type cells, the length of prophase was 162 ± 33.6 min (n = 14). In both spo11Δ and spo11–Y135F, prophase is substantially shorter, 114 ± 8.4 min (n = 2) and 132 ± 43.8 min (n = 7), respectively. These values are both significantly different from the wild-type values as assessed by Student's _t_-test (P < 0.005) and are not significantly different from one another (0.25 < P < 0.4). Thus, prophase lasts ∼160 min in wild-type cells and is 30–50 min shorter in both of the spo11 mutants. Interestingly, Jiao et al. (1999) report that the DSB-defective mutants that exhibit accelerated meiosis I fell into two categories according to whether the effect was more or less extreme. Perhaps the two groups correspond to the phenotypes observed for spo11Δ and spo11–Y135F mutants.
Deletion of REC8 prolongs S phase in wild-type and spo11Δ strains
In S. cerevisiae, a rec8Δ mutation has no discernible effect on vegetative growth of a diploid but has severe effects on sister chromatid cohesion, meiotic recombination, and progression through the meiotic divisions (Klein et al. 1999; J. Dekker, unpubl.). Parallel analysis of wild-type and rec8Δ meiotic cultures reveals an increase of ∼10% in the length of meiotic S phase in rec8Δ mutant strains (Fig. 4C; Table 1). An even stronger effect is observed in a spo11Δ background: the length of meiotic S phase in the rec8Δ spo11Δ double mutant is ∼30% longer than in the spo11Δ mutant. S phase in the double mutant is restored to a length that is intermediate between S-phase lengths in rec8Δ and wild-type strains (and statistically indistinguishable from both cases) (Fig. 4C; Table 1). Cumulative curve analysis shows, however, that the kinetics of entry into S phase is normal in both rec8Δ and rec8Δ spo11Δ strains.
These studies also reveal that spo11Δ rec8Δ double mutant cells have a reduced probability of entering meiosis at all. The percentage of such cells that complete meiotic S phase is lower in the double mutant (71.2 ± 10.8%; n = 4) than for wild type (85.1 ± 8.3%; n = 4), and the deficit of 4C cells is found instead in the 2C peak (data not shown). The double mutant also exhibits a corresponding reduction in sporulation efficiency (76.1 ± 7.1%; n = 2 vs. 87.3 ± 4.0%; n = 14). No such effect is observed in rec8Δ and spo11Δ single mutant strains, both of which exhibit normal percentages of 2C and 4C cells. spo11Δ sporulated with normal efficiencies (above) while rec8Δ arrests in prophase (Klein et al. 1999; data not shown).
Discussion
Meiotic interchromosomal interaction proteins participate in meiotic S-phase progression
The products of the SPO11 and REC8 genes are involved in determining the length of meiotic S phase, that is, in how the DNA replication process proceeds. Such involvement presumably reflects a need to directly integrate events important for interhomolog and intersister interactions with ongoing DNA replication and for spatial and temporal coordination of the two types of interactions with one another (introductory section).
Because deletion of SPO11 results in a shorter S phase, Spo11p is defined as a negative regulator of DNA replication; as deletion of REC8 results in a longer S phase, Rec8p is defined as a positive effector of replication progression. The finding that Rec8 is required for timely S-phase progression is analogous to the finding that its mitotic homolog Mcd1/Scc1 is required for timely S-phase progression during the mitotic cell cycle (Guacci et al. 1997). The identification of a molecule that makes S phase proceed more rapidly than normal is unique, although a negative regulator of DNA replication initiation has been identified in E. coli (Boye et al. 1996).
Among the genes examined in this study, only SPO11 and REC8 were involved in meiotic replication progression, implying that their roles in this process are relatively fundamental and unique.
Earlier and later roles for Spo11
The spo11–Y135F mutant exhibits normal S-phase length and normal early homolog pairing but is defective for DSB formation and formation of SC. The activities of Spo11p for the two early functions, replication control and homolog pairing, could be rather closely related, as a spo11Δ mutation eliminates both of these processes and spo11–Y135F, red1Δ, hop1Δ, and rec102Δ mutations leave both processes intact. Similarly, the two later functions are likely also related to one another, as the extent and timeliness of SC formation is closely correlated with the process of DSB formation (for review, see Zickler and Kleckner 1999). All activities of Spo11p could, however, involve the same basic ability of the protein to bind chromosomes and/or to localize to particular structural features of the chromosomes as they develop during S phase.
Regulation of DNA replication by interchromosomal interaction proteins
We like the idea that progression of replication through the genome is directly promoted by the development of specific chromosomal features in newly replicated regions. Interchromosomal interaction proteins could then directly influence the rate of S-phase progression via their involvement in the development of these features (introductory section). Formally, such a model requires the existence of a kinetic barrier(s) that impedes progression of replication and a feedback process in which proteins that influence the length of S phase alter the time required to get through the kinetic barrier. Rec8-mediated events should be required for satisfying progression requirements; when those requirements are not satisfied, progression is slower. Spo11-mediated events should be required for maximal robustness of the barrier, that is, for imposing additional requirements that must be met before the barrier is eliminated; in the absence of these additional requirements, progression is faster. Such a model also requires additional features, however, to ensure that the temporal program of origin firing is adjusted to accommodate the slower progression of replication from region to region through the genome.
Rec8 is involved in both intersister and interhomolog interactions, whereas Spo11 is thus far implicated only in interhomolog events. The proposed roles for Rec8 and Spo11 are consistent with a mechanism in which intersister requirements must be satisfied and interhomolog requirements are coupled to that underlying process. Specifically, Rec8 would act both to satisfy intersister requirements and to couple interhomolog-specific effects to the intersister-specific process; a Spo11-dependent process would then be layered on top of, and linked to, this Rec8-dependent process. The functional relationships between rec8Δ and spo11Δ mutations with respect to S-phase progression are also consistent with this model: A rec8Δ mutation increases the length of S phase in both the presence and absence of SPO11; a spo11Δ mutation, in contrast, shortens the length of S phase in the presence of REC8 but has little or no effect in a rec8Δ background.
Meiotic S-phase prolongation
Meiotic S phase is longer than mitotic S phase in all organisms examined to date (Holm 1977 and references therein; for S. cerevisiae, see above). Notably, also, the lengthening of meiotic S phase in yeast is not a “trivial” response to the nutrient deprivation conditions normally used to induce meiosis in this organism. Yeast cells can carry out normal meiosis in nitrogen-rich YPA growth medium under certain conditions, that is, when stable expression of the meiotic regulator IME1 is favored (Hayashi et al. 1998; Colomina et al. 1999). S-phase data obtained in the latter study clearly demonstrate a long (meiotic-like) S phase under such conditions.
Several types of comparisons have failed to reveal significant differences that could account for the difference in S-phase lengths between the mitotic and meiotic programs. For example, within the left-most 200 kb of yeast chromosome III, the same origins are utilized, with similar relative efficiencies, during meiosis as in vegetatively growing cells (Collins and Newlon 1994). Also in yeast, rates of fork movement, fork lengths, and Okazaki fragment sizes are similar or identical in both mitotic and meiotic S phases in yeast (Petes and Williamson 1975; Riven and Fangman 1980; Johnston et al. 1982; Williamson et al. 1983). Similarly, fork movement rates were found to be the same during mitotic and meiotic DNA replication in newt (Callan and Taylor 1968).
Three types of mechanisms for S-phase prolongation have generally been considered: (1) Fewer origins might be utilized during meiotic as compared to mitotic S phase; (2) the rate of DNA synthesis during the elongation phase might be reduced; and (3) the same origins may be used in both cases, but the time between successive origin firings is longer during meiosis than in mitotic cells. The first two models appear to be excluded by available data (above); the third model remains possible but carries with it an additional implicit requirement. It is known for yeast that if replication of one origin passes through another origin, firing of the latter origin is suppressed (Santocanale et al. 1999). Thus, in the absence of other effects, model 3 requires the presence of fork barriers to prevent the over-reaching of replication forks from early firing origins. Given the existence of fork barriers, feedback controls might modulate the rate of progression through those barriers, thereby permitting specific features of the chromosome to influence the length of S phase at that level. It would not be surprising if several types of effects are involved. For example, in lily, where replication of euchromatic sequences precedes replication of heterochromatic sequences in two distinct phases, meiosis has two distinct effects: (1) The length of time required for replication of each set of sequences is increased; and (2) the interval between replication of the two sets of sequences is also increased (Holm 1977).
Nonetheless, whatever mechanisms are responsible for meiotic S-phase prolongation, it seems likely that effects of spo11Δ and rec8Δ mutations on the length of meiotic S phase reflect the participation of Spo11 and Rec8 proteins in that process. And given such a linkage, it is more economical to invoke the participation of these proteins via a feedback process that senses the completion of functions relevant to their primary roles for meiosis than to assign them additional, separate roles directed at the status of replication origins. Therefore, although we do not rule out contributions from additional regulatory processes, the interchromosomal feedback model seems to be an attractive way of explaining much of the available information.
SPO11 and REC8 also affect meiotic progression at G1/S and during prophase
Null mutations in the SPO11 and REC8 genes synergistically affect the probability that a cell will begin DNA replication. We infer that during G1, both proteins are present and involved in functionally related process(es) required for entry into S phase. Rec8p is known from earlier studies to be present on meiotic chromosomes prior to S phase in both fission yeast (Watanabe and Nurse 1999) and budding yeast (Klein et al. 1999). Localization of Spo11p to chromosomes during meiosis has not been reported for any stage.
Also, the length of meiotic prophase is reduced by ∼25% in both spo11Δ and spo11–Y135F mutants. By implication, Spo11 is required for implementation of some process that acts to slow down meiotic progression. In essence, Spo11 is a negative regulator of meiotic prophase progression just as it is a negative regulator of S-phase progression, except that the prophase effect requires later functions of Spo11, those specifically dependent on spo11–Y135F, which are not required for its S-phase role. Moreover, although the spo11–Y135F mutation affects DSB formation by affecting a residue directly involved in DSB catalysis, the process that slows down meiotic prophase progression does not appear to require meiotic DSBs per se, as several DSB-negative mutants seem to exit prophase at the normal time (Jiao et al. 1999). Perhaps a conformational change in Spo11p that accompanies DSB formation is required to set up an intraprophase checkpoint system for monitoring ensuing events. It has been proposed that DSB-independent homolog pairing contacts are lost subsequent to, and dependent on, the formation of DSBs (Zickler and Kleckner 1999). Perhaps a Spo11-dependent checkpoint system monitors the post-DSB loss of interhomolog pairing contacts. Such a system could be related and/or identical to that which monitors the progression of recombination itself.
Materials and methods
Strains
All yeast strains are isogenic derivatives of SK1. NKY3000 is a self-diploidized spore clone of NKY730 (MATa/_MAT_α, HO/′′, lys2/′′, ura3::hisG/′′, leu2::hisG/′′). The following are isogenic derivatives of NKY3000: NKY3001 (spo11–Y135F–HA-URA/+), NKY3002 (spo11Δ::hisG/+), NKY3135 (rec102Δ::URA3/+), NKY3157 (rec8Δ::KanMX4/+), NKY3190 (red1Δ::KanMX4/+), NKY3191 (hop1Δ::URA3/+), NKY3194 (rec8Δ::KanMX4/+, spo11Δ::hisG–URA3–hisG/+), NKY3196 (spo11Δ::hisG–URA3–hisG/+), NKY3197 (SPO11–HA–KanMX4/+). Each strain was generated by transformation using allele replacement constructs (details available on request) as confirmed by Southern analysis. NKY3002 is a 5-FOA resistant isolate of NKY3196. NKY3194 was generated by mating NKY3001 and NKY3196 (below).
Other strains used are also SK1 derivatives isogenic with the above: NKY611 (MATa/_MAT_α, ho::LYS2/′′, ura3/′′, lys2/′′, leu2::hisG/′′), NKY648 (MATa/_MAT_α, ho::LYS2/′′, ura3/′′, lys2/′′, spo11Δ::hisG–URA3–hisG/′′), BWY119 (MATa/_MAT_α, ho::LYS2/′′, ura3/′′, lys2/′′, leu2::hisG/′′, spo11–Y135F/′′), BWY168A (MATa/_MAT_α, ho::LYS2/′′, ura3/′′, lys2/′′, leu2::hisG/′′, his4X–LEU2/′′, ade2::LK/+, spo11Δ::hisG–URA3–hisG/′′), and NKY1992 (MATa/_MAT_α, ho::LYS2/′′, ura3/′′, lys2/′′, leu2::hisG/′′, his4X/′′, ade2::LK/+).
Mating of homothallic strains
To mate two homothallic strains, spores of each type were brought into association and allowed to germinate, and the desired diploids selected using appropriate markers. Spores from each strain were prepared by treatment with Zymolyase and sonicated briefly; spores of the two strains were then mixed, gently pelleted, and resuspended in 10–20 μl of H2O. The mixture was placed onto a YPD plate overnight and replica plated onto a double selection plate on the following day.
Synchronous mitotic and meiotic time courses
Liquid mitotic media were YPD (1% yeast extract, 2% Bacto-Peptone, 2% glucose), YPA (1% yeast extract, 2% Bacto-Peptone, 1% potassium acetate), or YPG (1% yeast extract, 2% Bacto-Peptone, 2% glycerol). The strain of interest was patched from a −80°C glycerol stock onto a YPG plate and incubated overnight at 30°C. Cells were streaked for single colonies on YPD which, after 2 days growth, were used to inoculate a 5-ml overnight YPD culture. To obtain a synchronous G1/G0 cell population, cells from these cultures were diluted to a final OD600 of 0.2 ± 0.02 in 100 ml of YPA in a 1-liter flask, and grown with vigorous shaking for 13.5 hr at 30°C. Cultures with an OD600 of 1.15–1.4 and consisting of >85% large unbudded cells were chosen for setting up synchronous meiotic and/or mitotic cultures. To induce meiosis, cells were washed and transferred into the same volume of SPM (0.3% potassium acetate, 0.02% raffinose). For mitotic analysis, cells were diluted to OD600 of 0.2 in the desired medium with or without nocodazole (10 μg/ml). Bud morphology and nuclear morphology were examined by combined phase contrast and fluorescence microscopy. FISH assay was performed as described by Weiner and Kleckner (1994; details available on request).
FACS analysis of DNA replication
One-milliliter samples from meiotic or mitotic cultures were pelleted and resuspended in 40% ethanol, 0.1 m sorbitol. Samples were stored at −20°C until FACS analysis. A day before FACS analysis, cells were pelleted and resuspended in 0.3 ml of 50 mm Tris at pH 7.5 containing 100 μg/ml RNAse and left overnight at room temperature. Next, 0.3 ml of propidium iodide solution (38 mm sodium citrate, 6.9 mm propidium iodide) was added. Samples were briefly sonicated and analyzed on a Becton-Dickinson FACScan analyzer.
Quantitation of S-phase and G2/M-phase fractions
Quantification of the fraction of cells undergoing DNA replication and those that have completed bulk DNA replication is shown in Figure 1B. Two vertical lines (a,b) were drawn down the center of the 2C and the 4C peaks. A third line (c), which is tangent to the lowest point of the histogram between the 2C and 4C peaks, and parallel to the baseline (e) is drawn to define the rectangular area. Another line (d) is drawn to define the right triangle comprising roughly half of the 4C peak. The rectangle and triangle were cut and weighed, and divided by the total weight of the intact histogram. The percentage of weight in the rectangle and twice that of the triangle are defined to represent the fraction of cells undergoing DNA replication and those that have completed bulk DNA replication, respectively.
Analysis of FACS profiles
The total distribution of DNA contents provide by FACS is the sum of three individual curves, for G1, S, and G2/M subpopulations. The fractions of S and G2/M cells can be approximated without knowing the three component curves by the approach described in Figure 1B, which is derived from those of Slater et al. (1977) and Padmore et al. (1991). The S-phase lengths determined for wild-type meiotic and mitotic cells in the current study are very similar to those obtained by earlier studies using different methods (see text). Furthermore, we applied our approach to certain sample FACS profiles analyzed previously by a computer model that provides best-fit curves for the three subpopulations (Dien et al. 1994); the two methods give essentially the same fraction of S-phase cells (within 10%; data not shown). Most importantly, however, the conclusions of the current analysis pertain to relative, rather than absolute, S-phase lengths. Thus, the conclusions would be compromised only if there were some systematic difference in the types of FACS profiles obtained in different strains that would introduce bias into the calculated fractions of S-phase cells. We have seen no evidence of such effects. Finally, S-phase estimates are not compromised by unsuspected contributions from mitochondrial DNA replication. During meiosis, mitochondrial replication is uncoupled from nuclear replication, which occur, respectively, immediately after transfer to SPM and after a 1- to 2-hr delay (e.g., Piñon et al. 1974; Williamson et al. 1983; Padmore et al. 1991; this study). Because no replication is detectable during the first 1–2 hr, either by FACS (Padmore et al. 1991) or by incorporation of radioactive precursors (Williamson et al. 1983), the contribution of mitochondrial replication to FACS profiles is apparently insignificant.
Life spans and cumulative curves for DNA replication
The duration of S-phase is given by the area under the corresponding noncumulative curve (e.g., Figs. 1B and 3B; Padmore et al. 1991; Burgess et al. 1999), divided by the total fraction of nuclei progressing through the mitotic or meiotic cell cycle (i.e., active fraction) defined in the current study as the fraction of cells in the 4C peak measured at the termination of each experiment. Cumulative curves describe the fraction of cells that have either entered or completed S phase as a function of time; the curve is identical to the noncumulative curve plotted up to the first non-zero time point and one life span thereafter. Then, for any time point (t) after one life span has elapsed, the cumulative curve value at t is equal to the noncumulative curve value at t plus the cumulative curve value at t minus one life span (Padmore et al. 1991; Burgess et al. 1999). The exit curve for S phase is given by plotting the entry curve at a rightward displacement along the _x_-axis by one life span.
Acknowledgments
Research was supported by National Institute of Health grants RO1-GM44794 and R37-GM25326 to N.K and EMBO fellowship ALTF223-1998 to J.D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kleckner@fas.harvard.edu; FAX (617) 495-0758.
References
- Atcheson CL, Didomenico B, Frackman S, Esposito RE, Elder RT. Isolation, DNA sequence and regulation of a meiosis-specifc eukaryotic recombination gene. Proc Natl Acad Sci. 1987;84:8035–8039. doi: 10.1073/pnas.84.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. Synaptonemal complex morphogenesis and sister-chromatid cohesion requires Mek1-dependent phosphorylation of a meiotic chromosomal proteins. Genes & Dev. 1998;12:3551–3563. doi: 10.1101/gad.12.22.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Smith J. The effect of polyploidy on meiotic duration and pollen development in cereal anthers. Proc R Soc Lond B. 1972;181:81–107. [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bhargava J, Engebrecht JA, Roeder GSH. The rec102 mutant of yeast is defective in meiotic recombination and chromosome synapsis. Genetics. 1992;130:59–69. doi: 10.1093/genetics/130.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: Differential roles for DnaA and SeqA proteins. Proc Natl Acad Sci. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B, Chlebowicz-Sledziewska E, Fangman W. Cell cycle phases in the unequal mother/daughter cell cycles of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2529–2531. doi: 10.1128/mcb.4.11.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Kleckner N, Weiner B. Somatic pairing of homologs in budding yeast: Existence and modulation. Genes & Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan HG. DNA replication in the chromosome of eukaryotes. Cold Spring Harbor Symp Quant Biol. 1973;38:195–203. doi: 10.1101/sqb.1974.038.01.023. [DOI] [PubMed] [Google Scholar]
- Callan HG, Taylor JH. A radioautoragraphic study of the time course of male meiosis in the newt Triturus vulgaris. J Cell Sci. 1968;3:615–626. doi: 10.1242/jcs.3.4.615. [DOI] [PubMed] [Google Scholar]
- Collins I, Newlon CS. Chromosomal DNA replication initiates at the same origins in meiosis and mitosis. Mol Cell Biol. 1994;14:3524–3534. doi: 10.1128/mcb.14.5.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomina N, Gari E, Gallego C, Herrero E, Aldea M. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 1999;18:320–329. doi: 10.1093/emboj/18.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes & Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- Dien B, Peterson M, Sriene F. Cell-cycle analysis of Saccharomyces cerevisiae. Methods in Cell Bio. 1994;42:457–475. doi: 10.1016/s0091-679x(08)61090-0. [DOI] [PubMed] [Google Scholar]
- Fox CA, Ehrenhofer-Murray AE, Loo S, Rine J. The origin recognition complex, Sir1, and the S-phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Ohkuni K, Yamashita I. Control of division arrest and entry into meiosis by extracellular alkalisation in Saccharomyces cerevisiae. Yeast. 1998;14:905–913. doi: 10.1002/(SICI)1097-0061(199807)14:10<905::AID-YEA290>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Holm PB. The premeiotic DNA replication of euchromatin and heterochromatin in Lilium longiflorum (Thunb.) Carlsberg Res Commun. 1977;42:249–281. [Google Scholar]
- Jiao K, Bullard SA, Salem L, Malone RE. Coordination of the initiation of recombination and the reductional division in meiosis in Saccharomyces cerevisiae. Genetics. 1999;152:117–128. doi: 10.1093/genetics/152.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Williamson DH, Johnston AL, Fennell DJ. On the mechanism of premeiotic DNA synthesis in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1982;141:53–62. doi: 10.1016/0014-4827(82)90067-2. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Klapholz S, Waddell CS, Esposito RE. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Meiosis: How could it work? Proc Natl Acad Sci. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo S, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Krawchuck M, DeVeaux L, Wahls W. Meiotic chromosome dynamics dependent upon the rec8+, rec10+, and rec11+ genes of the fission yeast Schizosaccharomyces pombe. Genetics. 1999;153:57–68. doi: 10.1093/genetics/153.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl S, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Parisi S, McKay M, Molnar M, Thompson M, van der Spek P, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers J, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD, Williamson DH. Fiber autoradiography of replicating yeast DNA. Exp Cell Res. 1975;95:103–110. doi: 10.1016/0014-4827(75)90614-x. [DOI] [PubMed] [Google Scholar]
- Piñon R, Salts Y, Simchen G. Nuclear and mitochondrial DNA synthesis during yeast sporulation. Exp Cell Res. 1974;83:231–238. doi: 10.1016/0014-4827(74)90334-6. [DOI] [PubMed] [Google Scholar]
- Riven CJ, Fangman WL. Replication fork rate and origin activation during the S phase of Saccharomyces cerevisiae. J Cell Biol. 1980;85:108–115. doi: 10.1083/jcb.85.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Engebrecht JA, Scherthan H, Loidl J, Roeder GS. The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics. 1995;141:49–59. doi: 10.1093/genetics/141.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Sharma K, Diffley J. Activation of dormant origins of DNA replication in budding yeast. Genes & Dev. 1999;13:2360–2364. doi: 10.1101/gad.13.18.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Shibahara K-I, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:576–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes & Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M, Sharrow S, Gart J. Cell cycle of Saccharomyces cerevisiae in populations growing at different rates. Proc Natl Acad Sci. 1977;74:3850–3854. doi: 10.1073/pnas.74.9.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes & Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Johnston LH, Fennell DJ, Simchen G. The timing of the S phase and other nuclear events in yeast meiosis. Exp Cell Res. 1983;145:209–217. doi: 10.1016/s0014-4827(83)80022-6. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic Chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]