Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1 (original) (raw)
Abstract
In the early Caenorhabditis elegans embryo, maternally expressed PIE-1 protein is required in germ-line blastomeres to inhibit somatic differentiation, maintain an absence of mRNA transcription, and block phosphorylation of the RNA polymerase II large subunit (Pol II) carboxy-terminal domain (CTD). We have determined that PIE-1 can function as a transcriptional repressor in cell culture assays. By fusing PIE-1 sequences to the yeast GAL4 DNA-binding domain, we have identified a PIE-1 repression domain that appears to inhibit the transcriptional machinery directly. A sequence element that is required for this repressor activity is similar to the Pol II CTD heptapeptide repeat, suggesting that the PIE-1 repression domain might target a protein complex that can bind the CTD. An alteration of this sequence element that blocks repression also impairs the ability of a transgene to rescue a pie-1 mutation, suggesting that this repressor activity may be important for PIE-1 function in vivo.
Keywords: Transcription, repression, C. elegans, germ line, RNA Pol II CTD, zinc finger
Maternally expressed PIE-1 protein is required in Caenorhabditis elegans for specification of embryonic cells (termed blastomeres) that give rise to the germ line (Mello et al. 1992). PIE-1 protein is present in the oocyte, and after fertilization it is segregated predominantly to the germ-line blastomere lineage, in which it accumulates in nuclei (Mello et al. 1996; Tenenhaus et al. 1998). Whereas somatic blastomeres activate mRNA transcription by the four-cell stage, germline blastomeres do not appear to produce any mRNAs until PIE-1 disappears at approximately the 100-cell stage (Seydoux et al. 1996). In the absence of PIE-1, however, germ-line and somatic blastomeres initiate mRNA transcription at the same time (Seydoux et al. 1996), and germ-line blastomeres differentiate in response to intrinsic maternally expressed transcription factors, adopting fates similar to those of their somatic sisters (Mello et al. 1992). These findings suggest that PIE-1 blocks differentiation in the germ line by inhibiting gene transcription (Mello et al. 1996; Seydoux et al. 1996). Germ-line blastomeres also appear to be transcriptionally inactive in the Drosophila embryo (Lamb and Laird 1976; Zalokar 1976; Kobayashi et al. 1988; Seydoux and Dunn 1997; Van Doren et al. 1998), indicating that a general transcriptional block may be a conserved aspect of germ cell specification.
During eukaryotic mRNA transcription, the RNA polymerase II large subunit (Pol II) carboxy-terminal domain (CTD) is phosphorylated upon the transition from initiation to elongation (for review, see Dahmus 1996). The CTD consists of tandem repeats of the sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser (YSPTSPS), in which the serines at positions 2 and 5 are the major phosphorylation targets (Zhang and Corden 1991; West and Corden 1995; Patturajan et al. 1998). Multiple lines of evidence indicate that the CTD is bound by regulatory components of the general transcriptional machinery, as well as by protein complexes that mediate mRNA capping, processing, and termination, and that it thereby integrates transcription initiation with subsequent elongation and mRNA processing events (Thompson et al. 1993; Cho et al. 1997; McCracken et al. 1997a,b; Neugebauer and Roth 1997; Myers et al. 1998). In C. elegans embryos, an antibody epitope corresponding to CTD phosphoserine 2 appears in somatic cells when embryonic transcription begins (Seydoux and Dunn 1997). This phosphoepitope is not detected in germ-line blastomeres in the presence of PIE-1 but is expressed in these cells in pie-1 mutant embryos. In contrast, PIE-1 does not completely inhibit appearance of an epitope corresponding to phosphoserine 5 and does not appear to block transcription by RNA polymerase I (Seydoux and Dunn 1997). These findings suggest that transcription by Pol II is specifically prevented by PIE-1 and that this inhibition may occur prior to phosphorylation of CTD serine 2 (Seydoux and Dunn 1997).
To maintain transcriptional silencing in germ-line blastomeres, it is possible that PIE-1 inhibits the transcriptional machinery directly, a model that is consistent with its presence in nuclei. Alternatively, PIE-1 could be required for a signal that prevents the onset of embryonic transcription, or it might regulate expression or modification of an essential transcriptional machinery component. The PIE-1 protein contains two central Cys-Cys-Cys-His (C3H) zinc fingers that are separated by an Arg- and Ser-rich sequence that includes Arg–Ser (RS) dipeptide repeats (Fig. 1A; Mello et al. 1996). Other C3H zinc finger proteins have been implicated in mRNA binding, cleavage, or processing (Barabino et al. 1997; Murray et al. 1997; Rudner et al. 1998) or in post-transcriptional gene regulation (Guedes and Priess 1997; Carballo et al. 1998; Tabara et al. 1999). Proteins with RS repeats are associated with pre-mRNA splicing (for review, see Valcarcel and Green 1996). These features suggest that PIE-1 or associated proteins might bind RNA, but they do not discriminate among possible models for PIE-1 function.
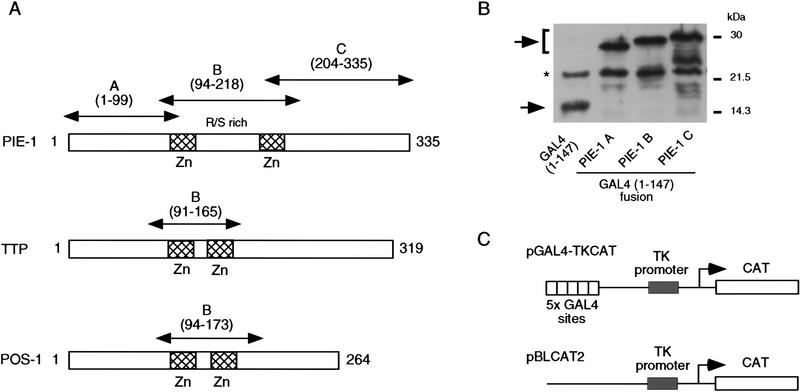
Figure 1.
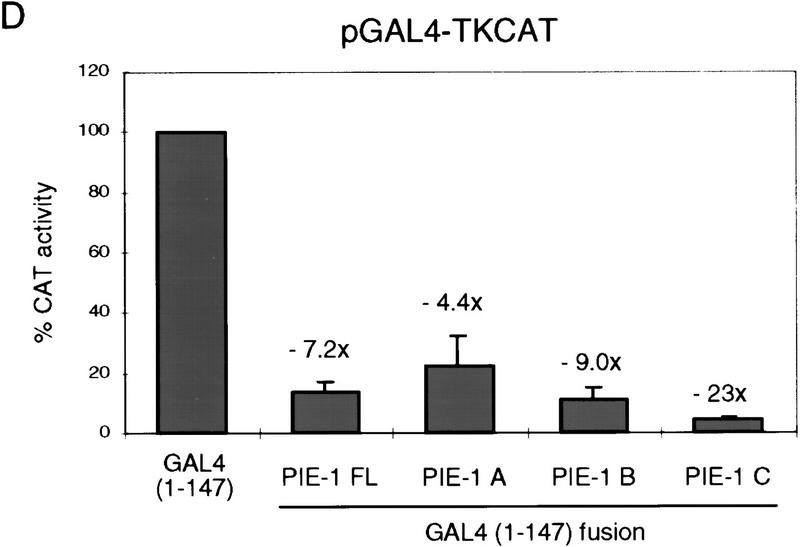
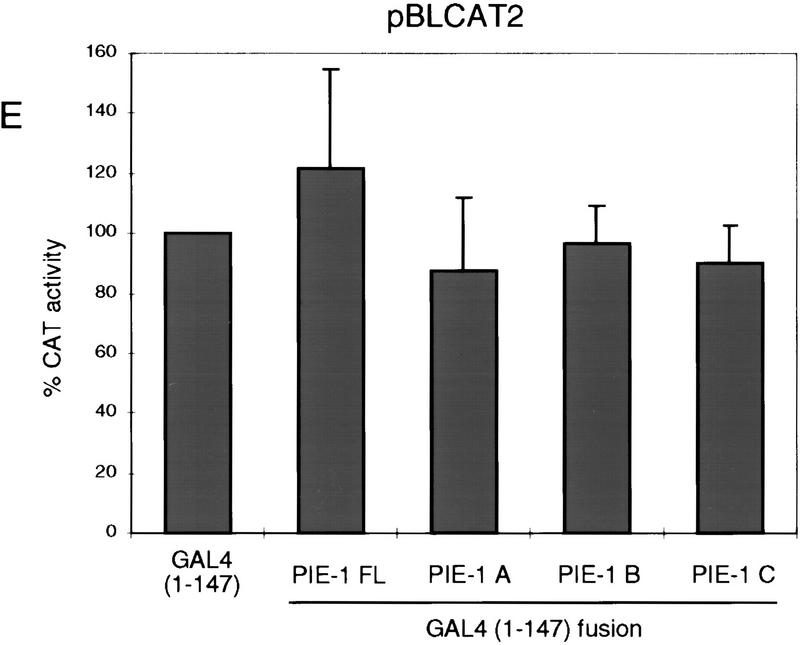
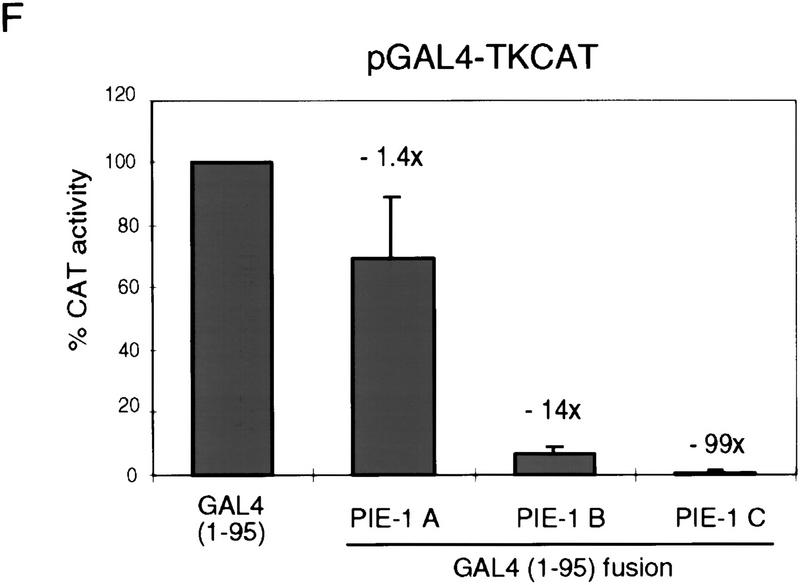
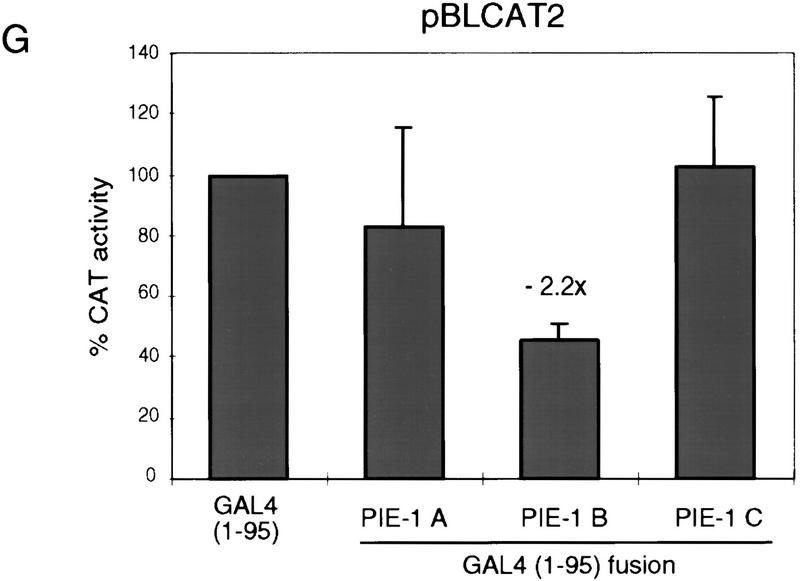
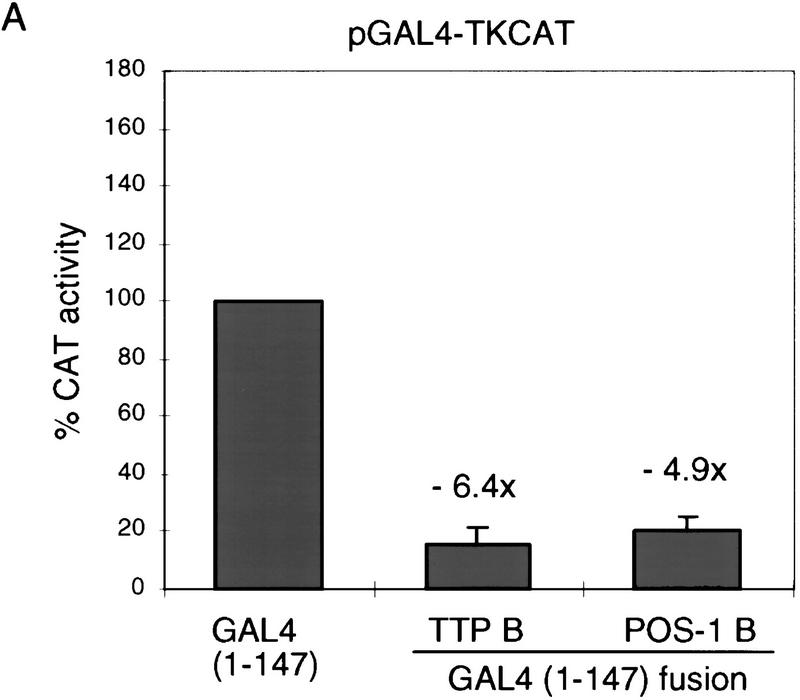
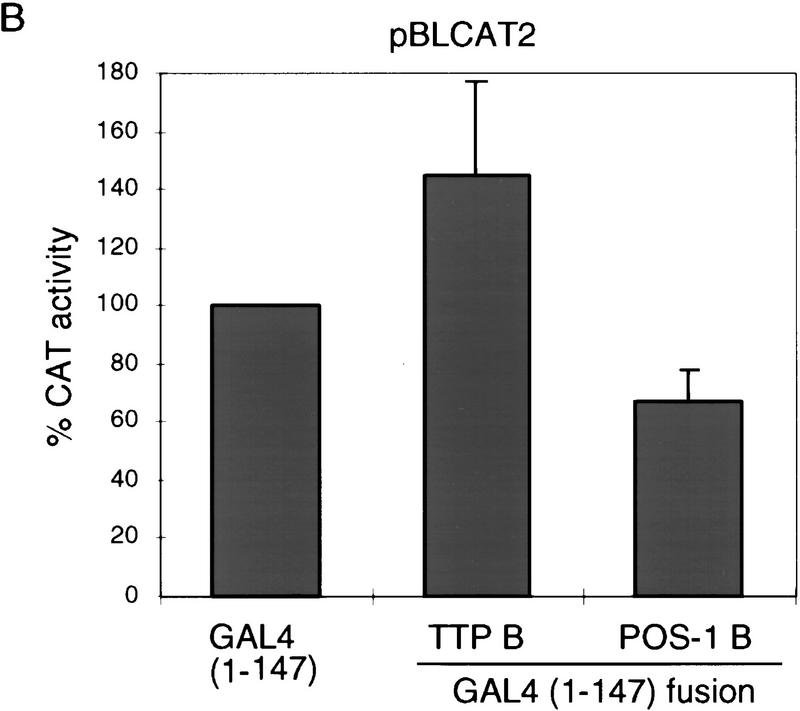
PIE-1 contains a powerful transcriptional repression domain. (A) Schematic diagram of the PIE-1 (Mello et al. 1996), mouse TTP (Lai et al. 1990), and C. elegans POS-1 (Tabara et al. 1999) C3H zinc finger proteins. Residues that are fused to the GAL4(1–147) DNA-binding domain are marked by arrows and indicated in parentheses. The boundaries of these regions were chosen on the basis of the location of the respective zinc fingers. The PIE-1 zinc finger regions span residues 99–123 and 185–208, those of TTP span 96–120 and 134–158, and those of POS-1 span 99–122 and 142–166. (B) Expression of GAL4(1–147) PIE-1 fusions in HeLa cells. Immunoblot of whole cell extracts from transfected HeLa cells probed with anti-GAL4(1–147) antibodies. Arrows indicate specific bands, and the asterisk a nonspecific band. Other GAL4(1–147) fusion proteins were expressed at similar levels (not shown). (C) Reporter constructs. pGAL4–TKCAT contains five GAL4 sites upstream of a HSV TK promoter that drives the CAT reporter gene (Shi et al. 1991). pBLCAT2 is an equivalent plasmid without GAL4 sites (Luckow and Schutz 1987). (D) Tethered repression by GAL4(1–147) PIE-1 fusions. HeLa cells were cotransfected with 1 μg of GAL4(1–147) effector plasmid and 5 μg of pGAL4–TKCAT reporter plasmid. Each data point represents the mean relative CAT activity compared to that of GAL4(1–147) alone (100%). Fold repression is indicated above each bar. (E) Assayed as in D, except with 5 μg of pBLCAT2 reporter plasmid. (F) Tethered repression by GAL4(1–95) PIE-1 fusions. HeLa cells were cotransfected with 1μg of GAL4(1–95) effector plasmid and 10 μg of pGAL4–TKCAT reporter plasmid. These fusion proteins were expressed at comparable levels (not shown). Each data point represents the mean relative CAT activity compared to that of GAL4(1–95) (100%). (G) As in F, except 10 μg of pBLCAT2 reporter plasmid was used. Error bars represent the standard deviation of three separate experiments.
If PIE-1 inhibits transcription directly, by acting on a component of the Pol II transcriptional machinery, it would be predicted to act on an evolutionarily conserved target, because the eukaryotic transcriptional apparatus is highly conserved (Orphanides et al. 1996; Hampsey 1998). This hypothesis also predicts that PIE-1 would contain regions that repress transcription when brought to a promoter. We have tested these predictions in human cell transfection assays and have determined that PIE-1 contains sequences that repress transcription when they are tethered to promoters through a yeast GAL4 DNA-binding domain. This repression domain is located outside of the PIE-1 zinc finger region, within the carboxy-terminal one-third of the protein. It appears to act directly on the Pol II transcriptional machinery and is composed of multiple sequence elements. One of these elements is similar to a CTD heptapeptide repeat and is required for the repressor activity, suggesting that the PIE-1 repression domain might target a protein complex that interacts with the CTD. A disruption of the CTD-like sequence motif that blocks repression also substantially decreases the efficiency of transgenic rescue of a pie-1 mutation, suggesting that this repressor activity may be a normal aspect of PIE-1 function in vivo.
Results
Repression of transcription by PIE-1 regions
To determine whether the PIE-1 protein has transcriptional repressor activity, we have linked full-length PIE-1 and three PIE-1 regions (PIE-1 A, B, and C; Fig. 1A) to the yeast GAL4 DNA-binding domain (residues 1–147), which can be targeted to heterologous promoters through its binding site. This approach has allowed us to test directly for repressor activity in cultured human cells, without requiring any species- or cell-type-specific factors that might otherwise be involved in targeting PIE-1 to transcription complexes. We have assayed the effects of these fusion proteins (Fig. 1B; not shown) on the pGAL4–TKCAT reporter, which contains five GAL4 sites adjacent to a herpes simplex virus thymidine kinase (HSV TK) promoter (Fig. 1C; Shi et al. 1991). Consistent with previous reports (Williams et al. 1995), GAL4(1–147) activates transcription of pGAL4–TKCAT approximately threefold in this assay (not shown). Compared to GAL4(1–147) alone, the GAL4(1–147) full-length PIE-1 fusion protein represses transcription from pGAL4–TKCAT approximately sevenfold (PIE-1 FL; Fig. 1D). GAL4(1–147) PIE-1 A represses this reporter by approximately fourfold, and GAL4(1–147) PIE-1 B by ninefold (Fig. 1D). Significantly, the GAL4(1–147) PIE-1 C fusion represses pGAL4–TKCAT by 23-fold (Fig. 1D), suggesting that in this assay, this more powerful repressor activity might be masked in the context of the GAL4(1–147) full-length PIE-1 fusion protein. These fusion proteins do not repress a control reporter (pBLCAT2) that lacks GAL4 sites (Fig. 1C,E), indicating that their repressor activities are dependent on tethering to the promoter.
We have also assayed for repression activity in the absence of any activation by GAL4, by fusing these three PIE-1 regions to the minimal GAL4 DNA-binding domain (residues 1–95). The GAL4(1–95) PIE-1 B and the GAL4(1–95) PIE-1 C fusions repress transcription from the pGAL4–TKCAT reporter by 14- and 99-fold, respectively (Fig. 1F). In contrast, the effect of GAL4(1–95) PIE-1 A is very weak (Fig. 1F), suggesting that the activity of PIE-1 A in Figure 1D derives from an effect on activation by GAL4(1–147). With the exception of a weak repression by GAL4(1–95) PIE-1 B, these fusion proteins do not not inhibit the pBLCAT2 control reporter (Fig. 1C,G), indicating that their effects are site dependent. These experiments demonstrate that the PIE-1 B and C regions each have a robust tetherable repressor activity.
To investigate whether these repressor activities might be a general characteristic of the C3H zinc finger protein family, we have tested whether two other C3H zinc finger proteins, mouse TTP (also known as Nup475 and TIS11; DuBois et al. 1990; Lai et al. 1990; Varnum et al. 1991) and C. elegans POS-1 (Tabara et al. 1999; Fig. 1A), have similar repressor activities in our assay. Neither of these proteins are predicted to function as transcriptional repressors: TTP is stimulated by growth factors (DuBois et al. 1990; Lai et al. 1990; Varnum et al. 1991) and is involved in regulating expression of the cytokine tumor necrosis factor-α at a post-transcriptional level (Carballo et al. 1998); POS-1 is a predominantly cytoplasmic protein involved in C. elegans germ-line development (Tabara et al. 1999). The PIE-1, TTP, and POS-1 zinc fingers are similar, but these last two proteins lack the RS-rich intervening region present in PIE-1 (Fig. 1A). Like the PIE-1 B region, the TTP and POS-1 zinc finger regions (Fig. 1A) each repress transcription in a site-dependent manner when fused to GAL4(1–147) (Figs. 1D,E, and 2A,B). This finding suggests that the repressor activity of these zinc finger regions should be interpreted cautiously, given that the latter two proteins are not predicted to be transcription factors. Unlike PIE-1, however, TTP and POS-1 lack any other regions that have repressor activity in this assay (not shown), indicating that the repressor activity in the C region is particular to PIE-1.
Analysis of the PIE-1 carboxy-terminal repression domain
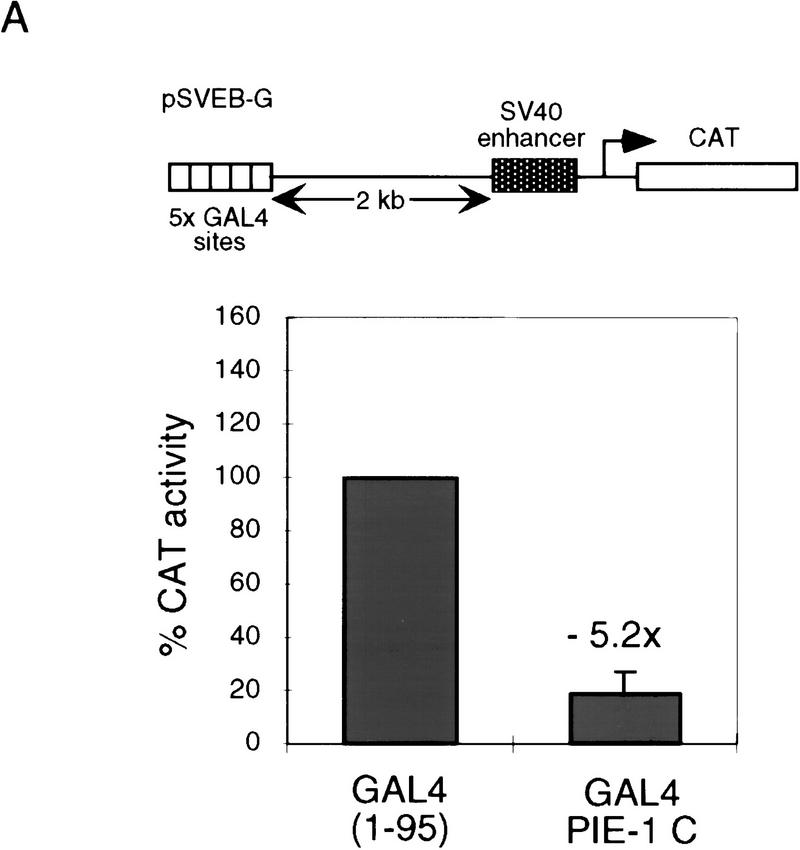
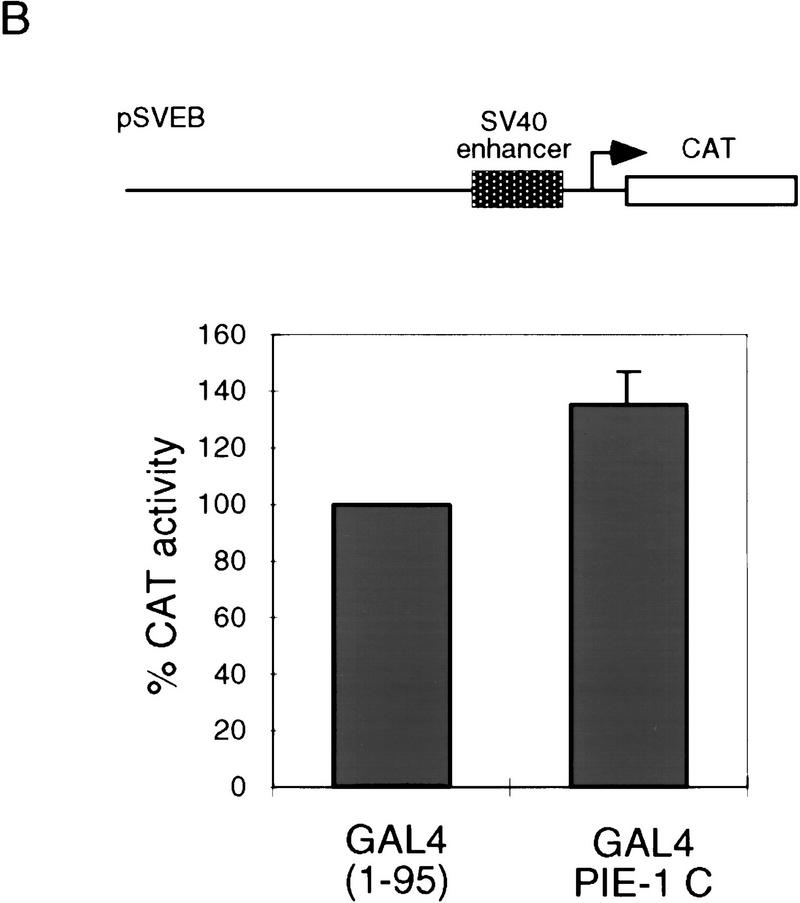
The repression domain in the PIE-1 C region (Fig. 1D,F) appears to be comparable in activity to strong repression domains, such as those of the Drosophila Knirps and Engrailed proteins (Han and Manley 1993; Gerwin et al. 1994). The GAL4(1–95) PIE-1 C fusion protein represses a reporter in which five GAL4 sites are located 2 kb from the SV40 enhancer/promoter (pSVEB-G; Fig. 3A) (Weintraub et al. 1995), but not the corresponding control reporter lacking GAL4 sites (pSVEB; Fig. 3B). This finding indicates that repression by the PIE-1 C region is neither derived simply from steric effects nor specific to the HSV TK promoter, and requires recruitment only to the general vicinity of a promoter.
Figure 3.
The tethered PIE-1 C region can repress transcription from a distance. (A) Repression of the pSVEB-G reporter (Weintraub et al. 1995). HeLa cells were cotransfected with 1 μg of GAL4(1–95) PIE-1 C and 5 μg of reporter. Data are displayed as in Fig. 1. (B) Effect of the PIE-1 C region on the pSVEB control reporter, assayed as in A.
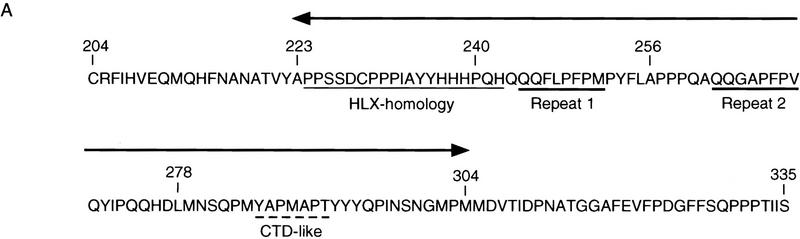
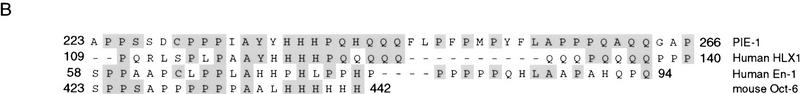
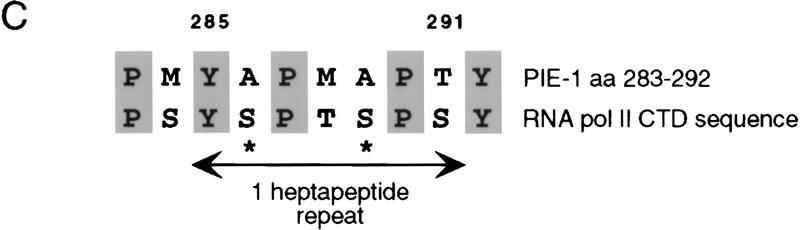
Within the PIE-1 C region (amino acids 204–335; Fig. 4A), positions 224–275 are rich in prolines and glutamines, residues that are often found in repressor domains (for review, see Cowell 1994; Hanna-Rose and Hansen 1996). The PIE-1 C region also contains three distinct sequence motifs. Residues 224–266 are similar to sequences in the transcription factors HLX1, En-1, and Oct-6 (Fig. 4B), all of which are involved in cell differentiation (Deguchi et al. 1992; Jaegle et al. 1996; Loomis et al. 1996; Anderson et al. 1997), but the function of this sequence in these proteins is not known. The PIE-1 residues that are related to all three of these proteins (224–242; Fig. 4A,B) are referred to as the HLX homology region. The area between residues 240 and 278 contains two repeats of the motif QQXZPFPZ, where Z is hydrophobic (Fig. 4A). Finally, residues 283–292 are similar to a portion of the Pol II CTD (Fig. 4A,C). This sequence contains a CTD-related heptapeptide motif (YAPMAPT), along with flanking residues, and differs from the CTD only by conservative substitutions, primarily at positions that would be phosphorylated. In particular, the central YAPMAPT sequence contains alanines in place of CTD serines 2 and 5 (Fig. 4C), which are important for transcription (West and Corden 1995; Dahmus 1996). Significantly, the tyrosines and prolines that are critical for recognition of the CTD repeat by kinases (Hengartner et al. 1998) are intact in the YAPMAPT motif and surrounding residues (Fig. 4C), suggesting that its structure is likely to be very similar to that of the CTD consensus.
Figure 4.
PIE-1 C region sequence motifs. (A) The PIE-1 C region (Fig. 1A). The HLX homology region, two repeated motifs, and the CTD-like YAPMAPT sequence are indicated. Residues that delimit some PIE-1 carboxy-terminal subregions are marked by residue number, and the complete C region repression domain (Fig. 5B) by arrows. (B) PIE-1 C region sequences aligned with the indicated transcription factors (Suzuki et al. 1990; Logan et al. 1992; Kennedy et al. 1994) using the MegAlign tool. (C) The YAPMAPT motif. This sequence is aligned with a consensus CTD repeat motif (YSPTSPS), which is in the context of adjacent repeats.
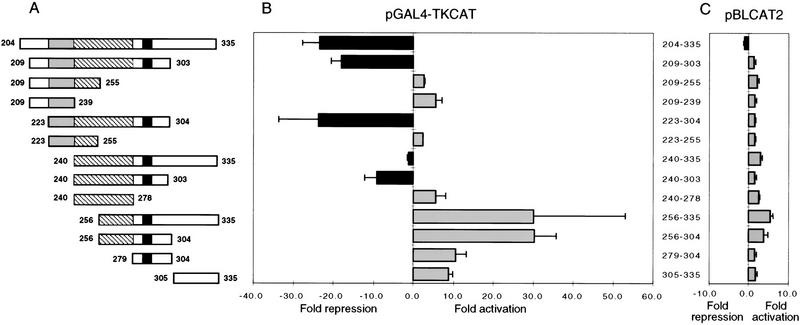
To identify important residues within the PIE-1 C region, we have assayed the repressor activities of GAL4(1–147) subregion fusion proteins that were constructed based on these sequence elements (Fig. 5A). All of these PIE-1 C subregion fusions are expressed at levels comparable to that of GAL4(1–147) PIE-1 C (Fig. 1B; not shown). Residues 223–304 repress pGAL4–TKCAT to an extent similar to PIE-1 C (>20-fold, Fig. 5B), indicating that residues 204–222 and 305–335 are not required for repression. Removal of the HLX homology region (Fig. 4A) decreases the degree of repression to approximately eightfold (residues 240–303; Fig. 5A,B), and further deletion from either end of this minimal repression domain abolishes repression entirely (240–278 and 256–304; Fig. 5A,B). In each case, repression is dependent on recruitment to the GAL4 sites (Fig. 5C). These experiments indicate that the minimal sequence capable of repression consists of the two QQXZPFPZ repeats, together with the region containing the YAPMAPT motif (residues 240–303; Fig. 5A,B). However, the complete PIE-1 repression domain, as defined by the smallest region capable of full activity (residues 223–304) also includes the HLX homology region (Figs. 4A and 5A,B).
Figure 5.
Dissection of the PIE-1 C region repression domain. (A) Schematic representation of PIE-1 C and its subregions, all tested as GAL4(1–147) fusions in B. The HLX homology region shaded, repeats (hatched areas), and YAPMAPT motif (solid areas) (Fig. 4A) are indicated. (B) Repression or activation by GAL4 PIE-1 C subregions. HeLa cells were cotransfected with 1 μg of GAL4(1–147) fusion construct and 5 μg of pGAL4–TKCAT reporter plasmid. (C) Effects of the PIE-1 C subregions on the pBLCAT2 reporter (5 μg), assayed as in B. Each bar represents the mean relative CAT activity, compared to that of GAL4(1–147) and reporter. Error bars indicate the standard deviation derived from three separate transfections.
Multiple subregions that constitute only part of, or that surround, the complete repression domain (residues 223–304) activate pGAL4–TKCAT transcription (Fig. 5A,B). For example, various fragments of the minimal repression domain (240–303) activate transcription through the GAL4 sites (240–278, 256–304, and 279–304), as do residues 305–335 (Fig. 5A,B). Similarly, although subregion 240–335 represses pGAL4–TKCAT slightly, residues 256–335 activate this reporter ∼30-fold (Fig. 5A,B). Given that recruitment of the transcriptional machinery is an important mechanism of activation (Ptashne and Gann 1997; Keaveney and Struhl 1998), the activator capability uncovered in these repressor domain fragments suggests that they can help recruit the transcriptional machinery to the reporter. This observation suggests that the PIE-1 C region interacts directly with the transcriptional machinery, and when it is intact is therefore more likely to inhibit a component of this machinery directly than to recruit a distinct inhibitory corepressor complex.
Importance of the YAPMAPT motif for PIE-1 function
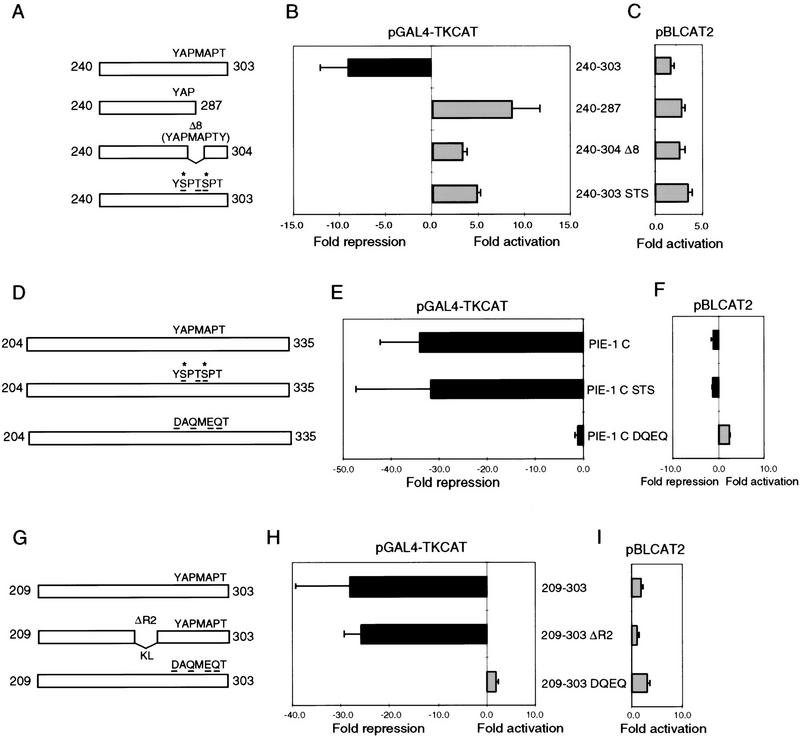
To test whether the CTD-related YAPMAPT motif might be important for repression, we have mutated or deleted this sequence (Fig. 4C) within both the minimal repression domain (residues 240–303; Fig. 6A), and the complete PIE-1 C region (residues 204–335; Fig. 6D). In the context of the minimal repression domain, which lacks the HLX homology region (Fig. 4A), either truncation of this motif at YAP(240–287; Fig. 6A), or its deletion together with the adjacent tyrosine (240–304 D8; Fig. 6A) abolishes repressor activity (Fig. 6B). Subtle mutation of the YAPMAPT motif to a ‘phosphorylatable’ CTD-like sequence (YSPTSPT) also abolishes the repression activity of the minimal domain (240–303 STS; Fig. 6A,B) but not the more robust activity of the complete PIE-1 C region (PIE-1 C STS; Fig. 6D,E). However, a nonconservative alteration of the YAPMAPT motif to DAQMEQT (PIE-1 C DQEQ; Fig. 6D) virtually eliminates repression by the complete PIE-1 C region (Fig. 6E). The repressor activity of residues 209–303, which include the complete repression domain (Figs. 4A and 5), is similarly impaired by conversion of YAPMAPT to DAQMEQT (209–303 DQEQ; Fig. 6G,H) but not by substitution of Lys–Leu (KL) for positions 256–268 (209–303 ΔR2; Fig. 6G,H), demonstrating that a major alteration can be tolerated within a different region of this domain. These results indicate that the YAPMAPT motif is important for activity of the PIE-1 repression domain.
Figure 6.
The YAPMAPT motif is required for the PIE-1 C region repressor activity. (A) Mutation of the YAPMAPT motif in the 240–303 subregion. Residues that correspond to phosphorylated CTD serines (Dahmus 1996) are marked by an asterisk. (B) Repression or activation by the GAL4(1–147) fusions described in A. HeLa cells were cotransfected with 1 μg of GAL4(1–147) fusion construct and 5 μg of pGAL4–TKCAT reporter. (C) Effects of the proteins described in A on the pBLCAT2 reporter plasmid, assayed as in B. (D) Mutation of the YAPMAPT sequence within the complete PIE-1 C region, diagrammed as in A. (E) Repression of pGAL4–TKCAT by the GAL4(1–147) fusions described in D, assayed as in B. In this experiment, repression by GAL4(1–147) PIE-1 C was greater than in Fig. 1D. (F) Effects of the proteins described in D on the pBLCAT2 reporter plasmid, assayed as in E. Data are graphed as in Fig. 5. (G) Mutations within the PIE-1 C 209–303 subregion (Fig. 5A). (H) Repression of pGAL4–TKCAT by the GAL4(1–147) fusions described in G, assayed as in B. (I) Effects of the proteins described in G on the pBLCAT2 reporter.
To determine whether this repressor activity is important for PIE-1 function in vivo, we investigated whether alteration of the YAPMAPT motif impairs the ability of a transgene to rescue a pie-1 mutation. We mutated this sequence to YSPMSPT and DAQMEQT within a full-length pie-1 transgene and assayed the ability of these mutants to rescue the maternal-effect lethality of a pie-1 null allele (zu154; Mello et al. 1992). Because standard techniques do not permit robust maternal expression of transgenes, we expressed these pie-1 transgenes using the recently developed complex array method (Kelly et al. 1997), with which transgene expression can be sustained over a few generations. In this assay, most F2 animals (81%) that carried a wild-type transgene produced multiple live progeny that included fertile animals (Table 1). The YSPMSPT transgene rescued at a comparable frequency (Table 1), a finding that is consistent with the repressor activity of the GAL4 PIE-1 C STS fusion protein (Fig. 6D,E). In contrast, only 25% of the DAQMEQT transgenic F2 animals produced any fertile progeny, and 44% produced only dead embryos (Table 1). Antibody staining indicated that these transgenic proteins were localized properly (not shown), but we were unable to compare levels of expression directly because of the transient nature of this in vivo assay. However, in parallel experiments in which these transgenes were tagged with green fluorescent protein (GFP), all three proteins were localized normally to the germ lineage and expressed at comparable levels (not shown). Although we cannot exclude that some differences in transgene expression might not have been detected in these assays, the finding that the DAQMEQT mutation impairs the frequency of rescue by a pie-1 transgene (Table 1) is consistent with the model that the C region repressor activity is important for PIE-1 function in vivo.
Table 1.
Transgenic rescue of pie-1 by YAPMAPT mutants
| Construct | Line | No. of F2s with fertile progeny/ total F2s tested | No. of F2s with all sterile progeny/ total F2s tested | No. of F2s with all dead progeny/total F2s tested |
|---|---|---|---|---|
| Wild-type transgene (YAPMAPT) | 1 | 2/4 | 0/4 | 2/4 |
| 2 | 1/2 | 1/2 | 0/2 | |
| 3 | 11/13 | 2/13 | 0/13 | |
| 4 | 4/4 | 0/4 | 0/4 | |
| 5 | 2/2 | 0/2 | 0/2 | |
| 6 | 6/7 | 0/7 | 1/7 | |
| Total | 26/32 (81%) | 3/32 (9%) | 3/32 (9%) | |
| YSPMSPT mutant | 1 | 1/1 | 0/1 | 0/1 |
| 2 | 2/3 | 0/3 | 1/3 | |
| 3 | 3/4 | 0/4 | 1/4 | |
| Total | 6/8 (75%) | 0/8 | 2/8 (25%) | |
| DAQMEQT mutant | 1 | 1/3 | 2/3 | 0/3 |
| 2 | 3/11 | 1/11 | 7/11 | |
| 3 | 3/5 | 0/5 | 2/5 | |
| 4 | 1/5 | 3/5 | 1/5 | |
| 5 | 0/8 | 5/8 | 3/8 | |
| 6 | 0/3 | 0/3 | 3/3 | |
| 7 | 1/1 | 0/1 | 0/1 | |
| Total | 9/36 (25%) | 11/36 (31%) | 16/36 (44%) |
Discussion
Previous studies have determined that the pie-1 gene product is required for specification of the C. elegans embryonic germ cell lineage and for the lack of mRNA transcription in these cells (Mello et al. 1992, 1996; Seydoux et al. 1996; Seydoux and Dunn 1997). We have now shown that the PIE-1 protein contains a transcriptional repression domain that, when tethered to promoters in mammalian cells, can inhibit a conserved aspect of the transcriptional machinery. The complete PIE-1 repression domain (residues 223–304; Fig. 4A) consists of the HLX homology region, two hydrophobic-rich repeats, and a region that contains the CTD-like YAPMAPT motif. Its repressor activity is decreased by removal of the HLX homology region and abolished by subsequent deletion of the first repeat element (Figs. 4A and 5). However, the second repeat element is not required (209–303 ΔR2; Fig. 6G,H), suggesting that these repeats may be redundant in the context of the full domain. In contrast, disruption of the YAPMAPT motif prevents repression (Fig. 6B,E,H). These data indicate that multiple distinct sequence elements are important for the repressor activity in the PIE-1 C region.
Transcriptional repression and PIE-1 function
To investigate whether this repressor activity might be required for PIE-1 function in vivo, we have mutated the YAPMAPT motif to YSPMSPT and DAQMEQT in the context of a pie-1 transgene, and assayed the capability of these transgenes to rescue a null pie-1 mutation (Table 1). Maternally expressed transgenes are highly susceptible to germ-line silencing in C. elegans, and our experiments represent the first application of a new technique (Kelly et al. 1997) to investigate how the molecular activity of a maternal protein might be linked to its developmental function. In these experiments, the DAQMEQT mutant transgene rescues this pie-1 mutation at a markedly decreased frequency compared to the wild-type and YSPMSPT transgenes (Table 1). The most direct interpretation of our findings is that this decrease derives from functional impairment of the PIE-1 repression domain, as observed in the GAL4 tethering assay (Fig. 6D,E), but it will be important to test this model in future in vivo studies. It should be noted that the DAQMEQT mutant transgene retains some rescuing activity (Table 1), suggesting that the YAPMAPT motif is not absolutely essential in this in vivo assay. It is possible that disruption of the YAPMAPT motif can be compensated for in vivo by other repressor elements present within the PIE-1 C region, even though such redundancy was not observed in the GAL4 fusion assay (Fig. 6D,E). Alternatively, this compensation could derive from other PIE-1 activities, such as the independent transcriptional repression capability identified in the PIE-1 zinc finger region (Fig. 1D,F). Given the importance of PIE-1 for germ cell specification and development of the embryo (Mello et al. 1992), it might be predicted that multiple mechanisms could contribute to its function.
Because our GAL4 fusion experiments indicate that the PIE-1 C region must be recruited to promoters to repress transcription, they raise the question of how this repressor activity might normally be brought to the transcriptional machinery. It is a reasonable model that the zinc finger region could be involved, particularly given its independent repressor activity (Fig. 1D,F). Binding of these PIE-1 zinc fingers to DNA at all potential target promoters seems unlikely, especially because other proteins that contain C3H zinc fingers appear to be involved in RNA binding or processing (Barabino et al. 1997; Murray et al. 1997; Carballo et al. 1998; Rudner et al. 1998). It seems more likely that the PIE-1 zinc fingers or other regions recruit this repression domain to the transcription complex through protein–protein interactions. Alternatively, or in addition, assuming that the PIE-1 zinc fingers and RS region indicate association with RNA-binding complexes, its recruitment could potentially involve binding to nascent RNA transcripts. The latter mode of recruitment has been described previously for transcriptional activators (Sit et al. 1998; Wei et al. 1998) but not for a repressor.
Potential targets of the PIE-1 repression domain
An interesting aspect of our findings is that multiple fragments of the PIE-1 repression domain activate transcription when tethered to a promoter, particularly fragments that contain the YAPMAPT motif (Fig. 5). This latter finding appears to be consistent with observations that CTD repeats can function as activators in this assay (Kim and Roeder 1994; Wendler et al. 1994; Xiao et al. 1994). Stimulation of transcription by these repressor domain fragments suggests that the tethered PIE-1 C region is more likely to bind and inhibit a positively acting component of the transcriptional apparatus than to recruit a negatively acting corepressor complex. Consistent with this model, our preliminary results indicate that treatment with the histone deacetylase inhibitor trichostatin A does not relieve repression by the GAL4 PIE-1 C region fusion protein (not shown). A ‘threshold’ level of PIE-1 expression appears to be required in vivo (Tenenhaus et al. 1998; T.H. Shin and C. Mello, unpubl.), also suggesting that PIE-1 may inhibit a positively acting factor directly.
Of the multiple sequence elements that contribute to the PIE-1 repression domain (Figs. 4 and 5), the YAPMAPT motif and surrounding sequences are particularly intriguing because of their apparent similarity to the YSPTSPS Pol II CTD repeat (Fig. 4C). Supporting the idea that these sequence motifs have similar structures, the YAPMAPT element is reminiscent of ‘pseudosubstrate’ kinase inhibitor sequences, in which serine phosphorylation targets are substituted with alanine (Scott et al. 1985; House and Kemp 1987; Graff et al. 1991; Cujec et al. 1997; Poteet-Smith et al. 1997). Replacement of YAPMAPT with the CTD-like sequence YSPTSPT does not impair repression by the complete PIE-1 C region (PIE-1 C STS; Fig. 6D,E) but abrogates activity of the minimal repression domain lacking the HLX homology region (240–303 STS; Fig. 6A,B). These findings suggest that a CTD-like sequence at these positions is compatible with repression, but also that the weaker minimal repression domain might be more sensitive to subtle differences between this sequence and YAPMAPT, or to its potential for phosphorylation. In contrast, both repression and in vivo rescue are impaired by the nonconservative DAQMEQT substitution (PIE-1 C DQEQ; Fig. 6D,E; Table 1). The repression domain can tolerate a major alteration within a different region, however, as indicated by the failure of the 209–303 ΔR2 mutation to prevent repression (Fig. 6G,H). These mutagenesis experiments suggest that the similarity of the YAPMAPT motif to the Pol II CTD repeat structure may be important for repression and, therefore, that the PIE-1 repression domain might target a protein complex that can recognize the Pol II CTD repeat. One possibility is that this repression domain might directly inhibit one of the kinase complexes that phosphorylate the CTD (Cisek and Corden 1989; Liao et al. 1995; Serizawa et al. 1995; Shiekhattar et al. 1995; Sterner et al. 1995; Jones 1997). However, the observation that repression can occur when the YAPMAPT alanines are substituted with serine is not consistent with a straightforward pseudosubstrate inhibition mechanism (PIE-1 C STS; Fig. 6E). Alternatively, the YAPMAPT element could contribute to the repression domain interacting with, and inhibiting or sequestering, any of the multiple other regulatory or enzymatic protein complexes that can interact with the Pol II CTD during an mRNA transcription and processing cycle (Thompson et al. 1993; Cho et al. 1997; Corden and Patturajan 1997; McCracken et al. 1997a,b; Neugebauer and Roth 1997; Myers et al. 1998). This model remains a working hypothesis until such a protein complex is identified, but it is attractive because it is consistent with both our repression data and our current understanding of PIE-1 function in vivo (Mello et al. 1996; Seydoux et al. 1996; Seydoux and Dunn 1997).
Materials and methods
Plasmid construction
GAL4(1–147) PIE-1 fusions were made in the pSG424 SV40-driven expression vector (Sadowski and Ptashne 1989) by cloning amplified PCR products into _Bam_HI and _Kpn_I sites. The PIE-1 A fusion then contained the linking residues S and A, and PIE-1 B and PIE-1 C contained the linker SAM. The PIE-1 C subregion fusions to GAL4(1–147) contained no linking region. TTP B and POS-1 B regions were similarly amplified and subcloned into pSG424 without linking regions. The TTP(Nup475) gene was a gift from Mark Worthington (Johns Hopkins, Baltimore, MD). The subcloning of PIE-1 A, B, and C regions into pCMV–GAL4 (1–95) created the following linking regions: SRSAM (A), SEFPGIRSAM (B), and SEFRSAM (C). The amplified coding regions of all constructs were checked by DNA sequencing. Site-directed mutagenesis was performed by the QuikChange method (Stratagene). Mutated sequences were confirmed by DNA sequencing and subcloned into backbone plasmid that had not undergone the thermal cycling reaction.
Transient transfections and CAT assays
HeLa cells grown in Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum, were plated at ∼1 × 106 per 100-mm tissue culture dish on the day before transfection. They were transfected by the BES calcium phosphate method (Chen and Okayama 1987) with 1 μg of effector plasmid, 5 or 10 μg of reporter plasmid and pBSKS+ (Stratagene) carrier DNA to 18 μg per dish. Harvested cells were lysed by freeze–thawing, and protein concentrations measured using the Bradford assay (Bio-Rad). The CAT assay was performed on 40 μg of protein using a scintillation overlay diffusion technique (Neumann et al. 1987), essentially as described by Sambrook et al. (1989). Reporter plasmids were not cotransfected to provide an internal reference for transfection efficiency, because some GAL4–PIE-1 subregion fusions affected transcription of several promoters that lack GAL4 sites (Fig. 5C; not shown). All transfection experiments were performed three times in duplicate.
Western blotting
HeLa cells were cotransfected with 10 μg of effector plasmid DNA and 8 μg pBSKS+ (Stratagene) carrier DNA. Harvested cells were resuspended in 100 μl of PBS and lysed by adding 6× sample buffer. The samples were boiled for 10 min, vortexed, and microcentrifuged at 10,000_g_ for 10 min to pellet cell debris. Proteins were separated by SDS-PAGE and blotted onto nitrocellulose (Schleicher & Schuell). The blot was probed with polyclonal antibodies raised to the GAL4(1–147) DNA-binding domain according to the manufacturer’s instructions (Upstate Biotechnology). A horseradish peroxidase-conjugated anti-rabbit secondary antibody (Promega) was used at a 1:2500 dilution and detected by enhanced chemiluminescence (Amersham).
In vivo rescue assay
The sequence of YAC Y49E10 (GenBank accession no. Z98866) was used to design primers to amplify a 7.8-kb genomic fragment containing the pie-1 gene (−2453 to +5373 relative to the pie-1 ATG). The resulting clone was engineered to contain a _Bam_HI site immediately after the pie-1 ATG. The YSPMSPT and DAQMEQT mutations were introduced by site-directed mutagenesis and recombinant PCR, respectively, and confirmed by DNA sequencing. Each clone was injected into unc-25(e156) pie-1(zu154)/qC1 hermaphrodites, along with 60 μg/ml _Pvu_II- or _Sca_I-digested N2 genomic DNA, 1 μg/ml linearized pRF4 (Rol-6D) DNA, and 1 μg/ml linearized pie-1 plasmid. This method creates complex arrays that allow maternal expression of the transgene in the first few generations after injection (Kelly et al. 1997). F2 Unc Rollers [unc-25(e156) pie-1(zu154) homozygotes containing the transgene] were cloned to individual plates and their progeny counted and examined for viability and fertility. For parallel analysis of transgenic protein expression and localization, transgenes were tagged with GFP by insertion of its coding region at the _Bam_HI site.
Figure 2.
Repressor activity of the TTP (Nup475, TIS11) and POS-1 C3H zinc finger regions. (A) HeLa cells were cotransfected with 1 μg of GAL4(1–147) TTP B or GAL4(1–147) POS-1 B and 5 μg of pGAL4–TKCAT reporter plasmid. (B) As in A, except 5 μg of pBLCAT2 reporter plasmid was used. Data are displayed as in Fig. 1. TTP regions that were tested and found to lack repressor activity in this assay were residues 1–96 and 159–319, and similarly inactive POS-1 regions were 1–99, and 167–264 (not shown).
Acknowledgments
We thank Grace Gill and Yang Shi for reagents, many helpful discussions, and critically reading this manuscript, for which we also thank Martin Highett, Steve Buratowski, Jeff Corden, and members of the Blackwell and Seydoux laboratories. We thank Wayne Waterman and Katherine Galvin for advice on transfections, and Bill Kelly and Andy Fire for advice on the complex array injection procedure. This work was supported by funding from the National Institutes of Health (T.K.B. and C.M.), the Searle Scholars Program/Chicago Community Trust (T.K.B. and G.S.), the Packard Foundation (G.S.), the Pew charitable trust (C.M.), a Schering Plough fellowship from the Life Sciences Research Foundation (T.H.S.), and the American Cancer Society, Massachusetts Division (T.K.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAILblackwell@cbr.med.harvard.edu; FAX (617) 278-3131.
References
- Andersen B, Weinberg WC, Rennekampff O, McEvilly RJ, Bermingham JR, Jr, Hooshmand F, Vasilyev V, Hansbrough JF, Pittelkow MR, Yuspa SH, Rosenfeld MG. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes & Dev. 1997;11:1873–1884. doi: 10.1101/gad.11.14.1873. [DOI] [PubMed] [Google Scholar]
- Barabino SMLX, Hübner W, Jenny A, Minivielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes & Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek LJ, Corden JL. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989;339:679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- Corden JL, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- Cowell IG. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Cujec TP, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan DO, Peterlin BM. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase. Genes & Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Deguchi Y, Thevenin C, Kehrl JH. Stable expression of HB24, a diverged human homeobox gene, in T lymphocytes induces genes involved in T cell activation and growth. J Biol Chem. 1992;267:8222–8229. [PubMed] [Google Scholar]
- DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J Biol Chem. 1990;265:19185–19191. [PubMed] [Google Scholar]
- Gerwin N, La Rosee A, Sauer F, Halbritter HP, Neumann M, Jäckle H, Nauber U. Functional and conserved domains of the Drosophila transcription factor encoded by the segmentation gene knirps. Mol Cell Biol. 1994;14:7899–7908. doi: 10.1128/mcb.14.12.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JM, Rajan RR, Randall RR, Nairn AC, Blackshear PJ. Protein kinase C substrate and inhibitor characteristics of peptides derived from the myristoylated alanine-rich C kinase substrate (MARCKS) protein phosphorylation site domain. J Biol Chem. 1991;266:14390–14398. [PubMed] [Google Scholar]
- Guedes S, Priess J. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development. 1997;124:731–739. doi: 10.1242/dev.124.3.731. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Manley JL. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao S-M, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- House C, Kemp BE. Protein kinase C contains a pseudosubstrate prototope in its regulatory domain. Science. 1987;238:1726–1728. doi: 10.1126/science.3686012. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- Jones KA. Taking a new TAK on TAT transactivation. Genes & Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MA, Rayner JC, Morris CM. Genomic structure, promoter sequence, and revised translation of human homeobox gene HLX1. Genomics. 1994;22:348–355. doi: 10.1006/geno.1994.1394. [DOI] [PubMed] [Google Scholar]
- Kim TK, Roeder RG. CTD-like sequences are important for transcriptional activation by the proline-rich activation domain of CTF1. Nucleic Acids Res. 1994;22:251. doi: 10.1093/nar/22.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Mizuno H, Okada M. Accumulation and spatial distribution of poly-A+ RNA in oocytes and early embryos of Drosophila melanogaster. Dev Growth Differ. 1988;30:251–260. doi: 10.1111/j.1440-169X.1988.00251.x. [DOI] [PubMed] [Google Scholar]
- Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- Lamb MM, Laird CD. Increase in nuclear poly(A)-containing RNA at syncytial blastoderm in Drosophila melanogaster embryos. Dev Biol. 1976;52:31–42. doi: 10.1016/0012-1606(76)90004-x. [DOI] [PubMed] [Google Scholar]
- Liao S-M, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJJ, Young RA. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- Logan C, Hanks MC, Noble-Topham S, Nallainathan D, Provart NJ, Joyner AL. Cloning and sequence comparison of the mouse, human, and chicken engrailed genes reveal potential functional domains and regulatory regions. Dev Genet. 1992;13:345–358. doi: 10.1002/dvg.1020130505. [DOI] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL. The mouse Engrailed-1 gene and ventral limb patterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. [DOI] [PubMed] [Google Scholar]
- Luckow B, Schutz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL Amgen EST program. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Murray MV, Turnage MA, Williamson KJ, Steinhauer WR, Searles LL. The Drosophila suppressor of sable protein binds to RNA and associates with a subset of polytene chromosome bands. Mol Cell Biol. 1997;17:2291–2300. doi: 10.1128/mcb.17.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. The Med proteins of yeast and their function through the RNA polymerase II carboxy terminal domain. Genes & Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. Transcription units as RNA processing units. Genes & Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- Neumann JR, Morency CA, Russian KO. A novel rapid assay for chloramphenicol acetyltransferase gene expression. BioTechniques. 1987;5:444–447. [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Poteet-Smith CE, Shabb JB, Francis SH, Corbin JD. Identification of critical determinants for autoinhibition in the pseudosubstrate region of type I alpha cAMP-dependent protein kinase. J Biol Chem. 1997;272:379–388. doi: 10.1074/jbc.272.1.379. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Breger KS, Rio DC. Molecular genetic analysis of the heterodimeric splicing factor U2AF: The RS domain on either the large or small drosophila subunit is dispensable in vivo. Genes & Dev. 1998;12:1010–1021. doi: 10.1101/gad.12.7.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Ptashne M. Vector for expressing GAL4(1–147) Nucleic Acids Res. 1989;17:7539–7544. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scott JD, Fischer EH, Demaille JG, Krebs EG. Identification of an inhibitory region of the heat-stable protein inhibitor of the cAMP-dependent protein kinase. Proc Natl Acad Sci. 1985;82:4379–4383. doi: 10.1073/pnas.82.13.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H, Makela TP, Conaway JW, Conaway RC, Weinberg RA, Young RA. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GL1-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Shiekhattar R, Mermelstein F, Fisher RP, Drapkin R, Dynlacht B, Wessling HC, Morgan DO, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- Sit TL, Vaewhongs AA, Lommel SA. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rohdewohld H, Neuman T, Gruss P, Scholer HR. Oct-6: A POU transcription factor expressed in embryonal stem cells and in the developing brain. EMBO J. 1990;9:3723–3732. doi: 10.1002/j.1460-2075.1990.tb07585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Schubert C, Seydoux G. Genetic requirements for inhibition of gene expression and PIE-1 localization in the embryonic germ lineage of Caenorhabditis elegans. Dev Biol. 1998;200:212–224. doi: 10.1006/dbio.1998.8940. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Varnum BC, Ma Q, Chi T, Fletcher B, Herschman HR. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol Cell Biol. 1991;11:1754–1758. doi: 10.1128/mcb.11.3.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang S-M, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity loop-specific binding to Tar RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- Wendler W, Altmann H, Ludwig-Winnacker E. Transcriptional activation of NFI/CTF1 depends on a sequence motif strongly related to the carboxy terminal domain of RNA polymerase II. Nucleic Acids Res. 1994;22:2601–2603. doi: 10.1093/nar/22.13.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Khachigian LM, Shows T, Collins T. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J Biol Chem. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- Xiao H, Lis JT, Xiao H, Greenblatt J, Friesen JD. The upstream activator CTF/NF1 and RNA polymerase II share a common element involved in transcriptional activation. Nucleic Acids Res. 1994;22:1966–1973. doi: 10.1093/nar/22.11.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol. 1976;49:425–437. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Corden JL. Identification of phosphorylation sites in the repetitive carboxyl- terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2290–2296. [PubMed] [Google Scholar]