Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae (original) (raw)
Abstract
Ca2+ signals regulate gene expression in animal and yeast cells through mechanisms involving calcineurin, a protein phosphatase activated by binding Ca2+ and calmodulin. Tcn1p, also named Crz1p, was identified as a transcription factor in yeast required for the calcineurin-dependent induction of PMC1, PMR1, PMR2A, and FKS2 which confer tolerance to high Ca2+, Mn2+, Na+, and cell wall damage, respectively. Tcn1p was not required for other calcineurin-dependent processes, such as inhibition of a vacuolar H+/Ca2+ exchanger and inhibition of a pheromone-stimulated Ca2+ uptake system, suggesting that Tcn1p functions downstream of calcineurin on a branch of the calcium signaling pathway leading to gene expression. Tcn1p contains three zinc finger motifs at its carboxyl terminus resembling the DNA-binding domains of Zif268, Swi5p, and other transcription factors. When fused to the transcription activation domain of Gal4p, the carboxy terminal domain of Tcn1p directed strong calcineurin-independent expression of PMC1–lacZ and other target genes. The amino-terminal domain of Tcn1p was found to function as a calcineurin-dependent transcription activation domain when fused to the DNA-binding domain of Gal4p. This amino-terminal domain also formed Ca2+-dependent and FK506-sensitive interactions with calcineurin in the yeast two-hybrid assay. These findings suggest that Tcn1p functions as a calcineurin-dependent transcription factor. Interestingly, induction of Tcn1p-dependent genes was found to be differentially controlled in response to physiological Ca2+ signals generated by treatment with mating pheromone and high salt. We propose that different promoters are sensitive to variations in the strength of Ca2+ signals generated by these stimuli and to effects of other signaling pathways.
Keywords: Calcineurin, transcription, signal transduction, calcium
Calcium signaling mechanisms are employed by all eukaryotic cells to regulate gene expression and a wide range of other cellular processes. During excitation of neurons, for example, calmodulin-dependent protein kinases differentially activate the transcription factors cAMP response element binding (CREB) and serum response factor (SRF) in response to subtle variations in the source or type of Ca2+ signal generated (for review, see Ginty 1997). Ca2+ signals can also regulate gene expression through the calmodulin-dependent protein phosphatase known as calcineurin. In human T cells, for example, calcineurin binds and dephosphorylates members of the NFAT family of transcription factors triggering their nuclear localization and stimulating gene expression (for review, see Rao et al. 1997). The immunosuppressive compounds cyclosporin A and FK506 are known as potent inhibitors of calcineurin (see Schreiber and Crabtree 1992) and have been used to reveal essential roles of calcineurin in T-cell activation and presumably in other cell types (see Collier 1990).
The budding yeast Saccharomyces cerevisiae maintains functional homologs of calmodulin (Davis et al. 1986), calcineurin (Cyert et al. 1991; Kuno et al. 1991; Liu et al. 1991; Cyert and Thorner 1992; Ye and Bretscher 1992) and calmodulin-dependent protein kinases (Ohya et al. 1991; Pausch et al. 1991). Although signaling by these factors is not required for vegetative growth (Geiser et al. 1991), it is required for long-term survival of cells responding to high doses of mating pheromones (Moser et al. 1996). Mating pheromones are diffusible peptide hormones secreted by the two haploid cell types that serve to prepare each other for sexual conjugation (for review, see Sprague and Thorner 1992). Binding of pheromones to specific receptors on the cell surface triggers a signaling cascade involving trimeric G proteins, a mitogen-activated protein (MAP) kinase cascade, and other associated factors that prepare the cell for conjugation. The effects of pheromone signaling can be categorized loosely into early and late responses, both of which are required for efficient mating. The early responses include induction of mating-specific genes and arrest in G1 phase of the cell division cycle. Late effects of pheromone signaling occur after prolonged intensive stimulation and include changes in cell morphology and polarity, down-regulation and desensitization of the signaling machinery, and resumption of mitotic growth. After at least 30 min of intensive pheromone signaling, the rate of Ca2+ influx is enhanced (Ohsumi and Anraku 1985) leading to an elevation of cytosolic-free Ca2+ (Iida et al. 1990; Nakajima-Shimada et al. 1991) and activation of the calmodulin-dependent kinases and calcineurin, which independently promote cell survival (Moser et al. 1996). The molecular mechanisms through which pheromone signaling stimulates Ca2+ influx and calcium signaling promotes cell survival are starting to be elucidated (Iida et al. 1994; Withee et al. 1997).
The calcium signaling pathway of yeast has also been implicated as a regulator of cation homeostasis. Ca2+/calmodulin appears to bind and stimulate members of the Pmr2p family of P-type ion pumps (Wieland et al. 1995), which are involved in Na+ and Li+ efflux (Rudolph et al. 1989; Haro et al. 1991). Additionally, maximal induction of the PMR2A/ENA1 gene in response to high environmental salt requires calcineurin activation by Ca2+/calmodulin (Garciadeblas et al. 1993; Cunningham and Fink 1996). Calcineurin may further promote Na+ tolerance through other mechanisms (Nakamura et al. 1993; Mendoza et al. 1994; Hirata et al. 1995; Danielsson et al. 1996; Mendoza et al. 1996) and also promotes Mn2+ tolerance (Farcasanu et al. 1995; Cunningham and Fink 1996; Pozos et al. 1996). In high Ca2+ conditions, activation of calcineurin by Ca2+/calmodulin induces the expression of PMC1 and PMR1 (Cunningham and Fink 1996), which respectively encode Ca2+-pumping ATPases in the vacuole and Golgi complex (Rudolph et al. 1989; Antebi and Fink 1992; Cunningham and Fink 1994; Sorin et al. 1997). Finally, calcineurin activation appears to strongly inhibit the function of Vcx1p (Cunningham and Fink 1996), a vacuolar H+/Ca2+ exchanger also known as Hum1p (Pozos et al. 1996). Additional roles of calcineurin have also been detected in mutants deficient in either the vacuolar or plasma membrane H+ ATPases (Hemenway et al. 1995; Tanida et al. 1995; Nass et al. 1997). These transcriptional and post-translational effects of calcineurin may be mediated by a number of unidentified factors.
This work aims to identify factors that mediate the calcineurin-dependent induction of PMC1 and to determine their roles in other calcineurin-dependent processes. Through genetic and molecular approaches, we have identified Tcn1p, also called Crz1p (Stathopoulos and Cyert, this issue), as a specific transcription factor required for calcineurin-dependent induction of all the previously reported target genes plus TCN1 itself. We also show that most Tcn1p-dependent genes can be differentially induced based on mechanisms sensitive to both strength of Ca2+ signals and other regulatory inputs.
Results
Differential expression of calcineurin-dependent reporter genes in response to various physiological stimuli
Previous work has demonstrated critical roles for calcineurin in the induction of PMC1, PMR1, PMR2A, and FKS2 in response to at least one of the three physiological conditions that generate Ca2+ signals (Garciadeblas et al. 1993; Mazur et al. 1995; Cunningham and Fink 1996). Treatment with high salt causes calcineurin-dependent induction of PMR2A, and treatment with high amounts of mating pheromone concentrations leads to induction of FKS2. Growth in high Ca2+ conditions induces all four genes. To determine whether all the calcineurin-dependent genes respond to these types of Ca2+ signals, we examined expression patterns of various lacZ reporter genes with and without FK506, a potent inhibitor of calcineurin in yeast (Foor et al. 1992). Treatment with high salt (750 mm NaCl) caused calcineurin-dependent (FK506-sensitive) induction of PMR2A–lacZ but not PMC1–lacZ or FKS2–lacZ. Similarly, treatment with high pheromone (20 μm α-mating factor) stimulated expression of the FKS2–lacZ reporter gene by a calcineurin-dependent mechanism but caused little or no calcineurin-dependent induction of either PMC1–lacZ or PMR2A–lacZ (Fig. 1). Thus, different physiological generators of Ca2+ signals can produce distinct transcriptional responses in yeast as in neurons (for review, see Ginty 1997).
Figure 1.
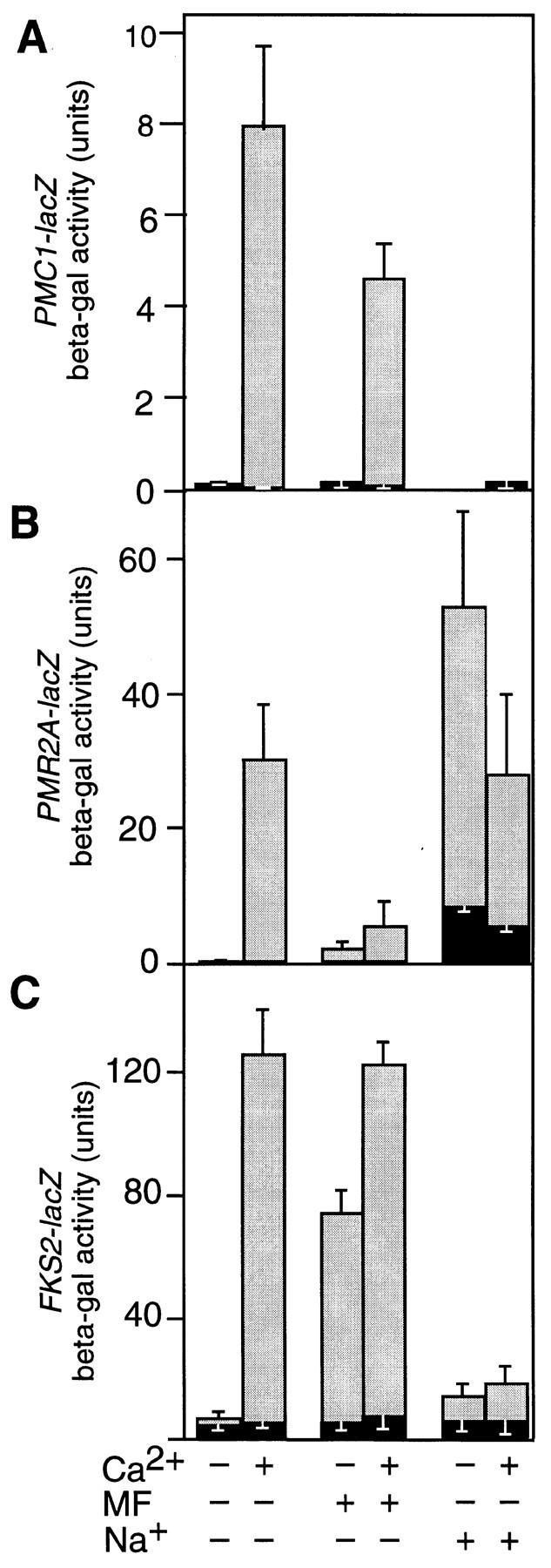
Treatments with CaCl2, pheromone, or high salt generate Ca2+ signals that differentially induce calcineurin-dependent reporter genes. Wild-type yeast (strain W303-1A) was transformed with either plasmid pKC190 carrying PMC1–lacZ (A), plasmid pKC201 carrying PMR2A–lacZ (B), or plasmid pDM5 containing FKS2–lacZ (C), grown to mid-log phase and treated for 4 hr at 30°C in YPD (pH 5.5) medium with 0.2 μg/ml of FK506 (solid bars) or without FK506 (shaded bars) with the additional supplements of Ca2+ (100 mm CaCl2), pheromone (20 μg/ml), Na+ (750 mm NaCl), or combinations thereof as indicated at the base of the plot. Each bar represents the average of three independent determinations of accumulated β-galactosidase activity (± s.d.).
To investigate this phenomenon further, the effects of high salt and pheromone on gene expression were examined during growth in high Ca2+ medium. High salt treatment blocked the normal calcineurin-dependent induction of PMC1–lacZ and FKS2–lacZ but not PMR2A–lacZ. Treatment with pheromone blocked the normal calcineurin-dependent induction of_PMR2A–lacZ_ but not FKS2–lacZ or PMC1–lacZ. These results indicate that in addition to generating Ca2+ signals that activate calcineurin, the response to high salt inhibits expression of both PMC1 and FKS2, whereas the pheromone response inhibits expression of PMR2A. One explanation for why pheromone treatment fails to modulate PMC1–lacZ expression emerges from the identification and characterization of a calcineurin-dependent transcription factor.
TCN1 encodes a zinc finger protein required for calcineurin-dependent induction of PMC1–lacZ
Differential expression of calcineurin-dependent genes may be accomplished using different sets of transcription factors or possibly using the same transcription factors and modulating their effectiveness toward specific targets. To help distinguish these possibilities, we sought to identify and characterize the calcineurin-dependent transcription factors regulating PMC1. A sensitive colony color assay was used to identify mutants of strains DMY62 and DMY63 (Table 1) that failed to express PMC1–lacZ during growth in high Ca2+ conditions (see Materials and Methods). Thirty-one independent recessive mutants were recovered from this genetic screen and placed into just three complementation groups. All members of the first group (six isolates) behaved like mutants lacking the regulatory B subunit of calcineurin (Cunningham and Fink 1994) and were allelic to cnb1 null mutants. As expected from earlier studies, no members of the second group (11 isolates) or third group (14 isolates) represented mutant alleles of calmodulin or the catalytic A subunit of calcineurin and, instead, defined two new components of the calcium signaling pathway. All members of the second group were complemented by low copy plasmids carrying MSN5/STE21, a previously characterized gene that functions in several processes unrelated to calcium signaling (Akada et al. 1996; P.M. Alepuz, D.P. Mathews, K.W. Cunningham, and F. Estruch, in prep). Members of the third group were complemented by plasmids carrying YNL027w, a previously uncharacterized gene on chromosome XIV, which we have named TCN1 for target of calcineurin.
Table 1.
Yeast strains used in this study
| Strain | Genotypea | Reference |
|---|---|---|
| W303-1A | + | Wallis et al. (1989) |
| YLIP179 | MATα fks1::HIS3 | Mazur et al. (1995) |
| YLIP183 | fks1::HIS3 | Mazur et al. (1995) |
| K601 | + | W303-1A |
| K473 | pmc1::LEU2 | Cunningham and Fink (1994) |
| K482 | MATα pmc1::TRP1 | Cunningham and Fink (1994) |
| K603 | cnb1::LEU2 | Cunningham and Fink (1994) |
| K605 | pmc1::TRP1 | Cunningham and Fink (1994) |
| K633 | pmr2::HIS3 | Cunningham and Fink (1996) |
| K661 | vcx1Δ | Cunningham and Fink (1996) |
| K665 | vcx1Δ pmc1::TRP1 | Cunningham and Fink (1996) |
| DMY14 | _tcn1::_G418 | |
| DMY18 | _tcn1::_G418 pmc1::TRP1 | |
| DMY20 | _tcn1::_G418 vcx1Δ | |
| DMY24 | _tcn1::_G418 vcx1Δ pmc1::TRP1 | |
| DMY44 | _tcn::_G418 pmr2::HIS3 | |
| DMY62 | _pmc1::_LEU2 PMC1–lacZ::URA3 | |
| DMY63 | MATα pmc1::TRP1 PMC1–lacZ::URA3 | |
| Y190b | gal4 gal80 GAL1–lacZ | Harpter et al. (1993) |
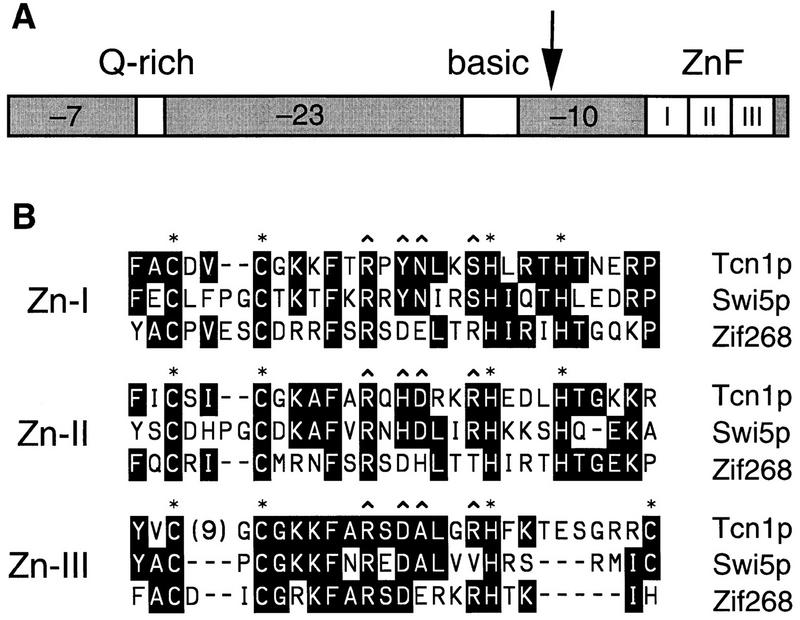
The predicted 678-amino-acid product of TCN1 contains three C2H2-type zinc finger motifs at the carboxyl terminus that strongly resemble the DNA-binding domains of numerous transcriptional regulators such as Swi5p from yeast and Zif268/early growth response (EGR)-1/Krox-24 from mammals (Fig. 2B). Outside of the zinc finger domain, Tcn1p shows no obvious sequence similarity to any proteins in current databases but contains sequence features found in many other transcriptional regulators, such as three acidic regions (net charges of −7, −23, and −10) separated by a glutamine-rich domain (residues 115–140) and a highly basic region (net charge of +13, residues 398–443) rich in serine and threonine residues (Fig. 2A). These sequence features suggest that Tcn1p may function as a specific transcriptional activator in the calcium signaling pathway.
Figure 2.
Sequence features of Tcn1p. (A) Predicted open reading frame of TCN1. Denoted are putative domains of Tcn1p: three acidic regions with net charges of −7, −23, and −10, respectively; a Q-rich domain where 24 of 27 amino acid residues are glutamine; a basic region containing a net charge of +13; and three putative zinc fingers. The arrow indicates the division between the amino and carboxyl termini used to assay functionality of these two domains. (B) Multiple sequence alignment of the three zinc finger motifs from Tcn1p, and the transcription factors Swi5p (residues 550–632) from yeast (Stillman et al. 1988) and Zif268/EGR-1 (residues 287–367) from mammals (Lemaire et al. 1988). Residues conserved in at least two of the three sequences are boxed and highlighted. Residues that coordinate zinc ions (asterisks) and that contact DNA (∧) in the crystal structure of Zif268 complexed with DNA (Pavletich and Pabo 1991) are indicated.
Distinct domains of Tcn1p interact specifically with calcineurin
Two functional domains of Tcn1p were defined by analysis of protein fusions constructed with the Gal4p transcription factor. Fusion of the carboxy-terminal zinc finger region of Tcn1p (residues 463–678) with the transcriptional activation domain of Gal4p yielded a functional hybrid protein that strongly induced PMC1–lacZ expression relative to controls in a fashion independent of Ca2+ and insensitive to FK506 (Fig. 3A). This constitutive Tcn1(C)::Gal4(AD) hybrid protein did not induce expression of reporter genes that are normally unresponsive to Ca2+ signals, such as GAL1–lacZ or CYC1–lacZ but stimulated expression of PMR2A–lacZ and FKS2–lacZ (data not shown). These results show that the carboxy-terminal zinc finger domain of Tcn1p retains the ability to form promoter-specific interactions.
Figure 3.
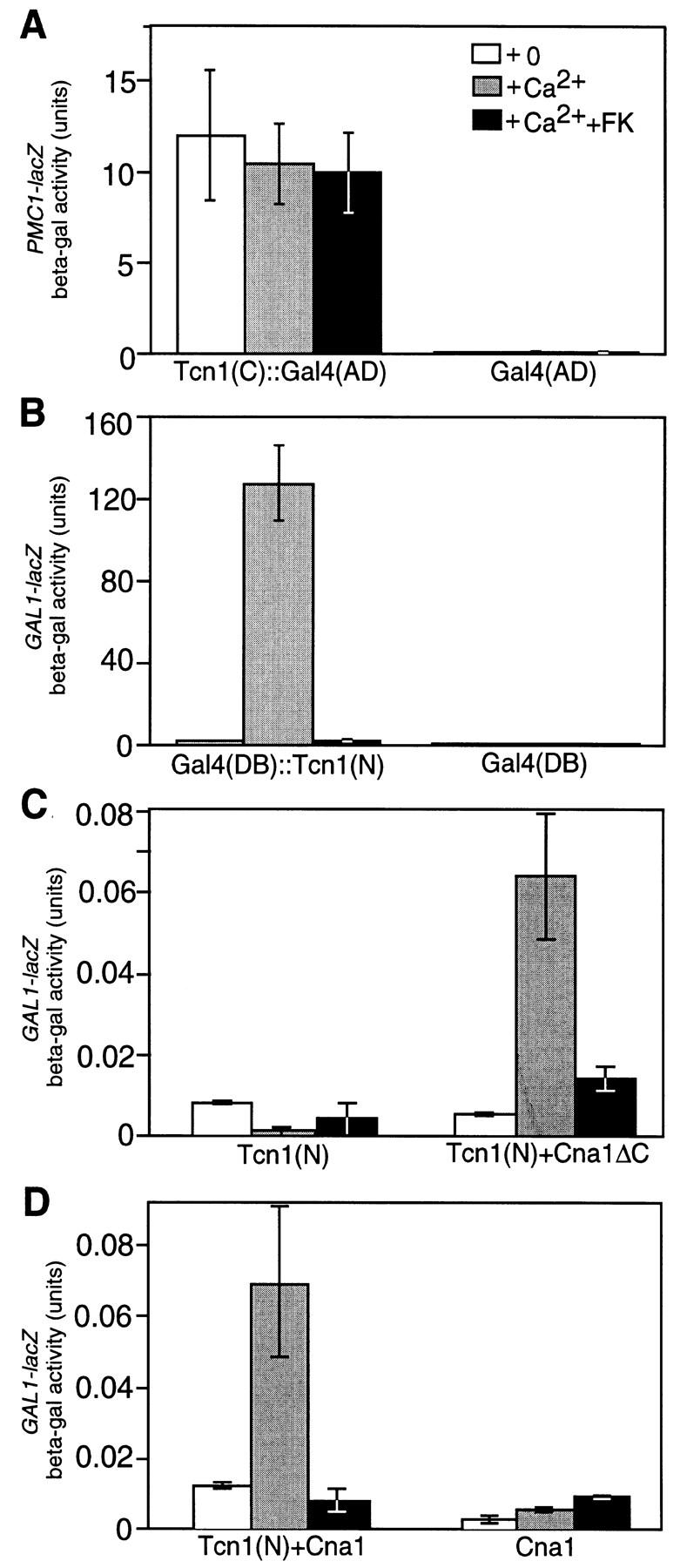
Functional domains of Tcn1p defined by fusions with Gal4p. (A) Expression of a PMC1–lacZ reporter gene on plasmid pKC190 in a tcn1 null mutant (strain DMY14) driven by either a Tcn1(C)::Gal4(AD) hybrid protein (plasmid pDM16) or a Gal4(AD) fragment (plasmid pPC86) as a control was measured after 4 hr growth in YPD (pH 5.5) medium supplemented as indicated with 200 mm CaCl2 and 0.2 μg/ml of FK506. Data are the averages of three independent transformants with standard deviation as indicated by error bars. Similar results were obtained using PMR2A–lacZ (pKC201) and FKS2–lacZ (pDM5) reporter genes. (B–D) Expression of a GAL1–lacZ reporter gene in a gal4 gal80 double mutant Y190 (Harper et al. 1993) was measured as above using plasmids expressing the following hybrid proteins: (B) Gal4(DB)::Tcn1(N) on plasmid pDM15 or Gal4(DB) on plasmid pPC97; (C) Tcn1(N)::Gal4(AD) on plasmid pTJK27 with either Gal4(DB) on plasmid pPC97 or Gal4(DB)::Cna1ΔC on plasmid pKC116; (D) Gal4(DB)::Cna1 on pKC115 with either Tcn1(N)::Gal4(AD) on plasmid pTJK27 or Gal4(AD) on plasmid pPC86. The results show the carboxyl terminus of Tcn1p interacts functionally with the promoter of PMC1 (A), whereas the amino-terminal region of Tcn1p functions as both a calcineurin-dependent transcription activation domain (B) and a calcineurin-interacting domain (C,D).
The amino-terminal region of Tcn1p contains a transcriptional activation domain responsive to calcineurin because fusion of residues 11–460 with the DNA-binding domain of Gal4p resulted in a Gal4(DB)::Tcn1(N) hybrid protein that conferred Ca2+-stimulated and FK506-sensitive induction to a GAL1–lacZ reporter gene (Fig. 3B). To test whether the amino-terminal domain of Tcn1p interacts directly or indirectly with calcineurin, a two-hybrid experiment was performed using fusions between the amino-terminal domain of Tcn1p and the catalytic subunit of calcineurin encoded by the yeast CNA1 gene (Cyert et al. 1991). Expression of a functional Gal4(DB)::Cna1 hybrid protein in which the DNA-binding domain of Gal4p was fused to calcineurin A failed to induce GAL1–lacZ in standard media or in media supplemented with Ca2+ or FK506 (Fig. 3D, right). Similarly, expression of a Tcn1(N)::Gal4(AD) hybrid protein, in which the activation domain of Gal4p was fused to the amino-terminal domain of Tcn1p, also exhibited no ability to induce GAL1–lacZ because this protein lacks the appropriate DNA-binding domain (Fig. 3C, left). Coexpression of both Tcn1(N)::Gal4(AD) and Gal4(DB)::Cna1 restored induction of GAL1–lacZ by added Ca2+, which was completely inhibited by FK506 (Fig. 3D, left). Similar results were obtained when the carboxy-terminal autoinhibitory domain of calcineurin A was deleted from the hybrid protein (Fig. 3C, right). Overexpression of the hybrid proteins using high-copy plasmid vectors increased the units but did not alter the patterns observed using the low-copy plasmids (data not shown). These results demonstrate that the amino-terminal domain of Tcn1p interacts functionally with activated calcineurin but not with inactive or FK506-inhibited calcineurin. Functional interactions detected using the two-hybrid assay usually reflect direct or indirect physical interactions (Fields and Sternglanz 1994).
Targets of Tcn1p
To determine whether Tcn1p mediates some or all of the effects of calcineurin on gene expression, both expression and function of the four known target genes were examined in a tcn1 null mutant in which the TCN1 coding sequence was deleted and replaced (see Materials and Methods). The tcn1 null mutant grew as well as wild-type strains in standard medium but completely failed to induce PMC1–lacZ in response to growth in high Ca2+ conditions (Table 2, line 1). Furthermore, the normal calcineurin-dependent induction of reporter genes for PMR1, PMR2A, and FKS2 was completely abolished in the tcn1 null mutant (Table 2, lines 2–4), whereas the control CYC1–lacZ reporter was not affected (line 5).
Table 2.
Expression of reporter genes in wild-type and tcn1 mutants
| β-Galactosidase (units) | Induction ratio | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Plasmid | Reporter | wild type | tcn1 mutant | wild type | tcn1 mutant | ||||
| +0+Ca+Ca+FK | +0+Ca+Ca+FK | (+Ca/+Ca+FK) | (+Ca/+Ca+FK) | ||||||
| 1. pKC190 | PMC1–lacZ | 0.2 | 19 | 0.1 | 0.1 | 0.1 | 0.1 | 270 ± 90 | 1.0 ± 0.0 |
| 2. pDM5 | FKS2–lacZ | 10 | 232 | 10 | 4.0 | 5.0 | 5.0 | 21 ± 7.0 | 1.1 ± 0.2 |
| 3. pKC199 | PMR1–lacZ | 11 | 28 | 12 | 5.0 | 5.0 | 6.0 | 2.3 ± 0.6 | 0.9 ± 0.1 |
| 4. pKC201 | PMR2A–lacZ | 0.1 | 59 | 4.5 | 0.1 | 0.1 | 0.8 | 14 ± 3.6 | 0.1 ± 0.05 |
| 5. pLGΔ312 | CYC1–lacZ | 424 | 483 | 466 | 358 | 483 | 415 | 1.0 ± 0.1 | 1.2 ± 0.1 |
| 6. pDM7 | TCN1–lacZ | 4.3 | 21 | 5.3 | 3.1 | 5.8 | 3.4 | 4.2 ± 1.4 | 1.7 ± 0.4 |
| +0+mf+mf+FK | +0+mf+mf+FK | (+mf/+mf+FK) | (+mf/+mf+FK) | ||||||
| 7. pDM5 | FKS2–lacZ | 39 | 166 | 50 | 17 | 26 | 24 | 3.4 ± 0.6 | 1.1 ± 0.1 |
| +0+Na+Na+FK | +0+Na+Na+FK | (+Na/+Na+FK) | (+Na/+Na+FK) | ||||||
| 8. pKC201 | PMR2A–lacZ | 3.3 | 163 | 39 | 1.4 | 53 | 38 | 4.2 ± 0.4 | 1.5 ± 0.2 |
To confirm that PMC1, PMR1, PMR2A, and FKS2 are physiological targets of Tcn1p, the function of each gene was compared in wild-type and tcn1 null mutants. The function of FKS2, for example, was measured qualitatively using a viability assay. Calcineurin-dependent expression of FKS2 is required for viability of cells lacking the homologous gene FKS1 (Parent et al. 1993; Eng et al. 1994; Garrett-Engele et al. 1995). We found that like calcineurin, Tcn1p is also required for functional expression of FKS2 because all fks1 tcn1 double mutants generated from 18 tetrads of a test cross (strain DMY14 crossed with strain YLIP179) were inviable. The functions of PMR1, PMR2A, and PMC1 in tcn1 mutants were assayed quantitatively using Mn2+, Na+, and Ca2+ tolerance tests, respectively. tcn1 mutants were significantly less tolerant than wild-type to these ions (Fig. 4). In the presence of FK506, wild-type strains and tcn1 mutants displayed approximately equal sensitivities to Mn2+, Na+, and Ca2+, suggesting that Tcn1p retains little or no activity in the absence of calcineurin function and that the two factors function in the same regulatory pathway.
Figure 4.
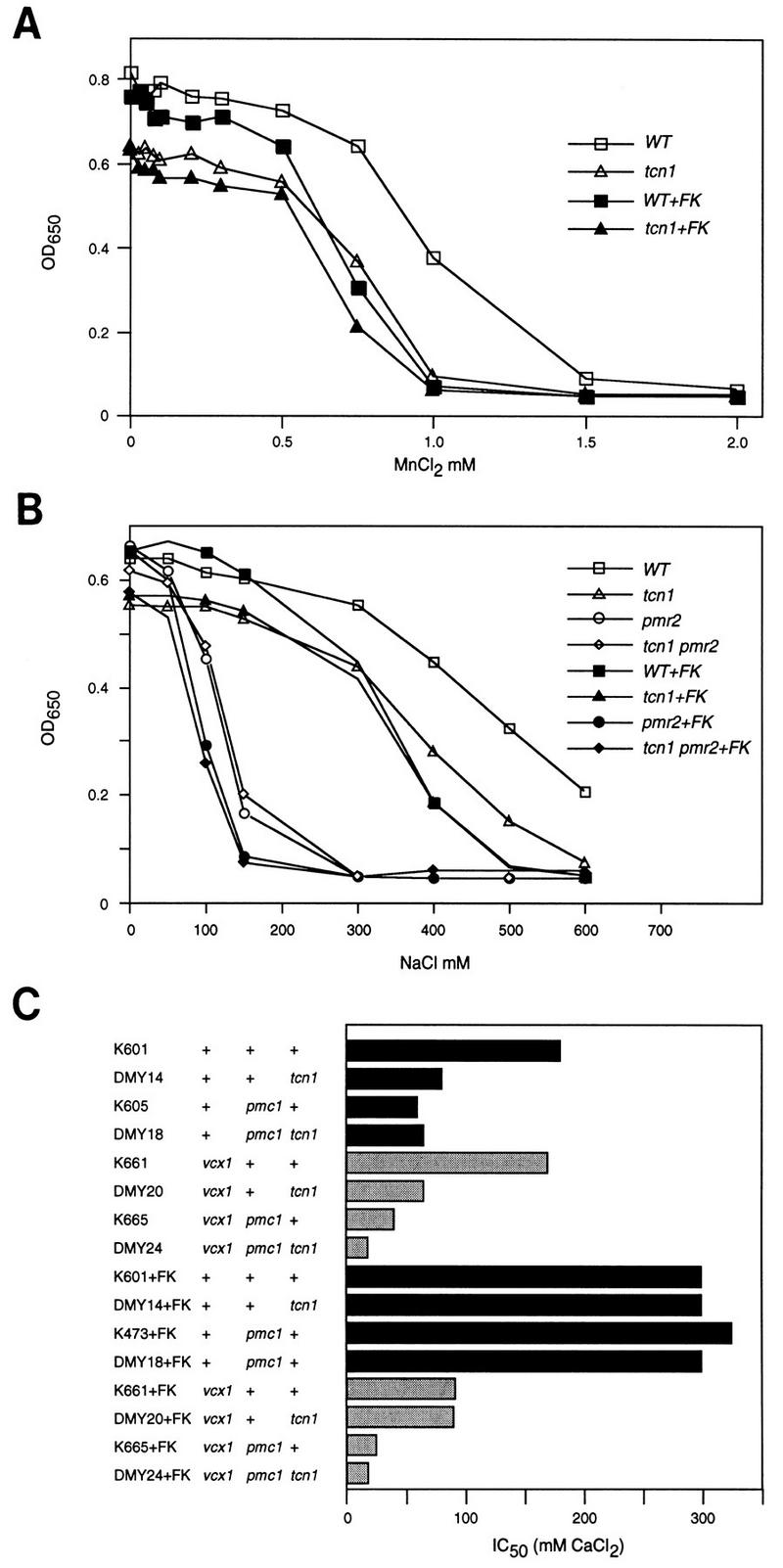
Mn2+, Na+, and Ca2+ tolerance assays of various yeast mutants showing roles of Tcn1p. All strains were grown to saturation in YPD medium at 30°C and diluted 1000-fold into fresh media containing a range of MnCl2, NaCl, or CaCl2 concentrations (with and without 0.2 μg/ml of FK506) and incubated for 1 day at 30°C in flat-bottom 96-well dishes (0.2 ml/well). Optical density at 650 nm was measured for each resuspended culture and plotted directly (A,B) or plotted and used to determine the 50% inhibitory concentration or IC50 (C) as described in Materials and Methods.
The ion tolerance assays shown in Figure 4 also reveal functions of calcineurin that are independent of Tcn1p function. For example, addition of FK506 to tcn1 mutants and to pmr2 tcn1 double mutants causes a further reduction in Na+ tolerance (Fig. 4B), suggesting that calcineurin affects other Na+ tolerance factors independently of Tcn1p. In tcn1 null mutants, weak effects of calcineurin on PMR2A–lacZ expression were also evident depending on the growth conditions (Table 2, lines 4 and 8). Calcineurin also inhibits the function of Vcx1p in Ca2+ tolerance assays by a Tcn1p-independent mechanism because FK506 addition increased Ca2+ tolerance of tcn1 mutants, pmc1 mutants, and pmc1 tcn1 double mutants but only when VCX1 was present. The finding that both pmc1 tcn1 double mutants and pmr1 tcn1 double mutants (not shown) are viable, whereas pmc1 pmr1 double mutants are inviable (Cunningham and Fink 1994), suggests that Tcn1p-independent basal expression of either Pmc1p or Pmr1p is sufficient for viability in standard media. In summary, the activity of Tcn1p on all known targets required calcineurin function, whereas the activity of calcineurin on at least two additional processes did not require Tcn1p function. These results suggest Tcn1p functions in a branch downstream of calcineurin in the calcium signaling pathway leading to gene expression.
Roles of Tcn1p and calcineurin in response to pheromone
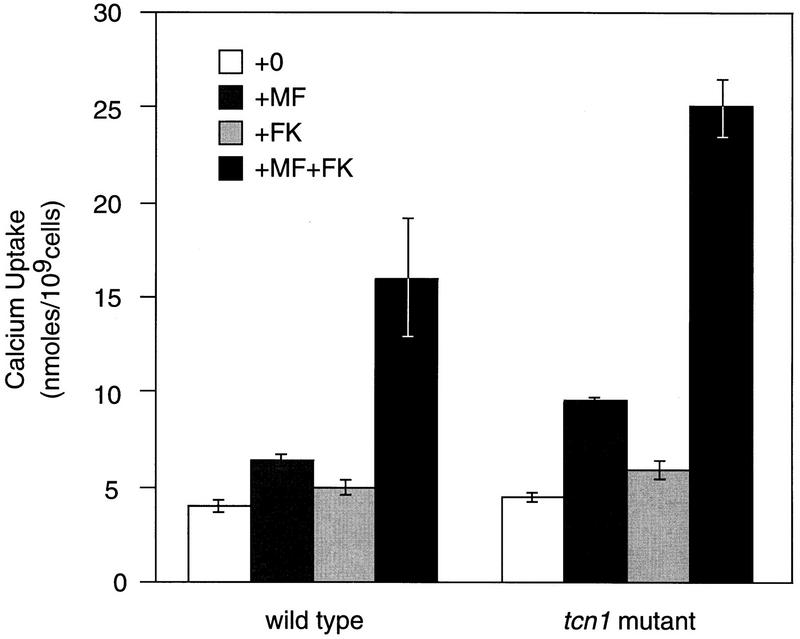
The Ca2+ signal generated in response to pheromone induces FKS2 through a calcineurin-dependent mechanism (Mazur et al. 1995). Induction of an FKS2–lacZ reporter gene was also observed after treatment of wild-type MATa cells with pheromone, and this induction was almost completely dependent on calcineurin and Tcn1p (Table 2, line 7). Calcineurin function is required for several additional responses to pheromone treatment, including maintenance of cell viability during prolonged pheromone stimulation (Cyert et al. 1991; Cyert and Thorner 1992; Moser et al. 1996) and for changes in cell morphology (Withee et al. 1997). Using similar methods, we found that tcn1 null mutants were indistinguishable from wild type (data not shown), suggesting that these effects of calcineurin are largely independent of Tcn1p. Another effect of calcineurin during the pheromone response, feedback inhibition of Ca2+ uptake, is illustrated in Figure 5. After 4-hr treatment with 20 μm pheromone, wild-type cells display a small increase in 45Ca2+ accumulation. Inhibition of calcineurin by addition of FK506 greatly potentiated the effect of pheromone but had little or no effect on untreated wild-type cells. In a parallel experiment, a tcn1 null mutant showed a pattern of 45Ca2+ uptake similar to wild type, indicating that Tcn1p is not required for the apparent calcineurin-dependent feedback inhibition of Ca2+ uptake after pheromone treatment. Although Tcn1p becomes activated during the pheromone response and induces genes such as FKS2, it plays no obvious role in several other calcineurin-dependent processes including increased Ca2+ uptake, morphological changes, and cell survival.
Figure 5.
Tcn1p is not required for calcineurin-dependent inhibition of Ca2+ uptake stimulated by pheromone. Log-phase cells were incubated for 4 hr at 30°C in YPD medium supplemented with 45Ca2+ tracer in the presence or absence of synthetic pheromone [10 μm of α-mating factor (MF)] and FK506 (1.0 μg/ml). Total cell-associated Ca2+ was determined as described in Materials and Methods, and the average of three independent experiments are shown (± s.d.). A large stimulatory effect of FK506 was observed for both the wild-type (strain W303-1A) and tcn1 null mutant (strain DMY14).
Dynamics of the calcium signaling pathway
Complementation tests performed during the characterization of isolated tcn1 mutants, showed that heterozygous tcn1/TCN1 diploids accumulated less than half as much β-galactosidase activity as homozygous TCN1/TCN1 diploids during growth in identical Ca2+ conditions (data not shown). This result implies that Tcn1p abundance may affect responsiveness to Ca2+ signals. Overexpression of Tcn1p by transforming a wild-type strain with the high dosage TCN1 plasmid (pLE66) resulted in higher induction of PMC1, FKS2, and PMR2A with lower doses of Ca2+ (Fig. 6B–D). For all reporters except FKS2, the maximum levels of expression were also elevated in the strain overexpressing Tcn1p (Fig. 6). A TCN1–lacZ reporter gene was constructed (see Materials and Methods) to determine whether Tcn1p levels might change in high Ca2+ conditions. In wild-type strains, TCN1–lacZ was induced about fourfold over basal levels by growth in high Ca2+ and this induction was largely dependent on calcineurin and Tcn1p (Table 2, line 6) and highly sensitive to dosage of Tcn1p (Fig. 6A). The apparent autoregulation of Tcn1p combined with the varying sensitivities of each target promoter to Tcn1p abundance and Ca2+ levels may permit differential gene expression in response to varying strength or intensity of Ca2+ signals.
Figure 6.
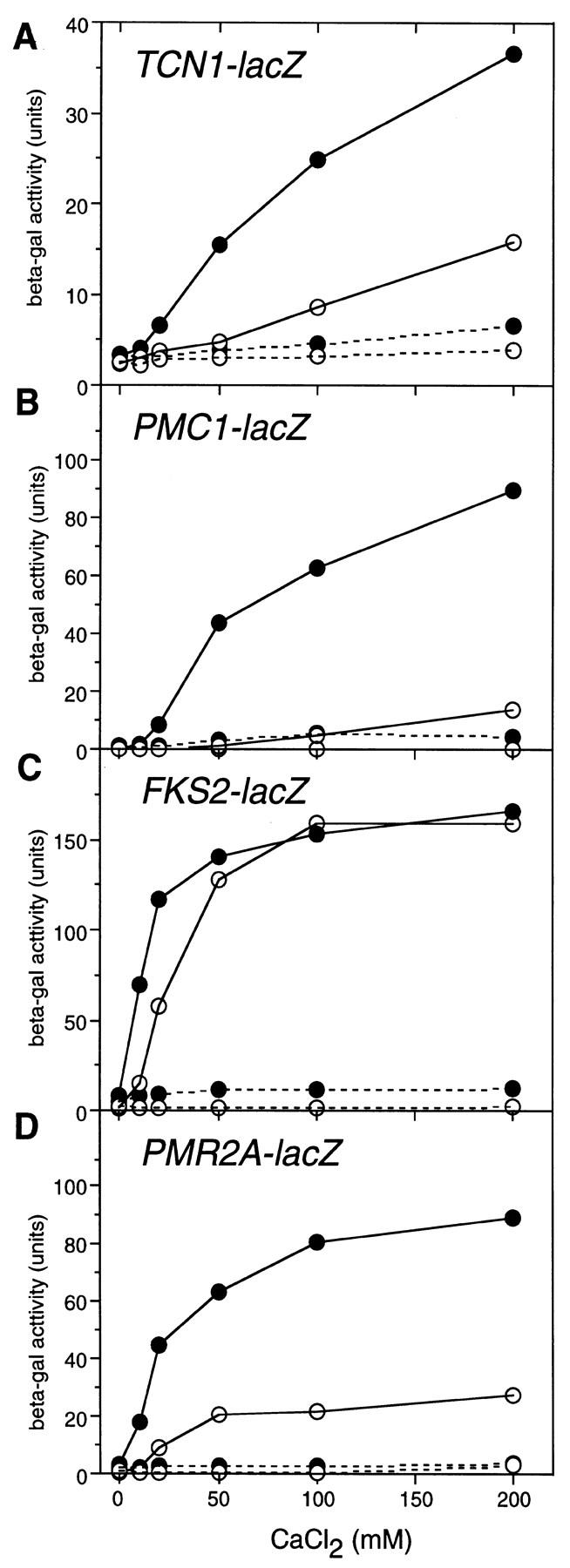
Calcineurin-dependent induction of TCN1–lacZ and other reporter genes is enhanced by overexpression of Tcn1p. Wild-type yeast (strain W303-1A) carrying the indicated reporter genes (plasmids pDM7, pKC190, pDM5, and pKC201, respectively) were transformed with either a control plasmid (YEp13, ○) or a similar high dosage plasmid containing TCN1 (pLE66, •) and grown to log phase in SC − ura − leu medium to maintain plasmid selection. After incubation for 4 hr at 30°C in YPD (pH 5.5) medium supplemented with CaCl2 as indicated and either with FK506 (0.4 μg/ml, broken lines) or without FK506 (solid lines), cells were collected and assayed for β-galactosidase accumulation.
Differential control of Tcn1p-dependent genes involves modulation of Ca2+ signal strength and other promoter-specific factors
One possible explanation for the failure of the calcium signaling pathway to induce PMC1–lacZ after pheromone treatment emerges from the above results: The response to pheromone may produce a relatively weak Ca2+ signal that is insufficient to induce low-sensitivity genes. In support of this hypothesis, overexpression of Tcn1p from a high dosage plasmid restored the calcineurin-dependent induction of PMC1–lacZ in response to pheromone treatment to ∼30% of maximal levels (Fig. 7A). Additionally, pheromone treatment caused a marked calcineurin-dependent induction of the high sensitivity GAL1–lacZ reporter driven by the Gal4(DB)::Tcn1(N) hybrid transcription factor amounting to ∼50% of the maximal induction observed upon treatment with both pheromone and high Ca2+ (Fig. 7B). These results suggest pheromone treatment generates a relatively weak Ca2+ signal that is ordinarily insufficient to induce low sensitivity genes such as PMC1 and possibly TCN1.
Figure 7.
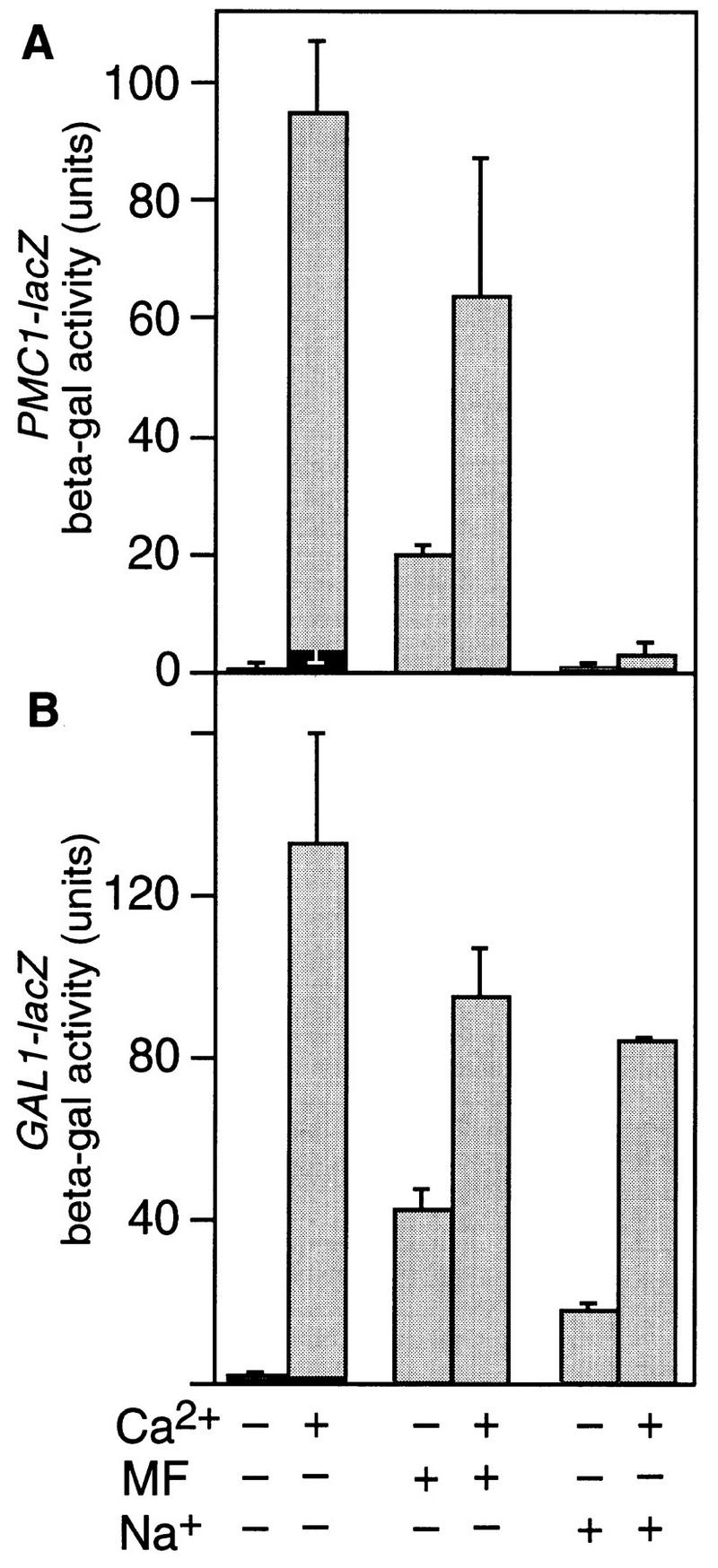
Differential expression of Tcn1p-dependent genes in response to strength of Ca2+ signals and other regulatory inputs. (A) Wild-type strain W303-1A was transformed with both plasmid pKC190 carrying PMC1–lacZ and plasmid pLE66 to increase dosage of Tcn1p, grown, and assayed for β-galactosidase activity as described in Fig. 1. (B) The MATa gal4 gal80 strain Y190 containing a GAL1–lacZ reporter gene was transformed with plasmid pDM15 expressing the Gal4(DB)::Tcn1(N) hybrid factor and then grown and assayed as above. The results suggest pheromone produces a weak Ca2+ signal that partially induces a high sensitivity reporter (B) but fails to induce a low sensitivity reporter unless Tcn1p is overexpressed (cf. A with Fig. 1A). High salt treatment induces the high sensitivity reporter and prevents induction of PMC1–lacZ by high Ca2+ treatment despite overexpression of Tcn1p.
Overexpression of Tcn1p failed to restore calcineurin-dependent induction to PMC1–lacZ by high salt treatment and failed to overcome the inhibitory effect of high salt on PMC1–lacZ induction by high Ca2+ treatment (Fig. 7A). In contrast, treatment with high salt caused ∼25% maximal calcineurin-dependent induction of GAL1–lacZ in cells expressing the Gal4(DB)::Tcn1(N) hybrid transcription factor and had only a slight inhibitory effect on induction by high Ca2+ (Fig. 7B). The simplest hypothesis consistent with these results is that the response to high salt includes both the promoter-specific blockers of gene expression and the production of relatively weak Ca2+ signals that are sensed by calcineurin and the amino-terminal domain of Tcn1p. Based on all these results, we conclude that differential expression of Tcn1p-dependent genes can be accomplished through mechanisms that distinguish both the strength of Ca2+ signals and inputs from other signaling pathways.
Discussion
The results reported here and elsewhere (Stathopoulos and Cyert 1997) propose that Tcn1p/Crz1p functions as an important part of a calcineurin-dependent transcription factor in yeast. Tcn1p contains within its amino-terminal region a domain that interacts functionally, and perhaps physically, with activated calcineurin. Genetic analyses of tcn1 null mutants suggest that Tcn1p functions downstream of calcineurin in the calcium signaling pathway on a branch leading to the expression of specific genes. Tcn1p contains three zinc finger motifs in its carboxy-terminal region resembling the DNA-binding domains of numerous transcription regulators and has been shown in vitro to bind a 24-bp element present in the promoter of at least one target gene (Stathopoulos and Cyert 1997). The simplest molecular mechanism consistent with these results would be that calcineurin directly dephosphorylates Tcn1p in response to Ca2+ signals and thereby stimulates either nuclear localization or transcriptional activation activity. The available data, however, do not rule out the involvement of unknown intermediary factors or additional steps in the mechanism. Regardless of the molecular mechanism, the analysis of tcn1 null mutants and its gain-of-function variants clarify the roles of specific factors in the calcium signaling pathway during the responses to high Ca2+, high salt, and mating pheromones.
Regulation of calcium transporters
The identification and characterization of Tcn1p confirms our previous model of Ca2+ homeostasis in yeast (Cunningham and Fink 1996) and extends our understanding of the feedback mechanisms controlling Ca2+ transporters. Briefly, we propose that as extracellular Ca2+ concentrations are increased from 0.1 mm to >100 mm the higher rates of Ca2+ influx elevate cytosolic Ca2+ to levels which activate calcineurin and Tcn1p, leading first to induction of PMR1 and eventually to induction of TCN1 and PMC1. Expression of the vacuolar Ca2+ ATPase Pmc1p correlates with Ca2+ tolerance and is consistent with a primary role of the vacuole in the sequestration of excess Ca2+ and precipitation with polyphosphate (Dunn et al. 1994). Previous work showed deletion of both PMC1 and PMR1 is lethal due to accumulation of toxic levels of Ca2+ in the cytoplasm (Cunningham and Fink 1994). Basal expression of either Ca2+ ATPase is sufficient for viability because tcn1 mutants, pmc1 tcn1 double mutants, and pmr1 tcn1 double mutants are all viable though highly sensitive to added Ca2+ (Fig. 4; data not shown). In high Ca2+ conditions, the essential function of Tcn1p appears to be the induction of PMC1 gene expression.
The analysis of tcn1 mutants also clarifies the role of calcineurin in Mn2+ tolerance. Pmr1p contributes strongly to Mn2+ tolerance but contributes much less than Pmc1p to Ca2+ tolerance (Cunningham and Fink 1994). Strains lacking calcineurin or Tcn1p function fail to induce PMR1–lacZ (Table 2) and are correspondingly less tolerant of added Mn2+ (Fig. 4A). This correlation suggests that Pmr1p levels directly determine Mn2+ tolerance just as Pmc1p levels directly determine Ca2+ tolerance levels. An alternative hypothesis proposed the role of calcineurin in Mn2+ tolerance was to limit Mn2+ influx by an unknown process (Farcasanu et al. 1995). However, a significant role for calcineurin in Mn2+ tolerance can only be detected when both Tcn1p and Pmr1p are functioning (Fig. 4A; Cunningham and Fink 1994). All of these results are consistent with a model in which calcineurin and Tcn1p induce expression of Pmr1p, which increases both Mn2+ sequestration in late compartments of the secretory pathway and Mn2+ export from the cell. Several other observations support this model. Mutants lacking PMR1 also display numerous secretory defects that can be attributed to insufficient Ca2+ and Mn2+ accumulation in compartments of the secretory pathway (Rudolph et al. 1989; Antebi and Fink 1992). Sufficient Ca2+ and/or Mn2+ is required for viability (Loukin and Kung 1995), and pmr1 mutants require much higher levels of these metals than wild type in spite of their increased uptake and sensitivity (Lapinskas et al. 1995). These findings are consistent with a model in which Pmr1p supplies compartments of the secretory pathway with Ca2+ and Mn2+ during standard conditions and promotes Mn2+ tolerance by sequestration and eventual export. Further analysis of tcn1 mutants and Mn2+ transport by Pmr1p and other factors (Supek et al. 1996) may resolve this issue.
Because calcineurin-dependent inhibition of the vacuolar H+/Ca2+ exchanger Vcx1p is independent of Tcn1p (Fig. 4C), the analysis of tcn1 mutants shed little light on the mechanisms regulating Vcx1p or on the physiological significance of this regulation. Previous work suggested that calcineurin may inhibit Vcx1p post-translationally, although other explanations were not ruled out (Cunningham and Fink 1996). Why yeast cells inhibit Vcx1p when this enzyme can greatly increase Ca2+ tolerance also remains unclear. However, analysis of constitutive mutant forms of Vcx1p that resist inhibition by calcineurin revealed inappropriate H+/Ca2+ exchange decreases the availability of cytosolic Ca2+ for calcium signaling (Cunningham and Fink 1996) and potentially for transport by Pmr1p into the secretory pathway.
Calcium signaling in response to salt stress
Evidence reported here and in previous studies all suggest that the response to high salt includes activation of the calcium signaling pathway, although no change in Ca2+ influx or accumulation in the cytosol has been reported. We observed submaximal induction of the Gal1–lacZ reporter by Gal4(DB)::Tcn1(N) after treatment with high salt (Fig. 7), and in tcn1 mutants we observed decreases in Na+ tolerance and PMR2A expression. These results suggest that high salt may generate a weak Ca2+ signal that mildly activates the calcium signaling pathway. In addition, calcineurin contributes to Na+ tolerance independently of Tcn1p and the Pmr2p ion pumps (Fig. 4B; Danielsson et al. 1996; Mendoza et al. 1996). Finally, Ca2+/calmodulin promotes Na+ tolerance by binding and activating Pmr2p ion pumps (Wieland et al. 1995). Together, these findings demonstrate multiple interactions between the calcium signaling pathway and Na+ tolerance factors. Another response to high salt appears to be negative regulation of genes such as PMC1 and FKS2 (Figs. 1 and 7). High salt may activate specific repressors or inhibit specific coactivators of transcription such as the MSN5/STE21 gene product, which is important together with Tcn1p for calcineurin-dependent induction of FKS2 and PMC1 (P.M. Alepuz, D.P. Matheos, K.W. Cunningham, and F. Estruch, in prep.).
Calcium signaling during the pheromone response
There is abundant evidence that Ca2+ signals are generated in yeast after treatment with high doses of mating pheromones (Ohsumi and Anraku 1985) and that the calcium signaling pathway becomes activated and induces genes such as FKS2 (Mazur et al. 1995). Here we show that induction of FKS2 in response to pheromone treatment also requires Tcn1p. The pattern of FKS2 expression contrasts with other Tcn1p-dependent genes and suggests that the pheromone response generates a relatively weak Ca2+ signal that is insufficient for induction of low-sensitivity genes such as PMC1, which required overexpression of Tcn1p before a significant response to pheromone could be observed (Fig. 7). In spite of this clear role for Tcn1p in the response to pheromone, we detected no significant role for Tcn1p in several other calcineurin-responsive phenomena, including feedback inhibition of Ca2+ uptake (Fig. 5), changes in cell morphology, or promoting cell survival during pheromone stimulation (not shown). Survival in pheromone was shown previously to depend on Ca2+ influx and the functions of calmodulin, calcineurin, and calmodulin-dependent protein kinases (Iida et al. 1990, 1994; Moser et al. 1996; Withee et al. 1997). The targets of the calcium signaling pathway involved in cell survival therefore remain to be identified.
Comparison of Tcn1p regulation with vertebrate systems
The calcineurin-dependent transcriptional activation domain of Tcn1p shows no significant sequence similarity to other proteins in current databases, so extrapolations to specific vertebrate mechanisms are not yet possible. Several parallels are noteworthy, nevertheless. Using the zinc finger domain of Tcn1p to search protein databases, the most similar vertebrate proteins are members of the EGR family of transcription factors. Zif268/EGR-1 is markedly induced by calcium signaling through the serum response factor SRF in many cell types (Cole et al. 1989; Ginty 1997), and induction of EGR-2 in B cells also depends on calcineurin function (Gottschalk et al. 1994). In these cases, the molecular mechanisms controlling EGR expression are not precisely known. We show evidence that Tcn1p may regulate its own expression by a positive feedback mechanism requiring calcineurin and that this autoregulation may affect expression of target genes such as PMC1. Remarkably, expression of vertebrate PMCA genes encoding the plasma membrane Ca2+ ATPases homologous to Pmc1p also appears to be regulated in response to Ca2+ signals and calcineurin activation in granule cells of the developing rat cerebellum (Carafoli et al. 1996).
An emerging question in understanding signal transduction networks is how cells utilize common signaling modules to generate distinct outputs depending on the type or source of input signal. In neurons, for example, Ca2+ signals generated by either activation of the NMDA receptor or activation of the L-type Ca2+ channel caused phosphorylation of the critical serine-133 in the CREB transcription factor but only the signal derived from the L-type channel could induce the c-fos gene (Bading et al. 1993). Differences in the spatial or temporal character of these Ca2+ signals have been proposed to accomplish this type of differential gene expression (Dolmetsch et al. 1997; Ginty 1997) although differences in Ca2+ signal strength or additional regulatory mechanisms analogous to those reported here may also be involved. Although the spatiotemporal characteristics of the Ca2+ signals in yeast caused by pheromone, high salt, and high Ca2+ are presently unknown, our results indicate that different promoters are sensitive to variations in the strength of Ca2+ signals and inputs from other types of signaling pathways. More work is needed to understand this phenomenon and to accurately compare the yeast mechanism with mammalian systems.
Materials and methods
Culture media and isolation/construction of tcn1 mutants
Synthetic complete (SC) and complex (YPD) media were prepared and supplemented with 2% glucose as described previously (Sherman et al. 1986) using reagents from Difco and Sigma Chemical Co. Where indicated, YPD medium was buffered to pH 5.5 by addition of 5 mm succinic acid and supplemented with various salt such as CaCl2, MnCl2, NaCl, or G418 sulfate. FK506 was generously provided by Fujisawa Corp. (Tokyo, Japan). The synthetic pheromone α-mating factor was obtained from Star Biochemicals.
All yeast strains listed in Table 2 were derived from W303-1A (Wallis et al. 1989) using standard methods of transformation and/or crossing (Sherman et al. 1986), and all strains except Y190 harbored the following genetic markers: MATa ade2-1 can1-100 his3-1 leu2-3,112 trp1-1 ura3-1. The tcn1::kanMX3 null mutation in which the chromosomal TCN1 gene was deleted and replaced was introduced into W303-1A by one-step gene replacement (Rothstein 1991) using a fragment of plasmid pKC287 generated by digestion with _Bgl_II plus _Xba_I. The resulting tcn1 null mutant (strain DMY14) was selected in YPD agar medium supplemented with 0.2 mg/ml of G418 sulfate (GIBCO BRL) and verified by PCR analysis of genomic DNA. Additional strains containing tcn1::kanMX3 were constructed by crosses between DMY14 and previously described derivatives of W303-1A (Cunningham and Fink 1996). Strains DMY62 and DMY63 were constructed by transformation of strains K473 and K482, respectively, with _Apa_I-digested plasmid pKC217, which integrates a PMC1–lacZ reporter gene at the ura3-1 locus. β-Galactosidase accumulation in strains DMY62 and DMY63 was very low during growth in YPD (pH 5.5) medium and very high after growth in YPD (pH 5.5) medium supplemented with CaCl2 as detected using the chromogenic substrates _O_-nitrophenyl-βd-galactopyranoside (ONPG) as described (Guarente 1983) or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal, see below).
To identify mutants unable to induce PMC1–lacZ, strains DMY62 and DMY63 were mutagenized to ∼30% viability using EMS, spread onto 40 plates of SC minus uracil agar medium at a density of ∼1000 colonies/plate, and grown at 30°C for 3 days. Colonies were then replica-plated onto paper filters (Whatman No. 3) placed on YPD (pH 5.5) agar plates supplemented with 0.3 mm adenine and 75 mm CaCl2, incubated for 1 day at 30°C, and assayed for β-galactosidase accumulation as follows. Filters were first soaked with 25 mm EGTA (pH 7.5) for ∼20 min at room temperature, plunged into liquid nitrogen for ∼1 min, and finally soaked ∼3 hr at room temperature with staining solution containing 0.3 mg/ml of X-gal dissolved in modified Z buffer [100 mm Na phosphate (pH 7.0), 10 mm KCl, 10 mm MgSO4, 0.1% SDS, 0.27% β-mercaptoethanol]. Most colonies stained dark blue after this protocol. All white or light blue colonies were collected, purified, retested, and then subjected to complementation testing where each DMY62 isolate was mated with each DMY63 isolate in all possible combinations and the resulting diploids were selected on SC − Leu–Trp agar medium and tested for β-galactosidase accumulation after calcium treatment as before. All recessive mutations were placed into one of three complementation groups—cnb1, msn5, and _tcn1_—which were identified by cloning of the functional loci by complementation and/or allelism tests using targeted disruption mutants.
Cloning of TCN1 and recombinant DNA
All recombinant DNA techniques were performed using standard techniques (Sambrook et al. 1989) with reagents supplied by Stratagene and New England Biolabs. To identify the TCN1 gene, a tcn1 mutant isolated in the screen was transformed with a low-copy genomic DNA plasmid library (kind gift of D. Levin, Johns Hopkins University, Baltimore, MD) based on the pRS313 shuttle vector (Sikorski and Hieter 1989), plated on SC − His agar medium, replica plated to filters placed on YPD (pH 5.5) agar medium containing 75 mm CaCl2, and stained with X-gal as described above to identify rare blue clones that regained ability to express PMC1–lacZ. Out of ∼10,000 independent transformants, 2 were found to yield plasmids that complemented the tcn1 defect. Restriction mapping and partial sequencing of the insert DNA from both plasmids (pDM1 and pDM2) demonstrated that the two plasmids contain distinct but overlapping inserts spanning two previously uncharacterized open reading frames from chromosome XIV termed YNL026w and YNL027w. Deletion of YNL026w by digestion of pDM1 with _Spe_I plus _Xba_I, followed by religation, resulted in plasmid pDM3, which retained the ability to complement tcn1 mutations. Prior to the release of the complete yeast genomic DNA sequence, A. Duesterhoeft and P. Phillippsen generously provided the sequence of the TCN1 locus and plasmid p678::lacZkanMX3, which was used to construct the tcn1::kanMX3 disruption plasmid pKC287 by digesting with _Spe_I plus _Bsr_GI to remove all lacZ sequences, generating blunt ends, and ligating (Philippsen et al. 1997). Null mutants obtained by gene replacement using tcn1::kanMX3 DNA failed to complement each of the tcn1 alleles isolated in the genetic screen, showing that the mutations in this group all resided within the TCN1/YNL027w gene.
Plasmids containing various reporter genes were constructed as follows. An integrating plasmid containing the PMC1–lacZ reporter gene (pKC217) was derived from pKC190 (Cunningham and Fink 1996) after digestion with _Spe_I and religation to remove the 2μ origin of replication. The FKS2–lacZ reporter plasmid (pDM5) containing the DNA segment from position −968 to +6 relative to the initiation codon of FKS2 fused in-frame to lacZ coding sequences was constructed by PCR amplification with oligonucleotides FKS2A (GGAGTCGACAGGGCTACTCAATCG) and FKS2B (GCCTCTAGAGGACATACCTATGACAG) followed by digestion with _Sal_I plus _Xba_I and cloning first into polylinker sites of YEp356R (Myers et al. 1986) and then subcloning into pLGΔ178 (Guarente and Mason 1983) after first digesting both plasmids with _Xho_I plus _Bam_HI. A TCN1–lacZ reporter plasmid (pDM7) was constructed by subcloning the DNA segment from −1485 to +42 relative to the TCN1 initiation codon from pDM3 (liberated with _Sal_I plus _Spe_I) first into YEp356R (digested with _Sal_I plus _Xba_I) and then subcloning the _Sal_I plus _Bam_HI fragment into pLGΔ178 digested with _Xho_I plus _Bam_HI.
All plasmids expressing Gal4 hybrid proteins were constructed from plasmids pPC97 and pPC86 (Chevray and Nathans 1992) as follows. The amino-terminal region of Tcn1p corresponding to nucleotides +33 to +1381 was amplified using oligonucleotides DB1 (GCCGCCAATATGGCGTCGACCATGACTAGTAGTAAT) and DB2 (GCCGCCATTGTCATCCTAGGCCCGATTATTGTCATT), purified on agarose gels, digested with _Sal_I plus _Avr_II, and cloned into pPC97 digested with _Sal_I plus _Spe_I to yield pDM15 containing Gal4(DB)::Tcn1(N). Plasmid pDM16 expressing Tcn1(C)::Gal4(AD) hybrid proteins was constructed by ligating into _Sal_I plus _Spe_I-digested pPC86 the _Sal_I plus _Avr_II-digested PCR product obtained using primers TA1 (GCCGCCAATCGGGAGTCGACTGACAATGATAGCAAA) and TA2 (GCCGCCTTAACTCCTAGGCTCTTGTCCCGATTTCTC), corresponding to nucleotides +1389 to +2033 of TCN1. Plasmids pKC116 and pKC117 containing Gal4(DB):: Cna1ΔC and Gal4(DB)::Cna1 hybrids, respectively, were constructed by subcloning the _Xho_I plus _Bam_HI fragments of pKC73 and pKC74 (Cunningham and Fink 1996) into pPC97 digested with _Sal_I plus _Bam_HI. Plasmid pTJK27 containing Tcn1(N)::Gal4(AD) was constructed by subcloning a _Sal_I plus _Sst_I fragment from pDM15 into pPC86 digested with _Sal_I plus _Sst_I.
β-Galactosidase assays
Strains were grown overnight in SC media lacking uracil to mid-log phase, harvested, and grown for an additional 4 hr in either YPD or YPD (pH 5.5) supplemented with CaCl2, NaCl, α-mating factor, and/or FK506 as indicated in the text. β-Galactosidase activity was assayed at room temperature using chloroform/SDS permeabilized cells as described previously (Guarente 1983).
Ion tolerance assay
Ion tolerance assays were performed as described previously (Cunningham and Fink 1996). For CaCl2, yeast strains were grown in YPD (pH 5.5), whereas MnCl2 and NaCl assays used YPD media. Cell density was measured at an OD650 with a Molecular Devices microplate reader. The concentration of cation resulting in a 50% decrease in cell growth relative to unsupplemented cultures (IC50) was interpolated from linear plots of the ion tolerance data.
Ca2+ uptake assays
Yeast cultures were grown to mid-log phase at 30°C in YPD media, harvested, and resuspended to an OD600 of 0.25 in 0.2 ml of YPD and supplemented with ∼10 mCi/ml of 45Ca2+ (Amersham Life Science). After incubation for 4 hr at 30°C with occasional mixing, 0.18 ml of culture was filtered through Whatman GFF filters and washed three times with buffer (5 mm Na HEPES, 10 mm CaCl2 at pH 6.5). Filters were dried and radioactivity quantitated using a scintillation counter. 45Ca2+ accumulation per 109 cells was calculated using the measured radioactivity retained on filters, the specific activity, and the cell density as determined by culture density using OD600.
Acknowledgments
We are indebted to Andreas Duesterhoeft and Peter Philippsen for generously providing DNA sequences prior to publication as well as yeast strains and plasmids. We are very grateful to Paula Alepuz, Trisha Davis, Cameron Douglas, Francisco Estruch, and David Levin for providing reagents and advice and to Fujisawa Corp. for gifts of FK506. This work was supported jointly by the Searle Scholars Program/The Chicago Community Trust, the Basil O’Connor Starter Scholar Research Award (FY96-1131) from the March of Dimes Birth Defects Foundation, and a Research Grant from National Institutes of Health (GM53082).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kwc@jhunix.hcf.jhu.edu; FAX (410) 516-5213.
References
- Akada R, Kallal L, Johnson DI, Kurjan J. Genetic relationships between the G protein β-γ complex, Ste5p, Ste20p, and Cdc42p: Investigation of effector roles in the yeast pheromone response pathway. Genetics. 1996;143:103–117. doi: 10.1093/genetics/143.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz PM, Cunningham KW, Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress activated ENA1 gene. Mol Microbio. 1997;26:91–98. doi: 10.1046/j.1365-2958.1997.5531917.x. [DOI] [PubMed] [Google Scholar]
- Antebi A, Fink GR. The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- Carafoli E, Garcia-Martin E, Guerini D. The plasma membrane calcium pump: recent developments and future perspectives. Experientia. 1996;52:1091–1100. doi: 10.1007/BF01952107. [DOI] [PubMed] [Google Scholar]
- Chevray PM, Nathans D. Protein interaction cloning in yeast: Identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- Collier SJ. Immunosuppressive drugs. Curr Opin Immunol. 1990;2:854–858. doi: 10.1016/0952-7915(89)90169-6. [DOI] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in yeast. Mol Cell Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS, Thorner J. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol Cell Biol. 1992;12:3460–3469. doi: 10.1128/mcb.12.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS, Kunisawa R, Kaim D, Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci. 1991;88:7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson A, Larsson C, Larsson K, Gustafsson L, Adler L. A genetic analysis of the role of calcineurin and calmodulin in Ca2+-dependent improvement of NaCl tolerance of Saccharomyces cerevisiae. Curr Genet. 1996;30:476–484. doi: 10.1007/s002940050159. [DOI] [PubMed] [Google Scholar]
- Davis TN, Urdea MS, Masiarz FR, Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986;47:423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- Dunn T, Gable K, Beeler T. Regulation of cellular Ca2+ by yeast vacuoles. J Biol Chem. 1994;269:7273–7278. [PubMed] [Google Scholar]
- Eng WK, Faucette L, McLaughlin MM, Cafferkey R, Koltin Y, Morris RA, Young PR, Johnson RK, Livi GP. The yeast FKS1 gene encodes a novel membrane protein, mutations in which confer FK506 and cyclosporin A hypersensitivity and calcineurin-dependent growth. Gene. 1994;151:61–71. doi: 10.1016/0378-1119(94)90633-5. [DOI] [PubMed] [Google Scholar]
- Farcasanu IC, Hirata D, Tsuchiya E, Nishiyama F, Miyakawa T. Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese, in blocking the entry of ions into the cells. Eur J Biochem. 1995;232:712–717. [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: An assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Foor F, Parent SA, Morin N, Dahl AM, Ramadan N, Chrebet G, Bostian KA, Nielsen JB. Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from alpha-factor arrest in yeast. Nature. 1992;360:682–684. doi: 10.1038/360682a0. [DOI] [PubMed] [Google Scholar]
- Garciadeblas B, Rubio F, Quintero FJ, Banuelos MA, Haro R, Rodriguez-Navarro A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol & Gen Genet. 1993;236:363–368. doi: 10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- Garrett-Engele P, Moilanen B, Cyert MS. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H+ ATPase. Mol Cell Biol. 1995;15:4103–4114. doi: 10.1128/mcb.15.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, van Tuinen D, Brockerhoff SE, Neff MM, Davis TN. Can calmodulin function without binding calcium? Cell. 1991;65:949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- Ginty DD. Calcium regulation of gene expression: Isn’t that spatial? Cell. 1997;18:183–186. doi: 10.1016/s0896-6273(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Gottschalk AR, Joseph LJ, Quintans J. Fc gamma RII cross-linking inhibits anti-Ig-induced Egr-1 and Egr-2 expression in BCL1. J Immunol. 1994;152:2115–2122. [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Haro R, Garciadeblas B, Rodriguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hemenway CS, Dolinski K, Cardenas ME, Hiller MA, Jones EW, Heitman J. vph6 mutants of Saccharomyces cerevisiae require calcineurin for growth and are defective in vacuolar H+/ATPase assembly. Genetics. 1995;141:833–844. doi: 10.1093/genetics/141.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Harada S, Namba H, Miyakawa T. Adaptation to high-salt stress in Saccharomyces cerevisiae is regulated by Ca2+/calmodulin-dependent phosphoprotein phosphatase (calcineurin) and cAMP-dependent protein kinase. Mol & Gen Genet. 1995;249:257–264. doi: 10.1007/BF00290525. [DOI] [PubMed] [Google Scholar]
- Iida H, Yagawa Y, Anraku Y. Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway. A study of [Ca2+]i in single Saccharomyces cerevisiae cells with imaging of fura-2. J Biol Chem. 1990;265:13391–13399. [PubMed] [Google Scholar]
- Iida H, Nakamura H, Ono T, Okumura MS, Anraku Y. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol Cell Biol. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno T, Tanaka H, Mukai H, Chang CD, Hiraga K, Miyakawa T, Tanaka C. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1991;180:1159–1163. doi: 10.1016/s0006-291x(05)81188-x. [DOI] [PubMed] [Google Scholar]
- Lapinskas PJ, Cunningham KW, Liu XF, Fink GR, Culotta VC. Mutations in PMR1 suppress oxidative damage in yeast lacking superoxide dismutase. Mol Cell Biol. 1995;15:1382–1388. doi: 10.1128/mcb.15.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P, Revelant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci. 1988;85:4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ishii S, Tokai M, Tsutsumi H, Ohki O, Akada R, Tanaka K, Tsuchiya E, Fukui S, Miyakawa T. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol & Gen Genet. 1991;227:52–59. doi: 10.1007/BF00260706. [DOI] [PubMed] [Google Scholar]
- Loukin S, Kung C. Manganese effectively supports yeast cell-cycle progression in place of calcium. J Cell Biol. 1995;131:1025–1037. doi: 10.1083/jcb.131.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P, Morin N, Baginsky W, El-Sherbeini M, Clemas JA, Nielsen JB, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-beta-D-glucan synthase. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo JM. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- Mendoza I, Quintero FJ, Bressan RA, Hasegawa PM, Pardo JM. Activated calcineurin confers high tolerance to ion stress and alters the budding pattern and cell morphology of yeast cells. J Biol Chem. 1996;271:23061–23067. doi: 10.1074/jbc.271.38.23061. [DOI] [PubMed] [Google Scholar]
- Moser MJ, Geiser JR, Davis TN. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol Cell Biol. 1996;16:4824–4831. doi: 10.1128/mcb.16.9.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Tzagoloff A, Kinney DM, Lusty CJ. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Nakajima-Shimada J, Iida H, Tsuji FI, Anraku Y. Monitoring of intracellular calcium in Saccharomyces cerevisiae with an apoaequorin cDNA expression system. Proc Natl Acad Sci. 1991;88:6878–6882. doi: 10.1073/pnas.88.15.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Ohmoto T, Hirata D, Tsuchiya E, Miyakawa T. Genetic evidence for the functional redundancy of the calcineurin- and Mpk1-mediated pathways in the regulation of cellular events important for growth in Saccharomyces cerevisiae. Mol & Gen Genet. 1996;251:211–219. doi: 10.1007/BF02172920. [DOI] [PubMed] [Google Scholar]
- Nass R, Cunningham KW, Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. J Biol Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Anraku Y. Specific induction of Ca2+ transport activity in MATa cells of Saccharomyces cerevisiae by a mating pheromone, α factor. J Biol Chem. 1985;260:10482–10486. [PubMed] [Google Scholar]
- Ohya Y, Kawasaki H, Suzuki K, Londesborough J, Anraku Y. Two yeast genes encoding calmodulin-dependent protein kinases. Isolation, sequencing and bacterial expressions of CMK1 and CMK2. J Biol Chem. 1991;266:12784–12794. [PubMed] [Google Scholar]
- Parent SA, Nielsen JB, Morin N, Chrebet G, Ramadan N, Dahl AM, Hsu J-J, Bostian KA, Foor F. Calcineurin-dependent growth of an FK506 and CsA hypersensitive mutant of Saccharomyces cerevisiae. J Gen Microbiol. 1993;139:2973–2984. doi: 10.1099/00221287-139-12-2973. [DOI] [PubMed] [Google Scholar]
- Pausch MH, Kaim D, Kunisawa R, Admon A, Thorner J. Multiple Ca2+/calmodulin-dependent protein kinase genes in a unicellular eukaryote. EMBO J. 1991;10:1511–1522. doi: 10.1002/j.1460-2075.1991.tb07671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Philippsen, P., K. Kleine, R. Pohlmann, A. Dusterhoft, K. Hamberg, J.H. Hegemann, B. Obermaier, L.A. Urrestarazu, R. Aert, K. Albermann et al. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature (Suppl.) 387: 93–98. [PubMed]
- Pozos TC, Sekler I, Cyert MS. The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol Cell Biol. 1996;16:3730–3741. doi: 10.1128/mcb.16.7.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao JI, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schreiber SL, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- Sherman F, Hicks JB, Fink GR. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin A, Rosas G, Rao R. PMR1, a Ca2+-ATPase in yeast Golgi, has properties distinct from sarco/endoplasmic reticulum and plasma membrane calcium pumps. J Biol Chem. 1997;272:9895–9901. doi: 10.1074/jbc.272.15.9895. [DOI] [PubMed] [Google Scholar]
- Sprague GF, Jr, Thorner JW. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones EW, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces: Gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- Stathopoulos, A.M. and M.S. Cyert. 1997. Calcineurin acts through the _CRZ1/TCN1_-encoded transcription factor to regulate gene expression in yeast. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Stillman DJ, Bankier AT, Seddon A, Groenhout EG, Nasmyth KA. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988;7:485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Hasegawa A, Iida H, Ohya Y, Anraku Y. Cooperation of calcineurin and vacuolar H+-ATPase in intracellular Ca2+ homeostasis of yeast cells. J Biol Chem. 1995;270:10113–10119. doi: 10.1074/jbc.270.17.10113. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wieland J, Nitsche AM, Strayle J, Steiner H, Rudolph HK. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee JL, Mulholland J, Jeng R, Cyert MS. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol Biol Cell. 1997;8:263–277. doi: 10.1091/mbc.8.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye RR, Bretscher A. Identification and molecular characterization of the calmodulin-binding subunit gene (CMP1) of protein phosphatase 2B from Saccharomyces cerevisiae. An α-factor inducible gene. Eur J Biochem. 1992;204:713–723. doi: 10.1111/j.1432-1033.1992.tb16686.x. [DOI] [PubMed] [Google Scholar]