The Inside-Out Mechanism of Dicers from Budding Yeasts (original) (raw)
. Author manuscript; available in PMC: 2012 Jan 22.
SUMMARY
The Dicer ribonuclease III (RNase III) enzymes process long double-stranded RNA (dsRNA) into small interfering RNAs (siRNAs) that direct RNA interference. Here, we describe the structure and activity of a catalytically active fragment of Kluyveromyces polysporus Dcr1, which represents the noncanonical Dicers found in budding yeasts. The crystal structure revealed a homodimer resembling that of bacterial RNase III but extended by a unique N-terminal domain, and it identified additional catalytic residues conserved throughout eukaryotic RNase III enzymes. Biochemical analyses showed that Dcr1 dimers bind cooperatively along the dsRNA substrate such that the distance between consecutive active sites determines the length of the siRNA products. Thus, unlike canonical Dicers, which successively remove siRNA duplexes from the dsRNA termini, budding-yeast Dicers initiate processing in the interior and work outward. The distinct mechanism of budding-yeast Dicers establishes a paradigm for natural molecular rulers and imparts substrate preferences with ramifications for biological function.
INTRODUCTION
RNA interference (RNAi) is a gene-silencing pathway triggered by dsRNA (Meister and Tuschl, 2004; Malone and Hannon, 2009). In this pathway, the RNase III enzyme Dicer first converts long dsRNA into 21–25 nucleotide (nt) siRNA duplexes bearing 2 nt 3' overhangs, a defining characteristic of RNase III cleavage products (MacRae and Doudna, 2007). The siRNA duplexes are then loaded into the effector protein Argonaute, and after one strand is discarded the remaining single-stranded RNA (ssRNA) pairs to RNA substrates, thereby guiding Argonaute to cleave these target transcripts (Tomari and Zamore, 2005; Wang et al., 2009b). Although RNAi or related silencing pathways are found in most eukaryotes, they are missing in Saccharomyces cerevisiae, which lacks both Dicer and Argonaute homologs. Nonetheless, other budding-yeast species, including Saccharomyces castellii and Kluyveromyces polysporus, contain a noncanonical Dicer, Dcr1, which fuels the Argonaute-mediated RNAi pathway in these organisms (Drinnenberg et al., 2009).
Dicer enzymes must generate siRNAs of a length compatible with loading into Argonaute. Canonical Dicers measure from the end of the dsRNA to determine product length, with siRNA length equaling the distance spanning the terminus-binding PAZ domain and RNase III active sites (Zhang et al., 2004; Macrae et al., 2006). The noncanonical Dicers of budding yeast have a very different domain architecture that lacks a PAZ domain (Figure 1A), raising the question of how they cleave dsRNA at precise intervals. Here we show how budding-yeast Dicers generate siRNAs of the correct length. Unlike canonical Dicers, which successively remove siRNA duplexes from the dsRNA termini, budding-yeast Dicers start in the interior and work outward. The distinct mechanisms of canonical and noncanonical Dicers confer different constraints on the types of substrates processed into guide RNAs and thus have important implications for RNAi function.
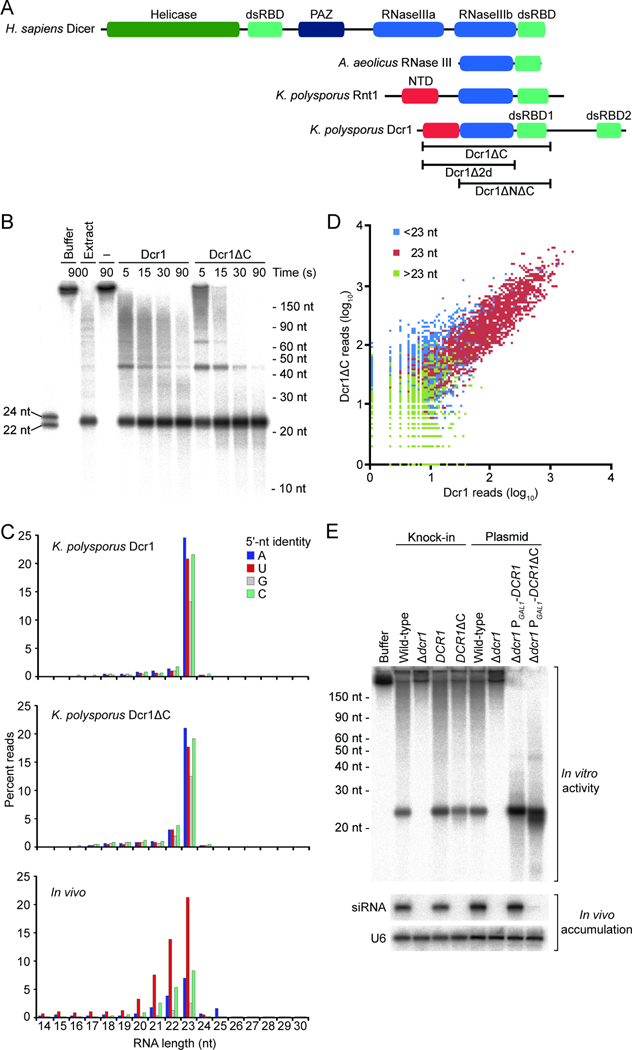
Figure 1. Activity of K. polysporus Dcr1ΔC.
(A) Domain architectures of representative RNase III proteins. The N-terminal domain (NTD) unique to budding-yeast RNase III enzymes is indicated.
(B) Activity of recombinant Dcr1 proteins under standard single-turnover conditions. Body-labeled 500 bp dsRNA was incubated with no enzyme (−), full-length K. polysporus Dcr1, or K. polysporus Dcr1ΔC for the indicated time. For comparison, the substrate was incubated with K. polysporus whole-cell extract (Extract) or with the buffer used for extracts (Buffer). Also shown are radiolabeled synthetic 22 and 24 nt RNAs (left) and the migration of other RNA standards (right).
(C) Length distributions of products with the indicated 5' nucleotides. Top and middle: Substrate-matching sequencing reads from analysis of in vitro reactions using the indicated purified proteins. Bottom: For reference, genome-matching small RNAs sequenced from K. polysporus, redrawn from Drinnenberg et al. (2009).
(D) Correlation between cleavage products generated by purified K. polysporus Dcr1 and Dcr1ΔC. Plotted is the read count (including a pseudocount of 1) for each sequenced product from the Dcr1ΔC reaction (y-axis) compared to the count for the corresponding product from the Dcr1 reaction (x-axis). Colors indicate the length of the sequenced products: 14–22 nt (blue), 23 nt (red), and 24–30 nt (green).
(E) Comparison of in vitro activity with product accumulation in strains expressing the corresponding Dcr1 variants. Top: Body-labeled 500 bp dsRNA was incubated with extracts from S. castellii strains with the indicated deletions, additions, and replacements. Bottom: An RNA blot with samples from the same strains was probed for an endogenous siRNA, then reprobed for U6 small nuclear RNA.
See also Figure S1 and Table S1.
RESULTS
Purified Dcr1 and Dcr1ΔC Retain Precise Cleavage Activity
One way that budding-yeast Dicer might have cleaved at precise intervals without a PAZ domain could have been through association with a protein cofactor that substituted for the PAZ domain. Arguing against this possibility, recombinant K. polysporus Dcr1 purified from E. coli converted body-labeled dsRNA into 23 nt products (Figure 1B). To facilitate subsequent biochemical and structural analyses, we identified a fragment of K. polysporus Dcr1 that could be purified from contaminating RNA (Figure S1A) and had enhanced stability and solubility. Spanning Ser15 to Glu355, this fragment lacked the extreme N-terminus and the C-terminal dsRBD (dsRBD2) and is referred to as Dcr1ΔC (Figure 1A). It had robust activity and generated 23 nt products indistinguishable from those of full-length enzyme or K. polysporus extracts (Figure 1B). Despite lacking one dsRBD, Dcr1ΔC retained specificity for dsRNA over ssRNA substrates (Figure S1B). In contrast, a fragment lacking both dsRBDs (Dcr1Δ2d) cleaved much less efficiently, failed to preferentially generate 23 nt products, and did not discriminate between dsRNA and ssRNA (Figure S1C). The requirement of dsRBD1 for dsRNA-specific cleavage activity was consistent with its requirement for dsRNA binding, as shown in a gel-shift assay (Figure S1D).
Sequencing of 18–30 nt RNAs isolated from processing reactions performed using full-length Dcr1 and Dcr1ΔC (Table S1) showed that products of both enzymes were predominantly 23 nt and displayed little 5'-nucleotide bias (Figure 1C). The abundances of 23 nt species in the two libraries were strongly correlated (_R_2 = 0.84), which showed that Dcr1ΔC faithfully recapitulated the siRNA-generating activity of full-length Dcr1 (Figure 1D). The length homogeneity of in vitro products exceeded that of endogenous siRNAs in K. polysporus (Drinnenberg et al., 2009) (Figure 1C). Perhaps length heterogeneity observed in vivo reflects subsequent events, such as 3'-end trimming of siRNAs that have been loaded into Argonaute (Halic and Moazed, 2010). In addition, the absence of a 5'-uridine bias among siRNAs generated in vitro suggested that the bias observed in vivo might arise from the binding specificity of Argonaute (Wang et al., 2009b; Frank et al., 2010).
Although dsRBD2 was dispensable for size-specific siRNA generation in vitro (Figures 1B–1D), its strict conservation among Dcr1 proteins and absence from Rnt1 proteins (RNase III enzymes involved in yeast ribosome biogenesis, Figure 1A) prompted us to examine its importance for siRNA accumulation in vivo using S. castellii, a species that is closely related to K. polysporus but has more tools available for molecular-genetic manipulation (Drinnenberg et al., 2009). The endogenous copy of S. castellii Dcr1 was replaced with S. castellii Dcr1ΔC expressed from the native promoter. Although extracts made from yeast expressing full-length Dcr1 and Dcr1ΔC had comparable in vitro dicing activity, endogenous siRNAs failed to accumulate in the Dcr1ΔC strain (Figure 1E). The absence of siRNAs in this strain could not be attributed to either lack of the extreme N-terminus or lower expression, as siRNA accumulation in vivo was not rescued by either restoration of the native N-terminus (data not shown) or overexpression of Dcr1ΔC in a Δ_dcr1_ strain (Figure 1E). In addition to size-specific cleavage, Dcr1 function in vivo might also involve appropriate protein localization, protection of product siRNAs from nucleases, and interaction with other proteins such as Argonaute. The C-terminal dsRBD is presumably involved in one or more of these additional aspects of Dcr1 function.
Crystal Structure of Dcr1ΔC
Having found that Dcr1ΔC recapitulates the siRNA-generating activity of the full-length enzyme, we determined its crystal structure at 2.3 Å resolution (Figure 2A and Table S2). Because RNase III domains work in pairs (MacRae and Doudna, 2007), Dcr1 has been proposed to act as a homodimer (Drinnenberg et al., 2009). Gel filtration and glutaraldehyde crosslinking confirmed that Dcr1ΔC behaves as a homodimer in solution (Figures S2A and S2C). The crystal structure had two homodimers in the asymmetric unit (Figure S2D), such that all four N-terminal domains (NTDs) and RNase III domains had well-defined electron density, whereas only one of the four dsRBD1 domains had traceable density, presumably because it had stabilizing crystal-packing interactions (Figure 2B, left panel). Dcr1ΔC forms a compact homodimer through the NTD and RNase III domains (Figure 2A), with the single observed dsRBD1, which exhibited variable density (Figure 2B), connected to the RNase III domain by a flexible linker. The buried surface within the dimer is 1,850 Å2, including 842 Å2 between the NTDs.
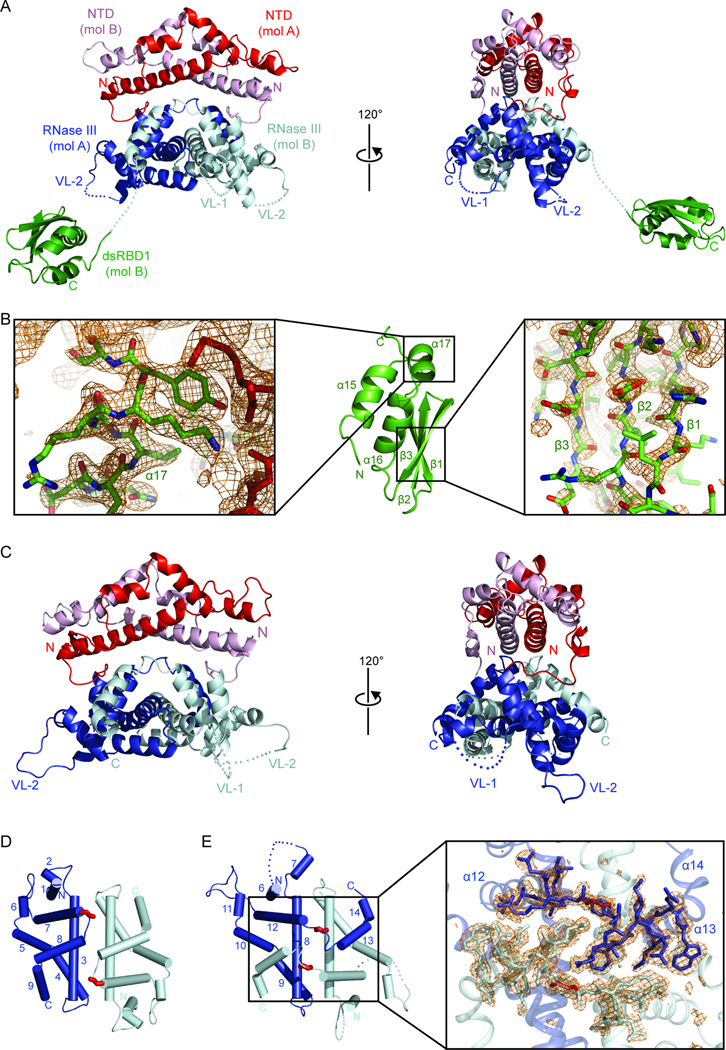
Figure 2. Crystal Structures of Dcr1ΔC and E224Q Dcr1Δ2d.
(A) Crystal structure of a Dcr1ΔC dimer at 2.3 Å resolution, showing a pair of NTDs (red and pink), a pair of RNase III domains (blue and silver) and a single dsRBD1 (green). The other dsRBD1 had poor density and is not shown. Disordered loops are shown as dotted lines, and VL-1 and VL-2 are labeled.
(B) Structure of Dcr1ΔC dsRBD (middle), flanked by close-up views of representative segments with good (left) or poor (right) electron density maps (2_F_o–_F_c). This domain is stabilized through interactions with a symmetry-related NTD of another Dcr1ΔC molecule (red sticks).
(C) Crystal structure of an E224Q Dcr1Δ2d dimer at 1.97 Å resolution. Domain designations and colors are as in (A).
(D) Topology of the A. aeolicus RNase III domain dimer (PDB code 2NUG). Individual monomers are colored in blue and silver, highlighting glycine residues at the kink point between helices α7 and α8 (red stick models).
(E) Topology of K. polysporus E224Q Dcr1Δ2d. Helices α13 and α14 (numbered according to the entire model) participate in a domain swap, with kink-point glycine residues indicated as in (D). The expanded region shows an _F_o–_F_c simulated annealing omit map of the loops between helices α12 and α13 contoured at 3.2σ (orange wire mesh). The α12–α13 loops are shown as stick models, and the other regions are shown as ribbon models.
See also Figure S2 and Table S2.
The NTD has a novel fold composed of five α-helices and forms a symmetric homodimer that packs against the RNase III domain homodimer (Figure 2A). Although a DALI search for structural similarity (Holm and Sander, 1993) failed to identify structures with significant homology to the NTD, sequence comparison of Dcr1 and Rnt1 N-terminal regions revealed a portion of Rnt1 with homology to the Dcr1 NTD (Figure 1A and Figure S2E), suggesting that both adopt a similar structure and have a common function and evolutionary origin. Indeed, the homodimeric nature of the Dcr1 NTD is consistent with the finding that the S. cerevisiae Rnt1 N-terminal region contains a dimerization signal (Lamontagne et al., 2000). Despite contributing an additional dimerization interface to the Dcr1 homodimer (Figure 2A), the NTD was not absolutely required for either dimerization (Figure S2F) or siRNA generation (Figure S1E and S1F), observations analogous to those for Rnt1 (Lamontagne et al., 2000).
The structure of the homodimeric Dcr1ΔC RNase III domain (Figure 2A) resembles structures of the homodimeric bacterial RNase III domain and the intramolecular heterodimeric RNase III domains of the canonical Dicer from Giardia intestinalis (_Gi_Dicer) (Gan et al., 2006; Macrae et al., 2006). One difference is that the Dcr1ΔC RNase III domains swap their terminal α-helices with each other (Figure 2A), which would presumably further strengthen the very stable interaction observed for RNase III homodimers (Meng and Nicholson, 2008). Our crystal structure of an active-site mutant (E224Q) of Dcr1Δ2d, which diffracted to higher (1.97 Å) resolution (Figure 2C, Table S2, and Figure S2B), confirmed this domain swap, as illustrated in a comparison with the Aquifex aeolicus RNase III (_Aa_RNase III) homodimer (Figures 2D and 2E). Another difference from the structures of _Aa_RNase III and _Gi_Dicer is that Dcr1ΔC has two long loops that correspond to the most variable regions in RNase III alignments (Figures 2A and 3A). We refer to the loops between α6 and α7 and between α11 and α12 as variable loops 1 and 2 (VL-1 and VL-2), respectively. α7, to which VL-1 is connected, is an additional helix that is not observed in other RNase III enzyme structures.
The disorder of the dsRBD1s in the Dcr1ΔC structure was consistent with reports of flexibility in the orientations between RNase III domains and dsRBDs in the absence of dsRNA (Akey and Berger, 2005). Nonetheless, the robust electron density observed for one dsRBD1 permitted confident determination of its structure (Figure 2B). Dcr1 dsRBD1 has the canonical α-β-β-β-α topology, with the pair of α-helices packed against an antiparallel β-sheet (Figure 2B). Among previously solved dsRBD structures, Dcr1 dsRBD1 most closely resembles Rnt1 dsRBD, in that these two dsRBDs are the only ones that include an α-helical extension (α17) beyond the canonical dsRBD fold (Leulliot et al., 2004). However, Dcr1 has not conserved the identities of residues that confer to Rnt1 a preference for AGNN tetraloops (Wu et al., 2004), which explains an absence of tetraloop specificity for Dcr1 (Figure S2G). In addition, the β1–β2 loop is especially short in Dcr1 dsRBD1 and thus would not be able to insert into the minor groove, which would presumably weaken affinity between this dsRBD and dsRNA (Figure S2H).
Sequence alignment of RNase III domains had suggested a close evolutionary relationship between Rnt1 and Dcr1 (Drinnenberg et al., 2009). Our results extend this relationship to two additional domains, showing that Rnt1 and Dcr1 have similar NTDs that expand the homodimer interface and that Dcr1 dsRBD1 shares many features with Rnt1 dsRBD but has lost otherwise conserved elements required for sequence specificity.
Metal-Ion Coordination in the Active Site
The RNase III domains of _Aa_RNase III and _Gi_Dicer form intermolecular and intramolecular dimers, respectively, to position a pair of active sites ~20 Å apart, which accommodates the width of the dsRNA minor groove with a 2 nt offset (Gan et al., 2006; Macrae et al., 2006). Within each active site, the most prominent metal-binding site (M1) is surrounded by four acidic residues (two Glu and two Asp), which are strictly conserved among RNase III enzymes and required for efficient catalysis (Takeshita et al., 2007). The RNase III domains of budding-yeast Dicers contain the four conserved acidic residues: Glu147, Asp151, Asp221, and Glu224 (Figure 3A). We observed clear octahedron electron density in the middle of these residues of Dcr1ΔC, indicating that each active site contained a metal ion (Figure 3B). Because the crystallization buffer contained 20 mM Mg2+, the ion presumably is Mg2+. The 20.6 Å distance between the two metals within each Dcr1ΔC dimer (Figure 3B) is consistent with that of other RNase III enzymes (Takeshita et al., 2007).
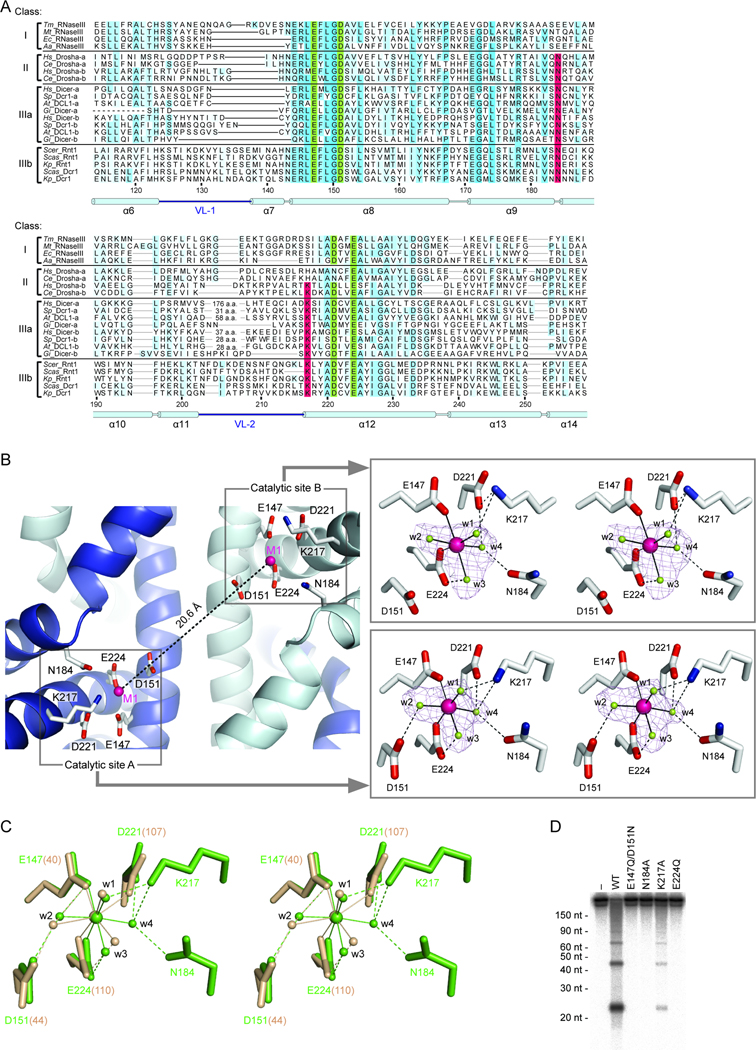
Figure 3. The Active Site of K. polysporus Dcr1.
(A) Sequence alignment of RNase III domains from each class of RNase III enzymes. Highlighted are the previously identified catalytic residues (green), two newly identified catalytic residues (red), and additional well-conserved amino acids (blue; intensity indicates degree of conservation). Residue numbers and the secondary structure of K. polysporus Dcr1 are indicated below the alignment. Tandem RNase III domains present in Drosha and canonical Dicer are designated a and b. Tm, Thermotoga maritoma; Mt, Mycoplasma tuberculosis; Ec, Escherichia coli; Aa, Aquifex aeolicus; Hs, Homo sapiens; Ce, Caenorhabditis elegans; Sp, Schizosaccharomyces pombe; At, Arabidopsis thaliana; Gi, Giardia intestinalis; Scer, Saccharomyces cerevisiae; Scas, S. castellii; and Kp, K. polysporus.
(B) Left: Close-up view of the Dcr1 catalytic sites showing RNase III domains (ribbons), conserved catalytic residues (sticks), and Mg2+ ions (spheres). Right: Metal-ion coordination in the active sites shown in stereo view. Octahedral coordination bonds and hydrogen bonds are drawn as solid and dotted lines, respectively. _F_o–_F_c simulated annealing omit maps of Mg2+ metal ion (magenta) and four water molecules (lime) are contoured at 4.0σ (purple wire mesh).
(C) Stereo view of the superimposed active sites of apo-Dcr1ΔC (green) and apo-_Aa_RNase III (tan; PDB code 1RC5). Bonds are drawn as in (B).
(D) Dicing activity of active-site mutants under standard single-turnover conditions. Body-labeled 500 bp dsRNA was incubated for 5 s with buffer only (−), wild-type Dcr1ΔC (WT), or Dcr1ΔC variants with the indicated mutations.
See also Figure S3.
In the structure of _Aa_RNase III (Blaszczyk et al., 2004), the metal ion at the M1 site is six-coordinated to Glu40, Asp107, and Glu110 (corresponding to Glu147, Asp221, and Glu224 of K. polysporus Dcr1, respectively) and three water molecules (w1–w3) (Figure 3C). The M1 site of Dcr1 is six-coordinated in a similar manner, with Glu147 and Glu224 participating in metal-ion binding in the same way as the corresponding residues in _Aa_RNase III (Figures 3B and 3C). In both structures, the metal-coordinated w3 is also hydrogen bonded to Glu110/224 (Figure 3C). Previous biochemical analyses of bacterial RNase III mutants demonstrated that Glu110 binds two Mg2+ ions and is essential for dsRNA cleavage, whereas the other active-site residues are not absolutely required for cleavage (Sun and Nicholson, 2001; Sun et al., 2004). As for the bacterial enzymes, replacement of Glu224 with Gln completely eliminated Dcr1ΔC activity, whereas a substitution of both Glu147 and Asp151 (with Gln and Asn, respectively) greatly diminished (Figure 3D) but did not eliminate (Figure S3) activity. As in most other apo-RNase III structures, no electron density was observed for a second metal ion in the active sites of our structure. By analogy to _Aa_RNase III (Gan et al., 2008), Glu224 of Dcr1 might coordinate two metal ions only after substrate RNA enters the active site.
An important difference between the M1 site of Dcr1 and that of bacterial RNase III is that in Dcr1 a fourth water molecule (w4), rather than Asp221, directly coordinates the Mg2+ ion (Figures 3B and 3C). In both active sites, this water is positioned by Asp221 as well as Asn184. An analogous Asn residue was proposed to hydrogen bond with a water molecule in a canonical Dicer RNase IIIb domain, which lacked a metal ion (Du et al., 2008). Our structure provides a function for this interaction, indicating that the water and thus Asn184 help position an active-site metal ion. In Dcr1, a sixth residue, Lys217, also contributes to metal-ion binding. In one of the active sites Lys217 forms hydrogen bonds with both w1 and w4, whereas in the other active site Lys217 forms hydrogen bonds with w1 and Asp221, which is associated with distortion of the octahedron (Figure 3B). Structural analysis of an _Aa_RNase III–product dsRNA complex indicates that two oxygen atoms of the scissile phosphate group replace w1 and w2 in the octahedral coordination (Gan et al., 2008). By analogy, Lys217 of Dcr1 would directly recognize the scissile phosphate group during the cleavage reaction.
The residues corresponding to Asn184 and Lys217 are not conserved in bacterial RNase III enzymes but are conserved throughout eukaryotic RNase III enzymes (with the exception of Lys217 in the RNaseIIIa domain of Drosha, Figure 3A), suggesting that they have an important catalytic role in eukaryotes. To test their importance for cleavage activity, we examined effects of Ala substitutions at Asn184 or Lys217. Replacement of Asn184 reduced in vitro cleavage activity to extremely low levels (Figure 3D and Figure S3). Replacement of Lys217 also reduced activity (Figure 3D and Figure S3), as previously observed for the corresponding residue in a canonical Dicer RNaseIIIb domain (Du et al., 2008). Thus, their positions in the structure, evolutionary conservation, and sensitivity to mutation all indicate that these two residues play important roles in the active sites of eukaryotic RNase III enzymes.
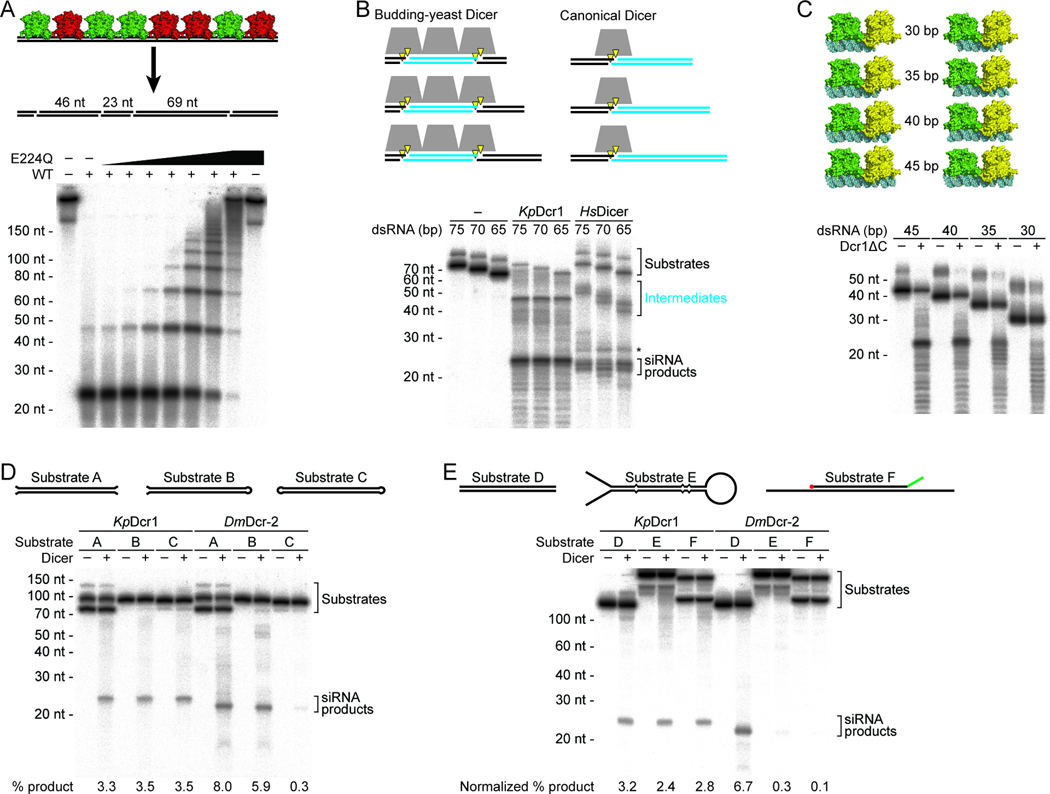
Closely Spaced Dcr1 Dimers Generate 23 nt Products from Internal dsRNA Regions
Because the NTD abuts the RNase III domain on the side opposite that of the catalytic surface (Figure 2A), a Dcr1ΔC dimer could interact with dsRNA in a manner analogous to that observed for _Aa_RNase III (Figure 4A). When apo-Dcr1ΔC and an _Aa_RNase III–dsRNA complex (Gan et al., 2006) are structurally aligned based on their RNase III domains, the modeled dsRNA only clashes with VL-1 of Dcr1, suggesting an induced fit wherein VL-1 extends along an RNA groove (Figure 4B). The alignment also suggested an induced fit of VL-2, because the corresponding loop in _Aa_RNase III (which constitutes RNA-binding motif 4) moves upon dsRNA binding (Gan et al., 2006). By using the loops VL-1 and VL-2 protruding from the sides, Dcr1ΔC would have an interface with dsRNA larger than that of _Aa_RNase III. To evaluate the importance of VL-1 and VL-2 for Dcr1 activity, we replaced these regions in Dcr1ΔC with the structurally analogous regions from the _Gi_Dicer RNase IIIb domain (Figure S4A). The VL-2 substitution abrogated cleavage activity, and the VL-1 substitution reduced activity such that even after extended incubation only heterogeneous products were observed (Figure 4C). Gel-shift assays indicated that the mutants had greatly reduced affinities for dsRNA substrate (Figure S1D).
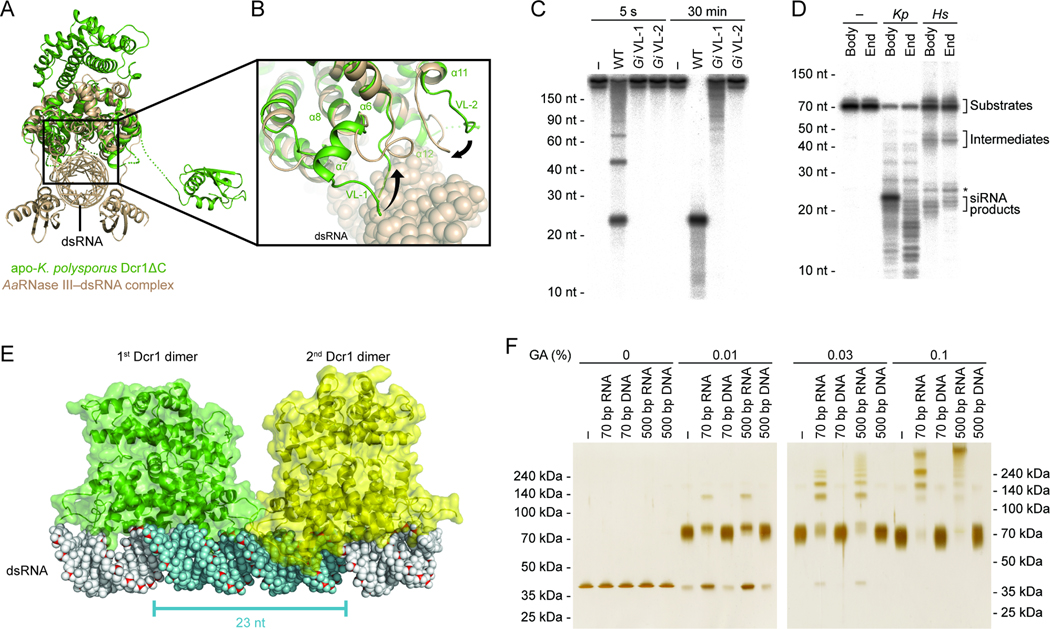
Figure 4. Binding and Cleavage of a dsRNA Duplex.
(A) Structural alignment of apo-Dcr1ΔC (green) and _Aa_RNase III–dsRNA complex (tan) based on their RNase III domains.
(B) Close-up view of the VL-1 and VL-2 loops. The dsRNA is represented with a sphere model. The numbering of secondary structure elements is as in Figure 3A.
(C) Dicing activity of variable-loop mutants under standard single-turnover conditions. Body-labeled 500 bp dsRNA was incubated for 5 s or 30 min with buffer only (−), wild-type Dcr1ΔC (WT), or Dcr1ΔC variants with the indicated _Gi_Dicer loop substitutions.
(D) Cleavage reactions following body- or end-labeled 70 bp dsRNA substrates. Substrates were incubated with buffer only (−), K. polysporus Dcr1ΔC (Kp; standard single-turnover conditions), or H. sapiens Dicer (Hs). _Hs_Dicer generates ~21–22 nt siRNA products and ~40–50 nt intermediates as indicated; a ~25 nt product of unknown origin is also observed (*).
(E) A pair of Dcr1ΔC dimers (green and yellow) modeled with dsRNA (sphere model). The anticipated 23 nt siRNA product is highlighted (cyan). The dsRBD is not shown for clarity.
(F) Protein cross-linking analyses of Dcr1ΔC oligomerization. E224Q Dcr1ΔC was incubated with buffer only (−) or the indicated nucleic acid before crosslinking with the indicated glutaraldehyde (GA) concentration. Reactions were analyzed by SDS-PAGE and visualized by silver staining. Also shown is the migration of protein standards with the indicated molecular weights.
See also Figure S4.
The structural alignment with _Aa_RNase III–dsRNA revealed that neither the NTD nor dsRBD of Dcr1ΔC are positioned to measure the length from the end of a dsRNA molecule to the active sites (Figure 4A). Therefore, we considered the possibility that unlike canonical Dicers, which sequentially remove siRNAs from termini of dsRNA, budding-yeast Dicers might generate 23 nt products from internal regions of dsRNA. To test this possibility, we compared the products of K. polysporus and canonical Dicers when acting on body- and end-labeled dsRNA substrates. Dcr1ΔC processing of body-labeled dsRNA yielded a predominant 23 nt radiolabeled product, whereas processing of end-labeled dsRNA yielded mostly shorter, variable-length radiolabeled fragments, without enrichment of 23 nt product (Figure 4D). In contrast, human Dicer generated radiolabeled siRNA products from both body- and end-labeled dsRNA substrates. These results indicate that internal regions of dsRNA but not the termini are incorporated into Dcr1-generated siRNAs.
The short lengths of end-labeled products generated by Dcr1ΔC suggested that they were terminal cut-off products. The length heterogeneity of Dcr1ΔC cut-off products (Figure 4D) suggested that the initiation of dsRNA processing from within the dsRNA molecule occurs at multiple potential sites (i.e., without a dominant register). Consistent with this interpretation, products deriving from a perfectly paired duplex were only weakly phased (Figure S4B). The stronger phasing observed for endogenous siRNAs (Drinnenberg et al., 2009) can be explained by loops and bulges present in many natural Dcr1 substrates, which could bias processing toward specific registers.
To understand how Dcr1ΔC might generate 23 nt products from internal regions of dsRNA, we manually docked two Dcr1ΔC dimers at sites 23 nt apart on a dsRNA substrate. The dimers aligned near to each other but without steric hindrance in a configuration that would generate 23 nt siRNA products with 2 nt 3' overhangs (Figure 4E). This docking model for Dcr1ΔC is remarkably similar to the crystal packing observed in the structure of _Aa_RNase III bound to a 22 nt dsRNA, in which RNase III dimers are adjacently positioned along a pseudo-continuous dsRNA generated by end-to-end stacking of short RNA helices (Figure S4C). Based on these structural observations, we hypothesized that Dcr1 dimers bind closely together along dsRNA and that the distance between adjacent active sites determines product length.
To test the first part of this hypothesis, we used glutaraldehyde crosslinking to determine whether the binding to dsRNA brings dimers sufficiently near to each other to be crosslinked into higher-order oligomers. For these experiments, we utilized a Dcr1ΔC mutant that contained an RNase III active-site substitution (E224Q) that prevents cleavage (Figure 3D) but does not affect substrate binding of E. coli RNase III (Sun and Nicholson, 2001). Although base-paired RNA does not react with glutaraldehyde (Hopwood, 1975), control experiments with dsDNA instead of dsRNA were used to identify any effects of double-stranded nucleic acid on crosslinking that were independent of Dicer binding. Inclusion of dsRNA shifted the distribution of species to molecular weights higher than those seen with no nucleic acid or dsDNA (Figure 4F). In lower glutaraldehyde (0.01% and 0.03%) the presence of dsRNA reduced intradimer crosslinking, as indicated by uncrosslinked monomer, but increased interdimer crosslinking, as indicated by higher molecular-weight bands. In 0.1% glutaraldehyde the addition of dsRNA caused nearly all of the protein to be crosslinked into higher-order oligomers. Larger species were observed when a 500 bp dsRNA was used compared to when a 70 bp dsRNA was used, consistent with the ability of the longer dsRNA to accommodate more dimers. Experiments following the fate of the dsRNA confirmed that the dsRNA-dependent crosslinked species represented protein–protein crosslinks rather than protein–RNA or RNA–RNA crosslinks (Figures S4D and S4E). Together, these results showed that Dcr1ΔC dimers can bind closely together along dsRNA.
Cleavage by Active Sites in Adjacent Dcr1 Dimers Generates 23 nt Products
To test the second part of our hypothesis, that the distance between adjacent active sites determines product length, we changed the distance between active sites to see if this caused a corresponding change in product length. To increase the distance between functional active sites, Dcr1ΔC with an active-site mutation (E224Q) was added to reactions containing wild-type Dcr1ΔC. If multiple Dcr1 dimers bind side-by-side along dsRNA, then binding of one or more E224Q dimers between two active dimers would increase the distances between active sites by multiples of 23 nt, with corresponding increases in product length (Figure 5A, top). Indeed, increasing E224Q Dcr1ΔC concentration in the presence of constant wild-type Dcr1ΔC produced progressively longer ladders with fragment lengths increasing in the predicted 23 nt increments (Figure 5A, bottom). A 46 nt fragment was also observed in the first few seconds of processing by wild-type Dcr1 (Figure 1B), which is explained by our model as a short-lived intermediate that is a consequence of cleavage occurring in the context of multiple Dcr1 active sites spaced along the dsRNA at 23 nt intervals. Likewise, larger intermediates with lengths in multiples of 23 nt were observed in reactions containing Dcr1ΔC active-site mutants (Figure 3D and Figure S3), as expected for rare cleavage events by closely packed dimers aligned on the dsRNA substrate.
Figure 5. The Relationship Between Active-Site Spacing and Product Length.
(A) Effects of changing active-site spacing. Top: The schematic depicts wild-type (green) and active-site mutant (red) Dcr1ΔC dimers assembled on dsRNA to generate different products with lengths varying by 23 nt increments. Bottom: Body-labeled 500 bp dsRNA present at 140 pM was incubated without (−) or with (+) 30 nM wild-type Dcr1ΔC in the absence (−) or presence of increasing concentrations of E224Q Dcr1ΔC (3-fold dilution series from 0.33 nM to 243 nM). Products were resolved by denaturing 10% PAGE.
(B) Length of dicing intermediates. Top: The schematic shows the predicted length variation of intermediates (cyan) for substrates of increasing length. Trapezoids represent dimers of RNase III domains. Bottom: Body-labeled dsRNA substrates of the indicated length (present at ~45 pg/µl) were incubated with buffer only (−), a mixture of 30 nM wild-type Dcr1ΔC and 3 nM E224Q Dcr1ΔC (_Kp_Dcr1), or H. sapiens Dicer (_Hs_Dicer).
(C) Substrate length requirements for preferentially generating 23 nt products. Top: Docking models depict a pair of dimers bound to the indicated substrates, either anchoring the first dimer at the dsRNA terminus (left) or centering the dimer pair (right). Bottom: Body-labeled dsRNA substrates of the indicated length were incubated for 2 min either without (−) or with (+) Dcr1ΔC under standard single-turnover conditions.
(D) Dicer activities on open and closed substrates. Top: Schematic of substrates, which contained 70 bp of dsRNA flanked by either short ssRNA overhangs or loops. Bottom: Body-labeled substrates were incubated without (−) or with (+) Dcr1ΔC (_Kp_Dcr1) or D. melanogaster Dcr-2 (_Dm_Dcr-2) under multiple-turnover conditions (30 nM substrate and 10 nM protein). Size markers were estimated based on RNA standards in Figure S5C.
(E) Dicer activities on substrates resembling endogenous yeast substrates. Top: Schematic of substrates, which contained 161 bp of dsRNA within either a perfect duplex (substrate D), a palindromic RNA (substrate E), or an internal duplex (substrate F; red, 7-methylguanosine cap; green, poly(A) tail). Bottom: Body-labeled substrates were reacted with Dicer enzymes as in (D). Percent product was normalized to account for radiolabeled phosphodiester linkages occurring outside of the dsRNA region of substrate E.
See also Figure S5.
Discrete processing intermediates separated by a constant interval equivalent to the dominant siRNA product length have been observed with canonical Dicers (Zhang et al., 2002; Macrae et al., 2006). These intermediates result from sequential removal of products from the ends, and thus their lengths are dependent on substrate length (Figure 5B, top right), whereas in our model for Dcr1 the lengths of intermediates should be independent of substrate length (Figure 5B, top left). Using 65, 70, and 75 bp substrates, a mixture of wild-type and E224Q Dcr1ΔC generated a 46 nt product irrespective of substrate length (Figure 5B, bottom). In contrast, human Dicer generated intermediates with lengths that varied predictably with substrate length, consistent with sequential removal of siRNA products from the ends. Thus, budding-yeast Dicer intermediates are fundamentally different from canonical Dicer intermediates.
According to our model for siRNA generation by budding-yeast Dicer, a substrate must be able to accommodate at least two dimers to preferentially generate a 23 nt product. Consequently, the shortest dsRNA substrate that could yield siRNA products should have a length corresponding to the length of dsRNA required for efficient binding of a pair of adjacent dimers. Structural modeling of Dcr1ΔC with dsRNA indicates that a pair of dimers spans 35–40 bp, excluding the outer VL-1 and VL-2 (Figure 5C). Indeed, reactions with substrates of at least 40 bp generated a predominant 23 nt product, whereas those with shorter substrates yielded little or no enrichment for 23 nt products and instead produced a collection of ≤23 nt products, which presumably derived from binding and cleavage by a single dimer (Figure 5C). Even 23 nt siRNA-like duplexes could be processed by Dcr1 into shorter fragments, albeit inefficiently (Figure S1G). The absence of >23 nt products in reactions with 30 bp substrates indicated that cleavage by a single dimer was more efficient when at least 7 bp flanked the cleavage site. The finding that the minimum substrate length for efficient siRNA production corresponded to the footprint of a pair of dimers further supported our model for the generation of 23 nt products.
Our model for budding-yeast Dicer predicts that even a dsRNA substrate that lacks termini can be processed into 23 nt siRNA products. In contrast, canonical Dicers cannot efficiently process substrates that lack free helical ends because of their reliance on the terminus-binding PAZ domain for measurement (Zhang et al., 2002; MacRae et al., 2007). To test these predictions, we compared the abilities of K. polysporus Dcr1ΔC and Drosophila melanogaster Dcr-2 (_Dm_Dcr-2)—a canonical Dicer specifically involved in siRNA biogenesis (Cenik et al., 2011)—to process dsRNA substrates with either two, one, or zero free termini (Figure 5D, top; substrates A, B, and C, respectively). Dcr1ΔC processed all substrates into siRNA products with similar efficiency, whereas _Dm_Dcr-2 efficiently processed only the substrates with at least one free end (Figure 5D), with the residual siRNA generation observed for the substrate without a free end at least partially attributed to contamination by a substrate with free ends (Figure S5B). _Dm_Dcr-2 was also more sensitive to the geometry of the substrate termini; addition of 3 nt overhangs to each end of a 70 bp substrate reduced processing by _Dm_Dcr-2 by ~4 fold with only minimal effect on processing by Dcr1ΔC (Figure S5C).
The terminus-independent processing by budding-yeast Dicer has implications for the generation of siRNAs in vivo because endogenous substrates of Dcr1 lack the dsRNA termini preferred by canonical Dicer and instead have long segments of single-stranded RNA extending from the duplex (Drinnenberg et al., 2009). To evaluate whether budding-yeast Dicer is uniquely suited to process these classes of substrates, we compared the abilities of Dcr1ΔC and _Dm_Dcr-2 to process three substrates: a perfect duplex; a palindromic RNA cloned from S. castellii, which forms a hairpin with 5' and 3' terminal extensions; and a sense-antisense pair comprised of a capped and polyadenylated RNA paired within the body of a longer transcript (Figure 5E; substrates D, E, and F, respectively). Dcr1ΔC efficiently processed all three substrates, whereas _Dm_Dcr-2 efficiently processed only the simple duplex (Figure 5E, bottom), suggesting that the termini of many endogenous Dcr1 substrates are not recognized by the PAZ domain. Although cofactors or pre-processing by nucleases might allow such non-ideal substrates to become processed by canonical Dicers, thereby explaining the phased siRNAs generated from hairpin precursors by _Dm_Dcr-2 in vivo (Czech et al., 2008; Okamura et al., 2008), these intrinsic differences in substrate specificity highlight the distinct mechanisms of budding-yeast and canonical Dicers.
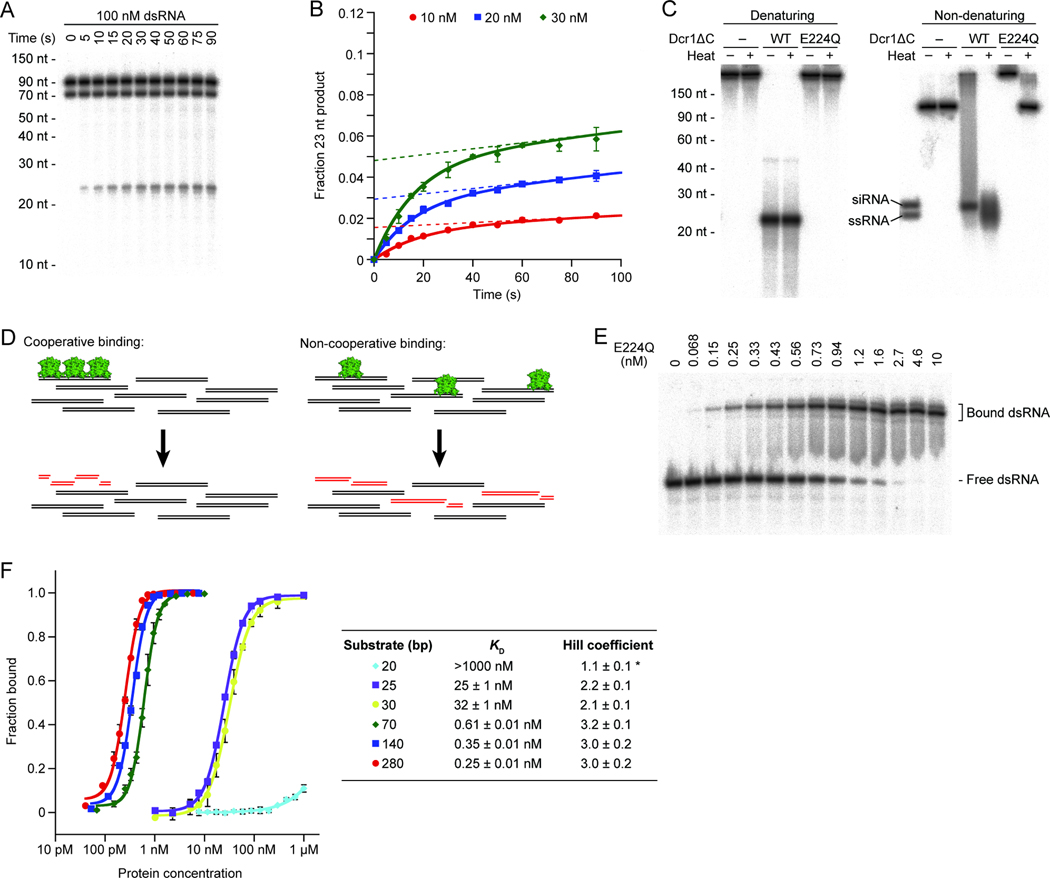
Slow Product Release and Cooperative Binding Contribute to siRNA Generation
Our model relies on a second dimer binding at the proper position before the first dimer has cleaved and released its products. This could be accomplished through cooperative binding along the length of the substrate and might be further favored by slow product release. To determine whether product release was slow relative to substrate binding and/or catalysis, we analyzed multiple-turnover kinetics. Two phases were observed, which corresponded to a pre-steady-state burst followed by slower steady-state turnover, with the amplitude of the burst increasing proportionately with the enzyme concentration (Figures 6A and 6B and Figure S6A). The simplest explanation for this behavior is that product release became rate limiting after the first round of cleavage.
Figure 6. Mechanisms for Preventing Off-Pathway Substrate Cleavage.
(A) Product accumulation under multiple-turnover conditions. Body-labeled 70 bp dsRNA present at 100 nM (implying a ~300 nM concentration of non-overlapping 23 bp binding sites) was incubated with 30 nM Dcr1ΔC for the indicated time.
(B) Quantitative analysis of multiple-turnover kinetics. Body-labeled 70 bp dsRNA present at 100 nM was incubated with the indicated concentration of Dcr1ΔC for the indicated time. Plotted are average values (n = 3; error bars, standard deviation). Solid lines, least-squares fit to the burst equation. Dotted lines, extrapolation of the steady-state rate to the y-axis.
(C) siRNA products bound to Dcr1ΔC. Body-labeled 500 bp dsRNA was incubated with buffer only (−), wild-type Dcr1ΔC (WT), or an active-site mutant of Dcr1ΔC (E224Q) under standard single-turnover conditions. Reactions were placed on ice (−) or heat-denatured at 90°C for 2 min (+) before fractionation. Products were resolved by denaturing 15% PAGE (left) or native 6% PAGE (right). siRNA, duplex of 23 nt siRNAs; ssRNA, 23 nt ssRNA.
(D) Schematic illustrating that for an enzyme that does not recognize the end of a duplex, siRNA generation under multiple-turnover conditions implies binding cooperativity.
(E) Representative gel-shift analysis of E224Q Dcr1ΔC binding to 70 bp dsRNA. Trace amounts of body-labeled 70 bp dsRNA were incubated with increasing amounts of protein.
(F) Binding isotherms for E224Q Dcr1ΔC. Data points represent average values (n = 3 for all substrates except 30 bp, for which n = 2; error bars, standard deviation). Solid lines show the best fit to the Hill equation, which produced the parameters shown to the right. For the 20 bp substrate, the maximum fraction bound was assumed to be 1.0 in order to obtain an estimate of the Hill coefficient (*).
See also Figure S6.
To visualize the protein-bound state of siRNA products, we analyzed single-turnover reactions on a non-denaturing gel. In reactions with wild-type Dcr1ΔC that had proceeded to completion, as shown on a denaturing gel (Figure 6C, left), RNA-containing species migrated as a smear on a non-denaturing gel (Figure 6C, right). After thermal denaturation, the smear resolved into unbound 23 nt siRNA and ssRNA, suggesting that it represented protein-bound RNA. Thus, a large fraction of siRNA products remained associated with Dcr1ΔC after cleavage occurred.
Together, our results indicate that slow product release contributes to siRNA generation by enabling additional dimers to bind adjacent to previously bound dimers before the previously bound dimers release their products. Slow product release might also protect the siRNAs from further nucleolytic cleavage, and it might enable Dcr1 to escort siRNAs to Argonaute and facilitate loading, as has been reported for canonical Dicers (Lee et al., 2004; Wang et al., 2009a).
The ability of sub-stoichiometric amounts of Dicer to preferentially generate 23 nt products in the presence of excess dsRNA-binding sites (Figure 6A) implied that binding of Dcr1ΔC to substrates was cooperative; otherwise, Dcr1ΔC dimers would bind randomly throughout the bodies of dsRNA molecules, only rarely positioning themselves precisely 23 nt apart to generate the proper product (Figure 6D). To examine binding of Dcr1 to dsRNA substrates, we performed gel-shift assays with E224Q Dcr1ΔC. Using a 70 bp dsRNA substrate, which can accommodate up to three dimers, increasing amounts of Dicer led to appearance of a single predominant gel-shifted band (Figure 6E). Although mobility in non-denaturing gels can be difficult to interpret, the observed pattern was consistent with the all-or-none behavior expected for strong cooperativity.
Binding curves for a series of dsRNA substrates showed that longer substrates had higher affinities (Figure 6F and Figures S6B and S6C). The affinities for 70, 140, and 280 bp substrates were similar but still correlated with substrate length, an observation consistent with Dicer binding to internal regions of dsRNA, as longer substrates contain more potential binding sites. Notably, the apparent _K_D for the 30 bp dsRNA—which could fully accommodate only a single dimer and did not preferentially generate 23 nt siRNA products—was ~50-fold weaker than that of the 70 bp dsRNA. The 30 and 25 bp substrates exhibited similar Dicer-binding behavior, but a 20 bp substrate failed to form significant amounts of stable complex detectable by gel shift, even when Dicer was present at concentrations as high as 1 µM (Figures 6F and S6). The striking sigmoidicity of the binding isotherms for all substrates ≥25 bp provided further evidence for cooperative binding, with Hill coefficients for substrates ≥70 bp reaching a plateau of ~3 (Figure 6F). Our experiments thus identified three types of dsRNA substrates with respect to binding affinity: long substrates (≥70 bp), which could fully accommodate at least 3 dimers and displayed pM affinities and Hill coefficients of ~3; shorter substrates (25–30 bp), which formed sufficient interactions with a pair of dimers to exhibit nM affinities and Hill coefficients of ~2; and a 20 bp substrate, which presumably bound only a single dimer with µM affinity and a Hill coefficient of ~1. Increasing monovalent ion concentrations from 41 mM to 150 mM reduced the binding affinity for the 140 bp dsRNA by ~5 fold and reduced the Hill coefficient to 1.6 (Figure S6D). However, Dcr1ΔC still behaved cooperatively under this more physiological salt concentration, as evidenced by both a Hill coefficient that exceeded unity and the preferential generation of 23 nt products under multiple-turnover conditions (Figure S6E). We conclude that binding cooperativity contributes to siRNA generation by precisely positioning dimers 23 nt apart even at limiting Dicer concentrations (Figure 6A), and this, supplemented by slow product release, helps prevent isolated dimers from enacting non-productive cleavage.
DISCUSSION
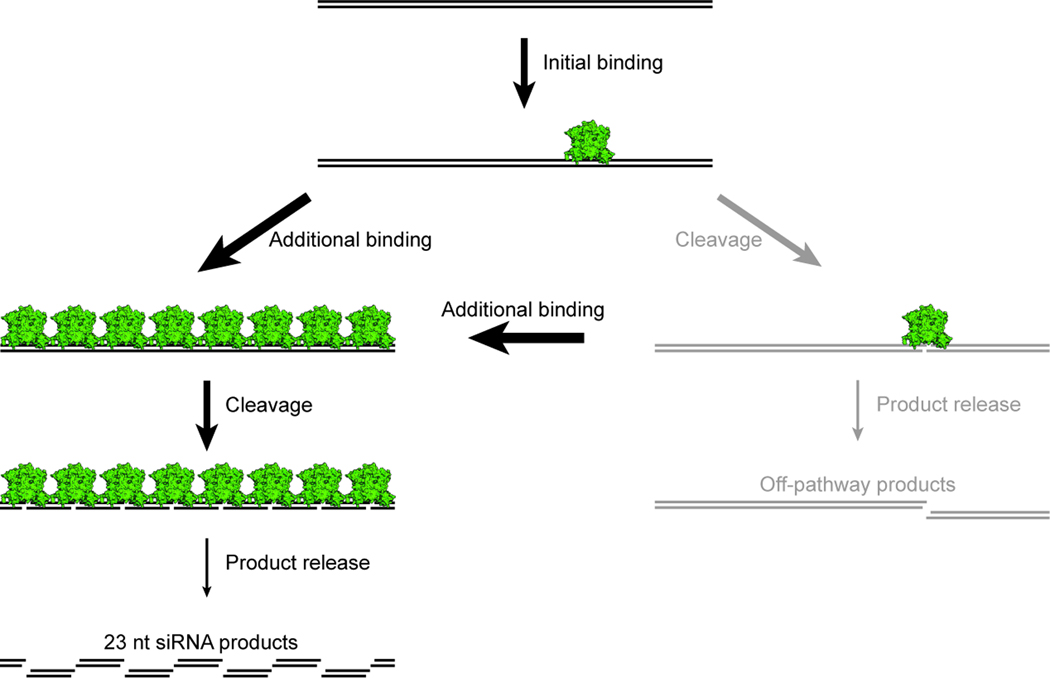
Our results show that budding-yeast Dicers produce siRNAs through a mechanism different from that of canonical Dicers. Instead of successively removing siRNA duplexes from the dsRNA termini, Dcr1 starts in the interior and works outward (Figure 7). This inside-out mechanism initiates with a dimer binding at an arbitrary position within the dsRNA, followed by the recruitment of additional dimers to adjacent sites. As binding propagates in both directions along the dsRNA, slow product release prevents cleavage events from disrupting maintenance of the phase. Cleavage by a collection of aligned dimers precisely generates 23 nt siRNA products paired to each other with 2 nt 3' overhangs.
Figure 7. Inside-Out Mechanism of Budding-Yeast Dicers.
See text for description. Off-pathway cleavage products (gray) are disfavored by both cooperative binding and slow product release.
See also Figure S7.
The mechanism for Dcr1-catalyzed siRNA production represents a natural example of a molecular ruler that is defined by the spacing of adjacent active sites. The concept of such a molecular ruler has been proposed but then rejected for other enzymes. The multimeric proteasome was hypothesized to generate short peptide products with a length determined by the distance between active sites (Wenzel et al., 1994), but subsequent experiments ruled out this model (Nussbaum et al., 1998). Ironically, models for product length determination based on active-site spacing were also proposed for both bacterial RNase III and canonical Dicers (Blaszczyk et al., 2001; Carmell and Hannon, 2004), but further study of these enzymes proved these models to be incorrect (Zhang et al., 2004). Nonetheless, under certain conditions E. coli RNase III can process long dsRNA into ~23 nt products in vitro by using a mechanism that might resemble the inside-out mechanism described here (Xiao et al., 2009). Products of this in vitro reaction act as potent siRNAs for mammalian gene knock-down (Yang et al., 2002; Xiao et al., 2009), as do siRNAs generated by budding-yeast Dicer (Figure S7).
In canonical siRNA-generating Dicers, the helicase domain uses ATP to facilitate complete processing of a duplex into siRNAs before beginning on the next duplex (Cenik et al., 2011; Welker et al., 2011). In budding-yeast Dicers, cooperativity could facilitate complete processing without requiring such a domain. dsRBD1 and VL-1/2 are candidates for forming cooperative interactions between adjacent Dcr1 dimers bound to dsRNA. Given their roles in dsRNA binding (Figure S1D), dissecting their potential contributions to cooperativity awaits a high-resolution view of the Dcr1–dsRNA complex, which would reveal dimer–dimer interactions that might be abolished without perturbing dimer–dsRNA interactions. In addition to mediating cooperativity, dimer–dimer interactions might allosterically activate adjacent dimers for cleavage, which would further favor productive cleavage.
In the current RNase III enzyme classification, which is based on domain architecture, class I includes both bacterial RNase III and yeast Rnt1 (MacRae and Doudna, 2007). We found that despite having similar domain architectures, bacterial RNase III and yeast Rnt1/Dcr1 use distinct active-site arrangements comprising four and six residues, respectively (Figures 3A and 3C). Adding this feature to the existing classification criteria would divide RNase III enzymes into four classes more parsimonious with their evolutionary relationships: bacterial RNase III, class I; Drosha, class II; canonical Dicer, class IIIa; and yeast RNase III, including both Rnt1 and Dcr1, class IIIb (Figure 3A). Despite its closer evolutionary relationship to Dcr1, Rnt1 behaves as a molecular ruler in a manner more analogous to canonical Dicer. Just as the canonical Dicer PAZ domain binds to the 2 nt 3' overhang of its substrate to position the RNase III active sites at a defined distance, the Rnt1 dsRBD recognizes the AGNN tetraloop of its substrate to position its active sites for precise cleavage (MacRae and Doudna, 2007). Thus, the terminus-independent measuring mechanism of Dcr1 departs from the principles operating in other class III RNase III enzymes.
The distinct mechanisms employed by canonical and budding-yeast Dicers to generate similarly sized siRNAs provide a striking example of convergent functional evolution. Although both mechanisms produce siRNAs, the canonical mechanism is more suitable for producing small RNAs that must be processed in a defined register, such as microRNAs and trans-acting siRNAs (Vaucheret, 2005). In contrast, the inside-out mechanism is more suitable for substrates that lack free helical ends, such as covalently closed molecules (e.g., viroids), dsRNA intermediates of rolling-circle replication, dsRNA with protected termini (e.g., viral ribonucleoproteins), and dsRNA with long single-stranded extensions, including endogenous Dcr1 substrates (Figure S5D). Thus, in a budding-yeast ancestor, the presence of dsRNA species resistant to processing by the canonical Dicer might have favored the evolution of an additional RNase III enzyme able to pre-process these substrates by cutting in their interior, thereby producing suitable substrates for canonical Dicer. After this enzyme acquired features that enabled it to produce siRNAs on its own, the absence of a phased small-RNA pathway might have allowed loss of the canonical Dicer without deleterious effects, thereby explaining the replacement of the canonical Dicer in the budding-yeast clade.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Detailed methods for expression and purification of His-Sumo-tagged proteins from bacterial plasmids (Table S4) are described in Extended Experimental Procedures.
Structure Determination and Refinement
Native and SeMet-substituted crystals of Dcr1ΔC were obtained by sitting-drop vapour diffusion at 20°C, and data sets were collected at Brookhaven NSLS beamline X29. Phasing was achieved using single-wavelength-anomalous-diffraction (SAD) with selenium anomalous signals. Native crystals of E224Q Dcr1Δ2d were grown under related conditions, data was collected at Argonne NE-CAT beamline 24-IDE, and the structure was solved by molecular replacement. For details on crystallization, structure calculation, and modeling, see Extended Experimental Procedures.
Dicer Activity Assays
Processing reactions using yeast whole-cell extracts were essentially as described (Drinnenberg et al., 2009). Reactions using recombinant Dcr1 variants were in 30 mM Tris-HCl (pH 7.5), 30 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 0.1 mM EDTA. Unless indicated otherwise, these reactions were for 90 s with 30 nM purified protein and ~45 pg/µl RNA, which corresponded to ~1 nM for 70 bp dsRNA (standard single-turnover conditions). Reactions using recombinant H. sapiens Dicer (Genlantis) and D. melanogaster Dcr-2 were in 30 mM Tris-HCl (pH 6.8), 25 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, and 1% glycerol, supplementing the Dcr-2 reactions with 1 mM ATP. Reactions were quenched by addition to ≥1 volume of Formamide Loading Buffer (90% formamide, 18 mM EDTA, 0.025% sodium dodecyl sulfate, 0.1% xylene cyanol, 0.1% bromophenol blue). RNA products were resolved by denaturing PAGE and visualized by phosphorimaging.
Dicer Binding Assays
Binding reactions were in 30 mM Tris-HCl (pH 7.5), 30 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, and 5% glycerol. Reactions were incubated at room temperature (22–24°C) for 10 min and then on ice for at least 20 min. Reactions were analyzed on native polyacrylamide gels run at 4°C, and RNA was visualized by phosphorimaging.
Glutaraldehyde Crosslinking
Crosslinking was in 30 mM HEPES-NaOH (pH 7.6), 30 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 300 nM protein, and indicated concentrations of nucleic acid and glutaraldehyde. After 10 min at room temperature, reactions were quenched by addition of an equal volume of 2X Laemmli Sample Buffer supplemented with 5% β-ME and 100 mM Tris-HCl (pH 8.0). Products were resolved by SDS-PAGE and visualized by silver staining and phosphorimaging.
Yeast Manipulations
S. castellii and K. polysporus culture, transformations, RNA isolation, and RNA blots were essentially as described (Drinnenberg et al., 2009) using strains and plasmids listed (Tables S3 and S4, respectively).
Small-RNA Sequencing
Single-turnover reactions were performed separately using dsRNA corresponding to fragments of mRNAs for Renilla luciferase and green fluorescent protein. After quenching, reactions were pooled, and total RNA was isolated by phenol extraction. Small-RNA cDNA libraries were prepared and sequenced (Drinnenberg et al., 2009). For a detailed description of data analysis, see Extended Experimental Procedures.
Supplementary Material
01
02
ACKNOWLEDGMENTS
We thank I.A. Drinnenberg, G. Fink, R. Fukunaga, C. Jan, R. Sauer, and D. Shechner for helpful discussions, D. Koppstein for performing mammalian gene knock-down experiments, the Whitehead Genome Technology Core for high-throughput sequencing, P. Zamore for providing _Dm_Dcr-2 enzyme, F. Roth for providing pAG416Gal-Dicer plasmid, Q. Yin in the H. Wu laboratory for assistance with light scattering experiments, the X29 beamline at Brookhaven National Laboratory, and the NE-CAT beamline at the Advanced Photon Source, Argonne National Laboratory. This work was supported by NIH grants GM061835 (D.P.B.) and AI121493 (D.J.P.), an NSF graduate research fellowship (D.E.W.), a Human Frontier Science Program Long-term Fellowship (K.N.), and a fellowship from the Japan Society for the Promotion of Science for Research Abroad (K.N.). D.P.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Small-RNA sequencing data were deposited in the Gene Expression Omnibus (GSE29168). X-ray coordinates of Dcr1ΔC and E224Q Dcr1Δ2d were deposited in the Protein Data Bank (3RV0 and 3RV1, respectively).
REFERENCES
- Akey DL, Berger JM. Structure of the nuclease domain of ribonuclease III from M. tuberculosis at 2.1 A. Protein Sci. 2005;14:2744–2750. doi: 10.1110/ps.051665905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk J, Gan J, Tropea JE, Court DL, Waugh DS, Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure. 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, Court DL, Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Hall TM, Zamore PD. Phosphate and R2D2 Restrict the Substrate Specificity of Dicer-2, an ATP-Driven Ribonuclease. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Lee JK, Tjhen R, Stroud RM, James TL. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc Natl Acad Sci U S A. 2008;105:2391–2396. doi: 10.1073/pnas.0711506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol Microbiol. 2008;67:143–154. doi: 10.1111/j.1365-2958.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Hopwood D. The reactions of glutaraldehyde with nucleic acids. Histochem J. 1975;7:267–276. doi: 10.1007/BF01003595. [DOI] [PubMed] [Google Scholar]
- Lamontagne B, Tremblay A, Abou Elela S. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol Cell Biol. 2000;20:1104–1115. doi: 10.1128/mcb.20.4.1104-1115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Leulliot N, Quevillon-Cheruel S, Graille M, van Tilbeurgh H, Leeper TC, Godin KS, Edwards TE, Sigurdsson ST, Rozenkrants N, Nagel RJ, et al. A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. EMBO J. 2004;23:2468–2477. doi: 10.1038/sj.emboj.7600260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meng W, Nicholson AW. Heterodimer-based analysis of subunit and domain contributions to double-stranded RNA processing by Escherichia coli RNase III in vitro. Biochem J. 2008;410:39–48. doi: 10.1042/BJ20071047. [DOI] [PubMed] [Google Scholar]
- Nussbaum AK, Dick TP, Keilholz W, Schirle M, Stevanovic S, Dietz K, Heinemeyer W, Groll M, Wolf DH, Huber R, et al. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc Natl Acad Sci U S A. 1998;95:12504–12509. doi: 10.1073/pnas.95.21.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li G, Nicholson AW. Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry. 2004;43:13054–13062. doi: 10.1021/bi049258i. [DOI] [PubMed] [Google Scholar]
- Sun W, Nicholson AW. Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant. Biochemistry. 2001;40:5102–5110. doi: 10.1021/bi010022d. [DOI] [PubMed] [Google Scholar]
- Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007;374:106–120. doi: 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. MicroRNA-dependent trans-acting siRNA production. Sci STKE. 2005;2005:pe43. doi: 10.1126/stke.3002005pe43. [DOI] [PubMed] [Google Scholar]
- Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009a;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009b;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer's Helicase Domain Discriminates dsRNA Termini to Promote an Altered Reaction Mode. Mol Cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W. Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett. 1994;349:205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- Wu H, Henras A, Chanfreau G, Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc Natl Acad Sci U S A. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Feehery CE, Tzertzinis G, Maina CV. E. coli RNase III(E38A) generates discrete-sized products from long dsRNA. RNA. 2009;15:984–991. doi: 10.1261/rna.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02