Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes (original) (raw)
Not only do molecular chaperones assist protein folding, they also facilitate the degradation of misfolded polypeptides. In coordinating with cochaperones CHIP and BAG3, Hsp70 can also target misfolded proteins to aggresomes, thereby protecting cells from proteotoxic stress.
Abstract
Protein misfolding is a common event in living cells. Molecular chaperones not only assist protein folding; they also facilitate the degradation of misfolded polypeptides. When the intracellular degradative capacity is exceeded, juxtanuclear aggresomes are formed to sequester misfolded proteins. Despite the well-established role of chaperones in both protein folding and degradation, how chaperones regulate the aggregation process remains controversial. Here we investigate the molecular mechanisms underlying aggresome formation in mammalian cells. Analysis of the chaperone requirements for the fate of misfolded proteins reveals an unexpected role of heat shock protein 70 (Hsp70) in promoting aggresome formation. This proaggregation function of Hsp70 relies on the interaction with the cochaperone ubiquitin ligase carboxyl terminal of Hsp70/Hsp90 interacting protein (CHIP). Disrupting Hsp70–CHIP interaction prevents the aggresome formation, whereas a dominant-negative CHIP mutant sensitizes the aggregation of misfolded protein. This accelerated aggresome formation also relies on the stress-induced cochaperone Bcl2-associated athanogene 3. Our results indicate that a hierarchy of cochaperone interaction controls different aspects of the intracellular protein triage decision, extending the function of Hsp70 from folding and degradation to aggregation.
INTRODUCTION
In living cells, both newly made and preexisting polypeptide chains are at constant risk for misfolding and aggregation (Goldberg, 2003). Aggregation of misfolded proteins is characteristic of various human diseases referred to as protein conformation disorders (Kopito, 2000; Chiti and Dobson, 2006; Morimoto, 2008). It is becoming evident that protein aggregation in cells is not an uncontrolled dead-end process driven by simple interaction among inappropriately exposed hydrophobic surfaces. Instead, directing protein aggregates to specific cellular sites is considered to be a second line of active cellular defense (Tyedmers et al., 2010). The sequestration of misfolded proteins into specific compartments can protect the cellular environment from potentially deleterious protein species (Arrasate et al., 2004; Lansbury and Lashuel, 2006). In addition, it allows for their subsequent clearance by alternative protein quality control systems such as the autophagy-lysosome pathway (Rubinsztein, 2006). Thus characterization of the pathways controlling protein aggregation is critical for elucidating the pathophysiology of a plethora of clinical diseases associated with misfolded proteins, including cystic fibrosis and Parkinson's disease.
Molecular chaperones and the ubiquitin-proteasome degradation pathway represent two main systems that protect eukaryotic cells against the buildup of misfolded polypeptides. Molecular chaperones assist the folding of newly translated and stress-denatured proteins (Frydman, 2001; Bukau et al., 2006; Hartl and Hayer-Hartl, 2009). However, some misfolded or “abnormal” polypeptides cannot fold correctly under any circumstance. The identification of ubiquitin ligases and chaperone cofactors that physically link chaperones to the ubiquitin-proteasome system indicates direct communication between the folding and degradation machineries (McClellan et al., 2005). In mammalian cells, the cochaperone ubiquitin ligase carboxyl terminal of heat shock protein 70 (Hsp70)/Hsp90 interacting protein (CHIP) acts to shift the quality control equilibrium from folding to ubiquitination and degradation (Cyr et al., 2002; Murata et al., 2003). CHIP interacts with the molecular chaperones Hsp70 and Hsp90 through its NH2-terminal tetratricopeptide repeat (TPR) domain, whereas its COOH-terminal U-box domain contains the E3 ubiquitin ligase activity (Ballinger et al., 1999; Connell et al., 2001). Cumulative data indicate that the majority of CHIP substrates are bona fide Hsp70/Hsp90 clients. Therefore CHIP acts as a quality control ligase to facilitate the clearance of misfolded proteins that cannot be rescued by chaperones.
Misfolded proteins aggregate in cells when the generation of misfolded proteins exceeds the intracellular refolding and degradative capacity. Compelling evidence suggests that protein aggregation is a much more organized process than previously thought (Tyedmers et al., 2010). In mammalian cells, the inclusion body called the aggresome is mainly localized to an indentation of the nuclear envelope at the microtubule-organizing center (Kopito, 2000). Although ubiquitination of the substrates is generally considered to be a prerequisite for its recognition and transport to aggresomes, it could not be shown for all the substrates found in aggresomes (Kawaguchi et al., 2003). Of interest, the intracellular quality control machinery partitions misfolded proteins between two distinct compartments (Kaganovich et al., 2008). Although soluble ubiquitinated misfolded proteins accumulate in a juxtanuclear compartment (JUNQ), terminally aggregated proteins are sequestered in an insoluble perivacuolor inclusion (IPOD). The interrelationships of aggresomes, JUNQ, and IPOD in mammalian cells remain to be established. In addition, little is known about the molecular mechanisms underlying diverse substrate protein recognition and specific aggregate deposition.
Given the strong correlation between the protein aggregates and the onset of several neurodegenerative diseases, more studies have focused on the reversal of protein aggregation (Tyedmers et al., 2010). Overwhelming experimental evidence indicates that molecular chaperones are crucial modulators of protein aggregation (Muchowski and Wacker, 2005). In line with the “holding and folding” function of chaperones, the frequent colocalization of chaperones with protein aggregates is suggestive of their role in “disaggregating” preexisting protein aggregates (Sherman and Goldberg, 2001). However, overexpression of chaperones in a diverse range of conformational disease models has yielded mixed results. For example, cotransfection of CHIP and Hsp70 facilitates tau ubiquitination and suppresses toxicity in cell culture but paradoxically enhances the levels of insoluble tau (Shimura et al., 2004). In a Drosophila melanogaster model of Parkinson's disease, coexpression of human Hsp70 prevents α-synuclein–mediated toxicity but has no visible effects on the inclusion body phenotype (Auluck et al., 2002). Moreover, unbiased genetic screens of protein aggregation modulators revealed that some chaperone molecules act as aggregation enhancers instead of suppressors (Zhang et al., 2010). Of interest, a recent study reported that the Hsc70/Hsp70 cochaperone network is essential in the formation of aggresome-like structures in dendritic cells for antigen presentation (Kettern et al., 2011). In addition, another recent study reported that the stress-induced cochaperone Bcl2-associated athanogene 3 (BAG3) mediates chaperone-based aggresome targeting (Gamerdinger et al., 2011). These observations strongly suggest that Hsp70 might have distinct roles in controlling protein aggregation. It remains open to speculation whether molecular chaperones act to solubilize protein aggregates or promote aggresome formation.
Here we sought to distinguish these contradictory roles for molecular chaperones in the aggregation of misfolded proteins in mammalian cells. To this end, we studied the chaperone and cochaperone requirements for the aggregation of several well-characterized misfolded proteins. We established that the stress-inducible molecular chaperone Hsp70, together with the cochaperone ubiquitin ligase CHIP, not only promotes the degradation of misfolded proteins, but also facilitates the aggregation of misfolded proteins when their degradation is impaired. The chaperone-based aggresome targeting also requires the stress-induced cochaperone BAG3 to deposit the Hsp70 substrates into the aggresome. These findings provide novel insights into the logic underlying triage decisions in the metabolism of misfolded proteins.
RESULTS
Degradation of misfolded protein cytosolic bovine serum albumin is CHIP dependent
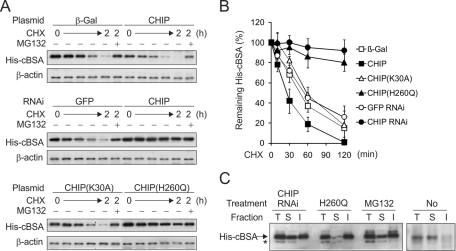
The cochaperone ubiquitin ligase CHIP facilitates the degradation of chaperone substrates including misfolded proteins (McDonough and Patterson, 2003). We initially determined the CHIP requirement for the degradation of a model misfolded protein—a cytosolic version of bovine serum albumin (cBSA; Qian et al., 2006a). cBSA could not fold correctly in the reducing cytoplasm environment and had t1/2 of ∼1 h (Figure 1, A, top, and B). Overexpression of CHIP further promoted the degradation of cBSA, resulting in t1/2 of <30 min (Figure 1, A and B). Remarkably, small interfering RNA (siRNA)–mediated CHIP knockdown largely stabilized cBSA and led to its accumulation in cells at levels similar to that after proteasome inhibition by MG132 (Figure 1, A, middle, and B). These results established the role of CHIP in the degradation of misfolded protein cBSA.
FIGURE 1:
Degradation of misfolded protein cBSA by CHIP. (A) HEK293 cells expressing His-cBSA were cotransfected with CHIP plasmids (top), siRNA targeting CHIP (middle), or CHIP mutants (bottom). Levels of cBSA were determined by immunoblotting after cycloheximide (CHX) chase up to 2 h. MG132 treatment was included at the last time point to serve as the control. (B) Quantitation of cBSA turnover by densitometry of immunoblotting results shown in A (mean ± SEM; n = 3). (C) Fractionation analysis of cBSA accumulated in cells with CHIP knockdown, CHIP(H260Q) overexpression, MG132 treatment, or no treatment. Whole-cell lysates were separated into 1% Triton X-100–soluble and –insoluble factions before immunoblotting. Arrow indicates the bands corresponding to cBSA, and asterisk indicates the nonspecific bands.
To further investigate the role of CHIP in the degradation of misfolded protein cBSA, we tested the effects of several CHIP mutants. CHIP(K30A) has a point mutation in the TPR domain and therefore is unable to bind chaperones (Jiang et al., 2003). Overexpression of CHIP(K30A) had a negligible effect on cBSA degradation (Figure 1A, bottom), suggesting that CHIP-mediated cBSA degradation relies on chaperone binding. CHIP(H260Q) has a point mutation in the U-box domain preventing its E3 ubiquitin ligase activity (Qian et al., 2006a). Indeed, an in vitro ubiquitination assay showed no ubiquitin conjugates in the presence of CHIP(H260Q) (Supplemental Figure S1). To our surprise, the intracellular degradation of cBSA was largely impaired in the presence of CHIP(H260Q) (Figure 1A, bottom). Notably, the steady-state level of cBSA was increased to a lesser extent than the turnover assay, suggesting a dose effect of CHIP(H260Q) during the course of plasmid transfection. This dominant-negative feature is likely due to the fact that CHIP(H260Q) maintains the intact chaperone-binding capacity that could potentially compete with the endogenous CHIP molecules in targeting misfolded proteins. Therefore either CHIP knockdown or CHIP(H260Q) overexpression stabilizes cytosolic misfolded proteins by preventing their ubiquitination.
Given the fact that ubiquitin conjugation may change the biochemical features of misfolded proteins, we next examined the detergent solubility of accumulated cBSA in cells with CHIP knockdown or CHIP(H260Q) overexpression or in the presence of MG132. Accumulated cBSA in cells with CHIP knockdown exhibited similar detergent solubility to that after proteasome inhibition, in which ∼50% of cBSA was detergent insoluble (Figure 1C). Of interest, the majority of accumulated cBSA in the presence of CHIP(H260Q) was detergent insoluble (Figure 1C). It was puzzling that a similar deficiency of ubiquitination led to different solubility of accumulated misfolded proteins in cells. It is likely that CHIP(H260Q) may interfere with other pathways that are unaffected by CHIP knockdown.
CHIP(H260Q) induces distinct aggregation of cBSA–green fluorescent protein
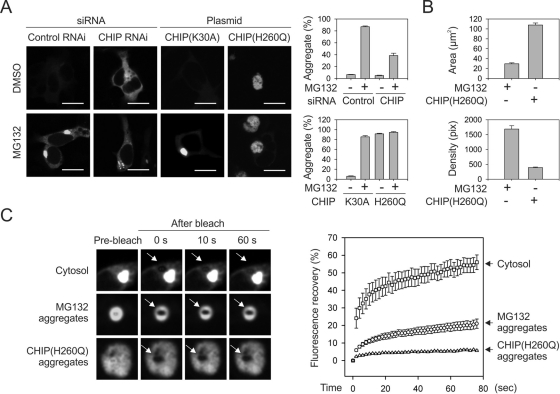
To determine the role of CHIP in the aggregation of misfolded proteins, we fused green fluorescent protein (GFP) to the COOH terminus of cBSA to facilitate evaluation by fluorescence microscopy. As expected, MG132 treatment not only increased the overall GFP fluorescence, but it also resulted in the formation of juxtanuclear aggresomes in HEK293 cells (Figure 2A, left). It is intriguing that cells with siRNA-mediated CHIP knockdown showed no obvious aggregate formation despite the stabilization of cBSA-GFP in these cells (Figure 2A, middle). Further MG132 treatment also failed to trigger the formation of typical aggresomes. It is likely that deficient ubiquitination in the absence of CHIP might prevent aggresome formation. However, reducing ubiquitin expression levels by siRNA-mediated knockdown did not prevent aggresome formation of cBSA-GFP in the presence of MG132 (Supplemental Figure S2). In any event, the stabilization of misfolded proteins in the absence of CHIP does not automatically lead to aggregate formation, further supporting the notion that protein aggregation is a regulated process in cells (Tyedmers et al., 2010).
FIGURE 2:
CHIP(H260Q) induces distinct protein aggregates of cBSA-GFP. (A) Fluorescence micrographs of HEK293 cells expressing cBSA-GFP after CHIP knockdown or overexpression of CHIP mutants. Twenty-four hours after transfection, cells were treated with 5 μM MG132 or dimethyl sulfoxide overnight. Bar, 10 μm. Aggregate formation in cells was determined as a single perinuclear GFP-positive inclusion and quantified using Nikon Elements imaging analysis (mean ± SEM; n = 4). (B) Quantitation of size and density of aggregates as shown in A using Nikon Elements imaging analysis (mean ± SEM; n = 30–36). (C) FRAP analysis of cBSA-GFP mobility in cytosol (top), MG132-induced aggregates (middle), or CHIP(H260Q)-induced inclusion body (bottom). After photobleaching, images were taken every 2 s up to 76 s. Arrow in the micrographs indicates the photobleach region. Percentage of fluorescence recovery was computed and plotted against the time (mean ± SEM; n = 6–11).
The dominant-negative U-box mutant CHIP(H260Q) also stabilizes the cBSA misfolded proteins in cells, as shown in Figure 1. Strikingly, overexpression of CHIP(H260Q) in HEK293 cells sensitized aggresome formation even in the absence of MG132 treatment (Figure 2A, right). Further MG132 treatment did not show any additive effects, suggesting that aggresome formation was maximal in the presence of CHIP(H260Q). Notably, the CHIP(H260Q)-induced aggregates were much larger but less dense than those induced by MG132. We measured both the averaged area and the pixel density of these aggregates and found that the cBSA-GFP aggregates in the presence of CHIP(H260Q) were about threefold larger but fourfold less dense (Figure 2B). To further characterize these distinct misfolded protein aggregates, we compared the dynamic properties of aggregates in the absence and presence of CHIP(H260Q) by applying fluorescence recovery after photobleaching (FRAP), a technique that measures the mobility of fluorescent molecules in living cells. Photobleaching a small section of cytosol had a rapid fluorescence recovery, whereas the recovery of MG132-induced aggregates was about threefold slower (Figure 2C). It is intriguing that the cBSA-GFP inclusion body in the presence of CHIP(H260Q) showed the slowest mobility, with <5% fluorescence recovery after photobleaching (Figure 2C, bottom). This suggests that CHIP(H260Q) induces distinct protein aggregates containing a large fraction of nondiffusing and insoluble substrates.
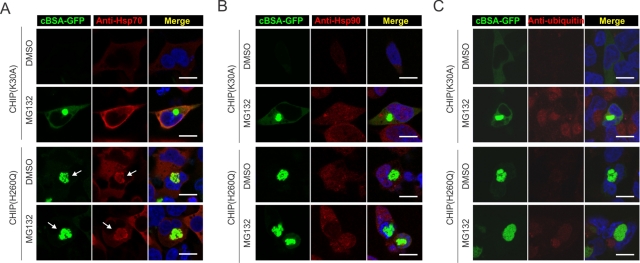
Because the CHIP(H260Q) mutant maintains intact chaperone-binding capacity, we then examined the colocalization of these cellular components with the aggregated cBSA-GFP via immune staining of endogenous chaperone molecules. As anticipated, MG132 treatment increased the expression levels of chaperone genes, including Hsp70 and Hsp90 (Figure 3, A and B, red channel). However, both chaperone molecules were largely excluded from the cBSA-GFP aggregates induced by proteasome inhibition. Cells expressing CHIP(H260Q) had a substantial induction of chaperone gene expression even in the absence of MG132, suggesting the existence of a potent stress response (Figure 3, A and B, and see later discussion). It is intriguing that a large fraction of Hsp70 molecules, but not Hsp90, were colocalized with the cBSA-GFP inclusion body in the presence of CHIP(H260Q) (Figure 3A, arrow). It is surprising to find that the colocalization of Hsp70 does not solubilize misfolded protein aggregates. It is likely that the dominant-negative CHIP(H260Q) mutant traps both the chaperone molecules and the associated misfolded protein substrates in the inclusion body because of deficient ubiquitination. Further supporting this notion, no positive ubiquitin signal was detected within the aggregates triggered by CHIP(H260Q) (Figure 3C). Therefore other E3s are unlikely involved in the aggregation process of misfolded proteins. To substantiate this point further, we knocked down another E3, Parkin, that collaborates with CHIP in the ubiquitination of substrates (Imai et al., 2002). Despite its positive role in the degradation of cBSA-GFP, reducing Parkin expression had little effects on the aggregation process (Supplemental Figure S3).
FIGURE 3:
Colocalization of cBSA-GFP aggregates with molecular chaperones. (A) HEK293 cells transfected with CHIP(K30A) or CHIP(H260Q) were fixed and stained with anti-Hsp70 antibody. Nuclei were stained with Hoechst. Arrow indicates the colocalization of cBSA-GFP with Hsp70. Bar, 10 μm. (B) HEK293 cells transfected with CHIP(K30A) or CHIP(H260Q) were fixed and stained with anti-Hsp90 antibody. Nuclei were counterstained with Hoechst. Bar, 10 μm. (C) HEK293 cells transfected with CHIP(K30A) or CHIP(H260Q) were fixed and stained with anti-ubiquitin antibody. Nuclei were counterstained with Hoechst. Bar, 10 μm.
CHIP(H260Q) elicits potent heat shock transcription factor activation without affecting 26S proteasome function
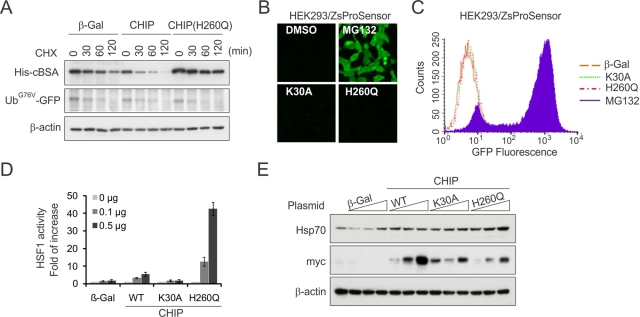
The different fate of misfolded proteins under either CHIP knockdown or in the presence of CHIP(H260Q) prompted us to investigate how the dominant-negative CHIP mutant sensitizes protein aggregation, whereas the CHIP knockdown prevents the aggresome formation. Because proteasome inhibition effectively triggers misfolded protein aggregation, we first asked whether the 26S proteasome function was impaired in cells expressing CHIP(H260Q). We examined the degradation of UbG76V-GFP, a proteasome substrate independent of chaperone function (Qian et al., 2002). Whereas the turnover of cBSA was significantly reduced in cells expressing CHIP(H260Q), the degradation of UbG76V-GFP was unchanged (Figure 4A). Thus CHIP(H260Q) has little effect on proteasome function in cells. To substantiate this finding further, we used a stable cell line expressing a 26S proteasome sensor, an ubiquitination-independent proteasome substrate fused with ZsGreen (Li et al., 1998; Qian et al., 2006b). In contrast to MG132 treatment, which caused a dramatic increase of ZsGreen fluorescence, both CHIP(H260Q) and CHIP(K30A) mutants had little effect on the fluorescence intensities as evidenced by both fluorescence microscopy and flow cytometry (Figure 4, B and C). Therefore CHIP(H260Q) does not interfere with 26S proteasome function in cells.
FIGURE 4:
CHIP(H260Q) elicits potent HSF1 activation without affecting the 26S proteasome function. (A) HEK293 cells were cotransfected with plasmids encoding CHIP or CHIP(H260Q) and substrates (His-cBSA or UbG76V-GFP) followed by CHX chase. Substrate levels were determined by immunoblotting. (B) A stable HEK293 cell clone expressing ZsProSensor were treated with MG132 or transfected with CHIP mutants as indicated. Live-cell images were taken using fluorescence microscope. (C) The mean fluorescence intensity of cells shown in B was determined by flow cytometry. (D) HEK293 cells were transfected with plasmids as indicated with 0, 0.1, and 0.5 μg/well in a six-well plate. HSF1 activity was measured by dual luciferase reporter assay using Hsp70-promoted firefly luciferase cotransfected with cytomegalovirus-promoted Renilla luciferase as an internal control (mean ± SEM; n = 3). (E) The levels of Hsp70 and myc-tagged CHIP in cells shown in D were determined by immunoblotting.
In our previous study, we demonstrated that wild-type CHIP stimulates the heat shock transcription factor (HSF1) transcriptional activity in a ubiquitination-independent manner (Dai et al., 2003). Consistently, the HSF1 activation was deficient in cells lacking CHIP (Qian et al., 2006a). To assess the effects of CHIP(H260Q) on HSF1 activity, we performed dual luciferase reporter assay in HEK293 cells. Remarkably, overexpression of CHIP(H260Q), but not CHIP(K30A), caused a dramatic increase of HSF1 activity in a dose-dependent manner (p < 0.01; Figure 4D). Consistent with the elevated HSF1 activity, there was an increase in the levels of Hsp70 in the presence of CHIP(H260Q) (Figure 4E). The potent HSF1 activation in the presence of CHIP(H260Q) further supports the idea that the U-box mutant of CHIP traps the chaperone molecules, which were occupied with misfolded proteins due to impaired ubiquitination.
CHIP(H260Q) relies on HSF1 in promoting misfolded protein aggregation
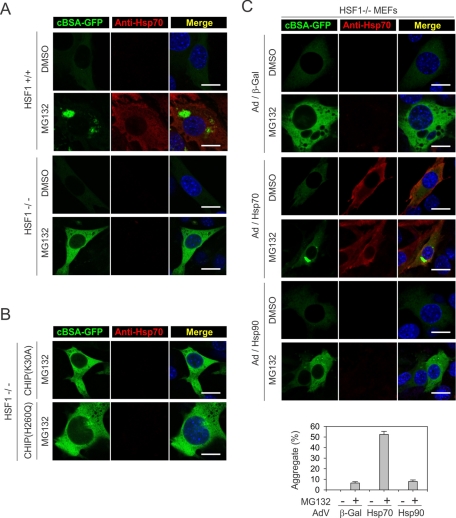
We next investigated whether it is the differential HSF1 activation that contributes to the distinct features of misfolded protein aggregation in the presence of CHIP(H260Q). To define the role of HSF1 in the aggregation of misfolded proteins, we used an immortalized fibroblast cell line derived from HSF1 −/− mice (McMillan et al., 1998). Both HSF1 +/+ and HSF1 −/− cells showed low basal levels of cBSA-GFP, suggesting that the degradation of cBSA-GFP is independent of HSF1 (Figure 5A). Proteasome inhibition by MG132 treatment led to stabilization of cBSA-GFP in both cell lines. However, only HSF1 +/+, and not HSF1 −/−, cells, showed juxtanuclear aggresome formation (Figure 5A). Furthermore, overexpressing CHIP(H260Q) in HSF1 −/− cells failed to induce cBSA-GFP aggregation even after MG132 treatment (Figure 5B). These results indicate that the dominant-negative CHIP mutant relies on HSF1 activation in promoting misfolded protein aggregation.
FIGURE 5:
CHIP(H260Q) relies on HSF1 in promoting misfolded protein aggregation. (A) HSF1+/+ and HSF1−/−− cells were treated with 5 μM MG132 overnight before immunostaining with anti-Hsp70 antibody. Nuclei were counterstained with Hoechst. Bar, 10 μm. (B) HSF1−/− cells were infected with recombinant adenovirus–expressing CHIP mutants, followed by MG132 treatment. Immunostaining was performed as in A. Bar, 10 μm. (C) HSF1−/− cells were infected with recombinant adenovirus–expressing molecular chaperones as indicated followed by MG132 treatment. Immunostaining was performed as in A. Bar, 10 μm. Aggregate formation in cells was quantified using Nikon Elements imaging analysis (mean ± SEM; n = 4).
Hsp70 is required for aggresome formation
HSF1 controls the expression of a large group of genes, including different classes of molecular chaperones (Hahn et al., 2004). Given the fact that CHIP associates with both Hsp70 and Hsp90, we focused on the roles of these chaperones in the process of protein aggregation. In contrast to HSF1 +/+ cells, which showed robust Hsp70 induction after proteasome inhibition, HSF1 −/− cells exhibited little Hsp70 expression in the presence of MG132 (Figure 5A). To specifically assess the role of Hsp70 in the fate of cBSA-GFP, we introduced Hsp70 into HSF1 −/− cells using recombinant adenovirus because of the low transfection efficiency of these mouse embryonic fibroblasts (MEFs). Remarkably, adding back Hsp70 alone robustly restored the aggregation of cBSA-GFP from <10% to >50% of the cells (Figure 5C, middle). In contrast, adenovirus-mediated Hsp90 expression had little effect on the fate of cBSA-GFP, as no aggregates were formed in these cells (Figure 5C and Supplemental Figure S4A).
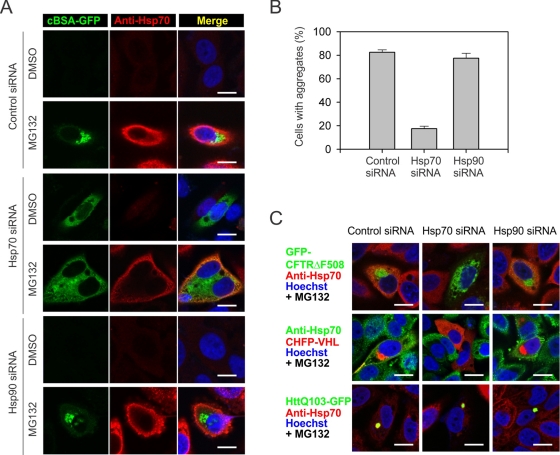
To further confirm the specific role of Hsp70 in the aggregation of misfolded proteins, we knocked down Hsp70 in HeLa cells using siRNA. The On-Target SmartPool was able to achieve more than 50% of Hsp70 knockdown (Supplemental Figure S4). Of interest, reducing Hsp70 expression alone stabilized the misfolded protein cBSA-GFP as reflected in the increased fluorescence (Figure 6, A and B). This result clearly indicates that Hsp70 contributes to the degradation of cBSA-GFP when the proteasome function is intact. However, the accumulated cBSA-GFP in cells with Hsp70 knockdown failed to form any apparent aggregates (Figure 6A). Further MG132 treatment also did not trigger the formation of juxtanuclear aggresomes. Notably, the knockdown of Hsp70 was not even complete after proteasome inhibition, presumably because of the stress-inducible feature of Hsp70 (Supplemental Figure S4B). Quantification of aggregates formation revealed that <20% of cells were able to form cBSA-GFP aggregates after partial Hsp70 knockdown, whereas ∼80% of cells showed aggregates in cells transfected with control siRNA (Figure 6B). Once again, Hsp90 knockdown had little effect on both the degradation and aggregation of cBSA-GFP, despite the high efficiency of knockdown (>90%; Supplemental Figure S4B). These results support the notion that Hsp70 not only facilitates the degradation of misfolded proteins but is also required to mediate the aggregation of misfolded proteins when their degradation is impaired.
FIGURE 6:
Hsp70 is required for misfolded protein aggregation. (A) HeLa cells were transfected with siRNA targeting molecular chaperones as indicated, followed by 5 μM MG132 treatment or overnight. Immunostaining was performed using anti-Hsp70 antibody. Nuclei were counterstained with Hoechst. Bar, 10 μm. (B) Aggregate formation in cells as in A was quantitated using Nikon Elements imaging analysis (mean ± SEM; n = 4). (C) HeLa cells expressing GFP-CFTRΔF508, CHFP-VHL, or HttQ103-GFP were transfected with siRNA targeting molecular chaperones as indicated. After 5 μM MG132 treatment overnight, cells were fixed and immunostained with anti-Hsp70 antibody. Nuclei were counterstained with Hoechst. Bar, 10 μm.
Hsp70 controls the aggregation of physiological substrates
Hsp70, among other molecular chaperones, has long been known to possess antiaggregation activity (Bukau et al., 2006; Kampinga and Craig, 2010). Therefore our finding that Hsp70 is indispensable for the aggregation of misfolded protein cBSA-GFP is quite surprising. Would Hsp70 also play a more generic role in the aggregation of other physiological substrates? One well-characterized CHIP substrate that readily forms aggresomes after proteasome inhibition is cystic fibrosis transmembrane conductance regulator (CFTR; Johnston et al., 1998; Meacham et al., 2001). Indeed, we observed a typical juxtanuclear aggresome of GFP-CFTRΔF508 in both HeLa and HEK293 cells after MG132 treatment (Figure 6C and Supplemental Figure S5). Consistent with the behavior of cBSA-GFP, knockdown of Hsp70, but not Hsp90, largely suppressed the aggresome formation of GFP-CFTRΔF508 (Figure 6C and Supplemental Figure S5). Thus Hsp70 plays a critical role in controlling the aggregation process of different misfolded proteins.
A recent study reported that the intracellular quality control machinery partitions misfolded proteins among two distinct quality control compartments—JUNQ and IPOD (Kaganovich et al., 2008). We then asked whether Hsp70 also controls the formation of JUNQ and IPOD in mammalian cells. We used a model JUNQ substrate CHFP-VHL, a physiological von Hippel–Lindau (VHL) protein fused with mCherry. Notably, the JUNQ formation of CHFP-VHL was largely colocalized with the aggresome of CFTRΔF508 and cBSA-GFP (Supplemental Figure S6). So, at least in mammalian cells, the JUNQ compartment resembles the aggresome structure. Similar to cBSA-GFP and GFP-CFTRΔF508, knocking down Hsp70 stabilized CHFP-VHL, but the accumulated CHFP-VHL failed to form aggregates (Supplemental Figure S7). It is significant that Hsp70 knockdown also prevented the aggresome formation in the presence of MG132 (Figure 6C and Supplemental Figure S7). In contrast, Hsp90 knockdown had little effect on the behavior of CHFP-VHL. We next examined the fate of a model IPOD substrate, HttQ103-GFP, a polyQ expanded Huntingtin fused with GFP. It is surprising that Hsp70 knockdown did not alter the aggregation of HttQ103-GFP in the absence or presence of MG132 (Figure 6C and Supplemental Figure S8). It appears that Hsp70 is involved in neither the degradation nor the aggregation of terminally misfolded proteins like HttQ103-GFP. These results suggest that distinct quality control pathways exist for different misfolded proteins.
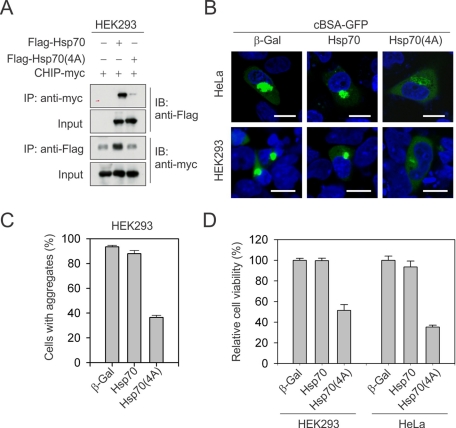
Hsp70–CHIP interaction is essential for the aggregation of misfolded proteins
The canonical function of Hsp70 is to assist protein folding and solubilize misfolded substrates. We then asked whether disrupting Hsp70–CHIP interaction would switch the functionality of Hsp70 from “proaggregase” to “disaggregase.” To this end, we mutated the COOH-terminal EEVD sequence of Hsp70 into four consecutive alanine residues. The resultant Hsp70(4A) mutant largely abolished the CHIP binding, as demonstrated by immunoprecipitation (Figure 7A). Despite the fact that most human cell lines such as HeLa have considerable high basal levels of endogenous Hsp70, overexpression of Hsp70(4A) largely suppressed the aggresome formation of cBSA-GFP in the presence of MG132 (Figure 7B). As a result, the majority of cBSA-GFP proteins were distributed evenly in the cytosol. Quantitative analysis further revealed that Hsp70(4A) suppressed the aggregate formation of cBSA-GFP by more than 50% in HEK293 cells as compared with the cells expressing either β-galactosidase (β-Gal) or wild-type Hsp70 (Figure 7B). Hsp70(4A) could also affect the binding of other cochaperones that rely on the EEVD motif of Hsp70. One example is Hsp70/Hsp90 organizing protein (HOP; Chen and Smith, 1998). HOP serves as an adaptor in connecting Hsp70 and Hsp90. Because Hsp90 showed little effect in aggresome targeting, we reasoned that the unique feature of Hsp70(4A) mutant was mainly due to the disruption of Hsp70–CHIP interaction. Although there are other TPR-domain–containing cochaperones, our results suggest that abolishing Hsp70–CHIP interaction converts the cellular function of Hsp70 from proaggregation to disaggregation. Taken together with the fact that CHIP knockdown alone prevented the aggresome formation of cBSA-GFP (Figure 2A), this indicates that Hsp70–CHIP interaction is essential for the aggregation of misfolded proteins.
FIGURE 7:
Hsp70-CHIP interaction is essential for the aggregation of misfolded proteins. (A) HEK293 cells were transfected with plasmids encoding CHIP-myc and FLAG-tagged Hsp70 or Hsp70(4A). Immunoprecipitation was performed using either anti-myc or anti-FLAG antibody, followed by immunoblotting. (B) HeLa and HEK293 cells were cotransfected with plasmids encoding cBSA-GFP and Hsp70 or Hsp70(4A). After 5 μM MG132 treatment overnight, cells were fixed. Nuclei were counterstained with Hoechst. Bar, 10 μm. (C) Aggregate formation in HEK293 cells was quantitated using Nikon Elements imaging analysis (mean ± SEM; n = 4). (D) HeLa and HEK293 cells were cotransfected with plasmids encoding cBSA-GFP and Hsp70 or Hsp70(4A). After 5 μM MG132 treatment for 18 h, viable cells were counted using trypan blue staining (mean ± SEM; n = 4).
The unique feature of Hsp70(4A) afforded us an opportunity to evaluate whether aggregate formation is cytotoxic or cytoprotective. We overexpressed misfolded protein cBSA-GFP in both HEK293 and HeLa cells, followed by MG132 treatment. We observed no appreciable protective effects of Hsp70 overexpression, presumably because both human cell lines have abundant endogenous Hsp70 molecules and can readily form misfolded protein aggregates after MG132 treatment (Figure 7, B and C). In contrast, overexpression of Hsp70(4A), which suppresses aggregates formation, reduced the cell viability to ∼50% as compared with cells expressing β-Gal (Figure 7C). Taken together, our results support the idea that Hsp70 exerts its cytoprotective effects by promoting aggregation of misfolded proteins in cells when their degradation is impaired.
Hsp70-based aggresome targeting relies on BAG3
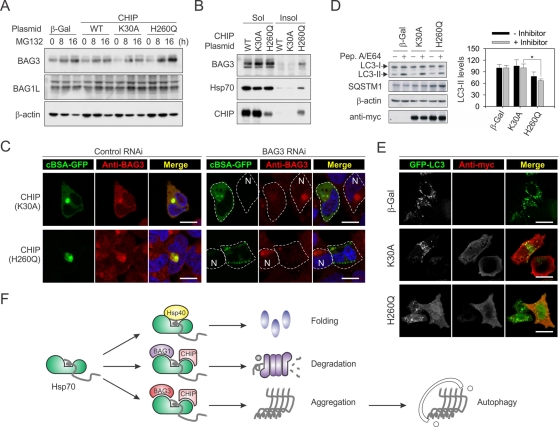
What is the molecular mechanism underlying Hsp70-mediated aggresome targeting? In particular, how does the dominant-negative CHIP mutant sensitize aggresome formation? A recent study reported that the stress-induced BAG3 interacts with the microtubule motor protein dynein and selectively directs Hsp70 substrates to the aggresome in a ubiquitination-independent manner (Gamerdinger et al., 2011). We examined the expression levels of BAG3 in cells expressing CHIP or CHIP mutants. Consistent with the previous report, proteasome impairment induced the up-regulation of BAG3 but not BAG1 (Figure 8A). It is remarkable that the dominant-negative CHIP mutant H260Q increased BAG3 expression to much higher levels. The clear time-course effects further support its dominant-negative nature and agree with our finding that CHIP(H260Q) triggers HSF1 activation (Figure 4, D and E). Because BAG3 binds the ATPase domain of Hsp70, CHIP(H260Q) could trap both BAG3 and Hsp70 together with their substrates. Indeed, considerable amounts of BAG3, Hsp70, and CHIP(H260Q) were recovered from the insoluble fraction (Figure 8B). In addition, BAG3 largely localized to the juxtanuclear aggresome after proteasome inhibition with an almost complete overlap with cBSA-GFP (Figure 8C). These results suggest a positive role of BAG3 in chaperone-based aggresome targeting.
FIGURE 8:
Hsp70/CHIP–mediated aggresome targeting relies on BAG3. (A) HEK293 cells were transfected with plasmids encoding CHIP or CHIP mutants, followed by MG132 treatment. BAG3 and BAG1L levels were determined by immunoblotting. (B) HEK293 cells were transfected with plasmids encoding CHIP or CHIP mutants followed by MG132 treatment. Forty-eight hours after transfection, whole-cell lysates were separated into 1% Triton X-100–soluble and –insoluble factions before immunoblotting. (C) HEK293 cells were transfected with siRNA targeting BAG3 before plasmid cotransfection to express both cBSA-GFP and CHIP mutants. After 5 μM MG132 treatment overnight, immunostaining was performed using anti-BAG3 antibody. Nuclei were counterstained with Hoechst. N, the neighboring control cells without BAG3 knockdown. Bar, 10 μm. (D) HeLa cells were transfected with plasmids encoding CHIP or CHIP mutants. Four hours before the sample harvest, cells were treated with 10 μg/ml pepstatin A and 10 μg/ml E64d. Whole-cell lysates were immunoblotted using antibodies as indicated. LC3-II levels were quantified and normalized against β-actin (mean ± SEM; n = 3; *p < 0.05). (E) HeLa cells were cotransfected with plasmids encoding GFP-LC3 and CHIP mutants, followed by rapamycin treatment for 24 h. Immunostaining was performed using anti-myc antibody to indicate CHIP expression. Bar, 10 μm. (F) Model for a hierarchy of cochaperone interaction controlling different aspects of the intracellular triage decision. Hsp70, coupled with cochaperones such as Hsp40, assists protein folding. When the successful folding cannot be achieved, the association of CHIP and BAG1 facilitates the degradation of chaperone substrates. When the degradative capacity in cells is exceeded or impaired, the binding of BAG3 directs the Hsp70/CHIP–associated substrates to the aggresome.
To further investigate the role of BAG3 in Hsp70/CHIP–mediated aggresome formation, we knocked down BAG3 in HEK293 cells using siRNA. In line with the notion that BAG3 promotes aggresome assembly by coupling chaperone substrates to the dynein motor (Gamerdinger et al., 2011), BAG3 knockdown largely prevented the aggresome formation of cBSA-GFP in cells expressing control plasmid CHIP(K30A) (Figure 8C). Of interest, the presence of CHIP(H260Q) caused formation of punctuated small aggregates dispersed in the cytoplasm after BAG3 knockdown. These preaggregates apparently failed to be delivered to the juxtanuclear inclusion body in the absence of BAG3. This unique pattern of cBSA-GFP aggregation could be readily observed in cells with BAG3 knockdown even without MG132 treatment, but only in the presence of CHIP(H260Q) (Supplemental Figure S9). Therefore BAG3 acts at a later stage than CHIP in targeting Hsp70 substrates to the aggresome, whereas Hsp70–CHIP interaction is essential in the primary aggregation process of misfolded proteins.
It has been reported that BAG3 promotes the turnover of aggregates via autophagy (Gamerdinger et al., 2009). Although CHIP(H260Q) sensitizes aggresome formation, it remains unknown how this dominant-negative CHIP mutant affects the autophagy pathway. A well-established marker for autophagy is LC3-II, and the autophagic flux can be measured by monitoring LC3-II accumulation in the presence of lysosomal inhibitors (Mizushima and Klionsky, 2007). In cells expressing CHIP(H260Q), the accumulation of LC3-II on lysosomal inhibition by pepstatin A and E64 was reduced to ∼35% (Figure 8D). This result suggests that the autophagy pathway was blunted in the presence of CHIP(H260Q). Further supporting this notion, one autophagic degradation substrate, SQSTM1, largely accumulated in cells expressing CHIP(H260Q) (Figure 8D). Next we expressed LC3 fused to GFP (GFP-LC3) to visualize autophagosomes in cells. CHIP(H260Q) overexpression led to a clear reduction in the number of GFP-LC3 puncta, whereas the neighboring nontransfected cells showed many typical GFP-LC3 puncta (Figure 8E). Together, our data indicate that CHIP(H260Q) sensitizes aggresome formation by suppressing the macroautophagy pathway.
DISCUSSION
The intracellular chaperone network plays a key role in maintaining protein homeostasis (Balch et al., 2008). Molecular chaperones are involved in a multitude of cellular functions, including de novo folding, refolding of stress-denatured proteins, oligomeric assembly, intracellular protein transport, and assistance in proteolytic degradation (Hartl and Hayer-Hartl, 2009). Hsp70-mediated folding is often linked to the prevention of misfolded protein aggregation. Here we provide evidence showing that Hsp70, coupled with cochaperone ubiquitin ligase CHIP, not only mediates the degradation of misfolded proteins, but also controls their aggregation when the proteasome function is impaired. Our finding challenges the belief that molecular chaperones solely act to solubilize protein aggregates in cells. We propose that a hierarchy of cochaperone interaction controls different aspects of the intracellular triage decision, thereby extending the function of Hsp70 from folding and degradation to aggregation.
The proaggregation function of Hsp70 is cytoprotective
Mounting evidence suggests that molecule chaperones assist protein folding and prevent protein aggregation. For example, protein disaggregation by the Hsp104–Hsp70 bichaperone system is well documented in Escherichia coli and Saccharomyces cerevisiae (Glover and Lindquist, 1998; Weibezahn et al., 2004). However, each chaperone component on its own has only limited disaggregation activity (Tyedmers et al., 2010). Of interest, no homologue of the Hsp100 family has been found in mammals (aside from mitochondria). A recent study reported the existence of a mammalian amyloid disaggregase, but its identity is ill characterized (Murray et al., 2010). Thus the existence of chaperone cooperation in the disassembly of protein aggregates in mammalian cells remains to be investigated. In our study, the seemingly contradictory activity of Hsp70 in the aggregation process of misfolded proteins is not incompatible with the protective role of Hsp70 under stress conditions. Instead, directing nondegradable misfolded protein substrates to specific cellular sites can be viewed as a second line of active cellular defense. Despite the strong correlation between the accumulation of aggregates proteins and the onset of several neurodegenerative diseases, a consensus is emerging that oligomeric, soluble states of the respective disease proteins are the primary cytotoxic species (Chiti and Dobson, 2006). Therefore the sequestration of misfolded proteins in the inclusion bodies is protective for cell function. Supporting this notion, overexpression of Hsp70(4A) not only prevented the aggresome formation, but also reduced cell viability (Figure 7D).
Among molecular chaperones, the effect of Hsp70 on the toxicity of disease proteins has been investigated intensely in a diverse range of models, including yeast, worms, flies, and mice. Overexpression of Hsp70 generally protects cells from the toxicity of disease proteins, including polyglutamine stretches, α-synuclein, and mutant ataxin (Muchowski and Wacker, 2005). However, the protective effects of Hsp70 do not always correlate with the suppression of aggregate formation. In some cases, Hsp70 overexpression even increased the size and number of inclusion bodies (Warrick et al., 1999; Auluck et al., 2002; Shimura et al., 2004). These results indicate that the protective effect of Hsp70 does not require inclusion body clearance. Our findings potentially solve the conundrum by demonstrating that Hsp70 promotes the spatial sequestration of small, potentially toxic misfolded assemblies into an inclusion body. Taken together, the data indicate that molecular chaperones facilitate neuroprotection by functioning at multiple levels that might not be linked exclusively to their refolding or solubilizing activities.
New insights into the logic of protein triage decisions
By investigating the fate of a misfolded protein cBSA, we confirmed that Hsp70 and CHIP promote the degradation of misfolded proteins. The collective activities of the molecular chaperones and the ubiquitin-proteasome system are normally sufficient to prevent the accumulation of misfolded proteins. However, when the degradative capacity of a cell is exceeded or impaired, protein aggregates accumulate. Despite typical aggresome formation of cBSA-GFP under proteasome inhibition, accumulated cBSA-GFP failed to form discernible aggregates in cells with either CHIP or Hsp70 knockdown. Both mouse and human cell lines exhibit similar features, supporting a generic mechanism of aggresome targeting involving CHIP–Hsp70 interaction. This result clearly supports the notion that misfolded protein aggregation is a regulated process rather than an unspecific and uncontrolled dead-end pathway (Tyedmers et al., 2010). More important, it suggests an essential role of Hsp70-CHIP interaction in the aggregation process of misfolded proteins. Directing of misfolded proteins to an aggresome in the cytoplasm of mammalian cells is believed to be mediated by histone deacetylase 6 (HDAC6), which functions as an adaptor that binds polyubiquitin chains of substrates and the microtubule protein dynein (Kawaguchi et al., 2003). This could explain the failed aggresome formation in the absence of CHIP because of the deficient ubiquitin conjugation on substrates. However, the dominant-negative U-box mutant CHIP(H260Q) potently sensitized the aggregation of misfolded proteins despite its inability to ubiquitinate the chaperone substrates. It is likely that ubiquitination is not the only signal involved in depositing misfolded proteins to aggresomes. Recent evidence indicates that BAG3-mediated aggresome targeting is independent of substrate ubiquitination. In particular, BAG3 coupled chaperone-associated substrates to the dynein motor complex and promoted aggresome assembly (Gamerdinger et al., 2011). Notably, proteasome inhibition up-regulates BAG3 expression in a HSF1-regulated manner (Du et al., 2009). On the basis of the exceptionally augmented BAG3 levels in the presence of the dominant-negative CHIP(H260Q) mutant, our results support the notion that the Hsp70-associated substrates can be selectively targeted to the aggresome independent of ubiquitination.
Despite the severe accumulation of misfolded proteins in cells, the presence of CHIP(H260Q) does not interfere with 26S proteasome function (Figure 4, A–C). One prominent feature of CHIP(H260Q) in cells is the potent activation of HSF1. In mammalian cells, HSF1 is the major transcription factor controlling chaperone gene expression (Akerfelt et al., 2010). It is perplexing that the increased chaperone levels in the presence of CHIP(H260Q) do not prevent the misfolded proteins from aggregation. In contrast, it largely sensitizes the aggregate formation of misfolded proteins. It is known that CHIP–Hsp70 interaction prolongs the substrate holding that facilitates the ubiquitination and subsequent degradation. When the substrate degradation is blocked by either deficient ubiquitination (in the case of CHIP(H260Q)) or proteasome inhibition, the continuous substrate holding might lead to local aggregation of Hsp70-associated substrates. The HSF1-mediated up-regulation of BAG3 then binds the ATPase domain of Hsp70 and loads its substrates to the dynein motor, followed by subsequent delivery to the juxtanuclear aggresome. Supporting this model, BAG3 knockdown resulted in distinct preaggregate formation in the cytosol (Figure 8C). Taken together with the finding that knocking down either Hsp70 or CHIP prevents the aggresome formation, we conclude that Hsp70 relies on different cochaperone binding to mediate both the aggregation process and the aggresome targeting.
Our analysis of the chaperone pathways required for protein aggregation provides novel insights into the logic underlying triage decisions in the mammalian cytosol (Figure 8D). The finding that Hsp70 is essential in directing misfolded proteins to aggresomes suggests that the fate of a given chaperone substrate may be controlled by a hierarchy of chaperone interactions (Kampinga and Craig, 2010). In association with cochaperones like Hsp40, Hsp70 enhances the refolding cycle by increased ATP hydrolysis (Bukau et al., 2006). If the refolding of substrates is not successful, the binding of cochaperone ubiquitin ligase CHIP facilitates substrate ubiquitination (Cyr et al., 2002). It is intriguing that ubiquitin-related BAG1 provides an additional link between the molecular chaperones and the proteasome, thereby facilitating the sorting of chaperone substrates to the proteasome for degradation (Alberti et al., 2002). The BAG family proteins are modulators of protein quality control, so association with different BAG proteins might lead to the distinct fate of Hsp70 substrates. A recent study reported that a switch from BAG1 to BAG3 in association with Hsp70 converted the degradation of chaperone substrates from proteasome to autophagy (Gamerdinger et al., 2009). In addition, functional interaction between CHIP and BAG3 was reported in muscle maintenance (Arndt et al., 2010). Among BAG family proteins, BAG3 is the only stress-inducible one. When substrate degradation is impaired, the up-regulated BAG3 might compete with BAG1 in Hsp70 binding and direct the deposition of associated substrates into aggresome. This functional partition not only allows the efficient sequestration of misfolded proteins in cellular compartments, but it also saves the chaperone molecules that are vital for other cellular functions.
The essential role of Hsp70 in misfolded protein aggregation is not limited to the artificial substrate cBSA-GFP. Physiological substrates like CFTRΔF508 and VHL had the same behavior as cBSA-GFP in response to the Hsp70–CHIP interaction. Notably, all of these substrates are sequestered at the same cellular compartments as aggresomes when their degradation is impaired. It is intriguing that Hsp70 had little effect on both the degradation and the aggregation of HttQ103, a polyglutamine model substrate. A recent study reported that HttQ103 differs from VHL by forming distinct protein aggregate IPODs (Kaganovich et al., 2008). Substrates at JUNQ are still mobile and exchange rapidly with the surrounding cytoplasm, whereas IPODs harbor terminal aggregates and insoluble proteins. It has been reported that polyQ-containing proteins are poor substrates for proteasomes (Venkatraman et al., 2004). Supporting this notion, HttQ103 can readily form IPODs in cells without proteasome inhibition. Therefore the fate of HttQ103 is less influenced by Hsp70–CHIP interaction once the aggregates have been preformed. However, Htt protein bearing shorter polyQ expansion could still be subject to regulation by chaperones (Zhang et al., 2010). Taken together, our results suggest that the degradative role of Hsp70–CHIP interaction can be tilted toward proaggregation when the degradation route is blocked. Hsp70, uniquely required for both folding and degradation, may play a key role in misfolded protein aggregation.
MATERIALS AND METHODS
Cell lines and reagents
HEK293 and HeLa cell lines were maintained in DMEM with 10% fetal bovine serum (FBS). HSF1+/+ and _HSF1_−/− MEFs were kindly provided by I. J. Benjamin (University of Utah, Salt Lake City, UT). Protein synthesis inhibitor CHX and proteasome inhibitor MG132 were purchased from Sigma-Aldrich (St. Louis, MO). Hoechst and Alexa Fluor 488– and 546–labeled secondary antibodies (donkey anti–mouse immunoglobulin G [H + L]) were purchased from Invitrogen (Carlsbad, CA). Anti-His antibody was purchased from Qiagen (Valencia, CA), β-actin monoclonal antibody from Sigma-Aldrich, anti-Hsp70 (SPA810), and anti-Hsp90 (SPA830) from StressGen (Victoria, Canada), anti-ubiquitin (P4D1), anti-BAG1 (CC9E8), and anti-myc (9E10) from Santa Cruz Biotechnology (Santa Cruz, CA), anti-BAG3 from Abcam (Cambridge, MA), and anti-Parkin (PRK8) from Millipore (Billerica, MA). The Dual Luciferase Kit was purchased from Promega (Madison, WI).
Plasmids, siRNAs, and recombinant adenoviruses
Mammalian expression constructs (pcDNA3) for β-Gal, cBSA, CHIP, and CHIP mutants were as described previously (Qian et al., 2006a). cBSA-GFP fusion gene was constructed by inserting PCR-amplified cBSA fragment into EGFP-N1 using _Hin_dIII and _Bam_HI sites. siRNAs targeting Hsp70, Hsp90, ubiquitin, CHIP, Parkin, and control siRNA were purchased from Dharmacon (Lafayette, CO). siRNAs targeting BAG3 were purchased from Santa Cruz Biotechnology. Plasmids encoding CHFP-VHL and HttQ103-GFP were kindly provided by D. Kaganovich (Hebrew University of Jerusalem, Jerusalem, Israel). Hsp70(4A) was created using a PCR-based recloning strategy. Recombinant adenoviruses expressing β-gal, Hsp70, and Hsp90 were purchased from Vector Laboratories (Burlingame, CA). Recombinant adenoviruses expressing cBSA-GFP were made using AdEasy system (Clontech, Mountain View, CA).
Transfections
Plasmid and siRNA transfections were performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
Immunoblotting
Cells were lysed on ice in TBS buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA) containing protease inhibitor cocktail tablet, 1% Triton X-100, and 2 U/ml DNase. After incubating on ice for 30 min, the lysates were heated for 10 min in SDS–PAGE sample buffer (50 mM Tris-HCl, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol). For fractionation analysis, the Triton X-100–soluble and –insoluble fractions were dissolved in samples buffer separately. Proteins were resolved on SDS–PAGE and transferred to Immobilon-P membranes (Millipore). Membranes were blocked for 1 h in TBS containing 5% BSA, followed by incubation with primary antibodies. After incubation with horseradish peroxidase–coupled secondary antibodies, immunoblots were developed using enhanced chemiluminescence (ECLPlus; GE Healthcare, Piscataway, NJ).
Immunoprecipitation
Cells were lysed on ice in TBS buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail tablet) containing 1% Triton. Cell lysates were cleared by centrifugation at 5000 × g for 5 min. Supernatants were incubated with anti-myc or anti-FLAG agarose beads at 4°C for 60 min. Immunoprecipitates were washed four times with lysis buffer, followed by elution using 1× SDS–PAGE sample buffer. Samples were resolved on SDS–PAGE, followed by immunoblotting.
Immunofluorescence microscopy, FRAP, and quantification
Cells were grown on glass coverslips, fixed in 4% paraformaldehyde, permeabilized by 0.2% Triton X-100, and blocked in 2% BSA in PBS. Primary antibody incubation was done at 4°C overnight, followed by 1 h of incubation at room temperature with Alexa Fluor–labeled secondary antibodies. Cells were washed and then incubated for 5 min in PBS supplemented with Hoechst to stain the nuclei. After final wash, cover slips were mounted onto slides and viewed using a confocal microscope (LSM710. Zeiss, Jena, Germany).
FRAP was performed at room temperature using an X40 oil objective with scan zoom of 4. A rectangular region of interest (ROI) was bleached with 900 iterations and 100% laser power (488-nm argon laser). One image was captured before bleaching. An image was taken every 2 s (488-nm argon laser at 4% power) after bleaching over a 76-s period. For each time point, the fluorescence intensity of the photobleached ROI was determined using LSM710 software.
For quantification analysis, five fields of each sample were randomly selected. The percentage of cells containing aggresomes was counted and averaged. For measurement of the area and the intensity of aggregates, 35 cells of each sample were randomly selected, followed by quantitative measurement using microscope software (NIS-Elements; Nikon, Melville, NY). Student's t test was used for statistical comparison.
Cell viability assays
HEK293 and HeLa cells were cotransfected with the plasmid expressing cBSA-GFP and Hsp70 or Hsp70(4A). Thirty hours after transfection, cells were treated with 5 μM MG132 for another 18 h, and then viability was determined via cell counting with trypan blue staining.
Supplementary Material
Supplemental Materials
Acknowledgments
We thank Sylvia Lee and members of the Qian laboratory for critical reading of the manuscript. We are also grateful to I. J. Benjamin (University of Utah) for providing HSF1+/+ and HSF1−/− MEFs and Daniel Kaganovich (Hebrew University of Jerusalem) for plasmids expressing CHFP-VHL and HttQ103-GFP. This work was supported by grants from the National Institutes of Health (1 DP2 OD006449-01) and the Ellison Medical Foundation (AG-NS-0605-09) to S.-B.Q.
Abbreviations used:
BAG3
Bcl2-associated athanogene 3
cBSA
cytosolic bovine serum albumin
CFTR
cystic fibrosis transmembrane conductance regulator
CHIP
carboxyl terminal of heat shock protein 70/heat shock protein 90 interacting protein
HSF1
heat shock transcription factor
IPOD
insoluble perivacuolor inclusion
JUNQ
juxtanuclear compartment
VHL
von Hippel–Lindau.
Footnotes
REFERENCES
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J Biol Chem. 2002;277:45920–45927. doi: 10.1074/jbc.M204196200. [DOI] [PubMed] [Google Scholar]
- Arndt V, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- Dai Q, et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZX, Zhang HY, Meng X, Gao YY, Zou RL, Liu BQ, Guan Y, Wang HQ. Proteasome inhibitor MG132 induces BAG3 expression through activation of heat shock factor 1. J Cell Physiol. 2009;218:631–637. doi: 10.1002/jcp.21634. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Jiang J, Cyr D, Babbitt RW, Sessa WC, Patterson C. Chaperone-dependent regulation of endothelial nitric-oxide synthase intracellular trafficking by the co-chaperone/ubiquitin ligase CHIP. J Biol Chem. 2003;278:49332–49341. doi: 10.1074/jbc.M304738200. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kettern N, Rogon C, Limmer A, Schild H, Hohfeld J. The Hsc/Hsp70 co-chaperone network controls antigen aggregation and presentation during maturation of professional antigen presenting cells. PLoS One. 2011;6:e16398. doi: 10.1371/journal.pone.0016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Murata S, Chiba T, Tanaka K. CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol. 2003;35:572–578. doi: 10.1016/s1357-2725(02)00394-1. [DOI] [PubMed] [Google Scholar]
- Murray AN, Solomon JP, Wang YJ, Balch WE, Kelly JW. Discovery and characterization of a mammalian amyloid disaggregation activity. Protein Sci. 2010;19:836–846. doi: 10.1002/pro.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006a;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian SB, Ott DE, Schubert U, Bennink JR, Yewdell JW. Fusion proteins with COOH-terminal ubiquitin are stable and maintain dual functionality in vivo. J Biol Chem. 2002;277:38818–38826. doi: 10.1074/jbc.M205547200. [DOI] [PubMed] [Google Scholar]
- Qian SB, Princiotta MF, Bennink JR, Yewdell JW. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J Biol Chem. 2006b;281:392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- Shimura H, Schwartz D, Gygi SP, Kosik KS. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Zhang S, Binari R, Zhou R, Perrimon N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics. 2010;184:1165–1179. doi: 10.1534/genetics.109.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials