Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB (original) (raw)
. Author manuscript; available in PMC: 2011 Sep 28.
Published in final edited form as: Nature. 2010 May 2;465(7295):243–247. doi: 10.1038/nature08966
Abstract
Polycomb group (PcG) proteins are transcriptional repressors that control processes ranging from the maintenance of cell fate decisions and stem cell pluripotency in animals to the control of flowering time in plants1–6. In Drosophila, genetic studies identified more than 15 different PcG proteins that are required to repress homeotic (HOX) and other developmental regulator genes in cells where they must stay inactive1,7,8. Biochemical analyses established that these PcG proteins exist in distinct multiprotein complexes that bind to and modify chromatin of target genes1–4. Among those, Polycomb repressive complex 1 (PRC1) and the related dRing-associated factors (dRAF) complex contain an E3 ligase activity for monoubiquitination of histone H2A (refs 1–4). Here we show that the uncharacterized Drosophila PcG gene calypso encodes the ubiquitin carboxy-terminal hydrolase BAP1. Biochemically purified Calypso exists in a complex with the PcG protein ASX, and this complex, named Polycomb repressive deubiquitinase (PR-DUB), is bound at PcG target genes in Drosophila. Reconstituted recombinant Drosophila and human PR-DUB complexes remove monoubiquitin from H2A but not from H2B in nucleosomes. Drosophila mutants lacking PR-DUB show a strong increase in the levels of monoubiquitinated H2A. A mutation that disrupts the catalytic activity of Calypso, or absence of the ASX subunit abolishes H2A deubiquitination in vitro and HOX gene repression in vivo. Polycomb gene silencing may thus entail a dynamic balance between H2A ubiquitination by PRC1 and dRAF, and H2A deubiquitination by PR-DUB.
A genetic screen for Drosophila mutants with PcG phenotypes recently identified calypso as a complementation group with two lethal alleles that complemented mutations in any other PcG gene8. We mapped the calypso mutations (see Methods), and the following findings established that calypso corresponds to the uncharacterized gene CG8445. First, calypso1 and calypso2, two independently isolated lethal calypso alleles8, both contained a cytosine to thymine mutation in CG8445 that creates a premature termination codon (Fig. 1a), whereas the parental chromosome on which these mutations had been induced contained a wild-type cytosine. Second, a transgene expressing a tandem affinity purification (TAP)-tagged form of the CG8445 protein under control of the α-tubulin1 promoter rescued calypso mutant animals into viable and fertile adults (see Methods). Third, calypso1 and calypso2 mutants did not express detectable levels of CG8445 protein (see later and Supplementary Fig. 5). We therefore named the CG8445 gene calypso.
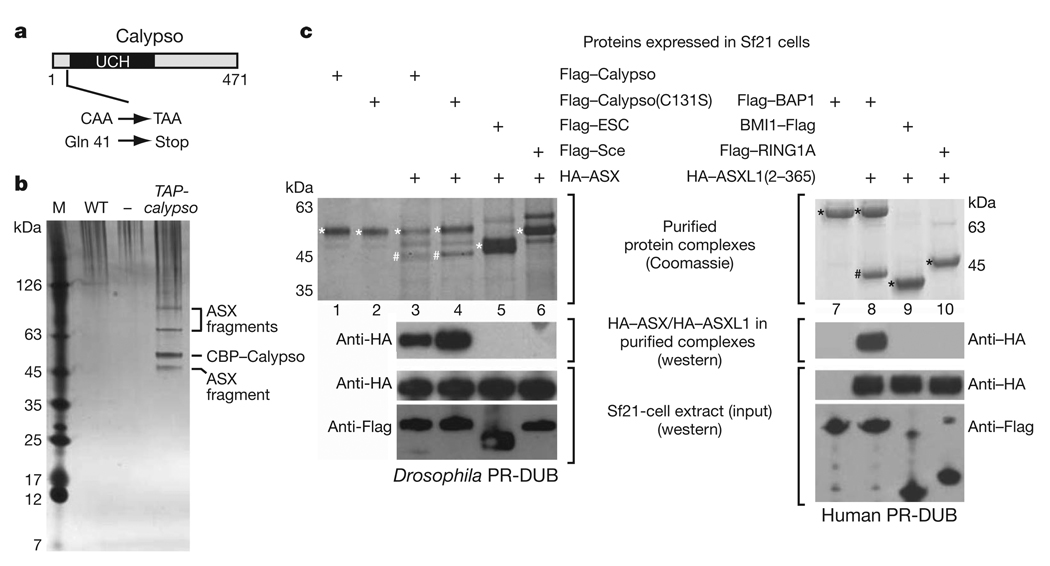
Figure 1. The Polycomb group proteins BAP1 and ASX form a conserved complex in vivo and in vitro.
a, Domain architecture of the Drosophila Calypso protein and molecular lesions in calypso mutant alleles. UCH, ubiquitin C-terminal hydrolase domain. b, Calypso complexes isolated by TAP26 from wild-type (WT) or TAP-calypso transgenic embryos. Input material for purification was normalized by protein concentration, and equivalent amounts of eluate from calmodulin-affinity resin were separated on a 4–12% polyacrylamide gel and visualized by silver staining together with a molecular mass marker (M). Calypso bait protein containing the calmodulin-binding tag (CBP–Calypso), and bands representing ASX fragments were identified by mass spectrometry (Supplementary Table 1 and Supplementary Fig. 2). No band corresponding to full-length ASX (180 kDa) was detected in several independent purifications, even though ASX is present as a single polypeptide of 180 kDa in total embryo extracts (Supplementary Fig. 5). This suggests that ASX is degraded during nuclear extract preparation or TAP purification. c, Reconstitution of recombinant Calypso–ASX and BAP1–ASXL1 complexes. Proteins were extracted by Flag-affinity purification from cell lysates containing the indicated Flag-tagged proteins and HA–ASX(2–337) (left) or HA–ASXL1(2–365) (right). Experiments with full-length ASX are shown in Supplementary Fig. 3. Proteins were visualized by Coomassie staining or western blotting analysis, as indicated. Input material for experiments in lanes 3–6 (left) and 7–10 (right) were probed by western blotting to ensure that comparable amounts of proteins were present in cell lysates. On the Coomassie-stained gel, Flag-tagged proteins are marked with an asterisk, HA–ASX(2–337) and HA–ASXL1(2–365) are marked with a hash symbol.
calypso encodes a polypeptide of 471 amino acids that is a member of the ubiquitin C-terminal hydrolase (UCH) subclass of deubiquitinating enzymes (Fig. 1a)9. UCH domains are cysteine proteases that hydrolyse the isopeptide bond between the C-terminal glycine of ubiquitin and the lysine side chain in the conjugated protein9–11. The closest human homologue of Calypso is BAP1 (Supplementary Fig. 1), a nuclear protein that possesses tumour suppressor activity9,12,13. The Calypso protein thus represents Drosophila BAP1.
Western blot analyses of Drosophila nuclear extracts and staining of imaginal discs with anti-Calypso antibodies showed that the Calypso protein is localized in nuclei (Supplementary Fig. 1c and see later). To identify interaction partners of Calypso, we purified proteins associated with a TAP–Calypso fusion protein from nuclear extracts of embryos that carried the α-tubulin1-TAP-calypso transgene. The purified material was separated on SDS-polyacrylamide gels and four major protein bands were identified (Fig. 1b). Sequencing of peptides from these bands by nanoelectrospray tandem mass spectrometry identified the 55-kDa band as the Calypso bait protein, whereas the other three bands all represented fragments of the PcG protein Additional sex combs (ASX) (Fig. 1b, Supplementary Fig. 2 and Supplementary Table 1). Analysis of other gel regions and liquid chromatography tandem mass spectrometry (LC–MS/MS) analysis of total purified material confirmed that ASX was the main protein co-purifying with TAP–Calypso (Supplementary Table 1). ASX is a PcG protein required for long-term repression of HOX genes during Drosophila development7,8,14, but it had not been identified in previously characterized PcG protein complexes and its molecular function has remained largely elusive. Calypso and ASX are thus components of a new, bona fide PcG protein complex that we named Polycomb repressive deubiquitinase (PR-DUB).
We tested whether Drosophila PR-DUB complexes could be reconstituted from recombinant Calypso and ASX proteins. Using baculovirus vectors, we expressed Flag–Calypso and haemagglutinin (HA)–ASX(1–1668) or HA–ASX(2–337) as individual proteins in Sf21 cells, mixed the cell lysates and performed Flag affinity purification. This strategy resulted in the isolation of stable Calypso–ASX complexes and showed that Calypso interacts with the amino-terminal 337 amino acids of ASX (Fig. 1c, left, lane 3 and Supplementary Fig. 3). ASX also formed stable complexes with Calypso(C131S), a mutant Calypso protein in which the predicted catalytic cysteine in the UCH domain10,15 had been substituted by serine (Fig. 1c, left, lane 4 and Supplementary Fig. 3). The interaction between Calypso and ASX(2–337) was specific because ASX(2–337) did not bind to the PcG proteins Flag–ESC or Flag–Sce under the same assay conditions (Fig. 1c, left, lanes 5 and 6). Using the same strategy, we found that human BAP1 also forms a stable complex with the N-terminal domain of human ASXL1 (ASXL1(2–365)) but not with the human PcG proteins BMI1 or RING1A (Fig. 1c, right, lanes 8–10). Like Drosophila Calypso and ASX, human BAP1 and ASXL1 proteins could thus also be assembled into a stable PR-DUB complex (Fig. 1c, right, lane 8), demonstrating the evolutionary conservation of this interaction.
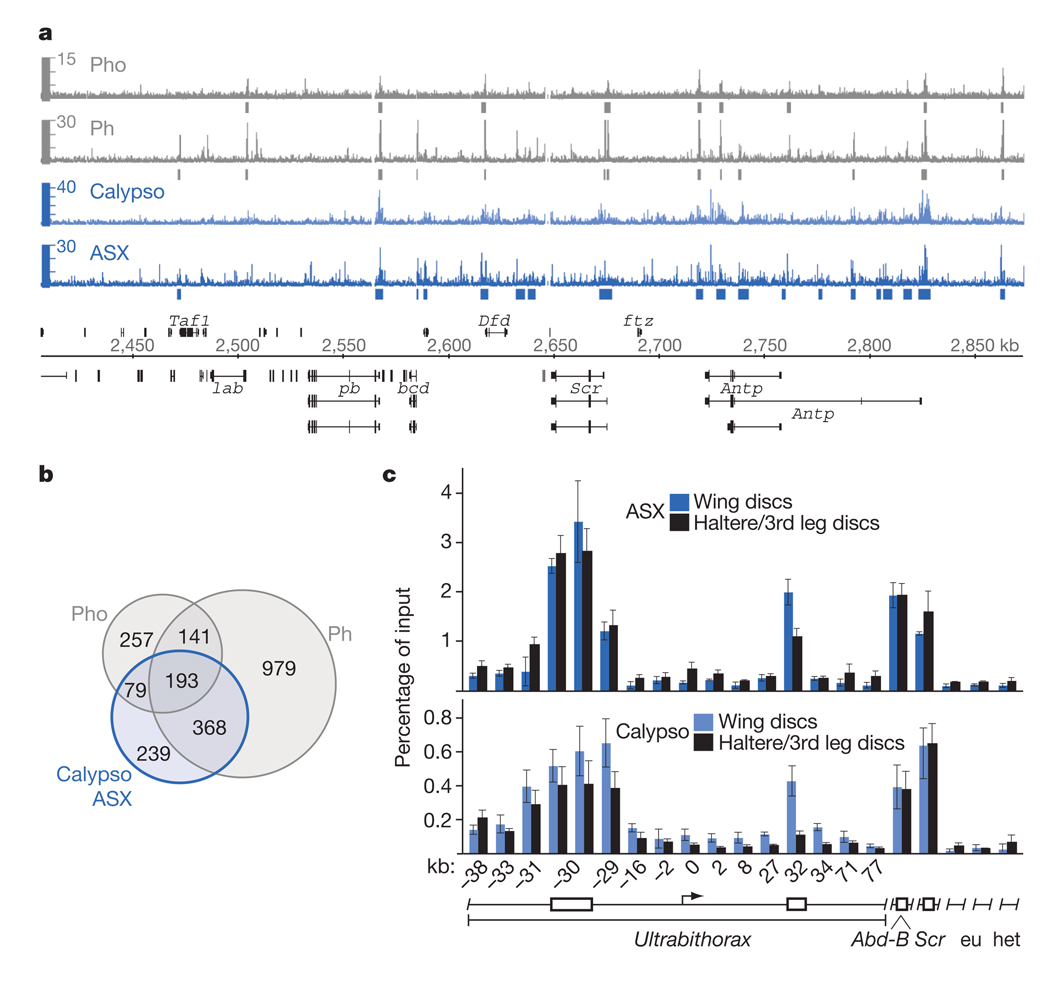
On polytene chromosomes, ASX protein binds at chromosome intervals encompassing the HOX genes and at many other chromosomal sites that co-map with binding sites for other PcG proteins14. We determined the genome-wide PR-DUB binding profile in the chromatin of Drosophila larvae by performing chromatin immunoprecipitation (ChIP) assays with antibodies against Calypso and ASX proteins. The precipitated material was hybridized to high-density whole-genome tiling arrays and analysed with TileMap16, using a stringent cutoff. We only considered genomic regions that were significantly enriched by both anti-Calypso and anti-ASX antibodies, and thus obtained a high-confidence set of 879 genomic sites bound by PR-DUB (Fig. 2a, b and Supplementary Table 2). We compared the PR-DUB binding profile with the profiles of the PRC1 subunit Ph and the PhoRC subunit Pho in imaginal disc cells17,18. PR-DUB is co-bound together with Ph and Pho at Polycomb response elements (PREs) of a large set of PcG target genes, such as the HOX genes (Fig. 2a, b and Supplementary Table 2). PR-DUB is thus a core PRE-binding complex, like PhoRC, PRC1 and PRC2. To extend these analyses, we compared the binding of Calypso and ASX in wing imaginal disc cells in which the HOX gene Ultrabithorax (Ubx) is inactive, and in haltere/third leg imaginal disc cells in which Ubx is expressed, at the same 16 locations across the Ubx transcription unit where we had previously analysed binding of the PcG protein complexes PhoRC, PRC1 and PRC2 (ref. 19). Like these other PcG protein complexes19, PR-DUB was bound at Ubx PREs both in cells where Ubx is repressed and in cells where it is active (Fig. 2c).
Figure 2. PR-DUB is bound at Polycomb target genes in Drosophila.
a, PR-DUB is bound at PREs of PcG target genes in Drosophila. ChIP profiles of PR-DUB subunits ASX (dark blue) and Calypso (light blue), and of Ph18 (grey) and Pho17 (grey) at the Antennapedia HOX gene cluster in imaginal disc and CNS tissues from third instar Drosophila larvae. Hybridization intensities for oligonucleotide probes are plotted as coloured bars above the genomic map (release 5, kilobase coordinates) of Drosophila melanogaster; significantly enriched regions are marked below plots. HOX genes labial (lab), proboscipedia (pb), Deformed (Dfd), Sex combs reduced (Scr), Antennapedia (Antp) and other genes on the plus (above) or minus (below) strand are represented with exons (black boxes) and introns (thin black lines). b, Venn diagrams showing the overlap of 879 PR-DUB-bound regions with 1,681 Ph-bound and 670 Pho-bound regions in larval cells. c, PR-DUB is bound at the inactive and at the active Ubx gene. ChIP analyses monitoring ASX and Calypso binding in wing and haltere/third leg imaginal discs from wild-type third instar Drosophila larvae. Graphs show results from independent ChIP reactions (n = 3 ChIP reactions) with ASX or Calypso antibodies. ChIP signals, measured by qPCR, are presented as the mean percentage of input chromatin precipitated at each region; error bars indicate±s.d. (see Methods). Locations of Ubx PREs (boxes) and other regions relative to the Ubx transcription start site are indicated in kilobases. As control, binding was monitored at two euchromatic (eu) and one heterochromatic (het) region elsewhere in the genome, and at the PREs of the HOX genes Abd-B and Scr that are both inactive in wing and haltere/third leg imaginal discs. Calypso and ASX are bound at Ubx PREs both in wing and in haltere/third leg disc cells; at the −30-kb PRE, Calypso and ASX ChIP signals were comparable in wing and haltere/third leg chromatin, at the kb PRE, the signal in haltere/third leg chromatin is about 2–4-fold lower than in wing chromatin, paralleling PRC1 and PRC2 binding at both these PREs19.
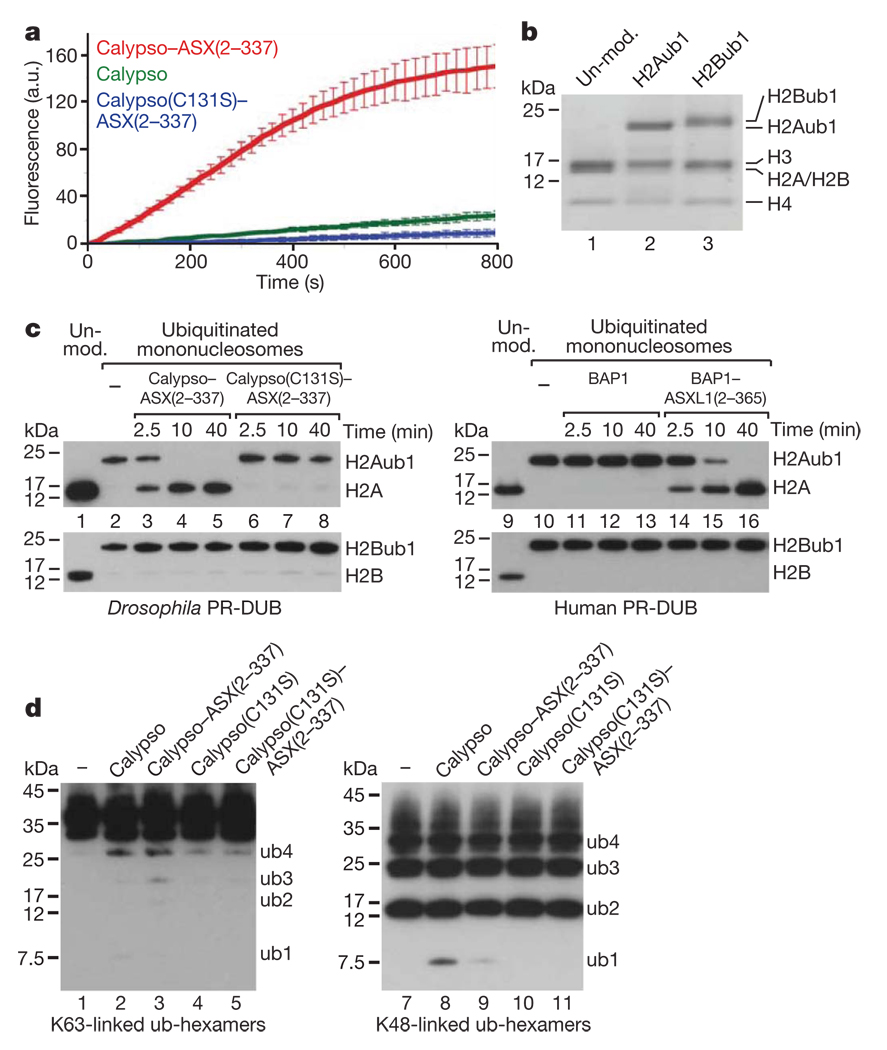
To characterize the deubiquitinase activity of PR-DUB, we tested whether the Drosophila complex could cleave the fluorogenic substrate ubiquitin-amidomethylcoumarin (Ub-AMC). Calypso alone hydrolysed the Ub-AMC bond, but the Calypso–ASX(2–337) complex was substantially more active in catalysing this reaction (Fig. 3a). In contrast, Calypso(C131S)–ASX(2–337) was virtually inactive (Fig. 3a). PR-DUB thus functions as a deubiquitinase in vitro and the catalytic activity of Calypso is strongly enhanced by association with the N-terminal domain of ASX. Because Drosophila PR-DUB associated with the chromatin of target genes, we then asked whether PR-DUB deubiquitinates histone H2A or H2B. Monoubiquitination of H2A (H2Aub1) at Lys 119 in vertebrates and Lys 118 in Drosophila by PRC1-like and dRAF, respectively, is thought to be critical for PcG repression20–22. Monoubiquitination of H2B (H2Bub1) at Lys 120 in vertebrates (corresponding to Lys 117 in Drosophila) is catalysed by a different E3 ligase, RNF20 (also known as BRE1), and has been implicated in transcriptional elongation22. We reconstituted recombinant mononucleosomes that contained either H2Aub1 or H2Bub1 (Fig. 3b) and used them as substrates in deubiquitination assays. Notably, the Drosophila Calypso–ASX(2–337) and Calypso–ASX(1–1668) complexes and the human BAP1–ASXL1(2–365) complex all deubiquitinated H2Aub1 but not H2Bub1 in nucleosomes (Fig. 3c, lanes 3–5, 14–16 and Supplementary Fig. 4c). Deubiquitination of H2Aub1 required both the presence of the catalytic cysteine in Calypso (Fig. 3c, lanes 6–8) and the association of ASX with Calypso (Supplementary Fig. 4) or of ASXL1 with BAP1, respectively (Fig. 3c, compare lanes 14–16 with 11–13). Moreover, PR-DUB showed only very poor activity for cleaving polyubiquitin chains that were linked through either Lys 63 or Lys 48 (Fig. 3d). PR-DUB thus specifically deubiquitinated H2Aub1 in nucleosomes in these assays.
Figure 3. Recombinant Drosophila and human PR-DUB deubiquitinate H2A in nucleosomes in vitro.
a, Cleavage of Ub-AMC by Calypso and Calypso–ASX(2–337) complexes. Reactions (n = 4) contained 25 pmol Ub-AMC and 10 pmol of the indicated protein (complex); release of AMC was monitored by fluorescence spectroscopy at 436 nm; error bars indicate ±s.d. a.u., arbitrary units. b, Mononucleosomes were reconstituted with recombinant Xenopus histone octamers and were unmodified (lane 1), monoubiquitinated at H2AK119 (lane 2) (see Methods) or monoubiquitinated at H2BK120 (lane 3) (see Methods). The material was analysed on a 4–12% polyacrylamide gradient gel and histones were visualized by Coomassie staining. c, Drosophila and human PR-DUB deubiquitinate H2Aub1 in nucleosomes. Xenopus mononucleosomes (15 pmol) containing 30 pmol of either H2Aub1 (top gels left and right) or H2Bub1 (bottom gels left and right) were incubated without (lanes 2 and 10) or with 30 pmol of the indicated Drosophila PR-DUB complexes (lanes 3–8) or human BAP1 or PR-DUB complex (lanes 11–16), respectively, and deubiquitination was monitored at indicated time points by western blot analysis with anti-H2A (top gels left and right) or anti-H2B (bottom gels left and right) antibody (5 pmol nucleosome per lane). Unmodified mononucleosomes (lanes 1 and 9) served as a control. Comparable results were obtained with Drosophila mononucleosomes containing H2Aub1 (Supplementary Fig. 4). PR-DUB containing full-length ASX(1–1668) also specifically deubiquitinated H2Aub1 but not H2Bub1 in nucleosomes (Supplementary Fig. 4). d, K48- or K63-linked hexameric polyubiquitin chains (160 ng; corresponding to maximally 17.5 pmol ubiquitin linkage bonds) were incubated for 40 min with 10 pmol of the indicated protein or protein complex under the same assay conditions as in c, followed by western blot analysis with an anti-ubiquitin antibody.
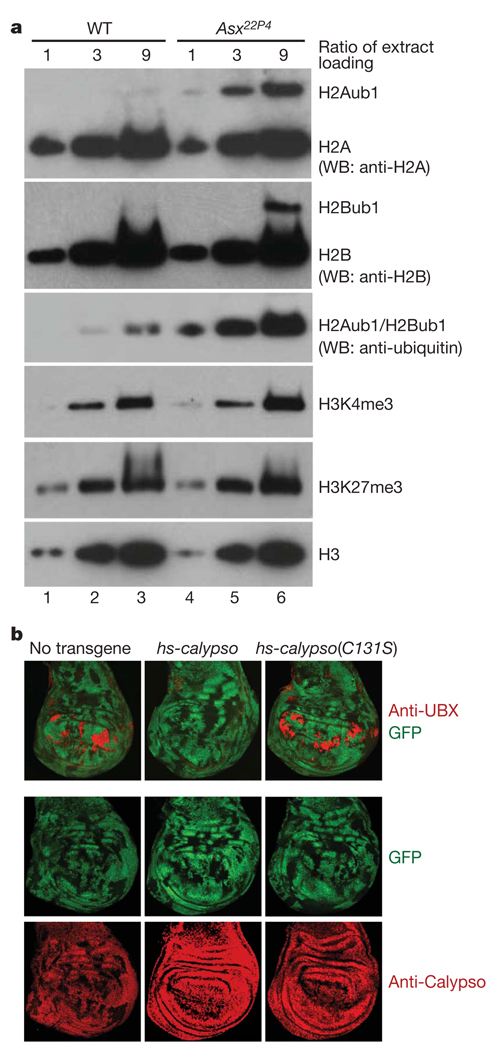
We next investigated how the lack of PR-DUB affects H2Aub1 levels in developing Drosophila. In embryos that are homozygous for Asx22P4, ASX protein is undetectable and Calypso protein levels are very drastically diminished (Supplementary Fig. 5). Asx22P4 mutant embryos thus have severely reduced levels of PR-DUB. We isolated bulk histones from wild-type and Asx22P4 homozygous embryos by acid extraction, and compared the levels of H2Aub1, H2Bub1, H3K27me3 and H4K4me3 in the two genotypes. Bulk H2Aub levels were almost tenfold increased in Asx22P4 mutant embryos (Fig. 4a). In contrast, the level of the PcG-specific histone tri-methylation mark H3K27me3 was comparable in Asx22P4 mutant and wild-type embryos (Fig. 4a). Unexpectedly, we also found a weak increase in H2Bub levels and a very slight concomitant increase in H3K4me3 levels (Fig. 4a). The higher H2Bub levels could be an indirect consequence of widespread global H2A ubiquitination, but it is also possible that, in vivo, PR-DUB deubiquitinates both H2A and H2B. Previous studies reported that the monoclonal antibody E6C5 specifically recognizes H2Aub1 in mammalian cells23, but we have not been able to specifically monitor H2Aub1 levels by ChIP in Drosophila using the commercially available E6C5 antibody (Supplementary Fig. 6 and Methods).
Figure 4. PR-DUB is required for H2A deubiquitination in Drosophila and its catalytic activity is essential for HOX gene repression.
a, PR-DUB is required for H2A deubiquitination in Drosophila embryos. Serial dilutions (1:3:9) of histone extracts from 16–18-h-old wild-type or Asx22P4 homozygous embryos were separated on 4–12% polyacrylamide gels and analysed by western blotting with the indicated antibodies. H2Aub1 levels in lanes 3 and 4 are comparable, suggesting that H2Aub1 levels are almost tenfold higher in Asx22P4 mutants than in wild type. H2Bub1 levels in Asx22P4 mutants are less than threefold increased compared to wild type (compare lane 3 with lanes 5 and 6). The band detected by an anti-ubiquitin antibody represents the combined signal of H2Aub1 and H2Bub1. H3K4me3 levels appear very slightly increased in Asx22P4 mutants. b, Calypso deubiquitinase activity is required for HOX gene repression. Wing imaginal discs with clones of calypso2 homozygous mutant cells from animals that carried no transgene or the indicated hsp70-calypso transgenes. calypso2 mutant cells are marked by the absence of GFP and discs were stained with antibodies against UBX or Calypso protein, as indicated. In all cases, clones were induced 96 h before analysis and larvae were repeatedly heat-shocked for 1 h every 12 h over a 96-h period to provide a continuous supply of Calypso protein from the transgene. In the absence of an hsp70-calypso transgene, Ubx is misexpressed in most calypso2 mutant clones in the pouch of the disc but remains repressed in the notum and hinge (left). Wild-type Calypso protein rescues repression of Ubx in mutant clones (middle), whereas the Calypso(C131S) protein fails to rescue (right), even though both transgene-encoded proteins are expressed at comparable levels and show nuclear localization like endogenous Calypso protein (bottom row).
Finally, we tested whether the deubiquitinase activity of PR-DUB is required for PcG repression. To this end we used a transgene rescue assay and asked whether the catalytically inactive Calypso(C131S) protein can repress the PcG target gene Ubx in Drosophila larvae, as follows. Clones of calypso2 mutant cells in larval imaginal discs lack detectable Calypso protein and fail to repress Ubx (Fig. 4b)8. However, a regular supply of wild-type Calypso protein from a heat-inducible hsp70-calypso transgene fully rescues repression of Ubx in such clones (Fig. 4b). In contrast, the catalytically inactive Calypso(C131S) protein expressed from a hsp70-calypso(C131S) transgene failed to rescue repression, and Ubx was misexpressed as in control animals lacking any hsp70-calypso transgene (Fig. 4b). PR-DUB deubiquitinase activity is thus critically required for repression of PcG target genes in Drosophila.
The following conclusions can be drawn from the work reported here: PR-DUB is a new PcG protein complex that comprises the Calypso and ASX proteins; PR-DUB is bound at the PREs of PcG target genes in Drosophila; reconstituted recombinant Drosophila or human PR-DUB deubiquitinate H2A in nucleosomes in vitro; Drosophila mutants lacking PR-DUB show an increase in global H2Aub1 levels; and a mutation in Calypso that disrupts H2A deubiquitinase activity in vitro impairs repression of HOX genes in Drosophila. Our analyses identified nucleosomal H2Aub1 as the preferred PR-DUB substrate; the complex failed to deubiquitinate nucleosomal H2Bub1 and showed only very poor activity for cleaving polyubiquitin chains. It is possible that PR-DUB also deubiquitinates other proteins but here we shall discuss its possible role in H2A deubiquitination. The observation that repression of PcG target genes in Drosophila requires not only the H2A ubiquitinase activity of PRC1 and dRAF but also PR-DUB may seem surprising. However, simultaneous depletion of Sce (that is, the H2A ubiquitinase subunit of PRC1 and dRAF20,21,24) and PR-DUB in embryos results in a more rapid loss of HOX gene repression and consequently more severe transformation of body segments than the depletion of Sce or PR-DUB alone (Supplementary Fig. 7). This suggests that appropriately balanced H2Aub1 levels in target gene chromatin may be critical for maintaining a Polycomb-repressed state. One possibility would be that PRC1/dRAF and PR-DUB act locally within target gene chromatin; the presence of H2Aub1 in some regions of a gene may be critical for repression but may be detrimental to it in others. Alternatively, H2A ubiquitination and deubiquitination may have to occur in a temporally regulated cycle to maintain repression, similar to what has been proposed for H2B ubiquitination and deubiquitination during transcriptional elongation25. Interestingly, calypso and Asx mutant embryos show not only derepression of HOX genes but also a partial loss of HOX gene expression in the central nervous system (Supplementary Fig. 8a). This loss of HOX gene expression seems to be restricted to the nervous system and we have not been able to detect a reduction of HOX gene expression in the embryonic epidermis or in imaginal disc cells (Supplementary Fig. 8b). Thus, even though PR-DUB is primarily required for repressing PcG target genes outside their expression domains, it might also be needed to fine-tune expression levels within these domains in certain tissues, perhaps by preventing repressive hyper-ubiquitination of H2A by PRC1 or dRAF complexes. It will be interesting to determine whether the mammalian complex has a similar prominent role in PcG repression during development and for maintenance of stem cell pluripotency, and to explore how the tumour suppressor activity of BAP1 (ref. 13) relates to the H2A deubiquitinase activity of human PR-DUB.
METHODS SUMMARY
Mapping and molecular cloning of the calypso gene
Detailed information can be found in the Methods.
ChIP assays and genome-wide Calypso and ASX profiling
X-ChIP in Drosophila imaginal disc cells, quantitative PCR (qPCR) analysis of immunoprecipitates and genome-wide profiling using Affymetrix whole genome tiling arrays were performed as described17,19.
Tandem-affinity purification of Calypso complexes
The α-tubulin1-TAP-calypso transgene fully rescued the viability and fertility of calypso2/Df(2R)Exel6063 animals. TAP was performed as described26.
Baculovirus expression of recombinant proteins
Flag-affinity purification of proteins was carried out as described26 with the only modification that protein complexes were reconstituted by infecting cells with individual viruses and then mixing cell lysates and incubating them for 2–3 h at 4 °C under mild agitation before Flag-affinity purification.
Recombinant mononucleosomes containing H2Aub1
Recombinant Drosophila and Xenopus octamers were assembled on a 5′ biotinylated 288-base-pair (bp) DNA fragment (601) by stepwise salt-dialysis. Nucleosomal H2A was ubiquitinated as described24. H2A-ubiquitinated nucleosomes were purified by binding to Streptavidin-coupled Dynabeads (Sigma), followed by washes and released by endonucleolytic cleavage with EcoRV. See the Methods for details.
Deubiquitination assays
Reactions were carried out at 25 °C in deubiquitination buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM ZnCl2, 1 mM dithiothreitol (DTT)) and were stopped by the addition of SDS sample loading buffer and incubation at 95 °C for 5 min.
Analysis of PR-DUB function in Drosophila
Induction of homozygous mutant cell clones in imaginal discs of Drosophila larvae, rescue experiments with hsp70-calypso transgenes and immunofluorescence staining were performed following previously established protocols27.
Generation of H2Bub1-containing mononucleosomes
Xenopus octamers containing H2Bub1 were generated as described28 and mononucleosomes were assembled as described earlier.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Supporting Material
02
Acknowledgements
We thank T. Sixma and G. Buchwald for the gift of proteins, H. W. Brock, R. E. Kingston, B. Korn and B. Turner for plasmids, baculoviruses and antibodies, V. Benes, J. de Graaf, S. Müller and A. Riddell for technical support, and W. Huber and J. Gagneur for discussions. T.W.M. is supported by NIH grant RC2CA148354. J.C.S., A.G.A.A., K.O., N.L.-H. and J.M. are supported by EMBL and by grants from the DFG.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions J.C.S., A.G.A.A., K.O., N.L.-H. and J.M. conceived the project, designed and carried out the experiments, discussed and interpreted the data and prepared the manuscript. R.K.M. synthesized H2Bub1 in the laboratory of T.W.M., S.F. performed the mass spectrometry analysis in the laboratory of M.W.
The authors declare no competing financial interests.
References
- 1.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 2.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Müller J, Verrijzer CP. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 5.Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Henderson IR, Dean C. Control of Arabidopsis flowering: the chill before the bloom. Development. 2004;131:3829–3838. doi: 10.1242/dev.01294. [DOI] [PubMed] [Google Scholar]
- 7.Jürgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985;316:153–155. [Google Scholar]
- 8.Gaytán de Ayala Alonso A, et al. A genetic screen identifies novel polycomb group genes in Drosophila. Genetics. 2007;176:2099–2108. doi: 10.1534/genetics.107.075739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijman SMB, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35:6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 11.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Jensen DE, et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 13.Ventii KH, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinclair DAR, et al. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development. 1998;125:1207–1216. doi: 10.1242/dev.125.7.1207. [DOI] [PubMed] [Google Scholar]
- 15.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 Å resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji H, Wong WH. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- 17.Oktaba K, et al. Dynamic regulation by Polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase Sxc/Ogt in Polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 19.Papp B, Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 21.Lagarou A, et al. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Vassilev AP, Rasmussen HH, Christensen EI, Nielsen S, Celis JE. The levels of ubiquitinated histone H2A are highly upregulated in transformed human cells: partial colocalization of uH2A clusters and PCNA/cyclin foci in a fraction of cells in S-phase. J. Cell Sci. 1995;108:1205–1215. doi: 10.1242/jcs.108.3.1205. [DOI] [PubMed] [Google Scholar]
- 24.Buchwald G, et al. Structure and E3-ligase activity of the Ring–Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry KW, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beuchle D, Struhl G, Müller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- 28.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Material
02