The Difficulty of Targeting Cancer Stem Cell Niches (original) (raw)
. Author manuscript; available in PMC: 2011 Sep 29.
Abstract
Normal stem cell niches typically are identified by their distinctive anatomical features and by association with tissue-specific stem cells. Identifying cancer stem cell (CSC) niches presents a special problem because there are few if any common anatomical features among tumors, and the physical phenotypes that reportedly describe the CSCs as entities may be subject to the host's microenvironment, sex, and tumor stage. Irrespective of a niche's location, the occupant's phenotype, or the precise molecular composition, all niches must do basically the same thing: maintain the activities in a stem cell that define it as such. Therefore, a potentially successful strategy, both for elaborating a molecular and cellular portrait of a CSC niche, and for therapeutically targeting them, is to identify components in the tumor microenvironment that are required for maintaining the functions of self-renewal, differentiation, and quiescence in the face of cytotoxic therapeutic regimens.
The cancer stem cell (CSC) hypothesis offers attractive explanations for generation of heterogeneity within tumors, metastatic dissemination, and resistance to therapy. The underlying logic is modeled on normal developmental hierarchies that are delineated for a number of adult tissues. Pools of undifferentiated stem cells give rise to less potent progenitors, which produce the most specialized cells of a given tissue. Only stem cells are thought capable of regenerating entire tissues in perpetuity. Analogously, only CSCs are thought capable of self-renewal, of initiating tumors at primary and distant locations, and of giving rise to more differentiated daughters that are incapable of reestablishing the tumor. Normal stem cell activity is maintained in niches; therefore, employing the same logic used for developmental hierarchies, niches that maintain CSCs, should also exist (see refs. 1–3 for additional reviews).
Niches are specialized microenvironments located within each tissue, wherein stem cells reside (Fig. 1; reviewed in refs. 4, 5). Microenvironments are defined as the sum total of cell-cell, -ECM, and -soluble factor interactions, and the physical states and geometric constraints that a cell may experience. Niche microenvironments can exert tremendous control over stem cell range of function. It was shown that progenitors both in skin and skeletal muscle could adopt residency in vacated stem cell niches, where they reacquired stem cell traits (6–8). Impressively, testis and neural stem cells from male mice were shown to give rise to lactating mammary glands when transplanted into the mammary fat pad (9, 10). Indeed, embryonic and adult stem and progenitor cell fate decisions are quantifiably flexible in response to combinatorial microenvironments (11–13). The ability of the niche to determine the functional spectrum of stem cell activities led us to hypothesize that stem cell niche microenvironments beget stem cell functions (14). Due to their role in maintaining stem cell activity, disrupting CSC-niche interactions may be crucial for overcoming barriers to therapeutic resistance.
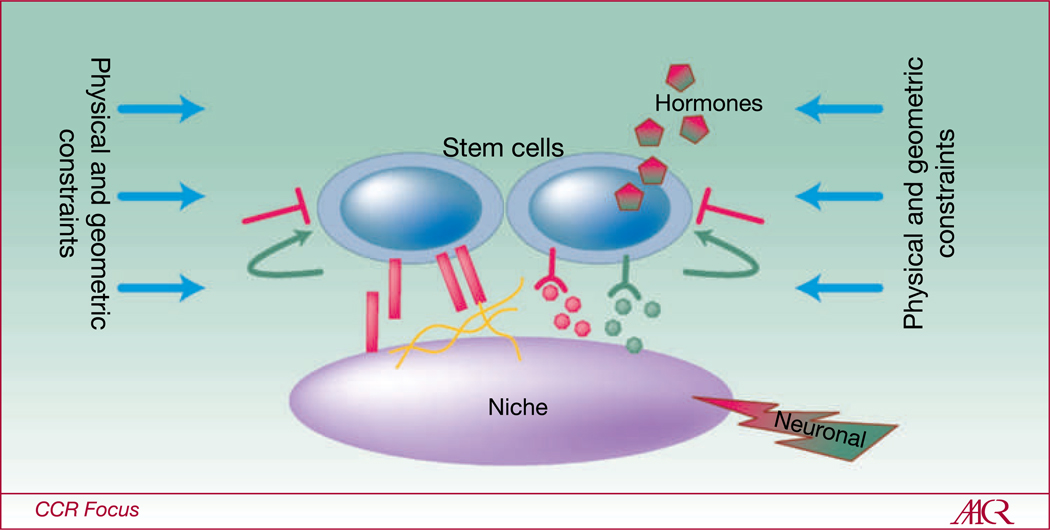
Fig. 1.
The basic concept of a stem cell niche. Niches are special microenvironments that maintain, or even endow cells with, stem cell activity. Cell-cell communication through adherens junctions, cell-ECM communication through integrins, paracrine, juxtacrine, and hormonal signaling, mechanical sensing of physical and geometric constraints, and integration of neural signals are all coordinated to enforce quiescence (red signals) and to tightly regulate self-renewal (green signals).
At least two possibilities exist for generating CSC niches: either they are manufactured as nascent domains by tumor cells, or CSCs usurp existing tissue-specific stem cell niches. Localization of nascent CSC niches is complicated because there are no common anatomical features among tumors (Fig. 2), and consensus is continually shifting about the identity of CSCs. On the other hand, usurped niches are most likely to be observed in early stages of cancer progression when there is some semblance of the original tissue's structure (e.g., ductal hyperplasias and possibly ductal carcinomas in situ in breast). In either scenario, the CSC niches and the CSC entities are linked like an amniotic sack and a fetus; one cannot exist without the other. Evidence suggests that the biochemical identity of CSCs is dependent on the host strain, sex, and tumor stage. Accordingly, the composition of CSC niches also will be dependent on host factors. Therefore, successful therapeutic strategies will target the functions that all CSC niches have in common: to mediate self-renewal and to maintain an undifferentiated state and CSC activity even in the presence of cytotoxic agents.
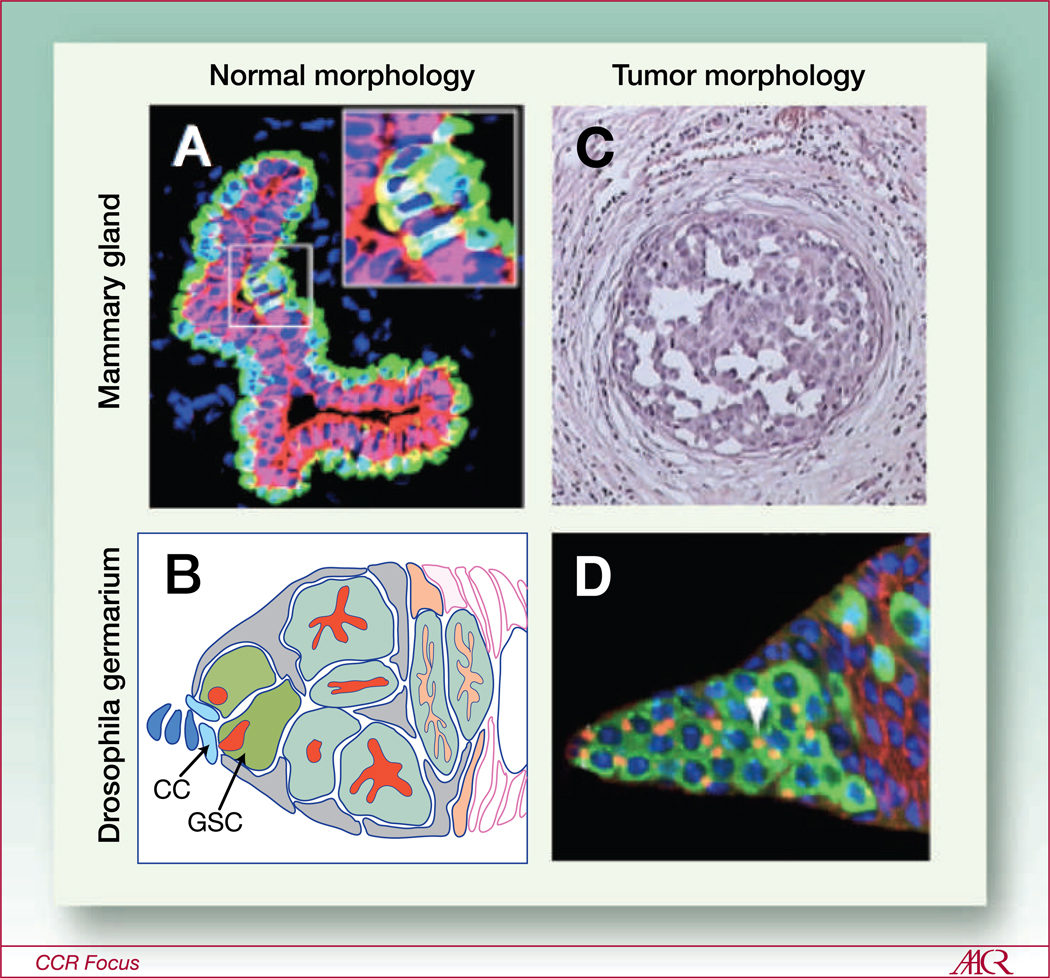
Fig. 2.
Loss of predictable tissue morphology in tumors presents challenges to identifying the CSC niche. A and B, stem cell niches are clearly identifiable in normal tissues. A, a cross-section of normal human mammary gland terminal duct shows the exquisite bilayered architecture of the tissue. The stem cells are nestled inside their suprabasal niche and express both keratins (K)14 and K19 (yellow, inset). They give rise to their differentiated progeny, the K14-expressing myoepithelial (green) and K19-expressing luminal cells. B, a depiction of the Drosophila germarium stem cell niche showing the stem cells (GSC, green) adjacent to the cap cells (CC, light blue). The stem cells give rise to the more differentiated progeny at their right. C and D, normal architecture is quickly lost in early stages of tumorigenesis. C, a cross section of human mammary duct, which is stricken with ductal carcinoma in situ, a noninvasive form of breast cancer that is thought to precede more invasive forms. D, shows a tumor laden germarial tip. The tumor in this case was derived from differentiation-defective bcgn mutant stem cells that outcompeted the normal stem cells for niche occupation due to upregulated E-cadherin expression, and subsequently filled the germarium with their mutant progeny. In both tumors, it is difficult or impossible to determine which of the tumor cells are the CSCs based on tissue morphology. A was adapted with permission from Villadsen et al. (71). B and D are adapted with permission from Jin et al. (1). C is reproduced from http://commons.wikimedia.org/wiki/File:DCIS.jpg
Have the Residents of CSC Niches Been Identified?
Unambiguous localization of the CSC niche will require characterization of the CSCs themselves. The basic algorithm used to identify and enrich for CSC activity is to first prepare malignant growths or tumors as single cell suspensions, which are divided into fractions by differential expression of marker proteins or by dye efflux ability. CSC activity is measured by introducing the fractions into living hosts, where their ability to reform the tumor is quantified. Frequencies of CSC within a population of tumor cells are calculated by doing limiting dilution studies, and the so called "gold standard" for identifying a stem cell, normal or cancer, is to do a single cell transplantation experiment. Since the first characterization of human leukemia initiating cells using severe combined immunodeficient (SCID) murine hosts (15), a number of reports have characterized CSCs that generate other heme malignancies (16–19) or solid tumors (20–29). Those studies either reported on murine tumor cells implanted into syngeneic inbred hosts, or human tumor cells xenografted into strains of SCID or nonobese diabetic (NOD)/SCID mice. With some notable exceptions (discussed below), the common trend suggested that CSC were rare within the total population, but were capable of self-renewal and were resistant to therapy. However, studies also have shown that the choice of host impacts the outcome of the experiment, and that host microenvironments will select for outgrowth of successful clones, suggesting that the host microenvironment will dictate the identity of the CSC.
Human melanomas xenografted for 8 weeks into SCID mice were reportedly generated from a CSC subpopulation that comprised ~ 1 of 1,000,000 of the tumor cells (24). A subsequent report similarly found that melanoma CSC occurred with a frequency of ~ 1 of 837,000 when xenografted in SCID mice for 8 weeks, but that the frequency increased to ~ 1 of 111,000 when measured after 32 weeks (30). Furthermore, 4,000 melanoma cells implanted into NOD/SCID mice and into NOD/SCID Il2rg−/− (NOG) mice, which are even more immunosuppressed hosts, showed that tumor formation was three fold more efficient in NOG mice. Indeed, 27% of random single melanoma cells were capable of forming tumors in the NOG strain (Table 1). The authors also showed that out of 50 distinct melanoma fractions, identified using antibodies recognizing previously identified CSC or melanocyte markers, none showed enrichment for tumor forming in NOG mice. Heterogeneity of expression with respect to one marker, CD133, was restored both from the CD133− and CD133+ fractions (30). In a different study, a similarly high frequency (~ 37%) of malignant engraftment was observed when single murine lymphoma cells were introduced into syngeneic hosts (31). These reports underscore that both time and host microenvironment are essential determinants of tumor formation in the context of tumor transplant assays.
Table 1.
Tumor formation from single cells
| Source tumor | Implant site | Host characteristics(Takes/Total animals) | Percent lethal takesfrom one cell | Citations |
|---|---|---|---|---|
| S2 Leukemia | Intravenous | Young mice (3 of 65) | 5 | Furth and Kahn (73)* |
| AKf5 | Intravenous | Young mice (2 of 32) | 6 | Furth and Kahn (73)* |
| Krebs2 carcinoma | Intraperitoneal | Adult mice (1 of 50) | 2 | Hauschka (32, 74, 75) |
| Krebs2 carcinoma | Intraperitoneal | Infant mice (10 of 62) | 16 | Hauschka (32, 74, 75) |
| Elrich near-tetraploid | Intraperitoneal | Adult mice (1 of 50) | 2 | Hauschka (32, 74, 75) |
| Elrich near-tetraploid | Intraperitoneal | Infant mice (7 of 50) | 14 | Hauschka (32, 74, 75) |
| Elrich near-tetraploid | Intraperitoneal | Infant mice (7 of 48) | 15 | Hauschka† |
| Elrich near-tetraploid | Intraperitoneal | Infant mice (13 of 53) | 25‡ | Hauschka† |
| Elrich near-tetraploid | Intraperitoneal | Infant mice (9 of 75) | 12 | Kaziwara† |
| Elrich near-tetraploid, clone 11 | Intraperitoneal | Infant mice (4 of 22) | 18 | Kaziwara† |
| Elrich hyperdiploid | Intraperitoneal | Infant mice (0 of 47) | 0 | Kaziwara† |
| Elrich hyperdiploid | Intraperitoneal | Infant mice (10 of 367) | 3 | Querner (76)* |
| Elrich hyperdiploid | Intraperitoneal | Infant mice (6 of 72) | 8 | Hansen-Melander† |
| 6C3HED lymphosarcoma | Intraperitoneal | Adult mice (0 of 24) | 0 | Hauschka (75) |
| S3A mammary carcinoma | Subcutaneous | Infant mice (3 of 44) | 7 | Klein (77) |
| Yoshida sarcoma | Intraperitoneal | Adult rats (1 of 3) | 33 | Ishibashi (78)* |
| Yoshida sarcoma | Intraperitoneal | Adult rats (69 of 150) | 46 | Hosokawa (79)* |
| Yoshida sarcoma | Subcutaneous | Adult rats (8 of 41) | 19 | Hosokawa§ |
| Hepatoma ascites | Intraperitoneal | Adult rats (0 of 15) | 0 | Hosokawa§ |
| Hepatoma ascites | Intraperitoneal | Adult rats (3 of 98) | 3 | Yoshida (80) |
| Hirosaki sarcoma | Intraperitoneal | Young rats (4 of 17) | 24 | Makino and Kano (81) |
| Melanoma from human | Subcutaneous | NOD/SCID Il2rg−/− (69 of 254) | 27‖ | Quintana et al (30) |
| Eμ-myc B Lymphoma | Intravenous | C57BL/6-Ly5.1+ (3 of 8) | 33‖ | Kelly et al (31) |
As detailed in studies done from 1937 to 1958, tumor formation from even one cell can be efficient in the contexts of particular hosts, and at particular developmental stages (Table 1). In 1958, Hauschka and Levan reported the results of a 5-year long study of tumor-forming efficiency from single cells of the Krebs-2 and Elrich Ascites tumors (32). The Krebs-2 tumor strain was derived from “either the skin or the mammary gland of a random-bred male mouse” and the notes pertaining to the exact origin of the Elrich strain were “lost in the war.” In a total of 212 infant and adult Swiss mice injected with single cells, 16% (Krebs) and 14% (Elrich) resulted in lethal tumors. Two clones derived from Krebs-2 cells were serially passaged 80 times through 564 Swiss mice (clone K2C) or passaged 70 times through 411 Swiss mice (clone K2D). Clone K2D generated a unimodal kill curve with a peak of activity between 5 to 7 days, whereas clone K2C generated a bimodal curve with peaks between 5 to 7 and 14 to 15 days. A second clone was isolated from K2C and it recapitulated the bimodal kill curve in Swiss mice, suggesting that one random cell was capable of making both types of tumors. Clone K2C then was used to form tumors in outbred Swiss agouti mice of three genotypes that differed by coat color (AA, Aa, and aa), and in eight different inbred strains. The bimodal kill curve was reproduced in the outbred Swiss strains and in one inbred strain (C57BL), whereas the other seven inbreds produced unimodal curves. The authors concluded the bimodal kill curves did not result from a “stem-line” origin, but that genetically heterogeneous agouti hosts selected for outgrowth of two clone types, whereas seven of the inbred strains selected only one. They proposed that the C57BL strain had some residual heterozygosity. Additionally, the authors noted that they originally preferred to use female mice due to their softer abdominal walls, which presumably pierced more easily with a glass pipette, but that tumor forming efficiency was only 2%, so the majority of their study was done in males. Because tumor forming was measured as the number of lethal “takes,” the variable of time could be omitted from the authors' interpretation, thus underscoring the importance of the host microenvironment in determining tumor-forming efficiency from single cells.
The examples above suggest that the methods commonly used to identify CSC activity are subject to a high degree of influence from tissue microenvironments, which differ according to time, host strain, and even sex. Therefore, any CSC phenotypes should be viewed as a product of the host strain used in the experiment. Using genetically heterogeneous outbred or wild-type strains of mice to identify CSC will generate more robust data, but may pose a problem for identifying human CSCs due to the need of immuno-compromised hosts. Outbred hosts will likely result in identifying more than one candidate CSC phenotype from the same tumor, as observed in Swiss agouti mice (32). However, those phenotypic differences may be used to advantage to identify common proteins essential to maintaining the functions that all CSC must have in common.
Are Niches and CSCs Tumor-Stage Specific?
In addition to maintaining the primary tumors, CSCs also are hypothesized to give rise to metastases. However, reports that identified different CSC subpopulations within primary and metastatic tumors, coupled with apparently stage-specific molecular requirements for progression, suggests that metastatic CSCs may be the children of de novo niches, which evolve during progression of the primary tumor. For instance, only the CD133hi fraction from human primary colon tumors, implanted subcutaneously (23) or under the kidney capsule (22), in NOD/SCID mice grew as xenografts. Whereas, both CD133hi and CD133lo fractions of human metastatic colon cancers isolated from livers and implanted subcutaneously in NOD/SCID mice formed tumors with similar efficiency (33). The tumors derived from the CD133lo fraction grew relatively faster, and those from the CD133hi fraction were the only ones to harbor both CD133+ and CD133− cells. Tumor cells from both subpopulations expressed human EpCam, which verified that the tumors were of epithelial origin (33). The discrepancy between the findings in primary and metastatic colon cancers may reflect experimental differences, but may also suggest that different cells are giving rise to the primary tumors and to the metastatic ones (reviewed in ref. 34).
Examination of the stage of tumor progression at which metastasis occurs, and the molecules that were involved, provided evidence for stage-specific CSCs. In a murine model of breast cancer, it was shown that the transcription factor GATA-3, a protein essential for differentiation into the luminal lineage (35, 36), was required by the tumor during the earliest hyperplastic stages of growth, but as the tumors progressed through early and late adenoma stages GATA-3 was silenced by promoter methylation (37). Metastases were observed only during the early and late adenoma stages when GATA-3 was lost, and GATA-3 expression was not detected at metastatic sites. Exogenous expression of GATA-3 in the early and late stage adenomas led to a reverted state reminiscent of early hyperplasia with no metastases, whereas knocking down GATA-3 in the early hyperplasia cells led to their death. Because there were distinctive stage-specific functional requirements for GATA-3, it seems unlikely that the same GATA-3+ CSC that established the primary tumor also established the GATA-3 null metastases. A second study also showed stage-specific requirements for growth of human and mouse breast cancer cell line-derived tumors in NOD/SCID and in syngenic immuno-competent mice, respectively. Growth of the primary tumors did not require the receptor tyrosine kinase Axl, but the protein was required for spontaneous metastasis (38). Moreover, Axl was highly expressed in patient metastases, but not by their primaries, and its high expression was found to be an independent predictor of recurrence. These reports serve as examples whereby the molecules GATA-3 and Axl are not merely markers that delineate potentially stage-specific CSCs in breast cancers, but they also play clear functional roles in the genesis of primary or metastatic tumors.
That different CSCs may give rise to primary and to metastatic tumors does not contradict the existence of CSCs per se, but does predict that the composition of metastatic and primary CSC niches will differ. Moreover, it suggests that either the original CSC evolves throughout tumor progression, or that there should be a mechanism to generate new metastatic CSCs de novo.
Microenvironments can impose specific cell fate decisions (12), thus a tumorigenic cell's location inside a tissue is a crucial determinant of its activity. Reacquisition of the stem cell phenotype has been observed in normal progenitors that populated vacant niches (6–8, 39); therefore, the possible consequence of CSCs usurping preexisting niches should be considered. In model systems of solid and heme malignancies, differentiation-defective stem cells were shown to be the etiological root of the diseases (16, 21, 40, 41). In Drosophila germaria, differentiation-defective hyperplastic stem cells outcompeted normal stem cells for occupancy of their niche (Fig. 2B and D; ref. 42). Those mutant stem cells exhibited upregulation of E-cadherin, an adherens junction receptor that mediates interaction between germinal stem cells and their niche. Malcom Steinberg's differential adhesion hypothesis (43) may explain why modulation of adherens proteins resulted in the reshuffling of CSC and normal stem cells for niche occupancy. He posited that different cell types self-organize into groups according to common levels of adherens junction proteins with an ultimate goal of reducing free-energy (43, 44). In the case of the germarium the mutant stem cells supplanted the normal by expressing relatively more E-cadherin, causing an energetically more favorable association with the niche. Germline mutations in E-cadherin are associated with diffuse gastric cancers, and cadherin expression is frequently misregulated in breast cancers (reviewed by Knudsen; ref. 45). Perhaps pathological modulation of cadherin expression can play a role in early disease stages by repositioning differentiation-defective CSCs into normal stem cell niches, where they receive self-renewal and survival signals (Fig. 2). Cadherin modulation may occur again in later tumor stages, at least in carcinomas, when tumor cells seem to lose epithelial characteristics (e.g., loss of E-cadherin) and acquire more mesenchymal ones.
Owing to genetic instability, the tumor microenvironment is a shifting landscape that may deleteriously impose stem-like phenotypes onto malignant cells. Normally, transforming growth factor β (TGF-β) is present in the mammary stroma in an inactive form, and is cleaved into its active form following exposure to radiation, wounding, and during tumorigenesis (46). Experiments with Rous Sarcoma virus-infected cells showed that active TGF-β at wound sites was necessary to realize the full malignant potential of a predisposed cell (47). Similarly, progression to frank neurfibromatosis required mutations both in the epithelial cells and in the nearby stromal cells, which disrupted TGF-β and receptor tyrosine kinase c-Kit regulation (48, 49). A CSC-like phenotype was reportedly induced in mammary epithelial cells exposed to active TGF-β, forcing them to undergo epithelial-to-mesenchymal transition (EMT; ref. 50), which included downregulation of E-cadherin. Induction of the stem cell-like program in nonmetastatic malignant mammary epithelial cells elicited invasive metastatic behavior (50). Other mechanisms that may drive evolution of cellular subtypes may include increased activity by proteases during tumorigenesis. Increased expression of matrix metalloproteinase-3, which is frequently observed in breast cancers, led to genomic instability and EMT in mammary epithelial cells (51, 52) and tumor formation in mice (53). Thus, a number of known tumor microenvironment components are sufficient to induce a CSC phenotype, and should be considered as potential CSC niche constituents.
Can the Elusive CSC Niche Be Therapeutically Targeted?
Because the biochemical identity of CSCs may shift as a tumor evolves, targeting specific entities for removal is unlikely to be effective. Instead, disrupting the functions that all stem cells have in common may yield better results. Accordingly, strategies that target the developmentally crucial Notch, Wnt, and Hedge Hog pathways are discussed in detail in this CCR Focus Series (see refs. 54–56, respectively), as well as strategies that target unchecked self-renewal (see ref. 57). For instance, activity in the Notch pathway is required for maintenance of mammary stem cell activity (12, 58), and a number of studies reported advantageous effects of down modulating the pathway in breast cancer models (59–62). When we impaired signaling in the Notch pathway in malignant mammary epithelial cells grown in three-dimensional culture, through use of gamma secretase inhibitors or of small hairpin RNAs directed against the four Notch receptors, a growth arrested polar phenotype was imposed (Fig. 3). Other candidate CSC niche constituents and their cognate receptors, which are not covered in detail in this series, include the ECM molecule, fibronectin, and the glucosaminoglycan, hyaluronic acid. Studies designed to understand a basis for therapeutic resistance suggested that those molecules facilitated a quiescent state in some cancer cells when they were under siege from chemotherapy. Antibodies against the fibronectin receptor VLA-4 (α4β1 integrin) prevented association of tumor cells with premetastatic niches (63), and reduced the incidence of minimal residual disease in a model of acute myelomonocytic leukemia (AML; ref. 64). Treatment of malignant breast cancer cells with fibronectin-(65) or β1 integrin-(66) blocking antibodies promoted phenotypic reversion to a polar, growth-arrested state in organotypic three-dimensional cultures. Finally, hyaluronic acid-rich substrates protect hematopoietic stem cells from the cytotoxic effects of 5′ fluoro-uracil (67), and antibodies against its receptor, CD44, reduced minimal residual disease in AML models (68). Thus ECM proteins and glucosaminoglycans that impose specific behaviors on stem cells also should be considered when formulating anti-CSC therapies.
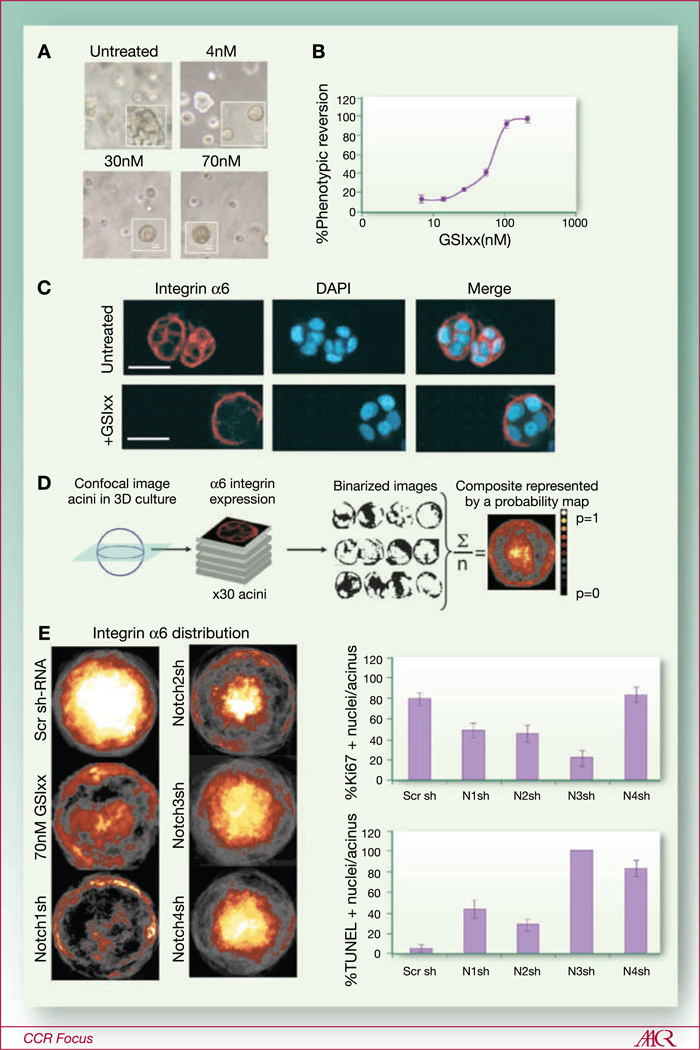
Fig. 3.
Modulating pathways involved in maintenance of the mammary stem cell state in malignant mammary epithelial cells may effectively halt disease progression. A, malignant mammary epithelial cells, HMT3522-T4-2 (T4-2), were embedded in Matrigel and grown for 72 hours in presence of the gamma secretase inhibitor (GSIxx), which blocks activation of the entire Notch pathway. Untreated cells formed disorganized and apolar colonies that do not growth-arrest, which appear characteristically rough under phase microscopy, whereas GSIxx treated cells growth-arrest after a few divisions and form smooth acini. B, GSIxx imposed the phenotypic reversion in a dose-dependent manner. C, representative confocal images of the hemispherical optical section from acini in three-dimensional culture. Basal polarity marker, integrin α6 (red), is expressed on all cell surfaces in colonies of the untreated malignant cells, whereas it was only expressed at the basal surfaces of the phenotypically reverted acini grown in presence of GSIxx. D, image analysis of three-dimensional cultures using NIH ImageJ was used to generate composite images, shown as probability of distribution heat maps, of integrin α6 distribution that occurs most frequently among the acini in a given three-dimensional culture condition. E, left side, Integrin α6 distribution maps are shown for cultures of GSIxx treated T4-2 cells, as well as cultures that were infected with retroviruses expressing small hairpin RNA (shRNA) to knock down each of the four Notch receptors (Notch1–4). E, right side, Bar graphs showing the impacts of each shRNA on apoptosis, via TUNEL stain, and proliferation, via Ki67 stain. Targeting different notch receptors elicited distinct phenotypes: Notch 1 and 2 shRNAs imposed normal polarity and had modest impact on apoptosis or proliferation, whereas Notch 3 and 4 shRNAs had little impact on restoring normal polarity and had a large impact on the apoptotic and proliferative phenotypes. (For more reading on the concept of phenotypic reversion see refs. 66, 72).
Summary
The presence of a CSC niche is dependent on the presence of a CSC. That tumors may be generated and maintained by CSCs, in a manner analogous to the developmental hierarchy of the hematopoietic system, is challenged by a number of reports that suggest the ability of any given subset of malignant cells to form tumors is determined by the host's microenvironment. Therefore, the biochemical identity of any putative CSC is likely to be host-specific to a large extent. Moreover, the ability of microenvironments to impose biochemical and functional phenotypes on cells may add additional complications, because tumors are constantly evolving by virtue of reciprocal and dynamic collaborations between patients' deteriorating genomes and twisting microenvironments (69, 70). Therefore, the cells that initiate tumor growth may differ from those involved in metastasis. However, it is clear that the functional abilities ascribed to stem cells are important to tumor progression, and it is suggested that those functions can be imposed upon malignant cells either by normal stem cell niches, or by tumor microenvironments. Accordingly, herein is presented a model whereby a malignant cell may first usurp preexisting tissue-specific stem cell niches, becoming the initial CSC that nurtures a tumor in its early stages. The evolved tumor microenvironment may then impose CSC-like functions onto other cells, which facilitate metastatic dissemination (Fig. 4). Targeting of the microenvironment molecules that can impose stem-like functions, or targeting of the signaling pathways in cells that mediate those functions, may represent worthwhile therapeutic paradigms.
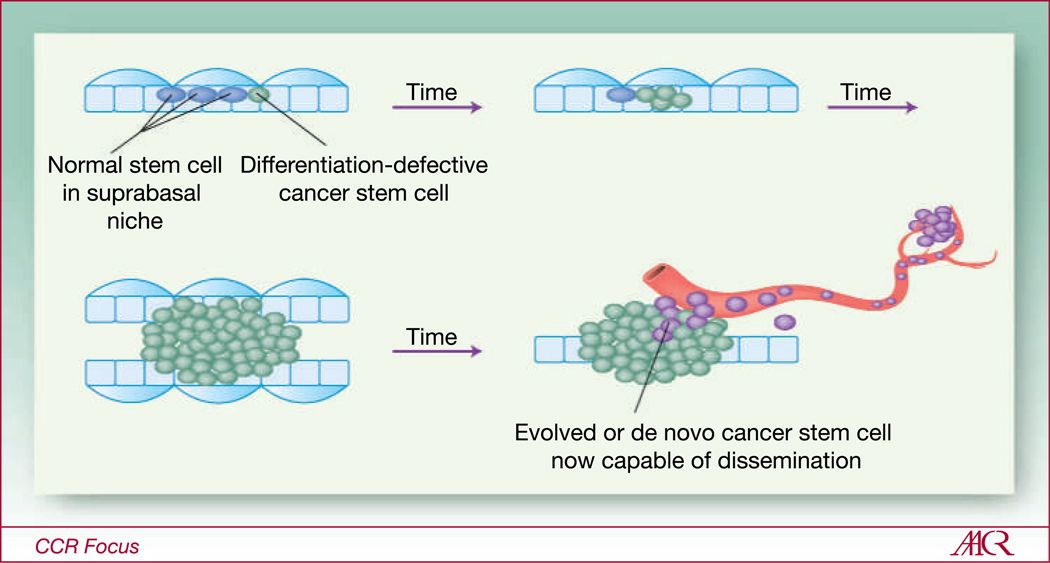
Fig. 4.
Do CSCs and their niches evolve in parallel with tumor progression? A depiction of a hypothetical bilayered, lumen-containing epithelial tissue (e.g., mammary gland). Normal stem cells (solid blue circles) are located inside their suprabasal niche, sandwiched between two differentiated cell types (represented by open rectangles and ovals). A differentiation defective CSC may outcompete the normal SCs for position within the niche. Because the CSCs are not bound by usual constraints on self-renewal, their mutant daughters begin to populate the tissue until normal organization and architecture are lost. Either the original CSC evolves, or the tumor microenvironment imposes stem cell-like properties onto random tumor cells to generate de novo CSCs, which accommodates tumor progression and eventual metastatic dissemination.
Acknowledgments
Thanks to Prof. George Klein for very useful discussions and correspondence, which led me to rediscover some oldies but goodies; they are real treasures.
Grant Support
M.A. LaBarge is supported by the National Institutes of Health (K99AG033176).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- 1.Sneddon JB, Werb Z. Location, location, location: the cancer stem cell niche. Cell Stem Cell. 2007;1:607–611. doi: 10.1016/j.stem.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang ZJ, Wechsler-Reya RJ. Hit 'em where they live: targeting the cancer stem cell niche. Cancer Cell. 2007;11:3–5. doi: 10.1016/j.ccr.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Flynn CM, Kaufman DS. Donor cell leukemia: insight into cancer stem cells and the stem cell niche. Blood. 2007;109:2688–2692. doi: 10.1182/blood-2006-07-021980. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 5.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 6.Collins CA, Olsen I, Zammit PS, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura EK, Jordan SA, Oshima H, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 8.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booth BW, Mack DL, Androutsellis-Theotokis A, McKay RD, Boulanger CA, Smith GH. The mammary microenvironment alters the differentiation repertoire of neural stem cells. Proc Natl Acad Sci U S A. 2008;105:14891–14896. doi: 10.1073/pnas.0803214105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulanger CA, Mack DL, Booth BW, Smith GH. Interaction with the mammary microenvironment redirects spermatogenic cell fate in vivo. Proc Natl Acad Sci U S A. 2007;104:3871–3876. doi: 10.1073/pnas.0611637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 12.LaBarge MA, Nelson CM, Villadsen R, et al. Human mammary progenitor cell fate decsions are products of interactions with combinatorial microenvironments. Integr Biol. 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaBarge MA, Petersen OW, Bissell MJ. Of microenvironments and mammary stem cells. Stem Cell Rev. 2007;3:137–146. doi: 10.1007/s12015-007-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 16.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 18.Hong D, Gupta R, Ancliff P, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 19.Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol. 2004;5:738–743. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- 20.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 22.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 23.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 24.Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 26.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 27.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Kruithof-de Julio M, Economides KD, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu L, Gibson P, Currle DS, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 32.Hauschka TS, Levan A. Cytologic and functional characterization of single cell clones isolated from the Krebs-2 and Ehrlich ascites tumors. J Natl Cancer Inst. 1958;21:77–135. [PubMed] [Google Scholar]
- 33.Shmelkov SV, Butler JM, Hooper AT, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaBarge MA, Bissell MJ. Is CD133 a marker of metastatic colon cancer stem cells? J Clin Invest. 2008;118:2021–2024. doi: 10.1172/JCI36046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 36.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kouros-Mehr H, Bechis SK, Slorach EM, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gjerdrum C, Tiron C, Høiby T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci U S A. 2010;107:1124–1129. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290:328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- 40.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 41.Yilmaz OH, Valdez R, Theisen BK, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 42.Jin Z, Kirilly D, Weng C, et al. Differentiation-defective stem cells out-compete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell. 2008;2:39–49. doi: 10.1016/j.stem.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinberg MS. Does differential adhesion govern self-assembly processes in histogenesis? Equilibrium configurations and the emergence of a hierarchy among populations of embryonic cells. J Exp Zool. 1970;173:395–433. doi: 10.1002/jez.1401730406. [DOI] [PubMed] [Google Scholar]
- 44.Steinberg MS. Differential adhesion in morphogenesis: a modern view. Curr Opin Genet Dev. 2007;17:281–286. doi: 10.1016/j.gde.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Knudsen KA, Wheelock MJ. Cadherins and the mammary gland. J Cell Biochem. 2005;95:488–496. doi: 10.1002/jcb.20419. [DOI] [PubMed] [Google Scholar]
- 46.Barcellos-Hoff MH, Derynck R, Tsang ML, Weatherbee JA. Transforming growth factor-β activation in irradiated murine mammary gland. J Clin Invest. 1994;93:892–899. doi: 10.1172/JCI117045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dolberg DS, Hollingsworth R, Hertle M, Bissell MJ. Wounding and its role in RSV-mediated tumor formation. Science. 1985;230:676–678. doi: 10.1126/science.2996144. [DOI] [PubMed] [Google Scholar]
- 48.Bhowmick NA, Chytil A, Plieth D, et al. TGF-β signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 49.Yang FC, Ingram DA, Chen S, et al. Nf1-dependent tumors require a microenvironment containing Nf1 ± - and c-kit-dependent bone marrow. Cell. 2008;135:437–448. doi: 10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radisky DC, Levy DD, Littlepage LE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sternlicht MD, Lochter A, Sympson CJ, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sternlicht MD, Bissell MJ, Werb Z. The matrix metalloproteinase stromelysin-1 acts as a natural mammary tumor promoter. Oncogene. 2000;19:1102–1113. doi: 10.1038/sj.onc.1203347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pannuti A, Foreman K, Rizzo P, et al. Targeting notch to target cancer stem cells. Clin Cancer Res. 2010;16:3141–3152. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi-Yanaga F, Kahn M. Targeting wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 56.Merchant A, Matsui W. Targeting hedgehog - a cancer stem cell pathway. Clin Cancer Res. 2010;16:3130–3140. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16:3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 58.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zang S, Chen F, Dai J, et al. RNAi-mediated knockdown of Notch-1 leads to cell growth inhibition and enhanced chemosensitivity in human breast cancer. Oncol Rep. 2010;23:893–899. doi: 10.3892/or_00000712. [DOI] [PubMed] [Google Scholar]
- 60.Efferson CL, Winkelmann CT, Ware C, et al. Downregulation of Notch pathway by a γ-secretase inhibitor attenuates AKT/mammalian target of rapamycin signaling and glucose uptake in an ERBB2 transgenic breast cancer model. Cancer Res. 2010;70:2476–2484. doi: 10.1158/0008-5472.CAN-09-3114. [DOI] [PubMed] [Google Scholar]
- 61.Rizzo P, Miao H, D'Souza G, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farnie G, Clarke RB. Mammary stem cells and breast cancer-role of Notch signalling. Stem Cell Rev. 2007;3:169–175. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- 63.Kaplan RN, Riba RD, Zacharoulis S, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsunaga T, Takemoto N, Sato T, et al. Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat Med. 2003;9:1158–1165. doi: 10.1038/nm909. [DOI] [PubMed] [Google Scholar]
- 65.Sandal T, Valyi-Nagy K, Spencer VA, Folberg R, Bissell MJ, Maniotis AJ. Epigenetic reversion of breast carcinoma phenotype is accompanied by changes in DNA sequestration as measured by AluI restriction enzyme. Am J Pathol. 2007;170:1739–1749. doi: 10.2353/ajpath.2007.060922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weaver VM, Petersen OW, Wang F, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matrosova VY, Orlovskaya IA, Serobyan N, Khaldoyanidi SK. Hyaluronic acid facilitates the recovery of hematopoiesis following 5-fluorouracil administration. Stem Cells. 2004;22:544–555. doi: 10.1634/stemcells.22-4-544. [DOI] [PubMed] [Google Scholar]
- 68.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 69.Bissell MJ. The differentiated state of normal and malignant cells or how to define a “normal” cell in culture. Int Rev Cytol. 1981;70:27–100. doi: 10.1016/s0074-7696(08)61130-4. [DOI] [PubMed] [Google Scholar]
- 70.Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 71.Villadsen R, Fridriksdottir AJ, Rønnov-Jessen L, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang F, Hansen RK, Radisky D, et al. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94:1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furth J, Kahn MC. The transmission of leukemia of mice with a single cell. Am J Cancer. 1937;31:276–282. [Google Scholar]
- 74.Hauschka TS. Methods of conditioning the graft in tumor transplantation. J Natl Cancer Inst. 1953;14:723–739. discussion 41–3. [PubMed] [Google Scholar]
- 75.Hauschka TS. Cell population studies on mouse ascites tumors. Trans N Y Acad Sci. 1953;16:64–73. doi: 10.1111/j.2164-0947.1953.tb01321.x. [DOI] [PubMed] [Google Scholar]
- 76.Querner H. Preparation and cytological properties of clones of Ehrlich's ascites tumor. Z Krebsforsch. 1955;60:307–315. [PubMed] [Google Scholar]
- 77.Klein E. Immediate transformation of solid into ascites tumors; studies on a mammary carcinoma of an inbred mouse strain. Exp Cell Res. 1955;8:213–225. doi: 10.1016/0014-4827(55)90057-4. [DOI] [PubMed] [Google Scholar]
- 78.Ishibashi K. Studies on the number of cells necessary for the transplantation of Yoshida sarcoma; transmission of the tumor with a single cell. Gann. 1950;41:1–14. [PubMed] [Google Scholar]
- 79.Hosokawa K. Further research on transplantation of Yoshida sarcoma with single cell and with cell-free tumor ascites. Gann. 1950;41:236–237. [PubMed] [Google Scholar]
- 80.Yoshida T. Contributions of the ascites hepatoma to the concept of malignancy of cancer. Ann N Y Acad Sci. 1956;63:852–881. doi: 10.1111/j.1749-6632.1956.tb50897.x. [DOI] [PubMed] [Google Scholar]
- 81.Makino S, Kano K. Cytological studies of tumors. XIV. Isolation of single-cell clones from a mixed-cell tumor of the rat. J Natl Cancer Inst. 1955;15:1165–1181. [PubMed] [Google Scholar]