Specific sequence determinants of miR-15/107 microRNA gene group targets (original) (raw)
Abstract
MicroRNAs (miRNAs) target mRNAs in human cells via complex mechanisms that are still incompletely understood. Using anti-Argonaute (anti-AGO) antibody co-immunoprecipitation, followed by microarray analyses and downstream bioinformatics, ‘RIP-Chip’ experiments enable direct analyses of miRNA targets. RIP-Chip studies (and parallel assessments of total input mRNA) were performed in cultured H4 cells after transfection with miRNAs corresponding to the miR-15/107 gene group (miR-103, miR-107, miR-16 and miR-195), and five control miRNAs. Three biological replicates were run for each condition with a total of 54 separate human Affymetrix Human Gene 1.0 ST array replicates. Computational analyses queried for determinants of miRNA:mRNA binding. The analyses support four major findings: (i) RIP-Chip studies correlated with total input mRNA profiling provides more comprehensive information than using either RIP-Chip or total mRNA profiling alone after miRNA transfections; (ii) new data confirm that miR-107 paralogs target coding sequence (CDS) of mRNA; (iii) biochemical and computational studies indicate that the 3′ portion of miRNAs plays a role in guiding miR-103/7 to the CDS of targets; and (iv) there are major sequence-specific targeting differences between miRNAs in terms of CDS versus 3′-untranslated region targeting, and stable AGO association versus mRNA knockdown. Future studies should take this important miRNA-to-miRNA variability into account.
INTRODUCTION
MicroRNAs (miRNAs) are non-coding regulatory RNA comprising ~22 nt. In all known animals, miRNAs guide Argonaute (AGO)-containing microribonucleoprotein (miRNP) complexes to target mRNAs (1). Important questions remain about how miRNAs’ nucleotide sequences relate to their biological functions. A handful of miRNAs are entirely conserved throughout all metazoan species and dozens of different miRNAs are conserved among vertebrates (2), which indicate that the entire length of these miRNAs must be serving important functions. However, the mechanisms for miRNA:mRNA binding are complex and incompletely understood. The importance of the miRNA ‘5′ seed’—nucleotides 2–7 from the 5′-end of the miRNA—in terms of mRNA targeting has been sustained consistently (3). Also, the 3′ untranslated region (3′-UTR) of mRNAs has been shown to be an important context for targeting by some miRNAs (4). However, it is mistaken to assume that every miRNA’s target repertoire is characterized by a 5′ seed of a miRNA interacting with target mRNA 3′-UTR (5–7).
A promising method for directly characterizing miRNPs is co-immunoprecipitation (co-IP) that pulls down AGO proteins along with associated molecules (8–10). Using AGO co-IP assays, researchers have isolated multiple proteins, miRNAs and mRNA targets from miRNPs (11–17). A subset of AGO co-IP experiments involve ‘RIP-Chip’ techniques (18,19) that integrate miRNP co-IP with downstream high-density microarrays. This assay enables high-throughput assessment of AGO-associated mRNA for the systematic study of target mRNAs.
RIP-Chip assays were used previously to study why particular mRNAs get recruited into miRNPs after miRNA transfections. The monoclonal antibody (‘anti-AGO’, which was also termed ‘2A8’) was raised against human AGO2 (product of the gene EIF2C2), and recognizes a C-terminal AGO2 epitope (20). Applying RIP-Chip to H4 glioneuronal cell line (21) enabled the evaluation of some of the sequence determinants of miRNA:mRNA interactions (19). Experimentally validated miR-107 targets, unlike targets of some other miRNAs, tend to have sequences complementary to miR-107 in the target mRNAs’ open reading frame/coding sequence (CDS), rather than the 3′-UTR (10,19,22). MiR-107 is a member of a group of genes that are highly expressed in virtually all known mammalian cells (23) and prior studies did not evaluate other members of this miRNA group systematically. Nor is it known whether the 3′ portion of miR-107 plays a role in guiding the miRNA to experimental-verified mRNA targets.
Here, we performed a series of new experiments studying the miR-15/107 group of miRNAs, attempting to better understand the implications of both 5′ and 3′ portions of miRNA in terms of mRNA targeting. We analyzed both the putative target mRNAs that are associated with AGO in the RIP-Chip, and also those mRNAs downregulated following miRNA transfections. These experiments have provided further data about miRNA:mRNA functions and targeting mechanisms.
MATERIALS AND METHODS
Transfections, Co-IP of miRNPs using anti-AGO antibodies
The Anti-AGO RIP-Chip methodology has been validated and described in detail (19). Transfections with ‘Pre-miR’ reagents (Ambion) were performed exactly as previously described in detail (24). RNA was extracted using Trizol LS (Invitrogen, Carlsbad, CA, USA) as described (25).
Transfections with miR-107 duplex RNA
Hsa-miR-107 guide and passenger strands sequences from mirBase (26) and the mature sequences were synthesized (Integrated DNA Technologies). For annealing, equimolar concentrations of the guide and passenger strands (100 pmol/μl) were mixed in annealing buffer, heated to 75°C for 5 min, and cooled at 37°C for 1 h and then stored at −20°C until needed. Transfections were performed identically as described above with the Ambion siRNA-like reagents. RT–qPCR reactions for BACE1 and GRN mRNA were performed exactly as described previously (24).
Microarray analysis and RT–qPCR and downstream data analyses
Microarray analysis of RNAs isolated from co-IP or from total lysates were performed using Affymetrix Human Gene 1.0 ST™ chip at the University of Kentucky Microarray Core Facility as described previously (19,22). Three biological replicates were carried out in each treatment. Similarity matrix data were prepared using the Partek Genomics Suite. Other figures show results of Log2 microarray values. Criteria for selecting PmiTs according to anti-AGO RIP-Chip data (based on the averaged results of the three biological replicates on the array data for each transfection condition) were as follows: (i) the mRNA was enriched in the AGO-miRNPs after the cells were transfected with a particular miRNA, relative to the negative control miRNA; and (ii) the same mRNA was not upregulated >2-fold in the lysate after transfection with the miRNA.
Following the identification of PmiT-AGOs using these criteria, the 5′-UTR, CDS, and 3′-UTR sequences were analyzed for 6-mer sequences correlating to portions of the miRNAs (in anti-sense and sense orientation), and compared to the control sequences as follows.
For each PmiT, the 5′-UTR, CDS and 3′-UTR sequence regions were analyzed separately. For each sequence region of a PmiT, 200 control sequences from non-PmiTs were selected based on sequence length. Specifically, the selected sequences (5′-UTR, CDS or 3′-UTR from non-PmiTs) had the closest length match to the corresponding sequence regions from the PmiT. Then from this pool of 200 sequences, 20 were randomly selected as negative controls for the PmiT in consideration. Thus, for 100 PmiTs, there were 2000 combined control sequences for 5′-UTR, CDS or 3′-UTR, respectively.
Identification of sequence motifs in PmiTs corresponding to the 3′ portion of miRNAs
We analyzed miR-107/103 and two mutated miRNAs derived from miR-107 (MUT1 and MUT2) to identify potentially enriched words in PmiTs corresponding to the 3′ portion of these miRNAs. In this analysis, putative transcript targets, as identified in either the anti-AGO co-IP or lysate fraction, were included to identify pentamer words that match to any pentamer sequence from the last 12 nt of the cognate miRNA. To reduce the background noise of unrelated pentamer matches from the whole-target transcript sequence, the matching pentamer words were required to reside within the putative target site, defined as 2–30-nt downstream of the predicted seed-binding site. For each miRNA, the number of target transcripts with a matching pentamer in the target site was identified and listed in Tables 2 or 3. Simulation tests were performed to evaluate the statistical significance of the observed motif enrichment. A separate simulation test was performed for each miRNA. In each simulation test, 10 000 runs were performed to identify pentamer words within the putative target sites that matched to a randomized 3′-end sequence of the cognate miRNA. The _P_-value was calculated by counting the percentage of simulation runs in which at least a threshold number of target transcripts had matching pentamers to the randomized 3′-end sequence.
Table 2.
Enrichment of the anti-sense miRNA 3′ portion sequences in the CDS of putative target transcripts
| IP fraction (PmiT-AGO) | Lysate fraction (PmiT-Lys) | |||||||
|---|---|---|---|---|---|---|---|---|
| miR-107 | MUT1 | MUT2 | miR-103 | miR-107 | MUT1 | MUT2 | miR-103 | |
| Number of genes | 21 | 6 | 13 | 25 | 10 | 5 | 8 | 11 |
| _P_-value | 0.04 | 0.24 | 0.27 | 0.09 | 0.22 | 0.26 | 0.50 | 0.16 |
Table 3.
Characteristics of the anti-sense miRNA 3′ sequence AGCCCUGU in miR-107 PmiT-AGO transcripts
| Number of occurrence | _P_-value | |
|---|---|---|
| AGCCC | 18 | 0.010 |
| CCCUG | 16 | 0.021 |
| GCCCU | 14 | 0.032 |
| CCUGU | 11 | 0.074 |
| One of the four | 33 | 0.008 |
RESULTS
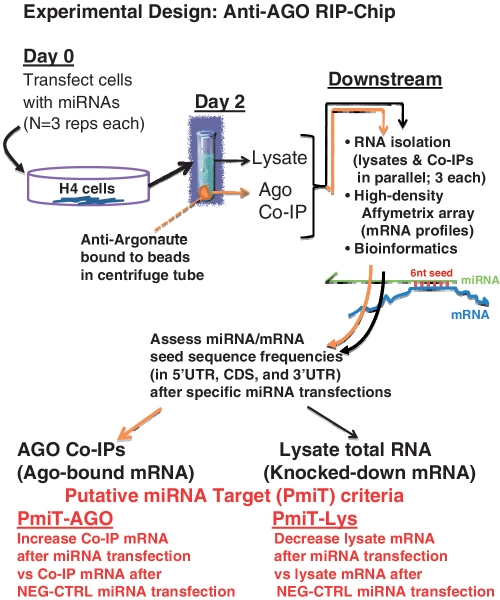
The study design is indicated schematically in Figure 1. The putative miRNA targets (PmiTs) were experimentally identified separately for mRNAs recruited to the Anti-AGO co-IP following miRNA transfections (PmiT-AGO) and mRNAs downregulated in the cell lysate following miRNA transfection (PmiT-Lys). Transfected miRNAs are shown in Figure 2. In this study, the ‘mRNA levels’ indicate the mean signal intensities read from the Affymetrix Human Gene 1.0 ST™ chip for each transcript (mean results from three different biological replicates each). After transfection with a specific miRNA, the designated PmiTs had increased mRNAs in the Anti-AGO RIP-Chip (PmiT-AGO), or decreased mRNA levels in the total RNA from cell lysates (PmiT-Lys), relative to the same mRNA levels following transfections with a negative control miRNA (NEG-CTRL, also transfected with three biological replicates, Figure 2). The data for PmiTs are presented in Supplementary Table S1.
Figure 1.
Experimental design of miRNA transfection experiments followed by anti-AGO co-immunoprecipitation (anti-AGO Co-IP) and microarray profiling of mRNAs from the anti-AGO Co-IP and lysates. Downstream bioinformatics allowed comparison between the mRNA ‘targets’ seen in the RIP-Chip experiments, relative to the mRNAs ‘knocked down’ in the lysates following miRNA transfections.
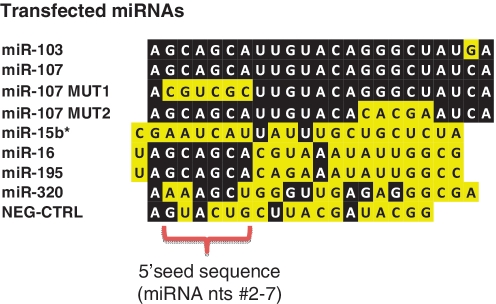
Figure 2.
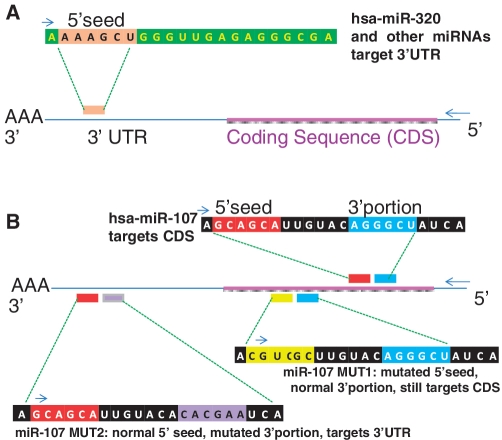
List of miRNAs transfected in the current study. The 5′ seed sequence (nucleotides 2–7) of the miRNAs is indicated. Note that there are three non-physiological controls—miR-107 MUT1, miR-107 MUT2 and NEG-CTRL. miR-15b* is an anti-sense control miRNA. Note that four members of the miR-15/107 gene family of miRNAs (miR-16, miR-195, miR-103 and miR-107) have similar 5′-seed sequences.
The microarray data indicated consistent results following miRNA transfections, both in the anti-AGO co-IP and in the cell lysates. A similarity matrix for the raw data analyzing the anti-AGO RIP-Chip microarray results are analyzed with a similarity matrix as shown in Supplementary Figure S1. Only a single sample, one of the control transfections (miR-15 b*) had a relatively low degree of similarity with the other samples. These data indicate a relatively high degree of correlation for all biological replicates, presumably indicating that most mRNAs are not miRNA targets for a given miRNA. Microarray data from the total lysate mRNAs had comparable between-replicate findings (data not shown). For complete data from all the experiments, see Supplementary Figure S1.
The data quality can be assessed further by analyzing the putative target (PmiT-AGO) mRNAs. The top 100 PmiT-AGO mRNAs for miR-107 and miR-103 are compared in Supplementary Figure S2. The potency of individual PmiT-AGO targets is judged based on the mean ratio of the mRNA signal on the microarray from anti-AGO RIP-Chip following transfection with each miRNA, to the signal for the same mRNA following transfection with the NEG-CTRL (Figure 2). The top 100 PmiT-AGO mRNAs of miR-103 and miR-107 were very similar (Supplementary Figure S2; _R_2 correlation coefficient is 0.769). In contrast, there is no similarity of top 100 PmiTs among those mRNAs enriched in the miRNP following miR-320 transfections (_R_2 < 0.001).
Another assessment of data quality was to ascertain whether the miR-107 results in these new experiments replicated prior studies (19,22). Previously, miR-107 transfection caused both BACE1 mRNA and GRN mRNA to be downregulated in cell lysates. However, of these two transcripts, only GRN mRNA was enriched in the anti-AGO co-IP after miR-107 transfection. Results in the current dataset are shown in Supplementary Figure S3. These data show the same findings as described previously. A potential confounder in both this and prior studies from our laboratory is that we use the Ambion ‘Precursor’ reagents. These RNA complexes are actually similar to an siRNA (perfect duplex) and raise the possibility that our data are epiphenomena related to the perfect (non-physiological) duplex conformation. To control for this, we performed an additional experiment with an imperfectly paired duplex that more faithfully recapitulates the physiological mature miR-107 duplex (Supplementary Figure S4). We then performed anti-AGO co-IP and then RT–qPCR for both BACE1 and GRN mRNA. As with the perfectly paired siRNA-like miRNA transfection reagents, the more physiological mature miRNAs had the same pattern with abundant enrichment for GRN, but not BACE1 mRNA in the anti-AGO RIP (Supplementary Figure S4D). An additional list is provided of the 20 mRNAs that are most knocked down in cell lysate following miR-107 transfection, but are not enriched in anti-AGO co-IP, as with BACE1 (Supplementary Table S2).
We next tested whether a sequence corresponding to the ‘5′ seed’ (Figure 2) for each transfected miRNA is enriched in the 5′-UTR, CDS and 3′-UTR from the miRNAs’ target mRNAs. The top 100 PmiT-AGOs of each miRNA were analyzed for miRNA seed sequences in 5′-UTR, CDS and 3′-UTR. The seed densities (number of anti-sense 5′ seed sequences per 1000 nt) for the top 100 PmiT-AGOs—in 5′-UTR, CDS and 3′-UTR—were compared to a control group of 2000 length-matched randomized non-PmiT mRNAs (see ‘Materials and Methods’ section for control sequence selection). The same comparative analysis was repeated 10 times by choosing different sets of length-matched randomized control sequences, and the results from the ten rounds were averaged and presented in Table 1. In addition, more details regarding sequence length and seed count of the PmiT-AGOs are summarized in Supplementary Table S3.
Table 1.
Presence of miRNA seed complementary sequence in the top 100 PmiT-AGO targets relative to controls
| miR-103 | miR-107 | miR-16 | miR-195 | miR-320 | MUT1 | MUT2 | miR-15b* | |
|---|---|---|---|---|---|---|---|---|
| Seed anti-sense | UGCUGC | UGCUGC | GCUGCU | GCUGCU | AGCUUU | GCGACG | UGCUGC | UGAUUC |
| 5′-UTR | 1.38 ± 0.10 | 0.57 ± 0.04 | 1.01 ± 0.06 | 0.85 ± 0.05 | 0.82 ± 0.06 | 2.58 ± 0.16 | 0.91 ± 0.07 | 0.90 ± 0.13 |
| CDS | 2.65 + 0.07 | 1.94 ± 0.04 | 1.38 ± 0.04 | 1.67 ± 0.05 | 1.31 ± 0.05 | 3.44 + 0.09 | 1.41 ± 0.03 | 1.76 ± 0.04 |
| 3′-UTR | 1.41 ± 0.05 | 1.20 ± 0.04 | 1.09 ± 0.04 | 1.18 ± 0.04 | 1.23 ± 0.03 | 1.42 ± 0.25 | 1.89 ± 0.06 | 1.63 ± 0.35 |
| Ratio CDS:3′-UTR | 1.88 | 1.62 | 1.26 | 1.41 | 1.07 | 2.43 | 0.75 | 1.08 |
Table 1 shows that among PmiT-AGO mRNAs, there is consistent enrichment of 6-mer sequences complementary to a transfected miRNAs’ 5′ seed portion, and these sequences tended to be present in the CDS and 3′-UTR. Intriguingly, the degree of enrichment in the CDS and 3′-UTR seems to be different for particular transfected miRNAs. For example, for both miR-103 and miR-107, the ratio of CDS enrichment to 3′-UTR enrichment (in terms of nucleotides 2–7 seed sequences/1000 nt) is above 1.6. In contrast, for miR-320, the ratio of CDS enrichment to 3′-UTR enrichment is 1.07. The difference was startling for the miR-107 mutant (non-physiological) miRNAs—the ratio of CDS enrichment: 3′-UTR enrichment was 2.43 for MUT1 (mutated 5′ seed), but only 0.75 for MUT2 (mutated 3′ portion).
The data in Table 1 indicate that the presence or absence of the 3′ portion of miR-107 altered the tendency of the miRNA to target the CDS versus the 3′-UTR. To convey the data more specifically, the 15 strongest AGO-enriched PmiTs of miR-107, miR-107 MUT1 and miR-107 MUT2 were further analyzed. Shown in Figure 3 are the number of 5′ seed (anti-sense) sequences for miR-107, MUT-1 and MUT-2 miRNAs, in the target mRNAs’ 5′-UTR, CDS and 3′-UTR. Note that as expected from Table 1, there are many miR-107 seed anti-sense sequences (UGCUGC) in the CDS of the strongest miR-107 PmiT-AGO targets. However, seed sequences were not enriched in the CDS of miR-107MUT2 targets; miR-107MUT2 has the same seed sequence as miR-107 but a mutated 3′ portion of the miRNA. In miR-107MUT-2, there are new mRNAs recruited into the anti-AGO RIP-Chip such as ASB1, KIAA0247 and FGFRL1 which have many miR-107 seed sequences in the 3′-UTR rather than the CDS. In contrast, for miR-107MUT1, which has a different seed sequence yet shares the same 3′ portion with miR-107, 8 of the top 15 PmiT-AGOs have MUT-1 seed sequences enriched in the CDS, and none in the 3′-UTR. This detailed analysis provides specific examples of how altering the 3′ portion of miR-107 leads to a change in the normal tendency of miR-107 to target mRNAs’ CDS.
Figure 3.
Top 15 targets (according to anti-AGO co-IP experiments) for miR-107, miR-107 MUT1 (5′ portion mutated) and miR-107 MUT2 (3′ portion mutated). Note that the miR-107 and miR-107 MUT2 have the same 5′-seed sequence (GCAGCA which is complementary to UGCUGC on the mRNA), but miR-107 MUT1 has a different 5′ seed (CGUCGC). The number of those seed sequences in the 5′-UTR, CDS/open reading frame and 3′-UTR for each of the targets is shown. Note that for the top 15 miR-107 targets, miR-107 seed sequences tend to be found in the CDS. For miR-107 MUT1, which has a completely different 5′-seed sequence, that seed sequence is found in the CDS of 8/15 top 15 targets (versus 0/15 for top 15 miR-107 targets). In contrast, miR-107 MUT2 has the same 5′-seed sequence as miR-107, but there is a remarkable tendency among the top 15 targets of miR-107 MUT2 to include mRNAs with the miR-107 seed sequence in the 3′-UTR instead of the CDS. These data appear to indicate that the 3′ portion of miR-107 may be directing the miRNA to target CDS rather than the 3′-UTR.
Whereas Table 1 and Figure 3 focus on the strongest PmiT-AGO mRNAs, we sought to better understand the tendency of miRNA seed sequences to be present in less strong targets, both for PmiT-AGOs and for PmiT-Lys mRNAs. We previously described RIP-Chip assays related to miR-107 (22), but the data presented here (including the miR-107 results) are the results of entirely different experiments. New analyses (Figures 5–7) show data for miR-103 and miR-320, a control physiological miRNA. Complete data referent to all the RIP-Chip and lysate RNA microarray results (a total of 54 microarrays) can be found in Supplementary Figure S1.
Figure 5.
Enrichment of 5′ miRNA seed sequences for miR-103 in 5′-UTR, CDS or 3′-UTR does not correlate with decreased mRNA in the lysate. Across all the evaluated mRNA, there is no evidence that for miR-103 (or miR-107, data not shown) that mRNAs ‘knocked down’ in the lysate after miRNA transfection correlate to those mRNAs with 5′-seed sequences enriched in 5′-UTR, CDS or 3′-UTR. In this case, the mRNAs are ordered (_x_-axis) based on the degree of decrease in the lysate following miR-103 transfection.
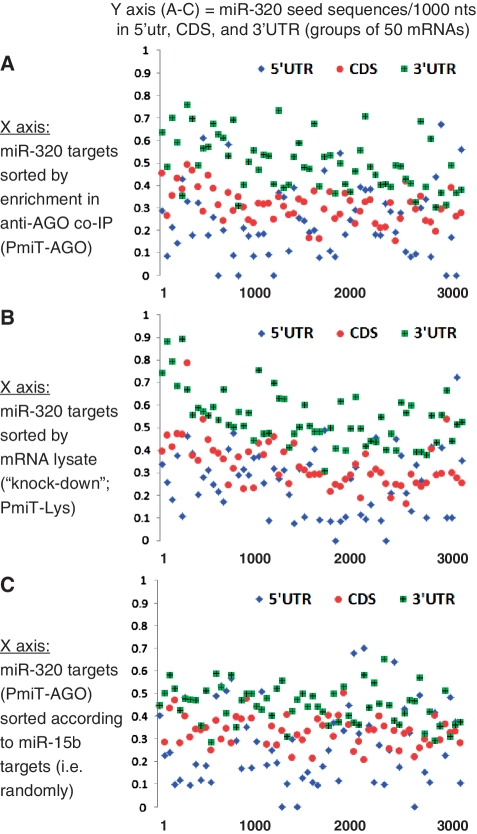
Figure 6.
Enrichment of 5′ miRNA seed sequences for miR-320 in the 3′-UTR of targets correlates with decreased mRNA levels for those targets in the cell lysates. For miR-320, unlike miR-103 or miR-107, there is robust evidence that the seed sequences in the 3′-UTR (as opposed to in the CDS) play a strong role in determining the miRNA/mRNA targeting. Also unlike miR-103 and miR-107, miR-320 transfections appear to result in target knockdown. Note that the seed sequences that are disproportionately found in the Anti-AGO Co-IP enriched miRNAs (A) and also the mRNAs knocked down in the lysate after miR-320 transfection (B), correspond to seed sequences in the 3′-UTR (green triangles). However, if sorted randomly, (C, where mRNAs are sorted according to the enrichment following miR-15b* transfection) the same tendencies are not seen. Thus, there is specific 3′-UTR enrichment of miR-320 complementary 5′ seeds that correspond specifically to those mRNAs that are most enriched in the Anti-AGO Co-IP and also the mRNAs that are most decreased in the lysate after miR-320 transfection.
Figure 7.
Depiction of some of the novel findings of the current study. (A) miR-320, like other miRNAs that we have evaluated previously (19,22), preferentially recognizes 3′-UTR sequences correlating to the miRNA 5′-seed sequence, and is more likely to induce decreased levels of mRNA targets in the lysates of cells transfected with miR-320. (B) In contrast, miR-107 targets the open reading frame/CDS of mRNAs leading to stable association between AGO and target mRNAs. The main determinant of target binding is the 5′-seed region of miR-107 (red rectangle). However, a miR-107 mutant (MUT-2) with altered 3′ portion (purple rectangle) targets the 3′-UTR sequences. A different miR-107 mutant (MUT-1) with altered 5′-seed portion still targets the CDS, but it targets sequences corresponding to the new 5′-seed portion (yellow rectangle). These data indicate the importance of the 5′-seed sequences in determining the particular mRNAs to target, but also indicate that the 3′ portion of miRNAs may help to select whether the miRNA targets the CDS or 3′-UTR.
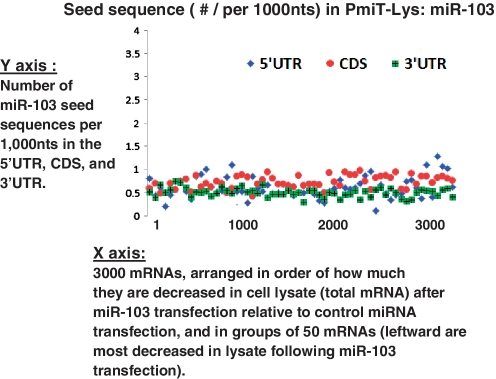
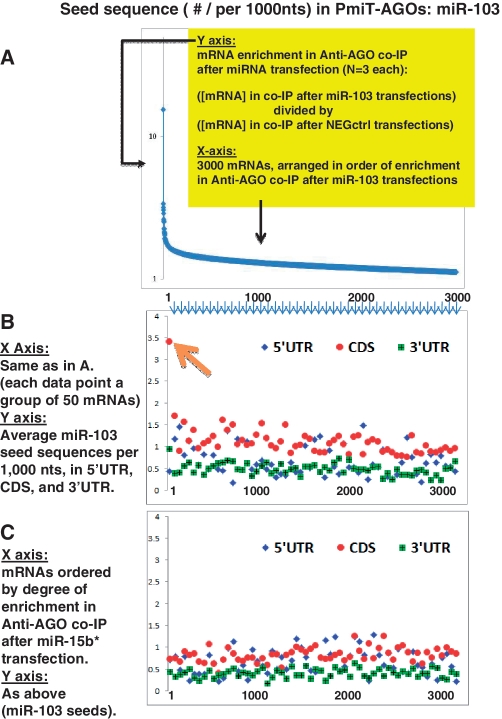
In a manner almost identical to miR-107, miR-103 PmiT-AGO mRNAs show increased density of 6-mer sequences corresponding to the miRNA nucleotides 2–7 in the CDS, but not in the 3′-UTR (Figure 4). There is also a tendency for other members of miR15/107 group to have enriched seeds in the CDS among PmiT-AGO mRNA targets (Table 1 and Supplementary Figure S5). In addition to stable association with AGO proteins, it is also known that many target mRNA are degraded following miRNA targeting (27). Enrichment was not seen in seed sequences among miR-103 (Figure 5) or miR-107 (data not shown) knockdown mRNAs (PmiT-Lys mRNAs) in total lysate. In contrast, miR-320 PmiT-Lys shows a tendency of seed sequence enrichment in 3′-UTR (Figure 6). In other words, unlike for miR-107, miR-320 tends to induce ‘knockdown’ of mRNAs that harbor miR-320 seed sequences in the 3′-UTR. One feature shared by miR-103, miR-320 and the other miRNAs is that seed sequences are enriched in thousands of lower strength PmiTs (Figures 4 and 6), confirming that a miRNA’s nucleotides 2–7 exert an important role in targeting for many hundreds of mRNAs per miRNA.
Figure 4.
miR-103 seed sequences are highly enriched in open reading frame (CDS) of targets. As for miR-103, the strongest miR-103 targets are those that are enriched for the complementary 5′-seed sequence in the CDS. These data correlate closely with results previously reported (22). To visualize the correlation between enrichment in the Anti-AGO Co-IP and the seed sequence densities in the 5′-UTR, CDS and 3′-UTR, we first arranged the mRNAs according to the enrichment in the anti-AGO Co-IP (A). Here one sees the top 3000 mRNAs in the order of the average expression levels detected on the microarray using RNA from the Anti-AGO Co-IP RNA, relative to the transfection using the NEG-CTRL miRNA (N = 3 each). This same order of mRNAs was used for the chart in B, which shows the miR-103 5′ complementary seed densities in 5′-UTR, CDS and 3′-UTR. The mRNAs are binned with each data point representing the average seed densities per 1000 nt for 50 different mRNAs. Note that the most-enriched 50 mRNAs (red arrow) have by far the highest miR-103 seed densities in the CDS. However, there remains some degree of enrichment for miR-103 5′-seed sequences even among the less-enriched mRNAs (orange arrow). In contrast, when the mRNAs are sorted randomly (C, where mRNAs are sorted according to the enrichment following miR-15b* transfection), there is lower miR-103 seed sequences in the 5′-UTR, CDS and 3′-UTR alike.
In contrast to many previously reported miRNAs such as miR-124 that predominantly target the 3′-UTR (22,28), the miR-103/107 family preferentially binds to the CDS of the target transcript. As stated above, there was no preference for CDS targeting when the 3′-end of miR-107 was mutated (i.e. MUT2). This indicates that the 3′ portion of miR-107/103 may play a role in CDS target determination. To test this hypothesis, word analyses were performed testing complementary sequences to the 3′ 12 nt of miR-103, miR-107 as well as two mutated miRNAs derived from miR-107 (MUT1 and MUT2), to assess whether there is any enriched 3′-end pairing sequences in the target sites in the CDS. The result is summarized in Table 2. In this sequence analysis, the strongest putative targets, as identified in either the anti-AGO co-IP or lysate fraction, were included. Following miR-107 transfection, 21 of the 50 selected transcripts from the IP fraction contain pentamer words in the target site that pair perfectly to the 3′ portion of miR-107, and this observed word enrichment was statistically significant (P = 0.04 by simulation test). The result was similar for miR-103 targeted transcripts from the IP fraction (trend with P = 0.09). In contrast, there was no significant enrichment of the 3′ portion matching sequences in the mutated miRNAs (MUT1 and MUT2) or the transcripts identified from the lysate fraction.
To further determine whether there are any specific sequence motifs in the 3′ portion of miR-107 that are involved in target recognition, we compared the 3′-end sequences of miR-107 and MUT2. To focus our analysis on the 3′ portion of miR-107, we only included 110 transcript sequences that are significantly enriched in the IP fraction of miR-107, but not that of MUT2. The enrichment of these genes is likely determined by the 3′-end of the miR-107 since the only difference between miR-107 and MUT2 is the mutated bases from the 3′-end. In the 3′ portion of miR-107, we found a sequence motif AGCCCUGU that was significantly enriched for these 110 genes. Within this motif, three adjacent pentamer words were significantly enriched, with anti-sense sequences found in the target sites in 14, 16 and 18 target transcripts for three pentamer words, respectively (Table 3). Combined together, 33 of the 110 target transcripts contain anti-sense pentamer words (any one of the four pentamers within AGCCCUGU) in the target sites, and this observation was statistically significant (P = 0.008 by simulation test).
DISCUSSION
RIP-Chip and miRNA transfection experiments focusing on miR-15/107 gene group members show that individual miRNAs follow idiosyncratic ‘binding rules’. We confirm that miRNA 5′ seeds appear to be critical targeting determinants. Yet, as shown previously (4), the 3′ portions of miRNAs also appear to contribute to target binding. We also provide word analyses showing pentamers recognized by miRNA 3′ sequence elements in strong miR-107 targets. The current study provides the first evidence that the 3′ portion of miR-103/7 appears to play a role in causing miRNAs to bind preferentially to CDS of target mRNAs (see Figure 7 for an overview). Transfections with a custom-synthesized miR-107 duplex produced mRNA profiles (BACE1 and GRN) in lysates and anti-AGO co-IPs that were similar to the commercial siRNA-like miRNA transfection reagents. Collectively, these data provide a large new repository of information about AGO-associated and ‘knocked down’ mRNA targets related to the miR-15/107 gene group (23). These data demonstrate the importance of evaluating individual miRNAs experimentally instead of relying on universal prediction algorithms before the full complexity of miRNA ‘binding rules’ is understood.
A focus of this study is the miR-15/107 group of miRNA genes (23). Although there are common sequence elements and overlapping functions among these vertebrate-specific genes, the evolutionary record is as yet inadequate to term them a true ‘gene family’. Members of this group of miRNA genes are highly expressed in every human cell evaluated to date, and the genes are increasingly appreciated to serve key functions including cell division, metabolism, stress response and angiogenesis (23). These genes have also been implicated in human cancers, cardiovascular disease and neurodegenerative diseases. For example, downregulation of multiple miR-15/107 gene group members has now been documented in Alzheimer’s disease (25,29–31).
We have previously validated our observation that miR-107 targets mRNAs through the CDS (19,22,23,32). In the current study, we find that multiple miR-107 paralogs target the GRN mRNA, which has implications for neurodegenerative diseases and cancer (24,33). It is hoped that the complete set of data will be of use to other researchers in better understanding the intriguing group of miRNAs that includes miR-16, miR-103, miR-107 and miR-195.
Research from many laboratories has now confirmed that the CDS of mRNAs can be, and often is, targeted by miRNAs as summarized by a recent review (6). In high-throughput, direct experimental evaluation of miRNA target with ‘HITS-CLIP’ in mouse brain, some 25% of miRNA target sequences resided in open reading frames of targeted mRNA (33). The biochemical determinants of CDS targeting are not firmly established. There has been indication that the tendency for particular miRNAs to target the 3′-UTR instead of the CDS was due to active translation in the CDS physically dislodging the miRNP (34), to the presence of upstream AUG sequences (35), or to the relative inability for CDS sequences to recruit cap-dependent translational inhibition (36). Just as the 3′-UTR is not the only part of an mRNA that a miRNA may target, it has also been shown that the 5′ seed portion of miRNAs is not the only determinant of targeting from the miRNA side. For example, Shin et al. (37) showed that the importance of central pairing miRNA sites for miRNA targeting. However, we know of no experimentally validated explanation as to why the 3′ portion of miRNAs helps to confer its tendency to bind to the CDS versus the 3′-UTR. The biochemical basis for this effect is a topic of active study in the Nelson laboratory.
Prior studies have concluded that miRNAs function through many different mechanisms. A recent paper showed data to indicate that regulation by three particular miRNAs (miR-1, miR-155 and miR-223) is achieved through knocking down the mRNA levels of these miRNAs’ targets in HeLa cells (27). These data do not include members of the miR-15/107 gene group and so our results are not in conflict with them. We found previously that for miRNAs outside the miR-15/107 gene group—for example, miR-124 and miR-320—the data is quite compatible with 3′-UTR targeting by miRNAs leading to mRNA degradation (22). We also continue to find evidence that some miR-107 targets (BACE1) may be targeted via the 3′-UTR and act through mRNA degradation: BACE1 mRNA is decreased in lysates following miR-107 transfection, protein levels are decreased, but the mRNA is not present in the anti-AGO co-IP (38). However, this mode of regulation does not appear to be the case for the large majority of miR-103/7 targets. Our high-throughput data provide compelling data that miR-103 and miR-107 seed sequences are neither enriched in the 3′-UTR of co-IP’d targets, nor in the 3′-UTR (or CDS) of mRNAs that are downregulated after miR-103 or miR-107 transfection. This is in marked contrast to miR-320 for which 3′-UTR seed sequences are enriched in both co-IP’d targets and also mRNAs that are downregulated in the lysate following miR-320 transfection.
There are limitations to the current study. We used cultured H4 cancer cells, derived from an aneuploid cell line (21). The miRNP is functional in H4 cells (19), but there may be currently uncharacterized idiosyncrasies. Aside from the inherent limitations of the cultured tumor cells, RIP-Chip is an assay that does not provide all the information about miRNA:mRNA interactions. AGO-miRNPs have been shown to exert gene expression regulation via multiple mechanisms (1,39). For the mRNA target to be detected by a microarray downstream of the anti-Ago co-IP, the target mRNA must be stably bound to the miRNP which is not necessarily expected in all modes of mRNA translational repression. We attempted to make up for this problem by also assessing mRNAs downregulated in the total lysate following miRNA transfection.
Another intriguing possibility is that the tendency of miR-15/107 family of miRNAs to target CDS systematically is related to sequence elements found most frequently in physiological CDSs. An exceptional example of how this might happen is provided by GRN. The CDS of GRN, across many animal species, contains many iterations of the sequence motif for cysteine–cysteine (UGCUGC), and this CDS motif is also targeted by the miR-107 seed sequence (24). We do not find a straightforward, systematic pattern of highly represented coding (versus UTR) sequences correlating with targeting by individual miRNAs, but this is an area that deserves further analyses.
In summary, analyses of RIP-Chip experiments in H4 cells provided the bases for detailed evaluation of miR-15/107 group member targets. Like previous studies, we find that the 5′ seed of miRNAs is important for target discrimination but the 3′ portion of miRNAs also contributes to miRNA binding. We also describe compelling data that for the first time indicates the 3′ portion of miR-103 and miR-107 contributes to these miRNAs’ tendency to target mRNAs’ CDS. These data do not deny or contradict findings with other miRNAs. However, these data surely provide new indications that miRNA biology defies simple formulas and doctrines. Apparently, members of the miR-15/107 gene family operate through some biochemical mechanisms that are somewhat distinct from the handful of miRNAs that were previously characterized in detail.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health, Bethesda, MD (grants R01 NS061933, R01 GM089784 and K08 NS050110); Alzheimer Association (NIRG-89917). Funding for open access charge: National Institutes of Health (grants R21AG036875, R01NS061933).
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
We thank Ms Willa Huang for technical and collegial assistance in the project. We thank the careful reviewers who provided us with ideas to improve the manuscript.
REFERENCES
- 1.Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. The deep evolution of metazoan microRNAs. Evol. Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 3.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 6.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 7.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson PT, Kiriakidou M, Mourelatos Z, Tan GS, Jennings MH, Xie K, Wang WX. High-throughput experimental studies to identify miRNA targets directly, with special focus on the mammalian brain. Brain Res. 2010;1338:122–130. doi: 10.1016/j.brainres.2010.03.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA. 2008;14:2440–2451. doi: 10.1261/rna.1139508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 13.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc. Natl Acad. Sci. USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 19.Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA. 2010;16:394–404. doi: 10.1261/rna.1905910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O’Doherty U, Mourelatos Z. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnstein P, Taylor DO, Nelson-Rees WA, Huebner RJ, Lennette EH. Propagation of human tumors in antithymocyte serum-treated mice. J. Nat Cancer Inst. 1974;52:71–84. doi: 10.1093/jnci/52.1.71. [DOI] [PubMed] [Google Scholar]
- 22.Wang WX, Wilfred BR, Xie K, Jennings MH, Hu Y, Stromberg AJ, Nelson PT. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol. 2010;7:373–380. doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J. Mol. Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am. J. Pathol. 2010;177:334–345. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2010;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS ONE. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Wang WX, Kyprianou N, Wang X, Nelson PT. Dysregulation of the mitogen granulin in human cancer through the miR-15/107 microRNA gene group. Cancer Res. 2010;70:9137–9142. doi: 10.1158/0008-5472.CAN-10-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ajay SS, Athey BD, Lee I. Unified translation repression mechanism for microRNAs and upstream AUGs. BMC Genomics. 2010;11:155. doi: 10.1186/1471-2164-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W-X, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. MiR-107 regulates Granulin/Progranulin with implicatins for traumatic brain injury and neurodegenerative disease. Am. J. Pathol. 2010;177:334–345. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Nelson PT, Tang G. Wiley Encyclopedia of Chemical Biology. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data