Balanced gene regulation by an embryonic brain non-coding RNA is critical for GABA circuitry in adult hippocampus (original) (raw)
. Author manuscript; available in PMC: 2011 Oct 28.
Published in final edited form as: Nat Neurosci. 2009 Jul 20;12(8):1020–1027. doi: 10.1038/nn.2371
Summary
Genomic studies demonstrate that while the majority of the mammalian genome is transcribed, only about 2% of these transcripts are protein coding. We have been investigating how the long, polyadenylated Evf2 non-coding RNA regulates transcription of homeodomain transcription factors DLX5 and DLX6 in the developing mouse forebrain. Here we show that in developing ventral forebrain, Evf2 recruits DLX and MECP2 transcription factors to key DNA regulatory elements in the Dlx 5/6 intergenic region and controls Dlx5, Dlx6, and GAD67 expression through trans and _cis_-acting mechanisms. Evf2 mouse mutants have reduced numbers of GABAergic interneurons in early post-natal hippocampus and dentate gyrus. Although the numbers of GABAergic interneurons and GAD67 RNA levels return to normal in Evf2 mutant adult hippocampus, reduced synaptic inhibition occurs. These results suggest that non-coding RNA-dependent balanced gene regulation in embryonic brain is critical for proper formation of GABA-dependent neuronal circuitry in adult brain.
Background
The potential of the genome to code for functional non-coding RNAs (ncRNAs) is only beginning to be uncovered1, 2. While many ncRNAs belong to classes of small regulatory RNAs, a subset are long, polyadenylated ncRNAs (lpncRNAs) that act cooperatively with protein partners3. We showed that Evf2, a lpncRNA target of sonic hedgehog (SHH) signaling in the developing telencephalon, exhibits _trans_-acting transcriptional cooperativity with DLX homeodomain proteins, increasing Dlx 5/6 enhancer activity in a neural stem cell line4. Identification of an ultraconserved Dlx 5/6 intergenic DNA regulatory element5 has led to discovery of greater than 1000 ultraconserved DNA sequences (UCRs), near key developmental regulators or transcription factors6–8. The necessary and sufficient domain of Evf2 transcription-regulating activity lies within this ultraconserved sequence at the 5′ end of Evf2 RNA4. The finding that Evf2 has transcription-regulating activity4 raised the possibility that subsets of UCRs are transcribed and functional. Recently, additional ultraconserved brain lpncRNAs have been identified9, supporting the possibility that ultraconserved ncRNAs constitute a new class of transcription-regulating ultraconserved ncRNAs (trucRNAs). Here we show for the first time that the Evf2 trucRNA plays a critical role in gene regulation and the development of interneurons that produce gamma-amino butyric acid (GABA), the major inhibitory neurotransmitter in the brain.
The balance between excitation and inhibition in the brain is critical for proper function and is maintained by two major classes of neurons: excitatory projection neurons and inhibitory local circuit interneurons. While excitation is primarily mediated by the neurotransmitter glutamate, GABA primarily mediates inhibition. Recently, multiple regulatory roles of GABAergic interneurons have been identified10. The dysfunction of GABA-regulated circuits has been implicated in different psychiatric disorders such as schizophrenia, autism, and Tourette’s syndrome, as well as epilepsy. In methyl CpG-binding protein (Mecp2) mutant mice11–13, a model for the human autism spectrum disorder (ASD) Rett syndrome, GABA-dependent inhibitory cortical activity decreases14. In dorsal lateral prefrontal cortex of schizophrenic patients, one of the most consistent findings is a reduction in GAD67, the enzyme responsible for GABA synthesis15. Therefore, multiple lines of evidence implicate alterations of GABAergic function in a variety of neurological diseases.
In this paper we show that a single non-coding RNA controls development of GABAergic interneurons and adult brain circuitry, making this mechanism a novel target for studying both development and disease.
Results
Trans and cis -gene regulation by Evf2 in vivo
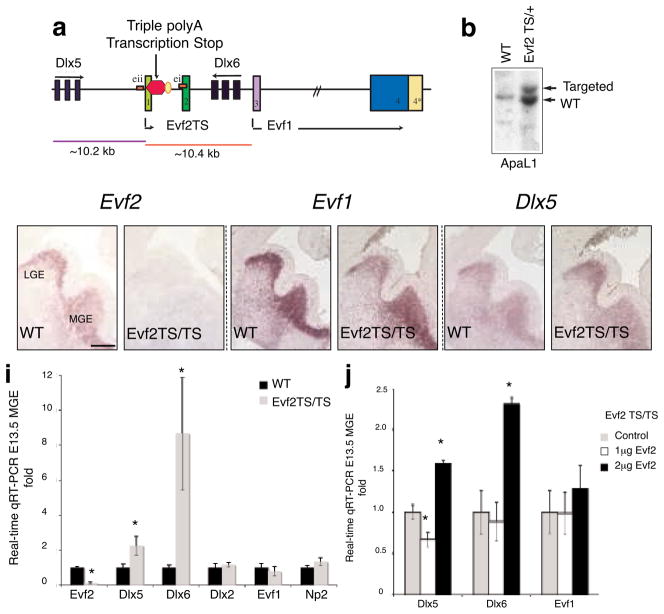
We designed Evf2 loss-of-function mice in order to determine the role of Evf2 in vivo. Because of the overlap of Evf2 with key Dlx 5/6 DNA regulatory elements, we inserted transcription termination sites rather than removing DNA fragments (Fig. 1a). We subcloned a 19.4 kb fragment spanning the Dlx 5/6 and Evf genes from a mouse BAC. We then introduced a triple polyadenylation signal16 into exon 1 (Evf2TS, Fig. 1a). Southern analysis verified correct targeting in ES cell lines (Fig. 1b) and mice (not shown). Evf2TS/TS mutant mice are fertile, lived for at least 1 year, and were morphologically indistinguishable from wild-type littermate controls (not shown). In situ hybridization analysis showed that transcription stop insertion into Evf exon1 eliminated Evf2, but not Evf1 or Dlx5 expression in E13.5 ventral telencephalon (Fig. 1c–h). Real-time qRT-PCR of E13.5 medial ganglionic eminence (MGE) tissue from Evf2TS/TS mice showed that Dlx6 and 5 transcripts increased by 8- and 2- fold, respectively (Fig. 1i). Despite the fact that Evf1 and Dlx5 transcription start sites are approximately equidistant from the TS insertion site (Fig. 1a), Dlx5, but not Evf1 transcription was affected in Evf2 mutants (Fig. 1i). Selective transcriptional effects supported that Evf2 loss rather than insertion of the TS sequence was responsible. If TS insertion were causing the observed transcriptional effects, all Dlx 5/6 enhancer activities would be expected to change, including Evf1.
Figure 1. Evf2TS/TS mice have increased Dlx5 and Dlx6 expression in the embryonic brain.
a. Schematic of targeting a triple polyadenylation transcription stop site into Evf exon1, truncating Evf2 transcripts from 3.7 kb to 101 bp (Evf2TS), but not Evf1, which is transcribed starting from exon3. Truncated Evf2TS transcript (101 bp) completely lacks the ultraconserved ei region. Only the distance from TS insertion to the Dlx5 (~10.2 kb) and Evf1 (~10.4 kb) transcription start sites are shown to scale. b. ES cells used for making Evf2TS mice contain correctly targeted transcription stop into Evf exon1, as verified by Southern analysis. c–h. RNA in situ hybridization of E13.5 coronal sections of wildtype (wt) and Evf2TS/TS mutant telencephalon, probed with anti-sense Evf2 (c, d), anti-sense Evf1 (e, f), or anti-sense Dlx5 (g, h). i. Quantitative real-time RT-PCR analysis of E13.5 MGE from wt and Evf2TS/TS mutants. Error bars: S.E.M. Statistical analysis: Mann Whitney U-test, p<.05: Evf2=0.0055, Dlx5=0.044, Dlx6=0.0055; n= 4 for wt, n=6 for Evf2TS/TS. j. Quantitative real-time RT-PCR analysis of E12.5 Evf2TS/TS mutant MGE electroporated with: control (pcDNA, 2μg), 1 μg pcDNA-Evf2 (+1 μg pcDNA), and 2μg pcDNA-Evf2. Error bars: S.E.M. Statistical analysis, ANOVA Dunnett’s two-sided test, p<.05 values: Dlx5, (0μg,1μg Evf2=0.05, 0μg,2μg Evf2=0.001), ANOVA Tukey test, p<.05 values, Dlx6 (0μg,2μg Evf2=0.041, 1μg Evf2, 2μg Evf2= .033); n=4.
* p < 0.05; _t_-test, Mann Whitney, or ANOVA
In order to distinguish between trans and _cis_-dependent Evf2 RNA regulatory effects, we performed Evf2 electroporation into E12.5 Evf2TS/TS brains at 2 different concentrations (Fig. 1j). At a lower Evf2 concentration (1μg) Dlx5 expression decreased, while Dlx6 and Evf1 remain unchanged. At a higher Evf2 concentration (2 μg), both Dlx5 and Dlx6 increased, while Evf1 did not change. The ability of Evf2 to partially rescue Dlx5 increase suggested that _Evf-2 trans_-regulatory mechanisms were involved in Dlx5 transcriptional control. The inability of ectopically expressed Evf2 to rescue Dlx6 increase in Evf2TS/TS mutants supported the idea that Evf2 reduced Dlx6 expression through anti-sense competition in cis, rather than by trans mechanisms. At higher concentrations of Evf2 (2 μg), both Dlx5 and 6 increased, supporting previously published results that Evf2 RNA can function as a transcriptional activator of Dlx 5/6 ei and eii when ectopically expressed4. Electroporation of an Evf2siRNA construct into E12.5 brains also increased levels of Dlx5 transcripts (Supplemental Fig. S1), further supporting that an Evf2 trans-acting mechanism regulated Dlx5 expression. Together, these data suggested that Evf2-mediated transcriptional control was concentration-dependent, utilizing both trans and cis regulatory mechanisms in vivo.
Evf2 recruitment of DLX and MECP2 to intergenic enhancers
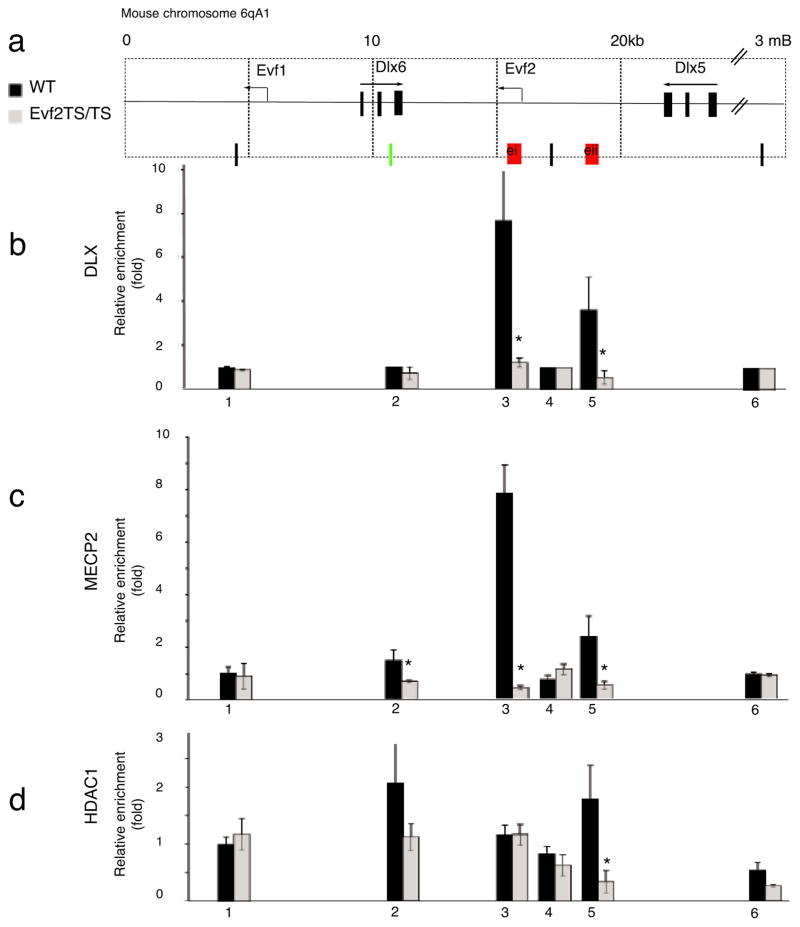
We recently showed that Evf2 forms a complex with DLX proteins in vivo and acts as a transcriptional co-activator of DLX activity with both target and homeodomain specificity in C17 cells4. In addition, we proposed a model in which Evf2 recruits DLX proteins to Dlx 5/6 intergenic enhancers. However, in the present study qRT-PCR analysis of mice lacking Evf2 (Fig. 1i) indicated that Dlx5 and 6 transcripts increased, suggesting that Evf2 played a negative, rather than positive, transcription-regulating role in vivo. Rescue experiments (Fig. 1j) suggested that increased Dlx6 in Evf2TS/TS mutants resulted from loss of anti-sense interference in cis, while more subtle repressive effects on Dlx5 transcription occurred in trans. In order to further investigate the mechanism of _Evf2_-dependent transcriptional control, we used chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qChIP-PCR) on wild-type and Evf2TS/TS mutant E13.5 MGE chromatin to determine whether Evf2 affects DLX binding to Dlx 5/6 ei and/or eii. Qualitative ChIP assays using anti-DLX1 and -2 antibodies previously identified Dlx 5/6 ei and eii as specific binding sites17. In wild type E13.5 MGE, pan-anti-DLX antibodies4, 18–21 recognized DLX protein-Dlx 5/6 enhancer (ei and eii) complexes (Fig. 2b, black bars, primers 3(ei) and 5(eii)). In Evf2TS/TS mutant chromatin, DLX did not bind to ei and eii (Fig. 2b, gray bars), showing that Evf2 was required for DLX protein recruitment to Dlx regulatory enhancers ei and eii.
Figure 2. Loss of Evf2 affects DLX and MECP2 binding to Dlx 5/6 intergenic enhancers in embryonic day 13.5 medial ganglionic eminence.
a. Schematic showing the region in mouse chromosome 6qA1 surrounding Dlx5/6 and Evf1/2. Quantitative chromatin immunoprecipitation of E13.5 MGE from wt (black bars) and Evf2TS/TS mutants (gray bars) using primers 1–6 across the Dlx 5/6 region, and antibodies to: b. anti-pan DLX, p <.05 values: 3=0.025, 5=0.025, c. anti-MECP2, p<.05 values: 2=0.025, 3=0.025, 5=0.025, d. anti-HDAC1, p <.05 values: 5=0.025, 6=0.025. qChIP-PCR was performed on optimized primer sets using previously defined primer sets (1=15, 2=24, 4=2723) and newly defined sets for ei (3- red), eii (5-red) and external primer (6). Primer 2 (green) was identified as a MECP2/HDAC binding site for transcriptional repression in adult cortex23. Error bars: S.E.M. Statistical analysis: Mann Whitney U-test. * p < 0.05 Mann Whitney U-test. The schematic in (a) is reversed from that shown for Evf2 transcription-stop insertion (Fig 1) in order to align with previously published primers23.
Given previous reports that mice lacking both Dlx1 and 2 show reduced expression of Dlx5 and 622, failure of DLX protein recruitment to Dlx 5/6 ei and eii would be expected to decrease Dlx5 and 6 transcription. However, Dlx5 and 6 transcripts increased and Dlx2 expression did not change (Fig. 1i). The report that a 2-fold increase in Dlx5 can occur in the adult prefrontal cortex upon loss of the transcriptional repressor Mecp223 led us to investigate whether loss of Evf2 in the embryonic brain can affect MECP2 binding in the Dlx 5/6 region. In the absence of Evf2, MECP211, a DNA methyl-binding protein associated with repressed chromatin, did not bind ei or eii (Fig. 2c). One mechanism proposed for MECP2-mediated transcriptional repression is its ability to recruit HDAC, a histone deacetylase responsible for chromatin inactivation24. However, loss of MECP2 binding to ei in Evf2 mutants did not alter HDAC binding to ei (Fig. 2d), suggesting that MECP2 binding functions through an alternate mechanism at this site and stage of development. Loss of MECP2 binding at eii did reduce HDAC binding, suggesting that ei and eii differ in how MECP2 function affects transcriptional regulatory activity by these sites. While site 2 was previously defined as an MECP2 and HDAC binding site in the adult prefrontal cortex23, MECP2 bound minimally to site 2 at this time during development. In addition, HDAC bound to site 2 in wild-type embryonic MGE, but reduction in Evf2TS/TS mutants was not statistically significant (Fig. 2d). Therefore, increased Dlx5 expression in Evf2 mutants may result from loss of MECP2 binding to ei and eii, and subsequent loss of HDAC from eii.
Evf2 recruited DLX and MECP2 to ei and eii, affecting Dlx 5/6 enhancer activities in trans on specific nearby genes such as Dlx5 but not Evf1, and regulating Dlx6 transcription through anti-sense inhibition in cis (Figs. 1 and 2). Several possibilities may explain how Dlx5, 6 and Evf1 are transcribed in the absence of DLX binding to ei and eii. First, DLX 1/2 may only be required for initial activation of Dlx 5/6 expression in an _Evf2_-independent manner; subsequent regulation of Dlx 5/6 levels by DLX and MECP2 may then require Evf2. Second, other DLX binding sites may compensate in the absence of DLX ei/eii interactions. Third, the major role of DLX1 and 2 may be to prevent MECP2 from binding ei/eii, acting through inhibition rather than as direct activators.
Evf2 did not control DLX or MECP2 nuclear localization
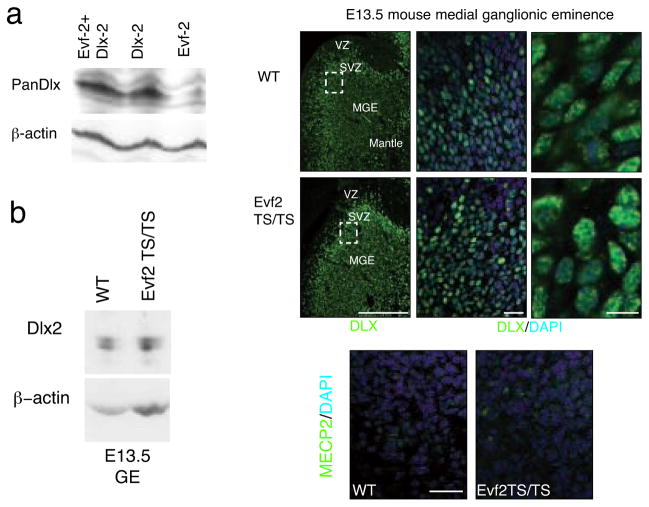
The absence of DLX and MECP2 protein binding to ei and eii in Evf2TS/TS mice raised the possibility that Evf2 influences protein stability and/or nuclear localization. In a direct test of the effect of Evf2 on DLX2 protein stability in C17 neural cells, DLX2 protein levels resulting from co-transfection in the presence or absence of Evf2 did not change (Fig. 3a). In addition, DLX2 protein levels in Evf2TS/TS mutant embryonic ganglionic eminences compared to wild-type did not change (Fig. 3b). By immunofluorescent visualization, DLX protein distribution in Evf2 mutant nuclei (Fig. 3f–h) was indistinguishable from wild-type (Fig. 3c–e). In addition, transcripts of Neuropilin 2 (NP2), a direct target of DLX1/225, did not change in Evf2TS/TS mutants (Fig. 1i). If Evf2 were controlling DLX protein stability, all DLX1/2 activities would be affected, including NP2, which is not. These data support a mechanism where Evf2 recruits DLX to ei and eii, rather than stabilizing DLX protein or directing DLX nuclear localization.
Figure 3. Evf2 does not affect DLX or MECP2 nuclear localization.
a. Western Blot analysis of DLX2 protein after transfection of a construct containing Dlx2 cDNA and/or Evf2 cDNA. Transfection was performed into C17 mouse neural stem cells in the presence or absence of Evf2 RNA, showing equal levels of DLX2 protein. That Evf2 RNA is stable after transfection was verified by RT-PCR (not shown). b. Western analysis of Evf2TS/TS and wt E13.5 ganglionic eminence extracts probed with anti-DLX2 specific antibody (gift of David Eisentstat) shows that loss of Evf2 does not affect DLX2 protein levels. c–h. Pan-anti-DLX antibody (green) stains nuclei counterstained with DAPI (blue) in both wt and Evf2TS/TS E13.5 MGE. MGE, medial ganglionic eminence; VZ, ventricular zone; SVZ, subventricular zone. c, f: scale bar= 200μm; d, g: scale bar=20μm; e, h: scale bar= 10μm. i–j. Immunohistochemistry using anti-MECP2 antibodies (green) and DAPI (blue nuclei) shows similar nuclear localization in both wt and Evf2TS/TS MGE cells, scale bar=20μm.
We next asked whether loss of MECP2 binding to ei/eii in Evf2 mutants resulted from effects on nuclear localization. Analysis of MECP2 localization in developing brain shows a gradient of increasing expression in neurons as they mature, with very little expression in immature neurons26. However, expression in E13.5 MGE has not been reported. Analysis of MECP2 in E13.5 MGE nuclei where Evf2 and DLX are normally expressed showed non-homogeneous, speckled MECP2 localization (Fig. 3i). In Evf2TS/TS mutants, MECP2 distribution is indistinguishable from wild-type (compare Fig. 3i and j), showing that failure of MECP2 recruitment to ei and eii in Evf2 mutants did not result from altered subcellular localization.
Reduced numbers of Evf2TS/TS hippocampal interneurons
The DLX homeodomain protein family is related to the Drosophila Distalless gene (dll)27. Mice lacking Dlx1 and 2 show significant loss of GABAergic interneurons in cortex (~75%) and hippocampus (~90%) due to defective migration from embryonic MGE28–30. Loss of Evf2 affected Dlx5/6 expression and DLX function (Figs. 1 and 2) in E13.5 MGE, the source of the majority of GABAergic interneuron precursors that will migrate to the hippocampus and dentate gyrus29, 31. Therefore, we asked whether GABAergic interneuron development in P2 hippocampus or dentate gyrus was affected in mice lacking Evf2.
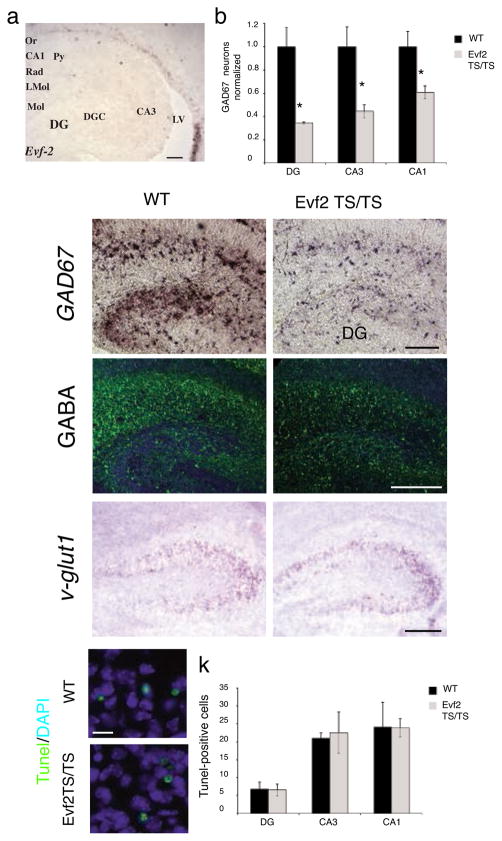
Expression of Evf2 in developing ventral telencephalon/subventricular zone persisted until birth in P2 subventricular zone and cells that appeared to be migrating to the hippocampus (Fig. 4a). However Evf2 was undetectable in wild-type postnatal day 2 (P2) hippocampus and dentate gyrus, as well as in adult subventricular zone or hippocampus (not shown). In situ hybridization using a probe against glutamate decarboxylase 67 (GAD67), an enzyme necessary for converting glutamate to GABA, showed that the number of GABAergic interneurons in Evf2 mutant dentate gyrus and hippocampal CA1 and CA3 layers are reduced by 40–65% (Fig. 4b–d). Reduced GAD67 expression in Evf2 mutants was accompanied by reduction in the neurotransmitter GABA, as shown by anti-GABA immunohistochemistry (Fig. 4e, f). In situ hybridization against vesicular glutamate transporter 1 (v-Glut1), a gene expressed specifically in glutamatergic neurons showed that v-Glut1 expression in Evf2TS/TS hippocampus and wild-type were similar (Fig. 4g, h). In addition, Tunel staining of Evf2TS/TS and wild-type hippocampus and dentate gyrus showed similar levels of cell death (Fig. 4i–k). Together, these data showed that Evf2 was required for proper GABAergic interneuron development, and that fate transformation or increased cell death were not responsible for this reduction.
Figure 4. GABAergic interneuron loss in the P2 hippocampus and dentate gyrus of Evf2TS/TS mutant mice.
a. RNA in situ hybridization analysis of Evf2 expression in the subventricular zone and cells lining the lateral ventricle as they migrate to the hippocampus and dentate gyrus. scale bar=130μm; Or, oriens; Py, pyramidal; Rad, radiatum; LMol, lacunosum molecular; Mol, molecular layer of the dentate gyrus; DGC, dentate granule cell layer; DG, dentate gyrus; LV, lateral ventricle. b. Quantification of the number of GAD67-expressing GABAergic interneurons in the dentate gyrus (DG), and hippocampal CA1 and CA3 regions. Error bars: S.E.M. Statistical analysis: Mann Whitney U-test, p =.025 for DG, CA3, CA1; n= 3 for wt, n=3 for Evf2TS/TS. c–d. RNA in situ hybridization of GAD67 expressing interneurons in the hippocampus, scale bar=135μm e–f. Immunohistochemistry, anti-GABA antibodies (green) and DAPI (blue nuclei), scale bar= 200 μm. g–h. RNA i_n situ_ hybridization of v-glut1 expressing neurons in the hippocampal CA3 region shows similar expression in wt and Evf2TS/TS, scale bar=80μm. i–j. Immunohistochemistry using the apoptosis TUNEL assay (green) and DAPI (blue nuclei) in the hippocampal CA3 region, scale bar= 10μm. k. Quantification of the number of TUNEL-positive cells in the dentate gyrus (DG), and hippocampal CA1 and CA3 regions shows no difference in cell death between wt and Evf2TS/TS mutants. Error bars: S.E.M. Statistical analysis: Student’s _t_-test, p >0.05 for DG, CA3, CA1; n= 3 for wt, n=3 for Evf2TS/TS. * p < 0.05; _t_-test or Mann Whitney U-test
Evf2 regulated GAD67 in embryonic, but not adult brain
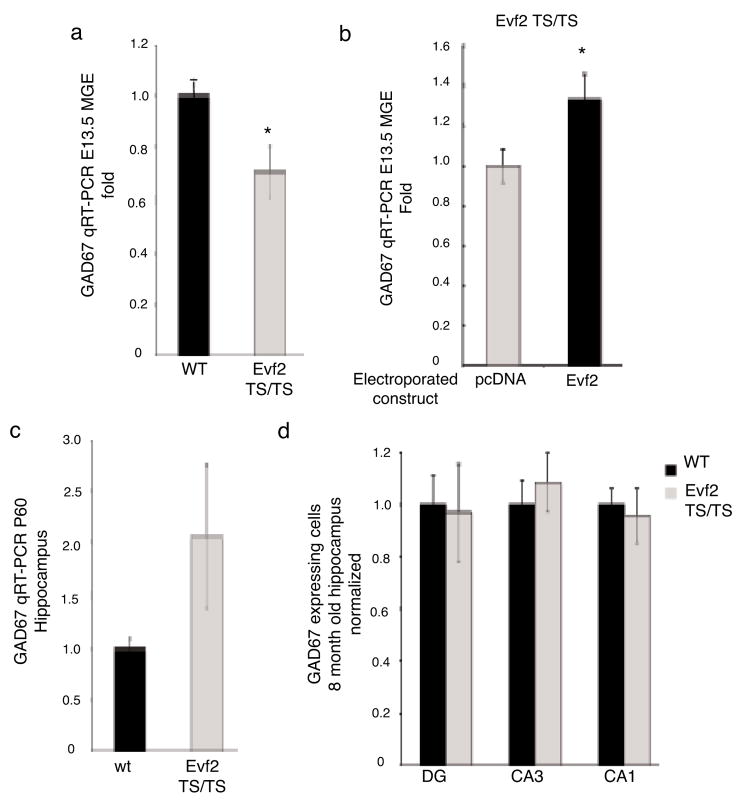
In order to further understand the role of Evf2 in GABAergic interneuron development, we analyzed GAD67 RNA levels in E13.5 MGE, where GABAergic interneuron precursors first arise. GAD67 transcripts decreased by ~30% in Evf2TS/TS compared to wt E13.5 MGE (Fig. 5a). Electroporation of Evf2 into Evf2TS/TS E12.5 MGE restored GAD67 levels by ~30% compared to pcDNA control (Fig. 5b). The ability of Evf2 to rescue decreased GAD67 in Evf2TS/TS mutants suggested that Evf2 activated GAD67 transcription through _trans_-acting mechanisms.
Figure 5. _Evf2 trans_-positively regulates GAD67 expression in E13.5 MGE, but not adult hippocampus.
a. Quantitative real-time RT-PCR analysis of GAD67 in E13.5 MGE from wt and Evf2TS/TS mutants. Error bars: S.E.M. Statistical analysis: Student’s _t_-test, p=0.031; n= 3 for wt, n=5 for Evf2TS/TS. b. Quantitative real-time RT-PCR analysis of GAD67 in E12.5 Evf2TS/TS mutant MGE electroporated with: control (pcDNA, 2μg), and 2μg pcDNA-Evf2. Error bars: S.E.M. Statistical analysis: Student’s _t_-test, p=0.0375; n=4. c. Quantitative real-time RT-PCR analysis of GAD67 in P60 hippocampus from wt and Evf2TS/TS mutants. Error bars: S.E.M. Statistical analysis: Student’s _t_-test, p=0.031; n=2 for wt, n=2 for Evf2TS/TS. d. Quantification of the number of GAD67-expressing GABAergic interneurons in 8-month old dentate gyrus (DG), and hippocampal CA3 and CA1 regions. Error bars: S.E.M. Statistical analysis: Student’s _t_-test, p>0.05 for DG, CA3, CA1; n=3 for wt, n=3 for Evf2TS/TS. * p < 0.05; _t_-test
In forebrain, Evf2 expression was limited to embryonic and early post-natal times of development (Figs. 1c and 4a), and undetectable in adult SVZ or hippocampus (not shown). We next asked whether reduced embryonic expression of GAD67 and reduction of GABAergic interneurons seen in early post-natal hippocampus persisted into adulthood. At P60 Evf2TS/TS GAD67 transcript levels were comparable to those in wild-type (Fig. 5c). In addition, the number of GABAergic interneurons in 8 month-old Evf2TS/TS and wt hippocampus and dentate gyrus were similar (Fig. 5d). This suggested that in Evf2 mutants GAD67 expression and GABAergic interneuron number recovered to normal levels sometime between early post-natal and 2 month-old hippocampal development, temporally correlating with timing of Evf2 down-regulation.
Reduced synaptic inhibition in Evf2TS/TS pyramidal neurons
Dlx1 −/− mice exhibit loss of a specific population of hippocampal GABAergic interneurons, which subsequently results in reduced synaptic inhibition32. Analysis of Evf2TS/TS hippocampus not only showed a greater percentage loss of total GABAergic interneurons in early postnatal hippocampus (Fig. 4b) than Dlx1−/− mice32, but also showed that GABAergic defects appeared earlier. Additionally, unlike Dlx1 −/− mice, Evf2TS/TS mice showed recovery of GABAergic interneurons in older animals (Fig. 5c, d). This led us to ask whether lower levels of embryonic and perinatal GAD67 can cause long-lasting effects on adult synaptic activity despite apparent transcriptional recovery.
In order to answer this question, we compared inhibitory post-synaptic currents (IPSCs) in CA1 pyramidal cells from Evf2TS/TS and wild-type littermates at ages older than P60, when GAD67 mRNA levels have recovered. First we analyzed spontaneous IPSPs (sIPSCs) and minimal IPSC (mIPSCs). In order to maximize outward inhibitory current in artificial cerebrospinal fluid (ACSF) containing glutamate receptor antagonists (20 μM CNQX and 50 μM D-AP5), we recorded at +20 mV. We added tetrodotoxin (2 μM) to isolate mIPSCs (Fig. 6a, b). We divided all mice into two age groups: adult (3–5 months old) and old (12 > months old) animals. The two age groups allowed us to distinguish any persistent differences between mutant and wild-type mice from those that might be attributable to specific developmental stages.
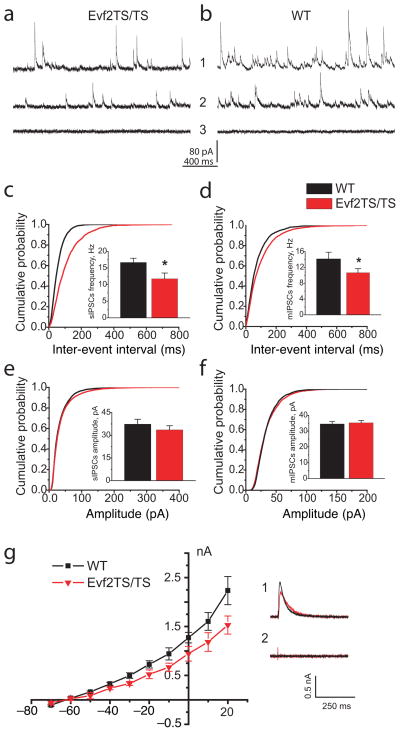
Figure 6. GABAergic synaptic inhibition is reduced in CA1 layer of the adult hippocampus of Evf2TS/TS mutant mice.
(a–b) Representative traces of sIPSCs (1) and mIPSCs (2) from CA1 pyramidal cells of wildtype (WT) and Evf2TS/TS mice. Picrotoxin (PTX) 0.25 mM blocks all IPSC activity in Evf2TS/TS and WT mice (3). (c–d) Averaged cumulative probability plots of sIPSC and mIPSC inter-event intervals from CA1 pyramidal cells (Evf2TS/TS, red; WT, black; Kolmogorov-Smirnov test, p<0.001). sIPSCs and mIPSCs event frequency (insert) in adult and old Evf2TS/TS mice (sIPSC adult: 11.75±1.73 Hz, sIPSC old: 9.17±0.71; mIPSCs adult: 10.65±1.05 Hz, mIPSCs old: 6.5±0.83 Hz) than in control WT mice (sIPSC adult: 16.66±1.26 Hz, sIPSC old: 12.62±0.92 Hz; mIPSCs adult: 14.16±1.67 Hz, mIPSCs old: 9.06±0.77 Hz). (e–f) Averaged cumulative probability plots of sIPSCs and mIPSCs amplitude. Evf2TS/TS, red; WT, black, sIPSC adult: 33.6±2.7 pA, sIPSC old: 48.50±3.06; mIPSCs adult: 35.26±1.41 pA, mIPSCs old: 34.87±3.76) and WT mice (sIPSC adult: 37.4±3.2 pA, sIPSC old: 53.78±3.98; mIPSCs adult: 34.54±1.64 pA, mIPSCs old: 37.03±4.63). (g) The I–V plot of evoked IPSCs of GABAergic inhibition in Evf2TS/TS mice (p<0.01; paired _t_-test). IPSC current was normalized to the amplitude of evoked EPSC in ACSF containing no drugs to suppress excitatory synaptic activity. (1) Representative IPSCs recordings at +20 mV in Evf2TS/TS (red) and WT (black) mice (average _n_=5) (2) PTX 0.25 mM blocks evoked IPSCs in CA1 neurons from both WT and Evf2TS/TS mice.
Error bars: S.E.M.; * p < 0.05; _t_-test.
Both Evf2TS/TS age groups demonstrated significant reduction in sIPSCs event frequency in CA1 pyramidal cells. sIPSC mean frequency in the adult group was lower in mutant mice by 42% compared with those in wild-type mice. In old animals, sIPSCs event frequency in mutant mice was lower by 38% compared to wild type. We observed similar significant reductions in mIPSCs event frequencies in adult and old Evf2TS/TS groups compared to wild-type. Cumulative probability plots and corresponding Kolmogorov-Smirnov statistical analysis further confirmed reduction in IPSC frequency in Evf2TS/TS mice (Fig. 6c, d). In contrast, we did not find significant differences in sIPSP amplitude or mIPSP amplitude between wild-type and mutant mice (Fig. 6e, f). This suggested that Evf2 did not control the property of synaptic contacts or GABA receptors formed by normally differentiated interneurons, but rather that reduced numbers of GABAergic synapses formed on CA1 pyramidal neurons.
Interestingly, the frequency of sIPSCs and mIPSCs were lower in old mice as compared to adult mice, but the comparable age-dependent changes occurred both in Evf2TS/TS and wild type mice. Thus the difference between mutant and wild-type mice persisted. Likewise, the amplitude of sIPSCs increased in old animals. These age differences are in line with those reported in the literature33, 34.
To further determine whether inhibitory input in CA1 pyramidal neurons were altered in the mutants, we measured the evoked IPSCs in CA1 pyramidal neurons from the adult mice group upon stimulation of the stratum radiatum. First, we recorded evoked excitatory postsynaptic current (EPSC) with amplitude 0.4–0.2 nA at −70mV adjusting the stimulus intensity as necessary. Glutamatergic antagonists blocked EPSCs (see above) and a series of evoked IPSCs were recorded at different holding potentials (−70 mV to +20 mV). The I–V plot constructed from these recordings was normalized to the amplitude of evoked EPSC at −70mV (Fig. 6g). We included only cells with similar biophysical characteristics in the analysis (wild-type n=5, mutant n=4). Evoked IPSCs were significantly smaller in mutant mice (p<0.01; paired _t_-test), further suggesting that CA1 pyramidal neurons received less GABAergic innervation.
Finally, we did not find significant changes in rise or decay time for spontaneous and evoked IPSCs. 0.25 mM picrotoxin when added in the recording ACSF abolished both spontaneous and evoked IPSCs in all our recordings confirming the evolvement of GABAA receptors. Therefore, GABAergic interneuron recovery in older animals did not result in the recovery of normal synaptic inhibition in the hippocampus.
Discussion
Non-coding RNA-dependent balanced gene regulation
This paper demonstrates that the Evf2 non-coding RNA is required for balanced gene regulation in developing ventral forebrain. In Evf2TS/TS mutants, Dlx6 transcripts increased by ~8-fold. The inability to rescue increased Dlx6 in Evf2TS/TS mutants supports the idea that Evf2 anti-sense transcriptional inhibition of Dlx6 occurs in cis through opposite strand transcription rather than anti-sense annealing. Data showing that higher Evf2 levels increase rather than decrease Dlx6 (Fig. 1j) supports opposite strand transcriptional inhibition, as would be expected with an anti-sense annealing mechanism. Although we cannot rule out that TS insertion rather than loss of Evf2 anti-sense interference resulted in Dlx6 increase, it is unlikely that TS insertion disrupted Dlx 5/6 ei or eii given that Evf1 transcription remained unaffected in Evf2TS/TS mutants. Furthermore, TS insertion in the 5′ of the Air non-coding RNA35 does not affect adjacent transcription. Together these data suggest that in vivo Evf2 transcription rather than Evf2 RNA negatively regulated Dlx6 transcription through competitive anti-sense inhibition in cis.
In contrast to Dlx6, the ability to rescue increased Dlx5 and decreased GAD67 indicated that Evf2 RNA negatively regulated Dlx5 and positively regulated GAD67 in trans. Therefore, _Evf2_-mediated _trans_-acting transcription-regulating effects were target-dependent and concentration-dependent: low levels of Evf2 repressed Dlx5 and higher levels activated Dlx5, Dlx6, and GAD67. These results also supported previous data that identified _trans_-acting transcriptional activation of Dlx 5/6 ei and eii by _Evf2_4. Further support that Evf2 acted in trans to repress Dlx5 stemmed from knockdown studies (Supplementary Fig. S1).
Studies show that SHH activates Dlx’s, Evf’s, and an embryonic form of GAD67 in embryonic forebrain19,4. In addition, ectopic expression of Dlx2 and Dlx5 activates GAD67 in embryonic forebrain slices 36. Together these data identify Evf2 and DLX as components of a signaling cascade that activates GAD67, supporting the idea that reduced GAD67 levels in Evf2TS/TS mutants may result from interference with Evf2/DLX regulation of GAD67. However, previous results and those from this paper do not distinguish whether loss of Evf2 directly or indirectly affects GAD67 transcription. Of the two known direct targets of DLX 1/2 signaling, Evf2 loss affected Dlx 5/65 but not Np225 (Fig. 1i), indicating that Evf2 did not affect all Dlx1/2 activities. Therefore, it is possible that unidentified targets of Evf2 may be responsible for GAD67 regulation. A key question raised by these experiments is whether Evf2/DLX directly or indirectly regulates GAD67 expression.
Transcription factor recruitment by non-coding RNAs
Experiments in this paper showed that the Evf2 trucRNA was required for positive (DLX) and negative (MECP2) transcription factor recruitment to ultraconserved DNA regulatory element Dlx 5/6 ei and Dlx 5/6 eii (Fig. 2) in developing ventral forebrain. Loss of Evf2 prevented recruitment of a known transcriptional activator (DLX) to positively-acting DNA regulatory elements (Dlx 5/6 ei and eii), with an unexpected increase, rather than decrease in Dlx5 transcription. We showed that _Evf-2_-mediated recruitment of MECP2, a known transcriptional repressor, was lost from both ei and eii and propose that MECP2 loss may explain Dlx5 deregulation. Despite differences between embryonic and adult MECP2 binding sites in the Dlx 5/6 region, 2-fold increase of Dlx5 in adult brain occurred upon loss of MECP223 and supported a repressive role for MECP2 on Dlx 5/6 ei activity. At eii MECP2 loss was accompanied by HDAC1 loss, suggesting that chromatin in eii was more active in Evf2 mutants, which in turn could increase Dlx5 transcription. However, HDAC1 binding to ei did not change, suggesting that an alternative regulation mechanism was employed at this site. Recent evidence suggests that in some cases MECP2 can act as a transcriptional activator, associating with CREB1 at activated targets37. What determines whether MECP2 acts as a transcriptional activator or repressor? Data from this paper demonstrated _Evf2_-dependent recruitment of MECP2, supporting the possibility that _trans_-acting RNAs may influence the choice between activator or repressor activities, either through co-recruitment of additional factors or by a novel mechanism.
Another question regarding the mechanism of _Evf2-_mediated recruitment of MECP2 and DLX proteins is whether recruitment occurs competitively, equally on both alleles, or in a mutually exclusive manner (Supplementary Fig. S2). Allelic imbalance causes both subtle (1.5-fold) and dramatic (9-fold) changes in gene regulation and occurs in 20–50% of tested genes38–40. Similarly, it is possible that the level of Evf2 localized at specific alleles may determine the extent or identity of the factor(s) recruited. Previous fluorescent RNA in situ hybridization data support the idea that E13.5 MGE nuclei display heterogeneous distribution of Evf2 between alleles; within an E13.5 MGE section Evf2 transcripts may be distributed on single, equal double, and unequal double alleles4. In this study, DLX and MECP2 were also distributed non-homogenously among different cells and within MGE nuclei (Fig. 3c–j). These data raise the possibility that unequal distribution of Evf2, DLX and MECP2 may play a role in regulating allelic imbalance within different populations of neuronal precursors, leading to neuronal diversity or phenotypic variation. Further studies to test the possible role of RNA-dependent control of allelic imbalance will be important to understanding how cellular diversity arises.
In humans, 15–25% of genes participate in anti-sense transcription41, however there are only a few reports of an in vivo mechanism that employs specific anti-sense transcriptional regulation1. Among these, in vivo roles of anti-sense transcripts have been described for Air35 and Kcnq1ot142 in imprinting control. In this paper, Evf2 mutant analysis combined with rescue experiments suggested that Evf2, an anti-sense transcript of Dlx6, negatively regulated Dlx6 transcription in cis through opposite strand transcriptional competition. However, the ability of Evf2 to regulate Dlx5 and GAD67 in trans raised the possibility that anti-sense transcripts may not be limited to _cis_-acting mechanisms or even to local regulation. Given the number of known anti-sense transcripts in non-imprinted regions, it will be important to determine how often anti-sense transcripts exhibit _trans_-regulatory effects, and the role of this regulation in various biological processes.
Non-coding RNA-dependent GABAergic interneuron development
The embryonic MGE is regulated by Dlx-dependent mechanisms and produces GABAergic interneuron precursors that will later populate the hippocampus29. In this paper, we show that loss of Evf2 resulted in imbalanced gene expression in the embryonic MGE, leading to decreased GABAergic interneurons in early post-natal (P2) hippocampus and dentate gyrus. Furthermore, this decrease in GABAergic interneurons in P2 Evf2 mutants did not result from increased cell death or a cell fate transformation in the hippocampus (Fig. 4g–k).
Why then are GABAergic interneurons decreased in P2 Evf2 mutant hippocampus? One possibility is that decreased GAD67 in E13.5 Evf2 mutant MGE reduced GABA levels in interneuron precursors, altering their tangential ventral to dorsal migration and reducing the number of GABAergic interneurons that reach their destination in the hippocampus. This is supported by reports that GABA affects neuronal migration in multiple contexts43 including tangential migration from MGE to cortex38, 44, 45. In addition, the interference with DLX 1/2 activity that was seen in Evf2 mutants would be expected to impair GABAergic interneuron migration from E13.5 MGE28,29. However, if the majority of GABAergic interneurons derive from embryonic and early post-natal ventral sources defective in Evf2 mutants, then how did GABAergic interneuron numbers recover in the adult Evf2 mutant hippocampus? One possibility is that the Evf2 mutant adult compensates by neurogenesis of GABAergic interneurons in either the hippocampal subgranular zone or rostral subventricular zone. If so, it is clear from the electrophysiology experiments that these neurons were not functionally equivalent at the synaptic level to their embryonically-generated counterparts. An alternate possibility is that GABAergic precursors in Evf2 mutants migrated at the appropriate time and to the proper destination, but produced lower levels of GAD67 in the absence of Evf2. If migration defects are found, future experiments will be needed to investigate why the numbers of GABAergic interneurons decrease at P2, how they eventually recover, and what is the basis for their synaptic defects.
Shh signaling in the embryonic ventral forebrain initiates a transcriptional cascade that requires DLX’s, Evf2, MECP2, and GAD67 for proper GABAergic interneuron development (Supplemental Fig. S3). While a large body of literature suggests that post-natal effects of MECP2 are likely to be critical for Rett syndrome46, 47 our results raise the possibility that Evf2 loss and MECP2 loss share a common mechanism where transcriptional effects in embryonic MGE cause adult GABAergic defects at the synaptic level. Efforts to identify single or multiple targets of MECP2 in the etiology of Rett syndrome have been inconclusive. Controversial evidence has raised the possibility that deregulation of Dlx5 in adult Mecp2−/− brains may be responsible for GABAergic defects23, 48. However, analysis of Evf2 mutants reveals that the correlation between MECP2 loss, increased Dlx5 and GABAergic defects may be indirect.
Given that DLX5 is a known activator of GAD6736, GABAergic interneuron loss would not be a predicted effect of the 2-fold Dlx5 increase in Evf2 mutants. In fact, Evf2 rescue experiments argued against Dlx5 upregulation as a cause for GABAergic defects in Evf2 mutants and suggested that the mechanism involved in Evf2 regulation of Dlx genes is separate from Evf2 regulation of GAD67. Rescue experiments suggested that Evf2 controls GAD67, Dlx5, and Dlx6 through distinct trans and _cis_- mechanisms. In addition, at the 2μg Evf2 rescue concentration, Dlx5 increased while GAD67 is rescued. If a 2-fold Dlx5 increase were the cause of GABAergic defects, GAD67 would not be expected to increase. These experiments suggest that GAD67 reduction rather than increased Dlx5 is more likely to cause GABAergic interneuron defects.
This paper describes the first demonstration that an ncRNA-dependent mechanism critical for early GABAergic interneuron development can determine GABA-dependent connectivity in the adult brain. The inability to recover proper connectivity, regardless of restored GAD67 and GABAergic interneuron number in adult brain, reinforces the idea that critical factors in the developing embryo influence GABAergic interneuron function in adult. This is especially important given the long-standing question of whether mental disorders, in the absence of apparent physiological adult deficits, can result from altered embryonic development.
Methods
Primer sequences
Gene targeting
Retrieval homology arms
- Cla I: GATGCGAATCGATCGGCTTAGGCCTCCAGGTTTC
- Hind III: AAACCCTAAGCTTGACTAGCGTGGCCCAAAGGT
- Hind III*: GATGCGAAAGCTTCTGTCAGTGCCAAAATGGAAGGACAT
- Nhe I: GATGCGAGCTAGCGGGGTTGGGACCTGGTTTTAGG
Targeting arms
- Sac II: TTAGTTCCGCGGCCTGGTCCTTTCTTCGTCTCAAGTC
- Not I: ATTTGCGGCCGCCTTAAGAGATATTCACCGGGGTAAGTTTTTATT
- Cla I: GATTTTATCGATCAATGATCAGGGTCTAGAAATCTATACTGAG
- Kpn: GATTTTGGTACCTTCAGGGTTTGATTTGATCGCTACTG
- 5′ ES Southern probe, mEvf5′.1: TGGTGAAGCTGGAGGAAGGAC
- mEvf5′.2: CACACTGACTTCTGAACACCCCTG
- 3′ Southern probe, mEvf3′.1: GGGGTGAAGGATGGTGATTAAAGAGC
- mEvf3′.1: GTGGCTGGCTGTCCTTTGGT
Quantitative Reverse Transcription PCR
SYBR Green:
The following concentration of primers:
- Evf2-F (0.2μM): 5′ CTCCCTCCGCTCAGTATAGATTTC 3′
- Evf2-R (0.2μM): 5′ CCTCCCCGGTGAATATCTCTT 3′
- Dlx2-F (0.15μM): 5′ CCCTACGGCACCAGTTCGT 3′
- Dlx2-R (0.15μM): 5′ TCGGATTTCAGGCTCAAGGT 3′
- Nrp2-F (0.5μM): 5′ ACTTTTCAGGACACGAAGTGAGAA 3′
- Nrp2-R (0.5μM): 5′ GCCAGCATCTTTGGAATTCAG3′
- Gad67-F (0.4uM): 5′ ACTCCTTCGCCTGCAACCT 3′
- Gad67-R (0.4uM): 5′ CGCCACACCAAGTATCATACGT 3′
- βActin-F (0.3μM): 5′ GCGAGCACAGCTTCTTTGC 3′
- βActin-R (0.3μM): 5′ TCGTCATCCATGGCGAACT 3′
TaqMan PCR:
- Dlx5-probe (0.1μM): 5′ CAAGCATCCGATCCGGCGACTTC 3′
- Dlx5-F (0.1μM): 5′ TATGACAGGAGTGTTTGACAGAAGAGT 3′
- Dlx5-R (0.1μM): 5′ ACGTCGGGAACGGAGCTT 3′
- Dlx6-probe (0.1μM): 5′ AACGCCTACGGAGCTTCTGAAGGAGACA 3′
- Dlx6-F (0.1μM): 5′ GAG ACCACAGATGATGTGACTTCTCT 3′
- Dlx6-R (0.1μM): 5′ CTGCCATGTTTGTGCAGATTCT 3′
- Evf1-probe (0.1μM): 5′ AGAGCTATGCGACTGTAGGCAAGCCAT 3′
- Evf1-F (0.1μM): 5′ GCATGGAAACTTTGATACCTTGGT 3′
- Evf1-R (0.1μM): 5′ GCCTTTCAGAACTAGAAGGGATTTAAA 3′
- βActin-probe (0.1μM): 5′ CAACGAGCGGTTCCGATGCCC T 3′
- βActin-F (0.1μM): 5′ ACGGCCAGGTCATCACTATTG 3′
- βActin-R (0.1μM): 5′ CAAGAAGGAAGGCTGGAAAAGA 3′
Quantitative PCR
SYBR Green PCR Core Reagents Kit were used for the following primers:
- 1-F (0.25μM): 5′ TATGAAAAGCCCAGGATTGC 3′
- 1-R (0.25μM): 5′ TGTCCCAGCTTCCTATCACC 3′
- 2-F (0.25μM): 5′ TGGTTTGAAAGAGGGGAATG 3′
- 2-R (0.25μM): 5′ AGAGCGCTTATTCTGAAACCA 3′
- 3-F (0.12μM): 5′ CCCAGGATCAATTCTGAACAAAG 3′
- 3-R (0.50μM): 5′ TCCCCAATGTCTGCTTCAAAT 3′
- 4-F (0.10μM): 5′ TGGATTCCCTGAACTCCAAG 3′
- 4-R (0.10μM): 5′ AGGGCTTGGGAACTCAAACT 3′
- 5-F (0.24μM): 5′ GGCGCATCTTTGCAAATTACA 3′
- 5-R (0.50μM): 5′ GCAGGCTGGATTAGGATGCTA 3′
- 6-F (1.0μM): 5′ TCGAAAGTATTGCGTGGATG 3′
- 6-R (1.0μM): 5′ GTGTGTACCAAGCGCATGTC 3′
- 7-F (0.25μM): 5′ GGCGTGTCAGCACCTGATTT 3′
- 7-R (0.25μM): 5′ GCCAAGTCACTGCCCATCTC 3′
Generation of Evf2TS/TS Mice
The Evf2 targeting construct was generated using lambda phage based recombineering in E.coli as described49. Using high fidelity Taq (Roche), homology arms of approximately 500bp were PCR amplified (with restriction sites added) from BAC DNA. Using a three-fragment ligation, homology arms were cloned into Cla I and Nhe I sites of PL253, with a Hind III site engineered between them. A 16.1kb region (corresponding to position 6,809,651–6825,742 on mouse chromosome 6, NCBI assembly) was retrieved from pBAC e3.6 M8 (M. Ekker) into the retrieval plasmid using recombination-induced EL250 cells 49. Further targeting was performed on the retrieved plasmid. The polyadenylation targeting vector was constructed in PL452, a floxed-Neo containing plasmid. The triple polyadenylation signal was cloned into EcoR I and Sal I sites of PL452. Approximately 500bp of targeting homology arms were cloned sequentially on either side of the polyA-floxed-Neo insert. Briefly, fragments were PCR amplified as above and cloned into either Cla I and Kpn I sites or Not I and Sac II sites. This triple polyA-floxed-Neo casette was targeted into the retrieved 16.1kb region using recombination-induced EL250 cells. Successful targeting was confirmed by Southern blot analysis of the completed construct using internal probes (NEBlot kit, NEB).
Mouse ES cells were targeted by homologous recombination using standard procedures. Successful targeting in ES cells was confirmed by Southern blot, verifying proper recombination at both the 5′ and 3′ ends. Probes were generated outside the 16.1kb homologous region. The 5′ probe was 499bp (Chr6 bases 6,808,430–6,808,928) and the 3′ probe was 991bp (Chr6 bases 6,825,821–6,826,811). Wildtype ApaL I sites are at Chr6 bases 6,828,765 and 6,817,913, yielding a 10.8 kb fragment that hybridizes with the 3′ probe. EL250 cells and recombineering plasmids PL253 and PL452 were provided by Dr. Neal Copeland.
Evf2TS (floxed neo)/+ heterozygotes were verified by Southern and crossed to EIIAcre (Jackson Labs) for two generations. Neo removal was verified by PCR (not shown). Mice are kept on a mixed 129/FVB/C57Bl6 background, and housed according to IACUC guidelines.
Chromatin Immunoprecipitation
MGE tissue was collected from E13.5 mouse embryos with genotype Evf2 +/+ or Evf2TS/TS (10–15 embryos per group). Tissue was broken into single cells by pipeting and spun through a 70μm filter. The DNA was crosslinked with 1% paraformaldehyde for 90 minutes on a rotator and then resuspended in SDS Lysis Buffer (1% SDS, 50mM Tris-HCl pH 8.1, 10mM EDTA) plus protease inhibitors: PMSF (170 μg/ml), pepstatin (0.7 μg/ml), leupeptin (10 μg/ml), and aprotinin (10 μg/ml). The crosslinked DNA was sonicated on a Microson Ultrasonic Cell Disruptor at Power 6 for 6 pulses, 10 seconds each. The sonicated mixture was spun down and the supernatant was used for ChIP.
For each IP condition, 20μg of chromatin was used in a total volume of 1000μl TES Buffer (50mM Tris-HCl pH 8.1, 1mM EDTA, 150mM NaCl) plus protease inhibitors. The chromatin was pre-cleared on a rotator at 4°C: 75μl washed Protein G-Agarose beads were added for 1 hour, the supernatant was incubated with 10μl rabbit preimmune serum for 1 hour, 75μl Protein G-Agarose were added for 1 hour, and the supernatant was split in half for an antibody condition and a rabbit preimmune condition. The chromatin was immunoprecipitated at 4°C: antibody (5μg) or rabbit preimmune serum (2μl) were added for 4 hours and then 100μl Protein G-Agarose was added overnight. The Protein G-Agarose beads were washed twice with Low Salt Buffer (0.1% SDS, 1% TritonX, 2mM EDTA, 20mM Tris-HCl pH 9.1, 150mM NaCl), once with 500mM NaCl, twice with LiCl Buffer (0.25M LiCl, 1% sodium deoxycholate, 1mM EDTA, 10mM Tris-HCl pH 8.1, 1% IGEPAL), and twice with TE Buffer (10mM Tris-HCl pH 8.1, 1mM EDTA). Then the chromatin was eluted off the Protein G-Agarose beads by incubation with 100μl Elution Buffer (1% SDS, 0.1M NaHCO3) twice. The DNA crosslinking was reversed by incubation in 0.5M NaCl for 5 hours at 65°C. Then the DNA was proteinase K treated by adding half a volume of 15mM EDTA/30mM Tris-HCl pH 8.1 and 750μg/ml proteinase K for 1 hour at 65°C. The uncrosslinked DNA was phenol extracted and ethanol precipitated using glycogen. Rabbit pan-anti-Dll antibody was produced in the laboratory. Pan anti-Dlx antibodies as originally described18, were made in our laboratory and extensively characterized4. Pan anti-Dlx antibodies were raised in rabbits against the 62 amino acid butterfly dll homeodomain containing fragment, affinity purified and tested by Western (dll fragment and zebrafish Dlx 1, 2 4, 6, mouse Dlx2) and immunohistochemistry (neural explants, embryonic forebrain sections). Pan anti-Dlx antibody recognizes zebrafish Dlx family members, including Dlx 1, 2, 4, and 64.
Monoclonal anti-Mecp2 antibodies were obtained commercially (Affinity Bioreagents, CO), and previously characterized26. We verified that these anti-Mecp2 antibodies bind only 2 bands in adult olfactory bulb extracts by Western analysis (data not shown). Anti-Hdac1 antibodies were obtained from SIGMA.
Primers were optimized to concentrations at which they were ~100% efficient with a standard curve slope of ~ −3.32.
Data Analysis
The enrichment of the antibodies and rabbit preimmune serum were determined by comparing the Ct values of the IP condition to the Ct value of input chromatin using the following formula: 2−(Ct(IP)−Ct(Input))
Once the enrichments were calculated, the relative enrichment of each antibody was calculated for Evf2 +/+ and Evf2TS/TS E13.5 MGE tissue using the following equation: Antibody Enrichment/Preimmune Enrichment
Immunohistochemistry
Brain tissue was fixed in 4% paraformaldehyde overnight, and then sunk consecutively in 15% and then 30% sucrose in PBS overnight. Tissue was cryostat sectioned at 18μm and air dried before staining. In situ hybridization was completed as previously published4. Immunohistochemistry was performed using tyramide amplification (TSA Kit #12, Invitrogen). Tissue was treated with 50% formamide/50% 2X SSC for 2 hours at 65°C and then washed in 2X SSC. Cells were permeabilized with 0.5% TritonX-100 in PBS and endogenous peroxidase activity was quenched by incubating the tissue in 1% hydrogen peroxide in PBS for 30 minutes. Tissue was blocked in 1% blocking solution (Component D) and labeled overnight with primary antibody, rabbit anti-GABA (Sigma, 1:500) or anti-pan-anti-dll (0.01 mg/ml, purified in the lab) in blocking solution. Tissue was washed in PBS and then incubated in secondary antibody, goat anti-rabbit IgG-HRP conjugate (Component C, 1:100) for 1 hour. Tissue was washed in PBS, and a working tyramide solution was applied for 15 minutes: Alexa Fluor 488 dye (Component A, 1:100) in amplification buffer (Component E) with 0.0015% hydrogen peroxide (Component F). Finally DAPI stain was applied for nuclear localization.
Western Blot
Evf2 +/+ and Evf2TS/TS E13.5 MGE tissue was homogenized in SDS-PAGE sample buffer, separated on a 12.5% SDS-PAGE gel, transferred to nitrocellulose and probed with primary rabbit anti-Dlx2 (1:2000) from Dr. David Eisenstat followed by secondary anti-rabbit peroxidase (1:5000 Sigma). Blot was re-probed with mouse anti-β-actin antibody (1:20,000, AC-15, SIGMA). Bands were visualized using a chemiluminescence kit (Perkin Elmer).
Electroporation
DNA was dissolved to a concentration of 0.1μg/ul in H2O + 0.04% trypan blue for visualization during injections. E12.5 Evf2TS/TS embryos were obtained from mating Evf2TS/TS males and females, and dissected in cold L-15 media. The heads were placed in cold PBS and 1.5 μl of DNA was injected into the lateral ventricles of the forebrain, under a dissection microscope. Electroporation using the square wave protocol (BioRad Gene Pulser XCell), delivered 5 X 50ms pulses of 30V with 1s intervals. Each embryo was electroporated twice using electroporation paddles (Protech) that were placed laterally on each side and then reversed in order to electroporate both left and right sides of the brain. The medial ganglionic eminence (MGE) was dissected and cultured in neurobasal medium (DMEM/F12 containing B-27 (Gibco), N2 supplement (Gibco), 200μM L-glutamine, 0.1μg/ml Penicillin/Streptomycin, and 1μg/ml mitomycin C on 0.02 μm filters (Nunc). After 24 hours in culture, RNA was isolated from pools of 4 MGE explants for qRT-PCR analysis.
Quantitative Reverse Transcription PCR
RNA Isolation and Reverse Transcription
Total RNA from pairs of E13.5 MGE tissue was isolated50 and treated with RNASE-free DNASE I (NEB), and reverse-transcribed using random hexamers (NEB) and MMoLV reverse-transcriptase (Invitrogen).
Quantitative PCR
SYBR Green PCR Core Reagents Kit (Applied Biosystems, 4304886) was used for all quantitative ChIP PCR on the 7500 Fast Real-Time PCR System (Applied Biosystems). The following PCR program was used: AMPErase UNG treatment at 50°C for 2 min., AmpliTaq Gold activation at 95°C for 10 min., then 40 cycles of denaturation at 95°C for 10 sec., anneal at 58°C for 5 sec., and elongate at 72°C for 32 sec. Each 25μl reaction included 5μl of a 1:50 IP dilution, 2.5μl 10x PCR Buffer, 3μl 25mM MgCl2, 2.0μl dNTP blend, 0.25μl AmpErase UNG, 0.25μl AmpliTaq Gold DNA polymerase. Primers were optimized to concentrations at which they were ~100% efficient or had a standard curve slope of ~ −3.32.
TaqMan PCR Core Reagents Kit (Applied Biosystems, N8080228) was used for the rest of the quantitative RT-PCR primers on the 7500 Fast Real-Time PCR System (Applied Biosystems). The following PCR program was used: AMPErase® UNG treatment at 50°C for 2 min., AmpliTaq Gold activation at 95°C for 15 min., then 40 cycles of denaturation at 95°C for 30 sec. and anneal/elongate at 59°C for 1 min. Each 25μl reaction included 50ng cDNA, 2.5μl 10x PCR Buffer, 5.0μl 25mM MgCl2, 0.75μl dATP, 0.75μl dGTP, 0.75μl dCTP, 0.75μl dUTP, 0.25μl AmpErase UNG, 0.25μl AmpliTaq Gold DNA polymerase.
Data Analysis
The fold gene expression of Evf2 +/+ and Evf2TS/TS E13.5 MGE tissue was determined by comparing the Ct values of the target gene to the Ct value of the control gene (β-actin) using the following formula: 2−(Ct(target)−Ct(βActin)).
Electrophysiology
Transverse hippocampal slices (300 μm) were prepared from 3–5 month old Evf2TS/TS mice. Slices were cut in ice-cold (~4 °C) oxygenated ACSF and then were allowed to recover for 1–2 hr before being transferred to the recording chamber where they were continuously superfused with solution heated to 32–34 °C and saturated with 95% O2/5% CO2. The standard extracellular solution contained (in mM) 124 NaCl, 3 KCl, 1.25 KH2PO4, 2.0 CaCl2, 1.3 MgCl2, 26 NaHCO3, 10 glucose. The concentration of MgCl2 was raised to 2.0 mM and that of CaCl2 decreased to 1.4 mM in the slicing solution. To isolate GABAergic current, 20 μM 6-ciano-7-dinitroquinoxaline-2,3-dione (CNQX) and 50 μM D-(−)-2-amino-5-phosphonovaleric acid (D-AP5) were added to the standard ACSF. Tetrodotoxin (TTX) (2 μM) was added to the drug solution to isolate minimal IPSCs.
Somatic whole-cell recordings were obtained from CA1 pyramidal neurons under visual guidance using infrared differential interference contrast (IR DIC) microscopy. Patch pipettes were pulled from borosilicate glass capillary tubing (A-M Systems, Inc). Internal solution for the patch pipettes contained (in mM): Cs methanesulphonate 120, CsCl 10, NaCl 5, HEPES 10, EGTA 10, TEA-Cl 5, Mg-ATP 4, GTP 0.3, QX-314 5, pH 7.3–7.4 adjusted with CsOH, osmolarity 290±10 mOsm.
Voltage-clamp recordings were performed using Axopatch-200B (Axon Instruments). In all our recordings series resistance was < 22 MΩ and was compensated at 80%. The recorded signal was low-pass filtered at 5 kHz, digitized at 10 kHz with a PCI-MIO-16E-4 board (National Instruments). All data were stored on a PC computer with custom software using C++ Builder 5.0 (Borland) and a NI DAQ 6.5 driver (National Instruments).
Evoked IPSCs were elicited at 0.1-0.07 Hz with a bipolar Pt/Ir electrode (2×25 μm) positioned on the stratum radiatum within 150 μm from the cell being recorded (Schaffer collateral-commisural fibres location site). At the beginning of each experiment, stimulus intensity was adjusted to evoke EPSCs with amplitude 0.4–0.2 nA. Evoked IPSCs were recorded at holding potential from −70 mV to +20 mV with 10 mV voltage steps after blocking EPSCs with a solution containing CNQX and AP5.
Spontaneous IPSCs were analyzed using MiniAnalysis 6.0.3 software (Synaptosoft). All events were manually selected based on their kinetics. Between 250 and 450 individual events were analyzed for each cell. Post-recording analyses of evoked IPSPs were done using Clampfit 10 (Axon Instruments). All statistical values were evaluated with Origin 8 (MicroCal Software). Values are presented as mean ± SEM. Statistical differences were established at p < 0.05 using the Student t-test, unless otherwise indicated.
Animal Use
Mice were handled under IACUC approval according to institutional guidelines.
Statistical Analysis
Statistical significance was determined using a variety of different tests, as indicated in individual figure legends, with *p<0.05, **p<0.01, and ***p<0.001. Error bars on graphs correspond to S.E.M.
Supplementary Material
1
Acknowledgments
We thank Kenny Campbell for providing early passage W4 embryonic stem cells, training and guidance in ES cell manipulation, Mark Ekker for Dlx 5/6 BAC, Pentao Lui and Neal Copeland for PL253, PL452, bacterial strains and protocols for BAC recombineering, Greg Taborn and Phil Iannaccone for blastocyst injections, David Eisentstat for anti-DLX2-specific antibody, Alex Joyner for triple polyA construct, John Rubenstein for GAD67 probe, and Qiufu Ma for v-Glut1 probe. We thank Kelly Jones (NUFSM) for establishing the conditions for embryonic tissue ChIP. This work was funded by NICHHD RO1 HD044745 and R21 HD049875, Illinois Regenerative Medicine Institute, and Illinois Excellence in Academic Medicine to J.D.K.
Footnotes
Author contributions
A.M.B performed experiments (Fig 1, 2, 3, 4, 5). M.GW.V. performed BAC recombineering, ES cell homologous targeting and screening for generating Evf2 TS mice. J.C.S. performed experiments (Fig 1, 3, 5). M.F.C. performed experiments (Fig 1, 5, S1), E.A.S. and J.F.D. contributed electrophysiology experiments and results for Fig 6. J.D.K. conceived of and directed experiments, wrote manuscript and assembled Fig 1–5, and S1-3.
References
- 1.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 3.Shamovsky I, Nudler E. Gene control by large noncoding RNAs. Sci STKE. 2006;2006:pe40. doi: 10.1126/stke.3552006pe40. [DOI] [PubMed] [Google Scholar]
- 4.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerucha T, et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilligan P, Brenner S, Venkatesh B. Fugu and human sequence comparison identifies novel human genes and conserved non-coding sequences. Gene. 2002;294:35–44. doi: 10.1016/s0378-1119(02)00793-x. [DOI] [PubMed] [Google Scholar]
- 7.Santini S, Boore JL, Meyer A. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 2003;13:1111–1122. doi: 10.1101/gr.700503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cristo G. Development of cortical GABAergic circuits and its implications for neurodevelopmental disorders. Clin Genet. 2007;72:1–8. doi: 10.1111/j.1399-0004.2007.00822.x. [DOI] [PubMed] [Google Scholar]
- 11.Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet. 2006;7:415–426. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- 12.Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16:276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Dani VS, et al. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 16.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 17.Zhou QP, et al. Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res. 2004;32:884–892. doi: 10.1093/nar/gkh233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panganiban G, Sebring A, Nagy L, Carroll S. The development of crustacean limbs and the evolution of arthropods. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- 19.Kohtz JD, Baker DP, Corte G, Fishell G. Regionalization within the mammalian telencephalon is mediated by changes in responsiveness to Sonic Hedgehog. Development. 1998;125:5079–5089. doi: 10.1242/dev.125.24.5079. [DOI] [PubMed] [Google Scholar]
- 20.Kohtz JD, et al. N-terminal fatty-acylation of sonic hedgehog enhances the induction of rodent ventral forebrain neurons. Development. 2001;128:2351–2363. doi: 10.1242/dev.128.12.2351. [DOI] [PubMed] [Google Scholar]
- 21.Feng J, et al. Synergistic and antagonistic roles of the Sonic hedgehog N- and C-terminal lipids. Development. 2004;131:4357–4370. doi: 10.1242/dev.01301. [DOI] [PubMed] [Google Scholar]
- 22.Anderson SA, et al. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 23.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 24.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 25.Le TN, et al. Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J Biol Chem. 2007;282:19071–19081. doi: 10.1074/jbc.M607486200. [DOI] [PubMed] [Google Scholar]
- 26.Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 28.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 29.Pleasure SJ, et al. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- 30.Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 31.Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- 32.Cobos I, et al. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- 33.Potier B, Jouvenceau A, Epelbaum J, Dutar P. Age-related alterations of GABAergic input to CA1 pyramidal neurons and its control by nicotinic acetylcholine receptors in rat hippocampus. Neuroscience. 2006;142:187–201. doi: 10.1016/j.neuroscience.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 34.Xu C, Cui C, Alkon DL. Age-dependent enhancement of inhibitory synaptic transmission in CA1 pyramidal neurons via GluR5 kainate receptors. Hippocampus. 2009 doi: 10.1002/hipo.20544. [DOI] [PubMed] [Google Scholar]
- 35.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 36.Stuhmer T, Anderson SA, Ekker M, Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- 37.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. Detection of regulatory variation in mouse genes. Nat Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- 39.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 40.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yelin R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 42.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heng JI, Moonen G, Nguyen L. Neurotransmitters regulate cell migration in the telencephalon. Eur J Neurosci. 2007;26:537–546. doi: 10.1111/j.1460-9568.2007.05694.x. [DOI] [PubMed] [Google Scholar]
- 44.Cuzon VC, Yeh PW, Cheng Q, Yeh HH. Ambient GABA promotes cortical entry of tangentially migrating cells derived from the medial ganglionic eminence. Cereb Cortex. 2006;16:1377–1388. doi: 10.1093/cercor/bhj084. [DOI] [PubMed] [Google Scholar]
- 45.Lopez-Bendito G, et al. Blockade of GABA(B) receptors alters the tangential migration of cortical neurons. Cereb Cortex. 2003;13:932–942. doi: 10.1093/cercor/13.9.932. [DOI] [PubMed] [Google Scholar]
- 46.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci U S A. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schule B, Li HH, Fisch-Kohl C, Purmann C, Francke U. DLX5 and DLX6 expression is biallelic and not modulated by MeCP2 deficiency. Am J Hum Genet. 2007;81:492–506. doi: 10.1086/520063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1