p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit (original) (raw)
Abstract
Accurate chromosome segregation requires the spindle assembly checkpoint to be active at the onset of mitosis, before being silenced following chromosome alignment. p31comet is a checkpoint antagonist in that its inhibition delays mitotic exit, whereas its overexpression overrides the checkpoint. How exactly p31comet antagonises the checkpoint is unclear. A prevalent model is that p31comet acts as a ‘cap’ by inhibiting recruitment of the open conformation form of Mad2 (O-Mad2) to the kinetochore-bound complex of Mad1–C-Mad2 (closed conformation Mad2), an essential step that is required for checkpoint activation. Here, we show that although p31comet localises to kinetochores in mitosis, modulation of its activity has no effect on recruitment of O-Mad2 to kinetochores. Rather, our observations support a checkpoint-silencing role for p31comet downstream of kinetochores. We show that p31comet binds Mad2 when it is bound to the mitotic checkpoint complex (MCC) components BubR1 and Cdc20. Furthermore, RNAi-mediated inhibition of p31comet results in more Mad2 bound to BubR1–Cdc20, and conversely, overexpression of p31comet results in less Mad2 bound to BubR1–Cdc20. Addition of recombinant p31comet to checkpoint-arrested extracts removes Mad2 from the MCC, whereas a p31comet mutant that cannot bind Mad2 has no effect. Significantly, expression of a Mad2 mutant that cannot bind p31comet prolongs the metaphase to anaphase transition. Taken together, our data support the notion that p31comet negatively regulates the spindle assembly checkpoint by extracting Mad2 from the MCC.
Key words: Mitosis, Kinetochore, Spindle assembly checkpoint, Anaphase promoting complex, Cdc20, BubR1
Introduction
The spindle assembly checkpoint (SAC) delays anaphase until all chromosomes are correctly attached to spindle microtubules through their kinetochores (Musacchio and Salmon, 2007). Unattached kinetochores activate the SAC, which leads to inhibition of the anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase that targets two anaphase inhibitors for proteolytic degradation, namely securin and cyclin B1 (Peters, 2006). The SAC inhibits the APC/C co-activator function of Cdc20, preventing ubiquitylation of securin and cyclin B1 (Musacchio and Salmon, 2007).
A key SAC component is Mad2, a small protein that can adopt two conformations, ‘open’ (O-Mad2) and ‘closed’ (C-Mad2) (Luo et al., 2004; Mapelli et al., 2007). Mad2 binds directly to Mad1, and, in doing so, adopts the closed conformation (Luo et al., 2002; Sironi et al., 2002). In interphase, the Mad1–C-Mad2 core complex localises to the nuclear envelope (Campbell et al., 2001), before relocalising to unattached kinetochores during mitosis (Chen et al., 1998). Because Mad2 can dimerise, C-Mad2 bound to Mad1 in turn recruits O-Mad2 and catalyses its conformational change to C-Mad2, thereby promoting its binding to Cdc20 (De Antoni et al., 2005). Thus, Mad1–C-Mad2 acts as a template, converting additional Mad2 molecules into Cdc20 inhibitors (Musacchio and Salmon, 2007). C-Mad2–Cdc20 then binds to the BubR1–Bub3 complex, forming the mitotic checkpoint complex (MCC), which in turn binds the APC/C (Sudakin et al., 2001). When bound by the MCC, APC/C activity towards securin and cyclin B1 is inhibited.
Once a cell correctly biorients its chromosomes, the SAC is satisfied and anaphase follows after a relatively constant period (Rieder et al., 1994). Several mechanisms are thought to contribute to checkpoint silencing. First, dynein-dependent stripping of SAC proteins from attached kinetochores prevents the generation of de novo signals (Howell et al., 2001). Second, activation of phosphatases counters SAC kinases such as Bub1 and Mps1 (Vanoosthuyse and Hardwick, 2009). Finally, p31comet has been shown to negatively regulate the SAC in mammalian cells (Habu et al., 2002; Xia et al., 2004).
p31comet (also known as MAD2L1-binding protein; SWISSPROT identifier Q15013) was originally identified as a Mad2 binding partner (Habu et al., 2002). Overexpression of p31comet overrides the SAC, whereas p31comet RNAi delays mitotic exit after nocodazole release (Habu et al., 2002; Xia et al., 2004). Taking these observations together with the fact that p31comet binds directly to C-Mad2 at its dimerization interface (Yang et al., 2007) and competes with O-Mad2 binding (Mapelli et al., 2006), it has been proposed that p31comet acts as an inhibitory ‘cap’ on the Mad1–C-Mad2 core complex (Mapelli et al., 2006; Musacchio and Salmon, 2007). According to this model, at unattached kinetochores, p31comet is somehow removed to allow recruitment of O-Mad2 to the Mad1–C-Mad2 core (Mapelli et al., 2006). Although this model is provocative, rigorous demonstration in cells is currently lacking. Alternative models have been proposed; one study suggested that p31comet contributes to SAC silencing by promoting Cdc20 ubiquitylation leading to disassembly of the MCC (Reddy et al., 2007). In addition, a recent report suggests that p31comet promotes dissociation of Cdc20 from BubR1 in an ATP-dependent manner (Teichner et al., 2011).
Our interest in p31comet was triggered by recent observations showing that Mps1 kinase activity is required to recruit O-Mad2 to the Mad1–C-Mad2 core complex during mitosis (Hewitt et al., 2010). If p31comet does act as an inhibitory cap, we reasoned that Mps1 might be required to release it, promoting O-Mad2 recruitment, and thereby kick-starting the Mad2-template reaction (Hewitt et al., 2010). Similarly, inactivation of Mps1 upon chromosome alignment might reactivate p31comet, capping the core and leading to checkpoint silencing. To test this possibility, we turned our attention to p31comet. However, although we could find no evidence supporting the capping model, we have uncovered a role for p31comet downstream of kinetochores. We show that p31comet promotes an early step in MCC disassembly, extracting Mad2 and leaving behind a BubR1–Bub3–Cdc20 complex that is still capable of inhibiting the APC/C.
Results
Novel reagents to study p31comet
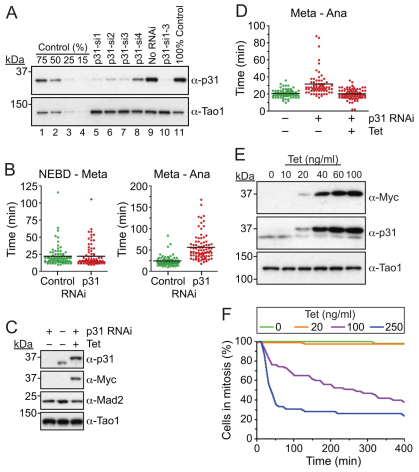
To study p31comet, we generated a novel anti-p31comet antibody. Anti-p31comet detected a single band, which diminished markedly when HeLa cells were transfected with siRNAs designed to repress p31comet (Fig. 1A). Of four siRNAs tested, three repressed p31comet by approximately 75% which, when pooled, routinely repressed p31comet by ~90% (Fig. 1A, lane 10). Previously, p31comet RNAi has been shown to delay mitotic exit following release from nocodazole (Xia et al., 2004). In our hands, p31comet inhibition delayed mitotic progression in an unperturbed mitosis. Time-lapse imaging of HeLa cells expressing a GFP-tagged Histone showed that p31comet RNAi had no effect on chromosome alignment (Fig. 1B). However, the time between metaphase and anaphase onset was extended from ~24 minutes in controls to ~56 minutes in p31comet-deficient cells (Fig. 1B). To confirm that this phenotype was not due to an off-target effect, we generated a HeLa line expressing a tetracycline-inducible Myc-tagged p31comet transgene rendered resistant to p31comet siRNA (Fig. 1C). When we repressed p31comet, the onset of anaphase was again delayed, but induction of the RNAi-resistant transgene restored normal anaphase timing (Fig. 1D).
Fig. 1.
Confirmation that p31comet negatively regulates the SAC. (A) Immunoblots of HeLa cells transfected with siRNA oligos targeting p31comet probed with sheep polyclonal antibodies against p31comet and Tao1. The negative control sample was titrated to quantify RNAi efficiency. (B) Time-lapse analysis of HeLa cells stably expressing GFP–Histone-H2B following p31comet RNAi. Scatter plots show the time from nuclear envelope breakdown (NEBD) to metaphase, and from metaphase to anaphase. Horizontal bars show the median. (C) Immunoblot of HeLa cells showing induction (20 ng/ml tetracycline) of an RNAi-resistant Myc-tagged p31comet transgene following p31comet RNAi. (D) Analysis of GFP–Histone-H2B HeLa cells showing that RNAi-resistant Myc–p31comet rescues the metaphase delay induced by p31comet RNAi. (E) Immunoblot showing overexpression of Myc-tagged p31comet. (F) Time-lapse analysis of GFP–Histone-H2B HeLa cells overexpressing p31comet treated with 0.66 μM nocodazole. The time taken from NEBD to mitotic exit was determined. The cumulative plot shows the number of cells remaining in mitosis.
Because overexpression of p31comet has been shown to override the SAC (Habu et al., 2002), we used low-level induction in the RNAi-rescue experiment to ensure that Myc–p31comet levels were similar to endogenous (Fig. 1C). To confirm that p31comet overexpression does override the SAC, we induced much higher expression levels (Fig. 1E). Under these conditions, the SAC was severely compromised, because cells could not arrest in mitosis when challenged with nocodazole (Fig. 1F). Taken together, these observations validate both the antibody and the siRNAs, and they confirm that p31comet is an inhibitor of the SAC (Habu et al., 2002; Xia et al., 2004).
p31comet localises to unattached kinetochores in a Mad2-dependent manner
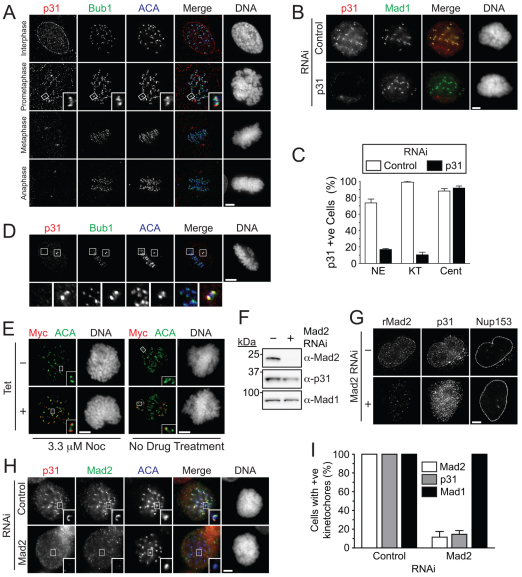
To better understand the localisation of p31comet during mitosis we stained asynchronous HeLa and RPE cells with the antibody against p31comet. During interphase and prophase, p31comet localised to the nuclear envelope (Fig. 2A), which is consistent with localisation of Mad1 and Mad2 (Campbell et al., 2001). In prometaphase, p31comet localised to kinetochores. By metaphase this signal was reduced, although centrosome staining was apparent (Fig. 2A).
Fig. 2.
p31comet localises to unattached kinetochores during mitosis. (A) Immunofluorescence images of hTERT RPE cells probed with the indicated antibodies, showing localisation of p31comet to kinetochores in prometaphase. (B) Images of p31comet RNAi HeLa cells treated with 3.3 μM nocodazole for 1 hour showing loss of kinetochore staining. (C) Quantification of cells in B, confirming loss of kinetochore staining following p31comet RNAi. (D) Images of a HeLa cell showing localisation of p31comet at the kinetochores of an uncongressed chromosome. Whereas kinetochores of congressed chromosomes stain poorly for p31comet (left inset), the kinetochores of an unaligned chromosome are enriched for p31comet (right inset). (E) Images of Myc-tagged p31comet-expressing (250 ng/ml tetracycline) HeLa cells treated with 3.3 μM nocodazole (left panel) and MG132 for 1 hour. (F) Western blot of an asynchronous HeLa cell extract showing Mad2 protein depletion after transfection of siRNAs targeting Mad2. (G) Representative immunofluorescence images showing diminished nuclear envelope localisation of p31comet in an interphase cell depleted of Mad2. (H) Images showing diminished p31comet kinetochore localisation in a mitotic cell depleted of Mad2, treated with 3.3 μM nocodazole. (I) Bar graph quantifying nocodazole-treated mitotic cells, displaying kinetochore localisation of the indicated proteins after Mad2 RNAi. Values represent the mean and s.e.m. of at least 90 cells counted across three independent experiments. Scale bars: 5 μm.
To verify this staining pattern, we repressed p31comet by RNAi: although kinetochore staining was clearly evident in control cells, p31comet RNAi diminished this markedly (Fig. 2B,C). By contrast the centrosome staining remained. Thus, the kinetochore localisation of p31comet appears to be bona fide, but the centrosome staining probably reflects a crossreactivity. Notably, in cells where the majority of chromosomes had aligned, p31comet was clearly visible at the kinetochores of unaligned chromosomes (Fig. 2D).
To confirm these observations, we analysed HeLa cells expressing Myc-tagged p31comet. When exposed to nocodazole, the majority of kinetochores were Myc positive (Fig. 2E). By contrast, in untreated cells, only kinetochores on unaligned chromosome stained for Myc. Thus, p31comet does indeed localise to kinetochores during mitosis, and similarly to other well-characterised SAC proteins, its abundance at kinetochores diminishes following chromosome alignment.
We were surprised that the localisation of p31comet was very similar to that of Mad2, i.e. abundant at unattached kinetochores (Waters et al., 1998). If the capping model is correct, one might expect p31comet levels at unattached kinetochores to be low, thereby allowing the Mad1–C-Mad2 core complex to recruit O-Mad2. However, it is conceivable that the localisation of p31comet is independent of Mad1–C-Mad2. Importantly, although Mad2 RNAi did not significantly affect p31comet protein levels (Fig. 2F), it did abolish p31comet nuclear envelope and kinetochore localisation (Fig. 2G–I). Because p31comet binds C-Mad2 (Xia et al., 2004), this suggests that p31comet targets these locations by binding Mad1–C-Mad2, which is consistent with p31comet capping the core complex. However, as we show later, an alternative explanation can account for how p31comet localises to kinetochores by binding C-Mad2.
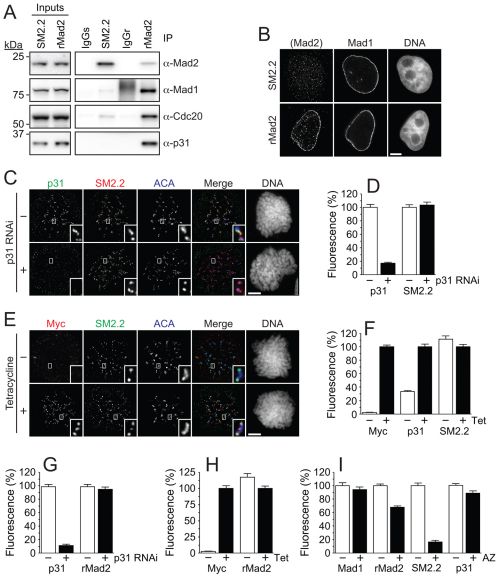
Testing the capping model
To test the capping model further, we asked whether modulation of p31comet levels affected the abundance of O-Mad2 at kinetochores. To do this, we used a sheep polyclonal antibody, SM2.2, which we recently showed preferentially recognises O-Mad2, at least in immunofluorescence experiments (Hewitt et al., 2010). To first confirm that SM2.2 does indeed selectively bind O-Mad2 in cells, we performed immunoprecipitations comparing SM2.2 with a rabbit polyclonal anti-Mad2 antibody. Importantly, rabbit anti-Mad2 efficiently co-precipitated Mad1 and Cdc20 along with Mad2 (Fig. 3A), suggesting that rabbit anti-Mad2 can bind C-Mad2. By contrast, although SM2.2 immunoprecipitated more Mad2, the amounts of co-precipitating Mad1 or Cdc20 were markedly reduced, which is consistent with the notion that SM2.2 preferentially binds O-Mad2 (Fig. 3A). Interestingly, rabbit anti-Mad2 co-precipitated p31comet but SM2.2 did not, confirming that p31comet binds C-Mad2 whereas SM2.2 does not.
Fig. 3.
Modulating p31comet levels has no detectable effect on kinetochore-bound O-Mad2. (A) Immunoblots of Mad2 immune complexes isolated from nocodazole-arrested HeLa cells using either SM2.2 or rMad2. SM2.2 poorly co-precipitates Mad1, Cdc20 and p31comet, which is consistent with it preferentially recognising O-Mad2. (B) Representative immunofluorescence images showing nuclear envelope localisation of Mad2 when probed with rMad2. Note that SM2.2 fails to detect the nuclear envelope. (C) Images of nocodazole-treated HeLa cells following p31comet RNAi showing no effect on kinetochore localisation of O-Mad2. (D) Bar graph quantifying pixel intensities of anti-p31comet and SM2.2 at kinetochores normalized to ACA in mitotic HeLa cells following p31comet RNAi. (E) Images of HeLa cells overexpressing Myc–p31comet (250 ng/ml tetracycline) showing no effect on kinetochore localisation of O-Mad2. (F) Bar graph quantifying pixel intensities of anti-Myc, anti-p31comet and SM2.2 at kinetochores normalised to ACA in mitotic HeLa cells following p31comet overexpression. (G,H) Bar graphs quantifying pixel intensities of anti-Myc, anti-p31comet and rabbit antiMad2 (rMad2) at kinetochores following p31comet RNAi (G) and overexpression (H). In all cases, cells were treated with 3.3 μM nocodazole 1 hour before fixation. (I) Bar graph quantifying Mad2 and p31comet pixel intensities at kinetochores in HeLa cells treated with the Mps1 inhibitor AZ3146 for 1 hour. The reduction in O-Mad2 is not accompanied by an increase in p31comet. Before treatment with AZ3146, cells were treated with 3.3 μM nocodazole and MG132 for 1 hour to ensure the cells were arrested in mitosis with unattached kinetochores. In all cases, bar graphs show the mean ± s.e.m. derived from three independent experiments, in which at least 50 kinetochores were quantified in five cells. Scale bars: 5 μm.
Interestingly, as alluded to above, Mad1 and Mad2 localise to the nuclear envelope (Campbell et al., 2001), as does p31comet (Hewitt et al., 2010) (Fig. 2A). The most likely explanation is that this reflects the existence of a Mad1–C-Mad2–p31comet complex bound to the nuclear envelope. If so, it is important to note that SM2.2 does not recognise the nuclear envelope of interphase cells, providing further evidence that SM2.2 does not efficiently recognise Mad2 when bound directly to Mad1 (Fig. 3B).
Having validated SM2.2 as a tool to recognise O-Mad2, we asked whether repressing p31comet increased O-Mad2 at kinetochores by uncapping additional Mad1–C-Mad2 core complexes. Although RNAi depleted p31comet from kinetochores, there was no effect on O-Mad2 binding (Fig. 3C,D). Conversely, when we overexpressed p31comet to a level that caused SAC override, there was no apparent reduction in the amount of O-Mad2 bound, despite p31comet levels at kinetochores increasing ~threefold (Fig. 3E,F). Thus, neither the ability of p31comet RNAi to delay mitotic exit nor the ability of overexpressed p31comet to override the SAC correlates with changes to recruitment of O-Mad2 to kinetochores. Note that modulating p31comet levels also had no effect on the level of kinetochore-bound Mad2 as measured using the rabbit anti-Mad2 antibody (Fig. 3G,H).
Previously, we speculated that Mps1 activity might be required to release p31comet, thereby allowing recruitment of O-Mad2 to the Mad1–C-Mad2 core complex (Hewitt et al., 2010). If true, p31comet levels should increase when Mps1 was inactivated. However, when we used AZ3146 to inhibit Mps1, we saw no increase in kinetochore-bound p31comet (Fig. 3I). Consistent with our previous observations (Hewitt et al., 2010), SM2.2 kinetochore staining diminished markedly upon Mps1 inhibition, reflecting a failure to recruit O-Mad2 (Fig. 3I). By contrast, the rabbit anti-Mad2 antibody still decorated kinetochores, although levels were reduced to ~70% of control cells (Fig. 3I), which is consistent with C-Mad2 still being bound to Mad1 (Hewitt et al., 2010).
In summary, we observed no changes in recruitment of O-Mad2 to kinetochores after inhibiting or overexpressing p31comet. Furthermore, when recruitment of O-Mad2 was blocked by inhibition of Mps1 activity, p31comet levels at kinetochores did not increase. Taken together, these data are difficult to reconcile with a role for p31comet acting as a cap on the Mad1–C-Mad2 complex. Therefore, we considered whether other aspects of the SAC might be regulated by p31comet.
p31comet interacts with SAC components downstream of kinetochores
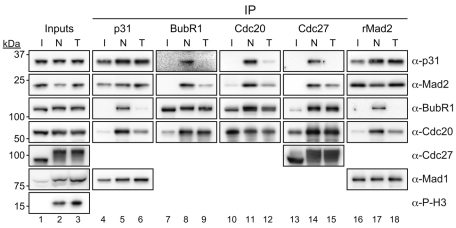
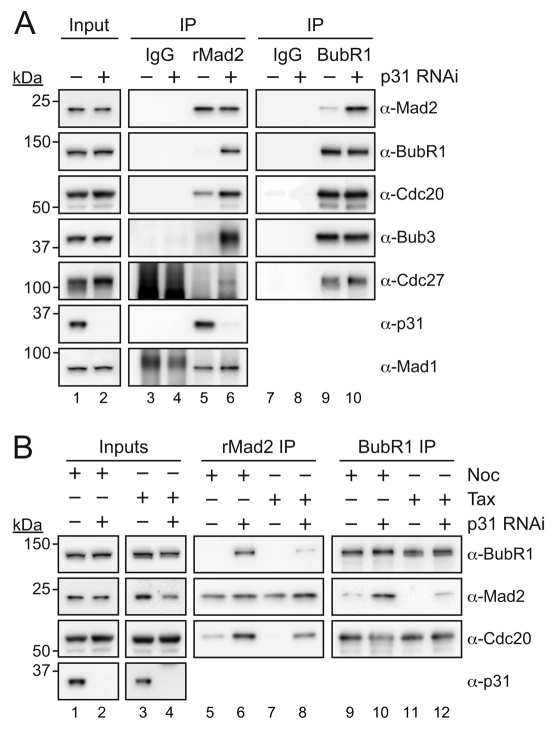
To explore the idea that p31 acts downstream of kinetochores (Reddy et al., 2007; Teichner et al., 2011), we asked whether p31comet binds other SAC components. When we immunoprecipitated p31comet from either interphase or mitotically arrested cells we could readily detect Mad1 and Mad2 (Fig. 4, lanes 4–6 and supplementary material Fig. S1A). In mitotic cells, p31comet also co-precipitated BubR1 and Cdc20 (Fig. 4, lanes 5 and 6). Consistently, when we immunoprecipitated BubR1 or Cdc20 from nocodazole-arrested cells, p31comet was detectable (Fig. 4, lanes 8 and 11). Furthermore, when we immunoprecipitated the APC/C from nocodazole-arrested cells using antibodies against Cdc27, p31comet was present (Fig. 4, lane 14). Thus, not only does p31comet interact with Mad2, but it is also present in complexes with other MCC components, namely BubR1 and Cdc20, and also the APC/C. This prompted us to investigate whether p31comet plays a role in regulating MCC function.
Fig. 4.
p31comet interacts with MCC components during mitosis. Immunoblots of immune complexes from interphase (I), nocodazole-arrested (N, 0.66 μM) or Taxol-arrested (T, 10 μM) HeLa cells probed with the indicated antibodies.
Less p31comet and Mad2 is bound to BubR1–Cdc20 in Taxol-arrested cells
During the course of the immunoprecipitation experiments described above, we were struck by a difference that manifested depending on whether the cells were arrested in mitosis with either nocodazole or taxol. Whereas nocodazole prevents microtubule polymerisation, leading to the vast majority of kinetochores being unattached, taxol stabilizes microtubules, thereby allowing kinetochore-microtubule interactions, albeit erroneous and unstable ones (Yang et al., 2009). Consequently, the number of kinetochores generating SAC signals is higher in nocodazole-treated cells compared with taxol-treated cells. However, because a single unattached kinetochore can prevent anaphase (Rieder et al., 1995), the SAC is ‘on’ in both cases because the APC/C is inhibited. Indeed, when either nocodazole or taxol was used, the extent of Cdc27 phosphorylation was the same, confirming that both samples were from mitotic-arrested cells (Fig. 4, compare lanes 2 and 3).
Although p31comet bound Mad1 and Mad2 equally in nocodazole and taxol, binding to BubR1, Cdc20 and Cdc27 was higher in nocodazole-treated cells (Fig. 4, compare lanes 5 and 6, 8 and 9, 11 and 12, and 14 and 15). Similarly, when we immunoprecipitated Mad2 using the rabbit polyclonal antibody, binding to BubR1 and Cdc20 was more prevalent in nocodazole-treated cells (Fig. 4, compare lanes 17 and 18). Thus, when many kinetochores are signalling, p31comet and Mad2 can be easily detected bound to BubR1, Cdc20 and the APC/C. However, when only a few unattached kinetochores are signalling, as in taxol-treated cells, less p31comet and Mad2 is bound to BubR1, Cdc20 and the APC/C. Importantly however, regardless of whether nocodazole or taxol were used, similar amounts of BubR1 and Cdc20 were bound to each other and to the APC/C (Fig. 4, compare lanes 8 and 9, 11 and 12, and 14 and 15), which is consistent with the fact that both drugs trigger a robust mitotic arrest. These data suggest therefore that the amount of Mad2 and p31comet bound to BubR1–Cdc20 is highly dependent on the number of unattached kinetochores.
Depletion of p31comet raises the level of Mad2 bound to BubR1–Cdc20
Because p31comet binds Mad2 when it forms part of the MCC, we asked whether modulating p31comet levels affected MCC composition. Significantly, upon p31comet RNAi more BubR1, Bub3 and Cdc20 were bound to Mad2, even though Mad1 binding was unaffected (Fig. 5A, compare lanes 5 and 6). Consistently, when we immunoprecipitated BubR1 from p31comet-depleted cells, significantly more Mad2 co-precipitated, whereas the amounts of Bub3 and Cdc20 were similar (Fig. 5A, lanes 9 and 10). Thus, p31comet appears to limit the amount of Mad2 bound to BubR1–Cdc20.
Fig. 5.
p31comet RNAi increases the level of Mad2 bound BubR1–Cdc20. (A) Immunoblots of Mad2 and BubR1 immune complexes isolated from nocodazole-arrested HeLa cells following p31comet RNAi. Mad2 was immunoprecipitated with a rabbit polyclonal antibody. (B) Immunoblots of Mad2 and BubR1 immune complexes isolated from p31comet RNAi HeLa cells arrested with either 0.66 μM nocodazole or 10 μM taxol. Note that the experiments in A and B were done in parallel, therefore the nocodazole ‘inputs’ in B are the same as those in A.
Because the amount of Mad2 bound to BubR1 and Cdc20 is higher in nocodazole-arrested cells compared with taxol-arrested cells (Fig. 4), we compared the effect of p31comet RNAi in these different conditions. Consistent with the data in Fig. 4, more Cdc20 co-precipitated with Mad2 from nocodazole-arrested cells, although BubR1 binding was difficult to detect (Fig. 5B, lanes 5 and 7). In both nocodazole- and taxol-treated cells, p31comet RNAi caused an increase in the amount of Cdc20 bound to Mad2, and BubR1 became detectable (Fig. 5B, lanes 6 and 8). In line with this, when we immunoprecipitated BubR1, p31comet RNAi increased the amount of co-purifying Mad2 in both nocodazole and taxol (Fig. 5B). In other words, p31comet RNAi had the same effect in either condition, namely increasing the amount of Mad2 bound to BubR1–Cdc20.
Together, these data suggest that the amount of Mad2 bound to BubR1–Cdc20 is variable during SAC signalling, and is positively regulated by unattached kinetochores and negatively regulated by p31comet: in the presence of nocodazole or in the absence of p31comet, the binding of Mad2 to BubR1–Cdc20 is enhanced.
p31comet extracts Mad2 from the MCC
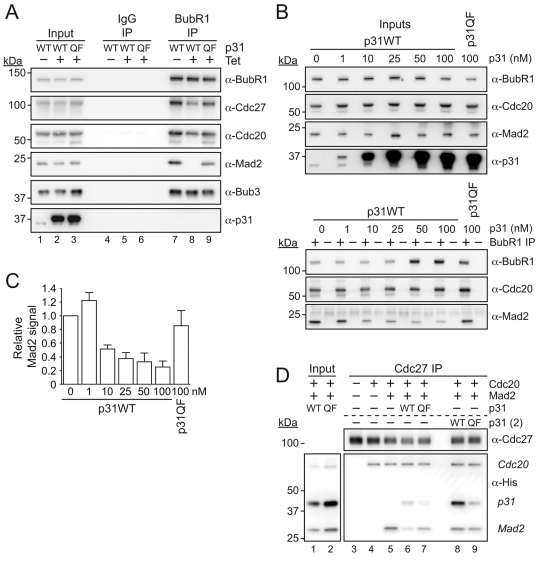
The observation that p31comet appears to regulate Mad2 binding to BubR1–Cdc20 during SAC signalling provides a novel explanation for why repressing p31comet delays anaphase onset. p31comet might contribute to efficient SAC silencing by limiting the amount of MCC that needs to be disassembled upon SAC satisfaction. If this model is correct, it might also explain why p31comet overexpression overrides the SAC; excess p31comet might reduce MCC levels below the threshold necessary for APC/C inhibition. In agreement, when p31comet was overexpressed in cells, the level of Mad2 present in BubR1 immunoprecipitates was drastically reduced (Fig. 6A, compare lanes 7 and 8). To ensure that this experiment was appropriately controlled, we wanted to ensure that this effect was dependent on the ability of p31comet to bind Mad2. Therefore, we generated a cell line expressing Myc-tagged p31comet Q83A F191A (p31QF), a mutant that has previously been shown to be deficient in Mad2 binding (Yang et al., 2007) (supplementary material Fig. S2). Consistent with previous observations, when we expressed p31QF in HeLa cells (supplementary material Fig. S2A) it did not bind Mad2 (supplementary material Fig. S2B). p31QF did not localise to kinetochores, suggesting that it is indeed recruited by Mad2 binding (supplementary material Fig. S2C). Furthermore, whereas p31comet overexpression overrode the SAC, overexpression of p31QF did not (supplementary material Fig. S2D). Interestingly, overexpression of p31comet had a potent effect in a colony formation assay, drastically inhibiting cell proliferation (supplementary material Fig. S2E). By contrast, p31QF was largely benign, demonstrating that the effect p31comet overexpression has on cell fate is absolutely dependent on its ability to bind Mad2. Importantly, when we overexpressed p31QF, the amount of Mad2 co-precipitating with BubR1 was similar to that in controls (Fig. 6A). Thus, the capacity of p31comet to limit the amount of Mad2 bound to BubR1 requires its ability to bind Mad2.
Fig. 6.
p31comet removes Mad2 from preassembled MCC. (A) Immunoblots of BubR1 immunocomplexes isolated from HeLa cells overexpressing p31comet or the p31QF mutant. Before harvesting, cells were released from an S-phase block into medium containing 0.66 μM nocodazole for 10.5 hours. MG132 was added for 1 hour before mitotic cells were isolated by selective detachment. (B) Immunoblots of BubR1 immunocomplexes isolated from nocodazole-arrested HeLa cell extracts after the addition of recombinant p31comet for 30 minutes at room temperature. (C) Bar graph quantifying the amount of Mad2 co-purifying with BubR1 following addition of recombinant p31comet. Values represent the mean ± s.e.m. derived from two independent experiments, each of which was analysed twice. (D) Immunoblots of Cdc27 immunocomplexes incubated with recombinant His-tagged Cdc20, Mad2 and p31comet as indicated. In lanes 3–7, 1 μM of the indicated His-tagged recombinant proteins were incubated together for 1 hour at room temperature, and added to the APC/C for 1 hour. In lanes 8 and 9, 1 μM p31comet protein was added after Mad2 and Cdc20 for 1 hour. Following protein incubation, the APC/C was eluted with a competitive peptide overnight and co-eluting proteins were analysed by western blotting.
The capacity of p31comet to reduce the amount of Mad2 in the MCC could arise by one of two mechanisms. One possibility is that p31comet prevents Mad2 from being assembled onto Cdc20, as predicted by the capping model. However, p31comet RNAi had little effect on the level of BubR1–Cdc20 (Fig. 5), and because BubR1–Cdc20 requires the prior formation of Mad2–Cdc20 (Davenport et al., 2006; Nilsson et al., 2008; Kulukian et al., 2009), this possibility seems unlikely. An alternative explanation is that p31comet extracts Mad2 from preassembled MCC, leaving behind an intact BubR1–Bub3–Cdc20 subcomplex.
To test this possibility, we prepared cell extracts from nocodazole-arrested HeLa cells and titrated in recombinant p31comet. After a brief incubation, we immunoprecipitated BubR1 and quantified the amount of bound Mad2. Interestingly, although addition of exogenous p31comet had no effect on Cdc20 binding to BubR1, it markedly diminished the amount of co-precipitating Mad2 (Fig. 6B,C). Importantly, addition of p31QF had no effect on Mad2 binding (Fig. 6B,C). Thus, our data suggest that p31comet silences the SAC by extracting Mad2 from the MCC complex.
When we repeated this experiment but immunoprecipitated Cdc27 instead of BubR1, we observed no effect on Mad2 (data not shown), suggesting that p31comet might not be able to extract Mad2 from the MCC when it is bound to the APC/C. To explore this further, we assessed the binding of recombinant Mad2 and Cdc20 to immunopurified APC/C in the presence of p31comet. When p31comet was incubated with Mad2 and Cdc20 before addition to the APC/C, it inhibited the association of Mad2 with the APC/C (Fig. 6D, compare lanes 5 and 6). However, if p31comet was added to Mad2-Cdc20 pre-assembled on the APC, it had no affect on Mad2 binding (Fig. 6D, compare lanes 5 and 8). Interestingly, we could detect p31comet bound to the APC/C in this instance, presumably through Mad2.
Taking together the data from p31comet RNAi, p31comet overexpression and the effect of adding exogenous p31comet to mitotic extracts, our observations support a model whereby p31comet negatively regulates the amount of Mad2 bound to BubR1–Cdc20, not by preventing the production of Mad2–Cdc20 complexes as predicted by the capping model, but rather by extracting Mad2 from the MCC, leaving behind an intact BubR1–Bub3–Cdc20 subcomplex.
The ability of Mad2 to bind p31comet is required for checkpoint silencing
If p31comet is required to remove Mad2 from the MCC, then a Mad2 mutant incapable of binding p31comet should stabilise the MCC, compromise SAC silencing, and in turn, prolong the metaphase to anaphase transition. It has previously been shown that Mad2 R133E,Q134A (Mad2RQ) does not bind p31comet (Mapelli et al., 2006). However, Mad2RQ is also dimerisation deficient (Mapelli et al., 2006). Thus, upon binding Mad1, Mad2RQ should form a core complex that is incapable of recruiting O-Mad2. This would be expected to override the SAC, thereby preventing us from probing the downstream role of p31comet. Bypassing this problem would require a Mad2 mutant that does not bind Mad1 but still binds Cdc20. Because Mad2 binds Mad1 and Cdc20 by a similar mechanism (Luo et al., 2002), we thought that such a mutant would not exist.
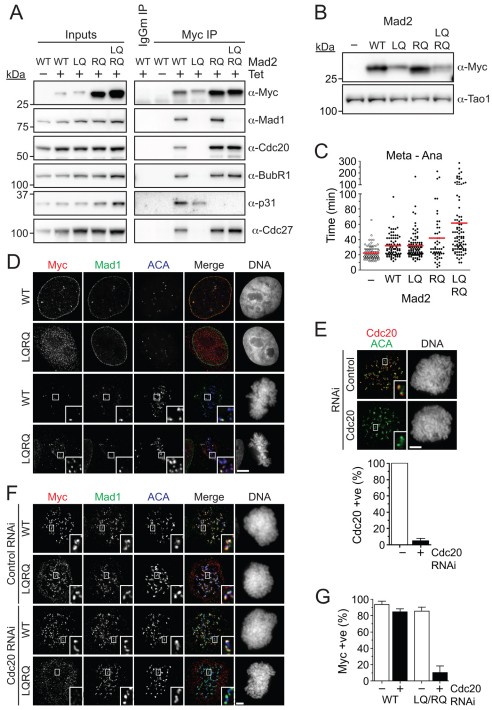
However, when we combined the RQ double mutant with the L13Q mutation, which promotes the folding of Mad2 into the closed conformation (Mapelli et al., 2007), this Mad2LQRQ triple mutant exhibited a very interesting behavior. When expressed in cells, Mad2LQRQ bound Cdc20, BubR1 and Cdc27, but not Mad1 or p31comet (Fig. 7A). In other words, Mad2LQRQ can assemble into the MCC and bind the APC/C, but cannot bind p31comet.
Fig. 7.
Overexpression of a Mad2 mutant that cannot bind p31comet delays anaphase. (A) Immunoblots of anti-Myc immune complexes isolated from nocodazole-arrested HeLa cells stably expressing the Myc-tagged Mad2 mutants indicated. (B) Immunoblot of HeLa cells transiently transfected with Myc-tagged Mad2 mutants. Note that Mad2LQ and Mad2LQRQ are expressed at similar levels. (C) Time-lapse analysis of HeLa cells co-transfected with Myc-tagged Mad2 mutants and GFP-Histone H2B measuring the time from metaphase to anaphase. (D) Immunofluorescence images of HeLa cells stably expressing wild-type Mad2 or Mad2LQRQ. Note that Mad2LQRQ does not localise to the nuclear envelope in interphase, but during mitosis it does localise to kinetochores that are negative for Mad1. (E) Representative immunofluorescence images and bar graph quantification showing Cdc20 is lost from kinetochores of nocodazole-treated cells after Cdc20 RNAi. (F) Immunofluorescence images showing that kinetochore localisation of Myc-tagged Mad2LQRQ (LQRQ) in nocodazole-treated cells is abolished following Cdc20 RNAi. (G) Bar graph quantifying the experiment in F, showing the number of cells that show a positive Myc signal at kinetochores. Quantifications in E and G are from three independent experiments with at least 50 cells counted in each. Scale bars: 5 μm.
To test whether Mad2 needs to bind p31comet for efficient checkpoint silencing, we transiently transfected Myc-tagged wild-type and mutant Mad2 into HeLa cells along with a GFP-tagged histone and analysed them by time-lapse microscopy. Note that Mad2LQ and Mad2LQRQ expressed to similar levels, whereas the wild-type protein was expressed at a slightly higher level (Fig. 7B). Consistent with data from yeast and Xenopus egg extracts (He et al., 1997; Li et al., 1997), overexpression of wild-type Mad2 extended the delay between metaphase and anaphase, from ~22 minutes to ~32 minutes (Fig. 7C). Cells expressing Mad2LQ also showed a delay in anaphase onset, taking ~33 minutes. Importantly, expression of Mad2LQRQ caused a significantly longer delay, doubling the mean time for the metaphase-anaphase transition to ~62 minutes when compared with Mad2LQ (Fig. 7C). Indeed, only ~1% of cells expressing Mad2LQ took over 100 minutes to initiate anaphase following metaphase alignment, whereas ~18% of cells expressing Mad2LQRQ did so. Thus, expression of a Mad2 mutant that cannot bind p31comet delays anaphase onset, which suggests that the Mad2–p31comet interaction is required for efficient checkpoint silencing.
Mad1-independent kinetochore localisation of Mad2
In line with Mad2LQRQ not binding Mad1, it did not localise to the nuclear envelope in interphase cells (Fig. 7D). Surprisingly however, it was readily detectable at kinetochores in nocodazole-treated cells (Fig. 7F). Furthermore, in unperturbed cells, Mad2LQRQ was evident at congressed kinetochores that were devoid of Mad1 (Fig. 7D). Together, these two observations raise the possibility that Mad2LQRQ localises to kinetochores independently of Mad1. Because Mad2LQRQ can bind Cdc20, which remains on kinetochores following microtubule attachment (Howell et al., 2004), we reasoned that kinetochore localisation of Mad2LQRQ might occur through Cdc20. To test this, we repressed Cdc20 by RNAi (Fig. 7E). Wild-type Mad2 was not affected by Cdc20 depletion, presumably because it binds Mad1 at kinetochores (Fig. 7F,G). However, as we predicted, kinetochore localisation of Mad2LQRQ was abolished in Cdc20-deficient cells. Thus, these data suggest that Mad2 can indeed bind kinetochores independently of Mad1, at least under these conditions where the p31comet -dependent extraction mechanism is inhibited as a result of the LQRQ mutant abolishing p31comet binding.
Discussion
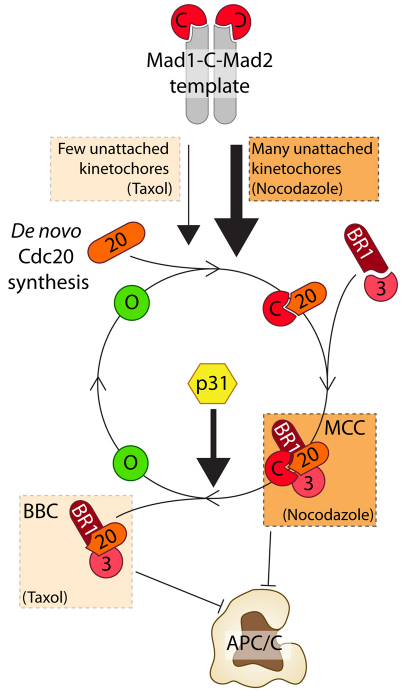
In this study, we confirm that p31comet is a spindle checkpoint antagonist and provide novel insight into how p31comet achieves this. Rather than capping Mad1–C-Mad2 templates, our data support a role for p31comet downstream of kinetochores. We show that p31comet associates with the MCC; based on elegant biochemical and structural studies, this interaction is most likely mediated by direct interaction with closed Mad2 (Xia et al., 2004;Mapelli et al., 2006). Our data show that p31comet limits the amount of Mad2 bound to BubR1–Cdc20 by promoting an early step in MCC disassembly. Indeed, addition of recombinant p31comet to preassembled MCC reduces the amount of Mad2 bound to BubR1–Cdc20, and importantly, this is dependent on the ability of p31comet to directly bind Mad2, because p31QF is benign in this assay. Significantly, upon Mad2 extraction, the BubR1–Cdc20 complex remains intact and capable of inhibiting the APC/C. In agreement with the notion that BubR1–Cdc20 complexes can inhibit the APC/C without Mad2, the amount of Mad2 bound to BubR1-Cdc20 is markedly lower in taxol-arrested cells compared with those arrested with nocodazole. Taking these observations together, we suggest a model whereby p31comet antagonises the SAC by extracting C-Mad2 from the MCC and recycling it back to free O-Mad2 (Fig. 8).
Fig. 8.
The Mad2 cycle. The Mad1–C-Mad2 core complex acts as a template at unattached kinetochores, recruiting O-Mad2 and Cdc20 to create C-Mad2–Cdc20 complexes. Binding to BubR1–Bub3 then results in production of tetrameric MCC. p31comet then removes Mad2, leaving a BubR1–Bub3–Cdc20 subcomplex. In nocodazole, the large number of unattached kinetochores generating C-Mad2–Cdc20 overwhelms the p31comet-dependent mechanism, leading to accumulation of MCC. In taxol, fewer kinetochores are unattached, therefore production of C-Mad2–Cdc20 complexes is reduced. In this case, the p31comet-dependent extraction of Mad2 dominates, reducing the amount of tetrameric MCC, but leaving behind a BubR1–Bub3–Cdc20 complex still capable of inhibiting the APC/C.
Our model builds upon the Mad2 template model, which posits that Mad1–C-Mad2 complexes at unattached kinetochores recruit O-Mad2, catalysing its conversion to C-Mad2 and concomitant binding to Cdc20. The C-Mad2–Cdc20 complex is then captured by BubR1–Bub3, thereby creating the MCC. This tetrameric complex can then bind and inhibit the APC/C. However, once the MCC is assembled, p31comet can extract Mad2 leaving behind a BubR1–Bub3–Cdc20 complex (Fig. 8). Importantly, because BubR1 remains bound to Cdc20, this complex can still inhibit the APC/C, despite the fact that Mad2 is no longer bound (Fig. 8). Importantly, when many kinetochores are unattached, the number of active Mad1–C-Mad2 core complexes is high so the Mad2 template mechanism outpaces p31comet-dependent extraction, and hence the MCC accumulates. However, when only a few kinetochores are unattached, p31comet-mediated recycling of Mad2 balances the production of new Mad2-Cdc20 complexes and hence the BubR1–Bub3–Cdc20 complex is the dominant species of APC/C inhibitor. This model provides a simple explanation for why the amount of Mad2 bound to BubR1–Cdc20 is variable depending on whether the cells are arrested with nocodazole or taxol (Fig. 8).
An immediate implication of this model is that the ultimate APC/C inhibitor is not the MCC, but rather the BBC, i.e. the BubR1–Bub3–Cdc20 complex. A corollary of this is that during a normal unperturbed mitosis, the balance of APC/C inhibitors in a cell might shift as chromosomes attach to the spindle, with the MCC dominating early on and the BBC dominating later. This model is consistent with recent data indicating that Mad2 is a substoichiometric MCC component, an observation which led to the suggestion that Mad2 acts catalytically to create BubR1–Cdc20 complexes (Nilsson et al., 2008; Kulukian et al., 2009). However, rather than Mad2 playing a catalytic role, we suggest that the MCC is indeed assembled from C-Mad2–Cdc20 and BubR1–Bub3 subcomplexes to create the tetrameric MCC, but then Mad2 is removed by p31comet to yield the BBC complex. Thus, the substoichiometric nature of Mad2 arises as a consequence of its extraction from the MCC by p31comet.
Although the model outlined above requires further testing, it raises a number of issues. First, how the BBC actually inhibits the APC/C still remains unclear. In vitro reconstitution experiments suggest that BubR1–Cdc20 might be sufficient for APC/C inhibition (Kulukian et al., 2009). However, Bub3 plays a key role in targeting BubR1 to kinetochores to efficiently capture C-Mad2–Cdc20 (P.L.G. and S.S.T., unpublished results), and in cells, Bub3 is clearly bound to BubR1–Cdc20. So, although Bub3 might be part of the ultimate inhibitor, it might not be required for inhibition per se. Similarly, the model implies that Mad2 is also not an actual APC/C inhibitor, but rather is required to promote the formation of the final inhibitor; perhaps upon Mad2 binding, Cdc20 adopts a conformation that allows it to be captured by BubR1–Bub3, but once BubR1 has bound Cdc20, Mad2 is no longer required to maintain the interaction (Nilsson et al., 2008; Kulukian et al., 2009).
Whether p31comet extracts Mad2 only when part of the MCC is unclear. Perhaps p31comet can recycle Mad2 from the C-Mad2–Cdc20 complex before BubR1 binding. Our preliminary observations suggest that p31comet is not sufficient to extract Mad2 from APC-bound MCC. Consistent with this, recent data indicate that ‘free’ MCC is disassembled before APC/C-bound MCC (Ma and Poon, 2011). It is unlikely that the APC/C masks Mad2 because we can detect p31comet bound to the APC/C. One possibility is that other factors in addition to p31comet are required to extract Mad2, and perhaps these are blocked when the MCC is bound to the APC/C. Alternatively, perhaps these additional factors are limiting in our in vitro experiments.
Although the exact mechanism by which p31comet recycles Mad2 off Cdc20 is unclear, several lines of evidence do support the notion that additional activities are involved in MCC disassembly. First, p31comet cannot be the only relevant factor because it is not essential for triggering anaphase: it is not conserved in yeast and even when potently repressed in human tumour cells, anaphase is only delayed. Second, an ATP-dependent process has been implicated in SAC silencing (Teichner et al., 2011), raising the possibility that an ATPase cooperates with p31comet in the disassembly process. In addition, CUEDC2 has very recently been implicated in dissociating Mad2 from Cdc20 (Gao et al., 2011). Finally, Cdc20 ubiquitylation and degradation also contributes to MCC turnover (Nilsson et al., 2008). Our preliminary observations suggest that p31comet is not required for Cdc20 ubiquitylation, but perhaps Mad2 extraction is required for ubiquitylated Cdc20 to be degraded, i.e. the p31-dependent Mad2 extraction mechanism might only be an early step of several involved in MCC disassembly.
We suggest that p31comet-mediated extraction of Mad2 is not a SAC silencing mechanism per se in that it is not ‘switched on’ following chromosome alignment. The fact that BBC is the predominant inhibitor in taxol-arrested cells indicates that Mad2 extraction occurs before the SAC has been satisfied. Thus, the p31comet mechanism appears to be a constitutive governor, dampening the amplitude of the SAC signal throughout mitosis, not just once the last chromosome aligns. Indeed, the notion that the MCC is continuously turned over when the SAC is on is supported by the demonstration that Cdc20 is ubiquitylated and degraded during a mitotic arrest (Nilsson et al., 2008). Why the spindle checkpoint is fine-tuned in this way remains unclear, but one possibility is that during an unperturbed mitosis, constant MCC turnover sensitises the SAC to the input signal at unattached kinetochores, thereby facilitating efficient anaphase onset irrespective of how long a cell has spent in prometaphase. Indeed, in both Ptk1 and HeLa cells, the onset of anaphase occurs after a relatively fixed time, regardless of how long it took to align the last chromosome (Rieder et al., 1994; Meraldi et al., 2004).
The capping model predicts that p31comet should compete with O-Mad2 for binding sites at kinetochores (Musacchio and Salmon, 2007). However, modulating p31comet levels had no discernible effect on kinetochore recruitment of O-Mad2. Indeed, the enrichment of p31comet at unattached kinetochores, which are presumably SAC active, was surprising. If p31comet is not capping Mad1–C-Mad2, then how is it being recruited to the kinetochore? p31comet certainly interacts with Mad1 in mitosis, and kinetochore localisation of p31comet is Mad2 dependent. Furthermore, because p31comet can only bind C-Mad2 (Xia et al., 2004), it seems reasonable to conclude that a Mad1–C-Mad2–p31comet complex might be present at kinetochores. However, our analysis of Mad2LQRQ opens up another possibility. This mutant assembles into the MCC because it readily binds BubR1 and Cdc20. Strikingly however, even though Mad2LQRQ cannot bind Mad1, it still localises to kinetochores, decorating aligned kinetochores negative for Mad1. This raises the possibility that C-Mad2 can bind to kinetochores through Cdc20 in addition to through Mad1. Indeed, RNAi experiments show that kinetochore localisation of Mad2LQRQ depends on Cdc20. In turn therefore, Mad2-dependent kinetochore localisation of p31comet might be due to it binding C-Mad2 bound to Cdc20, rather than C-Mad2 bound to Mad1. If this is the case, it suggests that p31comet-mediated disassembly of the MCC is occurring at kinetochores. Indeed, because p31comet is enriched on unaligned chromosomes, it would appear that unattached kinetochores are active sites for both assembly and disassembly of the MCC. However, it is important to stress that p31comet can be found bound to Mad1. This suggests that p31comet might cap Mad1–C-Mad2 complexes in the cytosol, or that the entire Mad2 cycle is taking place at the kinetochore, restricting the amount of inhibitor that diffuses away from the kinetochore to dampen the SAC signal and thereby ensure a timely anaphase.
Finally, although the p31comet mechanism might be a constitutive governor rather than a regulated silencing mechanism, the fact that overexpression of p31comet has such a potent effect on cell fate suggests that its levels must be carefully regulated. How this is achieved is unknown but clearly this issue has important implications for SAC control, the acquisition of chromosome instability, aneuploidy and cell fate.
Materials and Methods
Cell lines
All cell lines were cultured as described (Taylor et al., 2001). Stable HeLa cell lines were generated using Flp–FRT-mediated recombination (Tighe et al., 2004). Cells were synchronised in S-phase as described (Westhorpe et al., 2010). Nocodazole, Taxol and MG132 (Sigma) were used at a final concentrations of 0.66 μM, 10 μM and 20 μM, respectively, unless stated otherwise. AZ3146 (Tocris) was used at 2 μM.
RNAi and transient transfections
siRNA sequences (Dharmacon) were used at a final concentration of 50 nM (supplementary material Table S1) and transfected using Interferin™ (PolyPlus). For immunofluorescence and time-lapse analysis, cells were analysed 24 hours after siRNA transfection. For immunoprecipitation, cells were harvested 48 hours after transfection. Transient transfection of the Mad2 mutants was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Cell biology
The sheep polyclonal antibody Sp31 was raised and affinity purified against a GST fusion protein encoding amino acids 2–64 of p31comet using methods described previously (Taylor et al, 2001). All other antibodies used in this study are described in supplementary material Table S2. Immunofluorescence was performed as described (Hewitt et al, 2010). Immunofluorescence images were acquired either on an Axioskop2 Plus (Zeiss) fitted with CoolSNAP HQ CCD camera (Photometrics) or on a restoration microscope (Delta Vision RT; Applied Precision). Pixel intensity quantification was performed on deconvolved images using SoftWorx, as described previously (Hewitt et al., 2010). Graphs were produced in Prism (GraphPad). For time-lapse microscopy, HeLa cells were cultured in 24-well plates and imaged using an Axiovert 200 (Zeiss) microscope. Images were taken every 2 minutes and analysed using ImageJ.
Immunoprecipitation and immunoblotting
Cells were harvested and lysed for immunoprecipitation as previously described (Holland et al., 2007). After clearing by centrifugation, protein concentration was determined against a BSA standard using a Synergy HT plate reader (BIO-TEK). Normalised lysates were precleared with Protein-G–Sepharose beads and appropriate species of control IgG antibodies. After preclearing, cell lysates were transferred to fresh beads and the desired antibody added. The immunoprecipitate was washed in lysis buffer and isolated proteins resolved by SDS-PAGE, as previously described (Hewitt et al, 2010). In the case of membranes loaded with samples from immunoprecipitations, sheep primary antibodies were visualised using a recombinant Protein-G–HRP protein (Invitrogen). Immunoblotted proteins were visualised using chemiluminescence (SuperSignal; Thermo Fisher Scientific), either using film (Biomax MR; Kodak), or a Biospecturm 500 Imaging system controlled by VisionWorks LS software (UVP).
In vitro binding
Recombinant, 6×His-tagged wild-type Mad2, wild-type p31comet and p31QFprotein were produced by cloning respective cDNAs into the pET28a expression vector before transforming E.coli [BL21 (DE3) pLysS, Novagen]. Cells were induced overnight at 20°C with 0.4 mM IPTG, sonicated and centrifuged. 6×His–p31comet was purified using the TALON metal affinity resin system (BD Biosciences) essentially using the manufacturer's instructions, except that protein was bound, washed and eluted from beads using rounds of centrifugation rather than being column purified. Recombinant, 6×His-tagged Cdc20 was produced using a baculovirus expression system in Sf9 cells (P.L.G. and S.T., unpublished results). For the APC/C binding assay, 1 μM recombinant proteins were incubated together in PBS containing 10% glycerol and 4 mg/ml BSA for 1 hour at room temperature. Proteins were added to APC/C immunopurified from mitotic HeLa cells for 1 hour, and then washed in PBS containing 0.1% Triton X-100. Following washes, the APC/C was eluted using a competitive peptide as previously described (Herzog and Peters, 2005). Elutes were analysed by SDS-PAGE and immunoblotting.
Supplementary Material
Supplementary Material
Acknowledgements
The authors wish to thank Bill Earnshaw for the kind gift of ACA antibodies, and the rest of the Taylor lab for helpful discussions. We also thank Andrea Musacchio for helpful discussions.
Footnotes
Funding
F.G.W. is funded by the Wellcome Trust. P.L.G. is funded by CONICYT Chile. A.T. and S.S.T. are funded by a Senior Fellowship from Cancer Research UK. Deposited in PMC for release after 6 months.
References
- Campbell M. S., Chan G. K., Yen T. J. (2001). Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114, 953-963 [DOI] [PubMed] [Google Scholar]
- Chen R. H., Shevchenko A., Mann M., Murray A. W. (1998). Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143, 283-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J., Harris L. D., Goorha R. (2006). Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp. Cell Res. 312, 1831-1842 [DOI] [PubMed] [Google Scholar]
- De Antoni A., Pearson C. G., Cimini D., Canman J. C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E. D., et al. (2005). The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15, 214-225 [DOI] [PubMed] [Google Scholar]
- Gao Y. F., Li T., Chang Y., Wang Y. B., Zhang W. N., Li W. H., He K., Mu R., Zhen C., Man J. H., et al. (2011). Cdk1-phosphorylated CUEDC2 promotes spindle checkpoint inactivation and chromosomal instability. Nat. Cell Biol 13, 924-933 [DOI] [PubMed] [Google Scholar]
- Habu T., Kim S. H., Weinstein J., Matsumoto T. (2002). Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21, 6419-6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Patterson T. E., Sazer S. (1997). The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl. Acad. Sci. USA 94, 7965-7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F., Peters J. M. (2005). Large-scale purification of the vertebrate anaphase-promoting complex/cyclosome. Methods Enzymol. 398, 175-195 [DOI] [PubMed] [Google Scholar]
- Hewitt L., Tighe A., Santaguida S., White A. M., Jones C. D., Musacchio A., Green S., Taylor S. S. (2010). Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J. Cell Biol. 190, 25-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A. J., Bottger F., Stemmann O., Taylor S. S. (2007). Protein phosphatase 2A and separase form a complex regulated by separase autocleavage. J. Biol. Chem. 282, 24623-24632 [DOI] [PubMed] [Google Scholar]
- Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. (2001). Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155, 1159-1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. (2004). Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14, 953-964 [DOI] [PubMed] [Google Scholar]
- Hussein D., Taylor S. S. (2002). Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 115, 3403-3414 [DOI] [PubMed] [Google Scholar]
- Johnson V. L., Scott M. I., Holt S. V., Hussein D., Taylor S. S. (2004). Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117, 1577-1589 [DOI] [PubMed] [Google Scholar]
- Kulukian A., Han J. S., Cleveland D. W. (2009). Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell 16, 105-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gorbea C., Mahaffey D., Rechsteiner M., Benezra R. (1997). MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc. Natl. Acad. Sci. USA 94, 12431-12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H. (2002). The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 9, 59-71 [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Xia G., Wassmann K., Matsumoto T., Rizo J., Yu H. (2004). The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 11, 338-345 [DOI] [PubMed] [Google Scholar]
- Ma H. T., Poon R. Y. (2011). Orderly Inactivation of the Key Checkpoint Protein Mitotic Arrest Deficient 2 (MAD2) during Mitotic Progression. J. Biol. Chem. 286, 13052-13059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Filipp F. V., Rancati G., Massimiliano L., Nezi L., Stier G., Hagan R. S., Confalonieri S., Piatti S., Sattler M., et al. (2006). Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 25, 1273-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Massimiliano L., Santaguida S., Musacchio A. (2007). The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell 131, 730-743 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Draviam V. M., Sorger P. K. (2004). Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7, 45-60 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379-393 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Yekezare M., Minshull J., Pines J. (2008). The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10, 1411-1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N., Bastos R., McMorrow I., Burke B., Aebi U. (1994). Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126, 603-617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644-656 [DOI] [PubMed] [Google Scholar]
- Reddy S. K., Rape M., Margansky W. A., Kirschner M. W. (2007). Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446, 921-925 [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Schultz A., Cole R., Sluder G. (1994). Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127, 1301-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L., Cole R. W., Khodjakov A., Sluder G. (1995). The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130, 941-948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K. T., Musacchio A. (2002). Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21, 2496-2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G. K., Yen T. J. (2001). Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154, 925-936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., Hussein D., Wang Y., Elderkin S., Morrow C. J. (2001). Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114, 4385-4395 [DOI] [PubMed] [Google Scholar]
- Teichner A., Eytan E., Sitry-Shevah D., Miniowitz-Shemtov S., Dumin E., Gromis J., Hershko A. (2011). p31comet Promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc. Natl. Acad. Sci. USA 108, 3187-3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A., Johnson V. L., Taylor S. S. (2004). Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J. Cell Sci. 117, 6339-6353 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K. G. (2009). A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19, 1176-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Salmon E. D. (1998). Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141, 1181-1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F. G., Diez M. A., Gurden M. D., Tighe A., Taylor S. S. (2010). Re-evaluating the role of Tao1 in the spindle checkpoint. Chromosoma 119, 371-379 [DOI] [PubMed] [Google Scholar]
- Xia G., Luo X., Habu T., Rizo J., Matsumoto T., Yu H. (2004). Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23, 3133-3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li B., Tomchick D. R., Machius M., Rizo J., Yu H., Luo X. (2007). p31comet blocks Mad2 activation through structural mimicry. Cell 131, 744-755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Kenny A. E., Brito D. A., Rieder C. L. (2009). Cells satisfy the mitotic checkpoint in Taxol, and do so faster in concentrations that stabilize syntelic attachments. J. Cell Biol. 186, 675-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material