FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism (original) (raw)
. Author manuscript; available in PMC: 2012 Jan 22.
Summary
FMRP loss-of-function causes Fragile X Syndrome (FXS) and autistic features. FMRP is a polyribosome-associated neuronal RNA-binding protein, suggesting that it plays a key role in regulating neuronal translation, but there has been little consensus regarding either its RNA targets or mechanism of action. Here we use high throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) to identify FMRP interactions with mouse brain polyribosomal mRNAs. FMRP interacts with the coding region of transcripts encoding pre- and postsynaptic proteins, and transcripts implicated in autism spectrum disorders (ASD). We developed a brain polyribosome-programmed translation system, revealing that FMRP reversibly stalls ribosomes specifically on its target mRNAs. Our results indicate that loss of a translational brake on the synthesis of a subset of synaptic proteins may contribute to FXS. In addition, they provide insight into the molecular basis of the cognitive and allied defects in FXS and ASD, and suggest multiple targets for clinical intervention.
Introduction
Fragile X Syndrome (FXS) was the first genetic disorder to link RNA regulation to human cognitive function. Loss of function of the Fragile-X mental retardation protein (FMRP) causes FXS (Verkerk et al., 1991), the most common inherited form of mental retardation, which is further characterized by autistic behaviors, childhood seizures and abnormal dendritic spines (Hagerman and Hagerman, 2002; Hernandez et al., 2009). FMRP is an RNA binding protein (RNABP) whose function is incompletely understood, but is believed to be involved in translational regulation (Bassell and Warren, 2008; Gatto and Broadie, 2009; Costa-Mattioli et al., 2009; Zukin et al., 2009). This is of particular interest since new protein synthesis is required for long-term synaptic plasticity (Kelleher et al., 2004; Klann and Dever, 2004; Richter and Klann, 2009; Sutton and Schuman, 2006), a phenomenon thought to underlie the formation and persistence of memory (Malenka and Bear, 2004). Some activity-regulated translational control pathways have been identified, such as the ERK and mTOR pathways regulating initiation (Hoeffer and Klann, 2010), or eEF2 phosphorylation controlling elongation (Sutton et al., 2007), but in general these are thought to have broad effects on translation. A specific set of transcripts and the proteins regulating them to mediate synaptic plasticity remain to be defined.
FMRP is an excellent candidate for such a regulatory protein. In the brain, FMRP is present in the neuronal cell body, proximal dendrites and axons (Christie et al., 2009) and the majority of FMRP is associated with polyribosomes (Feng et al., 1997b; Khandjian et al., 2004; Stefani et al., 2004). Moreover, a missense mutation in the second RNA binding domain (I304N) abolishes FMRP polyribosome association (Zang et al., 2009; Feng et al., 1997a) and causes a Fragile X phenotype in mice (Zang et al., 2009) and humans (DeBoulle et al., 1993). Studies of Fmr1 knockout (KO) and I304N mice have documented a number of defects in synaptic plasticity (Pfeiffer and Huber, 2009; Zang et al., 2009). Together these observations suggest that FMRP regulates the translation of proteins important for proper synaptic function, yet there is no consensus as to how it might do so. In vitro, exogenous FMRP appears to repress translation of a variety of transcripts including luciferase reporter mRNA (Laggerbauer et al., 2001; Li et al., 2001). In dividing fibroblast cells, it has been suggested that transgenic Flag-tagged FMRP represses translation during elongation, but extrapolating this finding to endogenous FMRP in neurons is difficult (Ceman et al., 2003). Underscoring the uncertainty of such a connection, Napoli et al. found that non-polyribosomal FMRP in synaptoneurosomes can repress translation by inhibiting cap-dependent initiation through interaction with the novel eIF4E-BP, CYFIP1 (Napoli et al., 2008). However, this is likely to account for only a small fraction of FMRP function in vivo as the vast majority of the protein is polyribosome-associated. It has also been suggested that FMRP can activate translation (Bechara et al., 2009). The function of polyribosome-associated FMRP in neurons on endogenous mRNA target transcripts remains undefined.
A key to understanding FMRP function is to identify its RNA targets. FMRP binds to RNA in vitro (Siomi et al., 1993) with high affinity for kissing complex and G-quadruplex motifs mediated through its KH and RGG-type RNA binding domains, respectively (Darnell et al., 2005a; Darnell et al., 2001). Efforts have been made to identify specific FMRP target mRNAs by co-immunoprecipitation and microarray analysis (Brown et al., 2001), antibody positioned RNA amplification (APRA (Miyashiro et al., 2003)), and bioinformatic approaches (Brown et al., 2001; Darnell et al., 2001). However, these approaches do not identify RNA binding sites within transcripts, have inherent signal to noise issues (Mili and Steitz, 2004; Darnell et al., 2005b; Darnell, 2010), and are hampered by difficulties in bioinformatic prediction of complex RNA folding (Darnell et al., 2005a; Darnell et al., 2001). The net result is that they have met with limited success either in deriving a consensus set of FMRP mRNA targets, in identifying mRNAs that can be validated as targets in genetic systems, or in defining FMRP function.
We recently developed a general means of identifying RNA-protein interaction sites in vivo termed CLIP (crosslinking-immunoprecipitation; (Chi et al., 2009; Darnell, 2010; Licatalosi et al., 2008; Ule et al., 2003)). CLIP uses ultraviolet irradiation to penetrate tissue and create a covalent bond between proteins and RNA molecules that are in direct contact (within a bond-length). CLIP, particularly when combined with high throughput sequencing (HITS-CLIP), has been able to identify functional RNA-protein interaction sites in pre-mRNA that regulate alternative splicing, in 3′ UTRs that regulate alternative polyadenylation (Licatalosi et al., 2008; Katz et al., 2010) or miRNA-mediated translational regulation by Argonaute (Chi et al., 2009; Hafner et al., 2010; Zisoulis et al., 2010). Here we have applied HITS-CLIP to the mouse brain in order to identify FMRP-mRNA interactions. We have used the resulting set of robust FMRP targets to drive in vitro and in vivo functional assays that define the molecular role of FMRP in controlling translation.
Results
FMRP RNA targets in mouse brain polyribosomes
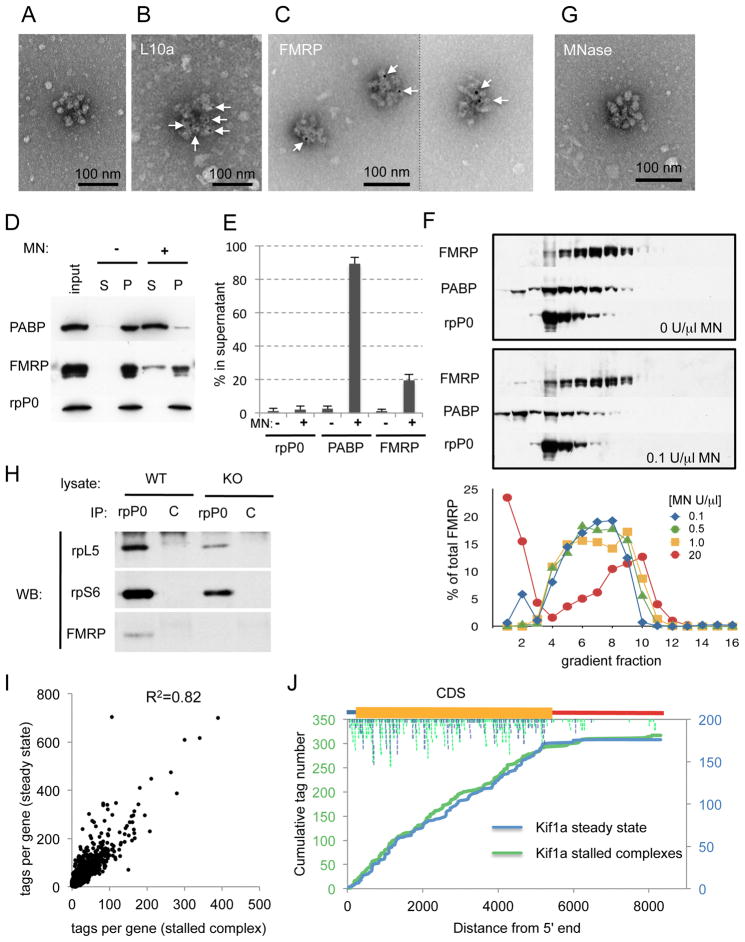
To develop FMRP CLIP (Figure 1A), we first took advantage of the observation that 85–90% of FMRP is associated with polyribosomes in the brain (Figure 1B–D) as a purification step (Stefani et al., 2004). These approaches eliminated detectable background RNA from immunoprecipitated FMRP-RNA crosslinked complexes (Figure 1E), and following RT-PCR amplification of crosslinked RNAs, products were readily detected from WT but not Fmr1 KO brain (Figure 1F). Using this protocol we performed 5 biologic replicates using two independent antibodies to FMRP. To assess the specificity and reproducibility of FMRP RNA binding we repeated CLIP in biologic replicate using a second protocol (Figure 1A) crosslinking intact brain slices, increasing the stringency of denaturation, and purifying Hu:RNA complexes (Darnell et al., 2009) from the same lysate as a control for specificity (Figure 1G–I).
Figure 1. FMRP CLIP on purified mouse brain polyribosomes.
(A) Schemes used for FMRP CLIP; steps specific to protocol 2 are indicated in green. Mouse brain post-mitochondrial supernatants (S2) were prepared as illustrated, and UV-crosslinked prior to loading on sucrose gradients for polyribosome purification. (B) Cell-equivalent aliquots of subcellular fractions from the purification steps indicated in (A) were analyzed by Western blotting for FMRP (with FMRP-specific ab17722); quantitation of three experimental replicates revealed 11.0% (standard deviation 1.4%) of FMRP is in P1. (C) The remaining ~90% of brain FMRP in S2 was applied to 20–50% sucrose gradients and gradient fractions analyzed by Western blot for FMRP. (D) Western blot comparison of the indicated fractions from (A) demonstrated that all the FMRP in pooled polyribosomes was pelleted at 300,000×g. (E) Autoradiogram of representative CLIP results from protocol 1. After dissociation of RNP complexes, samples were treated with RNAse T1, FMRP was IP’ed with either monoclonal or polyclonal antibodies, a 32P-labelled linker was ligated to the 3′ end of crosslinked RNA tags using T4 RNA ligase, and RNA protein-complexes run on denaturing PAGE, transferred to nitrocellulose and imaged by autoradiography. 32P-labeled RNA migrating at a modal size of 130kDa in IPs from WT but not Fmr1 KO brain (bracket) were taken for further workup; vertical line traces of each autoradiogram (blue (WT); red (KO); and green (non-crosslinked, not shown on autoradiogram) are shown to the right. (F) Following digestion of the radioactive RNA-protein complexes with proteinase K, a 5′ linker was added with T4 RNA ligase and products amplified by RT-PCR; product of the expected size, 60–100 nucleotides, was seen after 38 cycles (38X) from WT CLIP, but not from Fmr1 KO littermate CLIP. (G–H) To identify complexes crosslinked to RNA of an appropriate size using Protocol 2, aliquots of lysate were treated with a serial dilution of an RNAse A/T1 cocktail prior to IP of FMRP (G) or Hu (H). After 32P end-labeling with PNK, the RNA protein complexes were imaged by autoradiography. With no RNAse (0), complexes were of a wide range of sizes, most larger than desired, which progressively decreased as RNAse concentration increased, collapsing to bands close to the size of FMRP or Hu, as indicated (*). In the absence of crosslinking (no XL), only trace amounts of 32P-label were present. Protein-RNA conjugates were excised from the bracketed regions of the lanes indicated with blue arrows. (I) Final PCR products of CLIP tags of the expected size were obtained following 6–10 cycles of reamplification with sequencing primers, and the indicated samples (8 cycles) were used for Illumina sequencing. See also Table S2 and Figure S1.
We analyzed FMRP-crosslinked RNA tags from these seven independent experiments. 22 million tags were unambiguously mapped to the mouse genome, and after elimination of exact duplicates, 163,904 unique sequences were analyzed (Table S1). The results were very reproducible, with high correlations seen between exonic tag number per gene in CLIP experiments using different anti-FMRP antibodies, purification protocols and sequencing platforms (Figure S1A–B). The results were specific, as little correlation was seen between FMRP mRNA targets and either Hu targets or those of another neuronal RNABP, Nova (Figure S1C–D). Importantly, the number of FMRP tags per transcript showed little correlation with transcript abundance or length (Figure S1E–F). Taken together, these data demonstrate that FMRP reproducibly crosslinks to a subset of brain mRNAs in a manner distinct from other neuronal RNABPs.
Identity and functions encoded by FMRP target transcripts
We next defined the set of FMRP-mRNA interactions. We identified transcripts to which FMRP reproducibly crosslinked across the seven independent experiments, and used the number of unique tags per transcript in each experiment to rank the targets, allowing us to determine a chi-square score and false discovery rate (FDR) for each target based on rank and reproducibility. We identified a stringent set of 842 FMRP target transcripts (FDR <0.01) (Tables S2A–S2C). These FMRP targets were detectable with both antibodies (100%), using both CLIP protocols (100%), different sequencing platforms (100%), and were biologically reproducible (99% were detectable in at least 6 out of 7 experiments). It is likely that this analysis underestimates the true number of FMRP-regulated mRNAs; in addition to the conservative FDR threshold, some targets are likely to be rare, present in only a fraction of cells, or to interact with FMRP only under specific conditions. Therefore the absence of crosslinking to any one mRNA should be interpreted with caution.
Comparison of FMRP mRNA targets with a target list previously generated using FMRP-RNA co-immunoprecipitation (RIP-CHIP) from mouse brain (Brown et al., 2001) found significant overlap. 54% of RIP-CHIP targets (p=2.4 × 10−121; Tables S2C and S3A) were present in the FMRP CLIP target list, accounting for 24% of CLIP targets. However, despite the fact that our conditions were much more stringent than those used in RIP-CHIP, our data conservatively identified 661 novel FMRP mRNA targets not found previously. While our data overlaps and extends data from the RIP-CHIP study, comparison with 83 targets obtained using the APRA method (Miyashiro et al., 2003) revealed only 6 in common (p=0.28; Tables S2C and S3A).
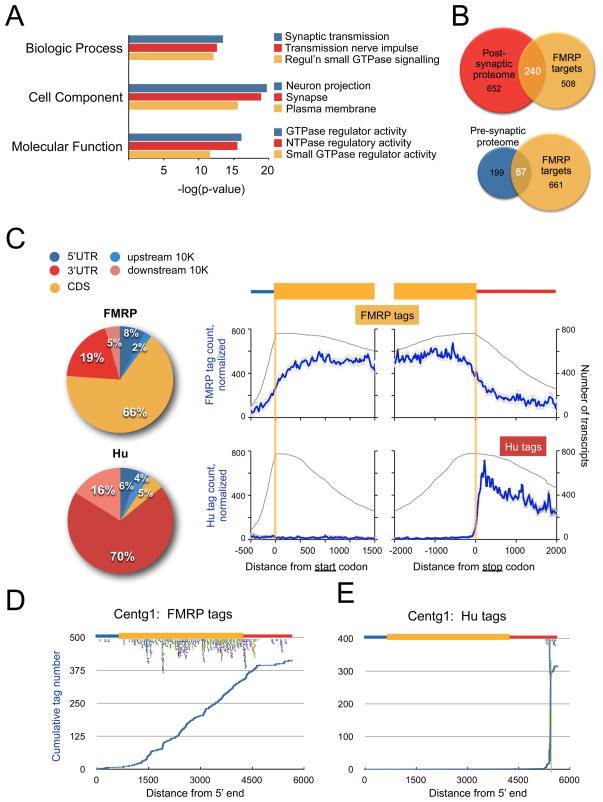
To gain further insight into the biologic functions encoded by the FMRP target transcripts, we used the DAVID Bioinformatics database to analyze the gene ontology (GO) terms assigned to them. The 842 FMRP targets were compared with the total mRNA population present within the polyribosomal fractions from which FMRP:RNA complexes were captured, and independently with a database of the transcriptome of neurons purified from mouse brain at similar ages (Cahoy et al., 2008) (Figure 2A; Table S3B). In both analyses, proteins encoded by FMRP-bound transcripts were enriched in those related to neuronal and synaptic transmission and regulation of small GTPase mediated signaling. FMRP was crosslinked to multiple members of these gene families (Table S2B).
Figure 2. Distribution of FMRP binding sites in target mRNAs.
(A) The top 3 Gene Ontology (GO) terms enriched in FMRP target transcripts in indicated GO categories, by p-value. (B) Overlap between the postsynaptic proteome (PSP) of the Genes2Cognition (G2C) database, the presynaptic proteome, and FMRP targets. Venn diagrams are drawn to scale and show the overlap between FMRP target transcripts and the indicated proteomic categories, with the absolute number of evaluable transcripts and p-values shown. (C) Distribution of unique tags among FMRP or Hu target transcripts represented by pie chart or graphically (blue line; error bars represent standard deviation), normalized to the total number of FMRP or Hu target transcripts (grey line), showing a predominance of FMRP tags within the coding sequence (gold, CDS). In contrast, Hu tags were predominantly present within the 3′ UTR (red) or mapped within 10,000 nt downstream of annotated genes (pink, downstream 10K), a region rich in unannotated 3′ UTR sequences. (D–E), Distribution of the cumulative number of FMRP or Hu tags, as indicated, across the length of a representative transcript. Centg1 is a target of both RNABPs and illustrates the different mechanisms of transcript association (Hu has a specific binding site, while FMRP is evenly distributed in CDS) within one starting pool. The positions of individual tags are also plotted below the cartoon of the mRNA structure; colors represent independent experiments. See also Tables S3-S5 and Figure S2.
We also compared FMRP target mRNAs to the curated dataset of mouse brain synaptic proteins (Genes to Cognition database, G2Cdb (Croning et al., 2009)). This identified an especially strong overlap with the postsynaptic proteome (32% of FMRP targets, p=2.1 × 10−83, Figure 2B and Tables S3A and S3C). Strikingly, a significant fraction of the mGluR5 (62%, p=5.8 × 10−22; Tables S3A and S3D) and NMDAR (34%, p=7.4 × 10−23; Tables S3A and S3E) receptor complex proteins were FMRP targets. Moreover, comparison of FMRP target mRNAs with a comprehensive presynaptic proteome database (Abul-Husn et al., 2009) revealed that FMRP also binds mRNAs encoding approximately one-third of the proteins in the presynaptic proteome (p=6.4 × 10−33; Figure 2B, Tables S3A and S3F). We used Ingenuity software to identify the top pathways enriched in FMRP targets (Figure S2A and Table S4). The most significant overlaps included synaptic signaling pathways: synaptic long-term potentiation, glutamate receptor signaling, neuropathic pain signaling, GABA receptor signaling, synaptic LTD and CREB signaling in neurons. Intracellular signaling pathways enriched in FMRP targets (p<0.05) included calcium, PKA, PKC, G-protein coupled receptor, RhoA, cAMP and PI3K/Akt signaling pathways. Taken together, the nature of the FMRP target transcripts suggests a direct role for FMRP in regulating translational control of the pre- and post-synaptic proteome and synaptic plasticity, likely underlying cognitive and behavioral deficits in children with FXS, as well as epilepsy and defects in pain sensitivity (Price et al., 2007; Symons et al., 2003).
Overlap of FXS and autism spectrum disorders (ASDs)
FXS is the leading monogenic cause of ASDs, accounting for up to 5% of all cases (Kelleher and Bear, 2008), and 90% of affected males show autistic behaviors (Hernandez et al., 2009). Comparison of the 842 FMRP target genes with 117 evaluable candidate genes from the SFARI database of autism candidate genes (http://gene.sfari.org (Basu et al., 2009)) revealed a highly significant overlap with 28 FMRP targets (24%, p=1.0 × 10−8; Table S3A), including several well-studied autism candidate genes such NLGN3, NRXN1, SHANK3, PTEN, TSC2, and NF1 (Table S5).
The ability of FMRP to repress translation of target mRNAs (see below) suggests that FXS, and by extension, some cases of ASD, may result from the overexpression of specific dosage-sensitive genes through loss of translational suppression. Gene overexpression can also occur due to copy number variations (CNVs) caused by de novo segmental duplications, and several CNV association studies have established links to ASD (Pinto et al., 2010; Sebat et al., 2007), therefore we looked for overlap between such genes and FMRP targets. Restricting our analysis to loci containing 5 or fewer protein-coding genes, we found a significant number (25/196) of candidate autism susceptibility genes in duplicated loci are FMRP targets (Tables S3A and S5, p=0.001). CNVs due to deletions showed less overlap and did not reach significance (12/121, p=0.28). Taken together, the overlap between FMRP target transcripts and genes linked to the ASDs, particularly overexpressed genes, provides a new connection between loss-of-function of FMRP and the development of autistic symptoms in patients.
FMRP binds coding sequences and stalls ribosomes
HITS-CLIP identifies RNABP binding sites within transcripts (Chi et al., 2009; Darnell, 2010; Licatalosi et al., 2008; Ule et al., 2003). Strikingly, 66% of FMRP mRNA binding was within the coding sequence (CDS), with no specific position relative to the start and stop codons, and with lower tag density in the 3′ or 5′ UTRs; in comparison, RNA tags of the neuronal Hu proteins mapped predominantly to the 3′ UTR of target transcripts (Figure 2C). This distribution of FMRP binding could arise in a population analysis from the sum of binding at unique positions in different transcripts. Examination of FMRP binding to the top-ranked individual targets also showed an even distribution of tags along the CDS (Figure 2D; Figure S2B–G), while in contrast, Hu binding to its top-ranked targets was restricted to very specific binding sites in 3′ UTRs (Figure 2E).
The distribution of FMRP binding was unexpected. FMRP crosslinking to the CDS is not easily reconciled with findings that most FMRP appears to be associated with actively translating polyribosomes, and that this association is sensitive to EDTA and RNAse (Stefani et al., 2004), sodium azide (Ceman et al., 2003; Feng et al., 1997a), which blocks initiation, and puromycin (Stefani et al., 2004), an aminoacyl-tRNA analog that causes translocating ribosomes to be released. We extended these results by treating N2a cells with puromycin or hippuristanol, an inhibitor of translation initiation, confirming that after ribosomal “run-off”, FMRP was associated with mRNAs still loaded with several ribosomes, in contrast to ribosomal protein P0 (rpP0) or poly(A) binding protein (PABP) (Figure S3A). Thus FMRP appears to associate with the CDS of transcripts on which some of the ribosomes may be stalled, suggesting that FMRP might play a role in regulating translation at the level of elongation.
Loss of FMRP function relieves ribosome stalling
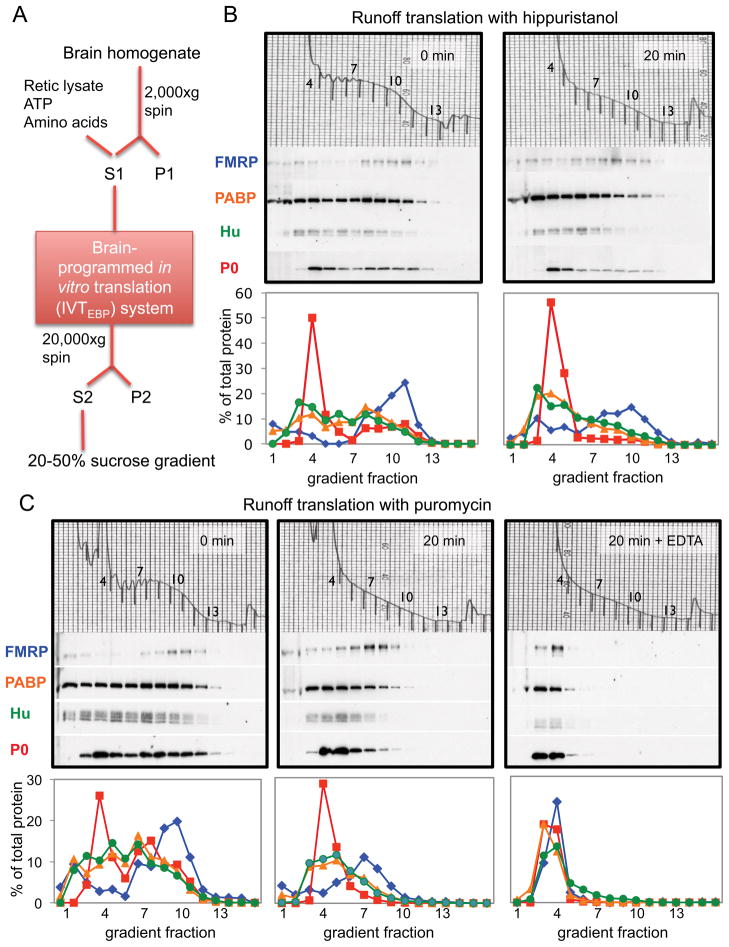
To explore this hypothesis, we developed an in vitro translation system programmed with endogenous brain polyribosomes (the “IVTEBP system”) to assess the role of endogenous FMRP on translation of individual neuronal mRNAs (Figure 3A). This system maintains normal RNA-protein stoichiometry, which is significant as FMRP may nonspecifically repress translation when overexpressed (Li et al., 2001; Laggerbauer et al., 2001). The IVTEBP system was translationally competent (Figure S3); in the presence of puromycin at 30°C, ribosomal run-off was evident within 2 minutes, reached a plateau by 15 minutes, and was saturable, as neither a ten-fold increase in puromycin nor incubation for up to 45 minutes caused additional run-off. No loss of polyribosome integrity was observed in parallel reactions incubated with cycloheximide (CHX) at 30°C for up to 45 minutes, indicating that ribosome run-off could not be explained by nuclease activity. Ribosomal run-off could be detected in the presence of hippuristanol, reflecting natural ribosome release at the stop codon (Figure 3B). This run-off was accompanied by 35S-methionine incorporation in a time- and brain polyribosome-dependent manner that was not increased if hippuristanol was omitted (Figure S3B–C), indicating that elongation can occur on pre-existing brain polyribosome-associated mRNAs in the absence of initiation in the IVTEBP system.
Figure 3. FMRP is associated with stalled polyribosomes in elongation-competent brain extracts.
(A) Schematic of the preparation of the brain-programmed in vitro translation (IVTEBP) system, in which S1 supernatants supplemented with amino acids, ATP and rabbit reticulocyte lysate allow ribosomal runoff, detected by analysis on sucrose gradients and Western blot of polyribosome-associated RNA binding proteins. (B) gradient fractions before run-off (0 minutes, left panel), or after 20 minutes of elongation in the presence of hippuristanol (right panel). Western blot analysis (middle panels) and their quantitation (bottom graphs) were used to compare the distribution of FMRP (blue diamonds), PABP (orange triangles), neuronal Hu isoforms (green circles) and ribosomal protein P0 (red squares) in 20–50% sucrose gradients. A254 traces of total RNA distribution are shown (top panels) and gradient fractions indicated. (C) gradient fractions analyzed as in (B), from ribosomal run-offs performed before (left panels, 0 min) or after run-off in puromycin (middle panels, 20 min) or puromycin followed by 30 mM EDTA treatment to release all ribosomes (right panels). See also Figure S3.
After ribosomal run-off in the presence of puromycin, FMRP shifted in a time-dependent manner to fractions containing ~5–8 ribosomes, recapitulating the behavior of FMRP in cells (Figure 3C; S3A; (Stefani et al., 2004)). In contrast, PABP or Hu shifted to lighter fractions, as did FMRP when EDTA was added to dissociate ribosomes (Figure 3C). In addition, natural run-off occurring in the presence of hippuristanol also resulted in retention of FMRP in large complexes in the IVTEBP system (Figure 3B). Based on the results of multiple experiments, the total amount of FMRP present on polyribosomes was not significantly changed before and after run-off. These independent approaches again suggest that FMRP may be associated with mRNAs harboring run-off resistant “stalled” ribosomes detectable in the IVTEBP system as well as in vivo.
If FMRP regulates the translation of its mRNA targets one might anticipate that the number of ribosomes associated with these target transcripts would be altered in its absence. We estimated the steady-state number of ribosomes associated with 9 FMRP target mRNAs or 9 non-target mRNAs by analyzing transcript distribution on polyribosome gradients. We found no reproducible differences in the sedimentation profile of any of these mRNAs in the presence or absence of functional FMRP in two different mouse FXS models (Figure 4, first column; Figure S4A). Moreover, global analysis of total and polyribosome-associated mRNA levels identified no statistically significant changes between 6 pairs of Fmr1 KO and WT littermates (other than the Fmr1 transcript itself; described in Extended Experimental Procedures; deposited in GEO database accession number GSE26809). These results were unexpected, given prior reports of FMRP-dependent changes in mRNA polyribosome distribution. While we cannot rule out that differences such as the cell types (e.g. human lymphoblastoid cell lines; (Brown et al., 2001)) or subcellular fractions (e.g. synaptoneurosomes; (Zalfa et al., 2003; Muddashetty et al., 2007)) used in these studies may account for this discrepancy, our results clearly demonstrate that on a cell population level there is no FMRP-dependent difference in steady state mRNA polysome distribution in the P8-P25 mouse brain.
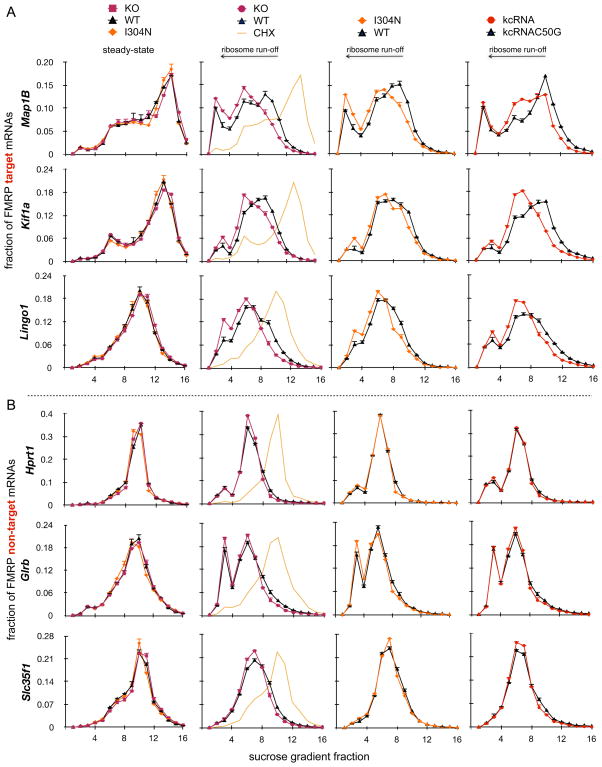
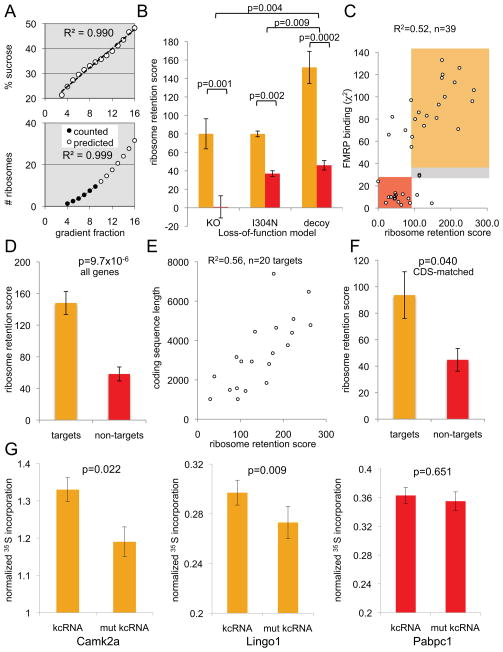
Figure 4. Ribosomal stalling on FMRP target transcripts is relieved in three FMRP loss-of-function models.
(A–B) FMRP target (A, Map1b, Lingo1, and Kif1a) or non-target (B, Hprt1, Glrb, and Slc35f1) mRNA distribution in each of 16 polyribosome sucrose gradient fractions was analyzed by qRT-PCR (see also Figure S4A). Prior to run-off (first column; CHX treated (“steady-state”) polyribosomes and reproduced in the second column, yellow line), no changes in mRNA distribution were evident between WT (black triangles), Fmr1 KO (red squares) or I304N knock-in (orange diamonds) brain polyribosomes. The same mRNAs were then analyzed in the IVTEBP system in puromycin to achieve run-off separating translocating from stalled ribosomes in three distinct FMRP loss-of-function systems (Fmr1 KO (second column, red circles versus WT in black), I304N (third column, orange diamonds versus WT in black), and from WT polyribosomes treated with kcRNA decoy to acutely disrupt FMRP polyribosome association (fourth column, red circles) compared with a non-functional kcRNA point mutant (kcRNAC50G, black)). Data are plotted as a fraction of total mRNA on the gradient; error bars represent standard deviation from three technical replicates. See also Figure S4.
The steady state number of ribosomes on a transcript is not a measure of active translation. There are numerous instances described in which significant inhibition of the synthesis of specific proteins is not accompanied by the expected decrease in the number of ribosomes associated with their encoding mRNAs (Olsen and Ambros, 1999; Clark et al., 2000; Braat et al., 2004; Nottrott et al., 2006; Maroney et al., 2006; Petersen et al., 2006; Lytle et al., 2007). Therefore, we separated translocating from stalled ribosomes by puromycin run-off in the IVTEBP system and assessed the number of puromycin-insensitive ribosomes remaining on target and non-target transcripts. In the presence of FMRP, target transcripts were associated with more residual ribosomes after run-off than in the absence of functional FMRP (WT vs. Fmr1 KO or I304N brain; Figure 4A, columns 2–3). Importantly, such differences were specific to FMRP-target transcripts (Figure 4B), suggesting that ribosomes are stalled on FMRP-bound mRNAs in an FMRP-dependent manner in vivo, with stalling relieved in mice harboring either of two different FMRP loss-of function mutations.
All three FMRP paralogs (“FXRP” proteins FMRP, FXR1P and FXR2P) show significant functional redundancy in binding to RNA and polyribosomes (Darnell et al., 2009), including the ability to be fully displaced from polyribosomes by a high affinity in vitro selected RNA ligand (kissing complex RNA (kcRNA); Figure S4B) (Darnell et al., 2005a). We compared the mRNA distribution of FMRP target and non-target mRNAs when WT brain polyribosomes were acutely incubated with kcRNA during puromycin run-off. Removal of FXRPs with kcRNA but not a non-binding point mutant (kcRNAC50G) also resulted in apparent relief of ribosome stalling specifically on FMRP target transcripts (Figure 4, column 4). Significantly, addition of kcRNA after run-off did not cause a shift in the distribution of mRNAs, ruling out the possibility that observed changes in mRNA distributions were due to the loss of the mass of FXRPs alone (Figure S4C). To confirm that FXRPs stall ribosomes in vivo, we repeated the puromycin run-off assay in N2A cells, comparing untreated cells with loss-of-function by knockdown of FXR1P, FXR2P and FMRP. qRT-PCR was used to determine the distribution of two FMRP target mRNAs (Map1b and Huwe1) and a non-target transcript (Hprt1) on sucrose gradients after puromycin run-off (Figure S4D–E). The FXRP-dependent association of target mRNAs with puromycin-insensitive ribosomes was remarkably similar to that seen in the brain polyribosome-programmed translation assay, underscoring the relevance of the in vitro system to living cells. Finally, addition of EDTA to remove all ribosomes caused some mRNAs to migrate to lighter fractions, consistent with the presence of residual ribosomes on some transcripts (those with more ribosomes initially, unrelated to whether the transcripts were FMRP targets; Figure S5). Taken together, these results demonstrate that acute loss of polyribosome-associated FMRP yields the same relief of translational repression as long-term loss of function in mouse models, indicating that ribosome stalling is likely to be caused directly and reversibly by FMRP.
We weighted the mRNA distribution on these plots according to the estimated number of ribosomes in each fraction (Figure 5; Figure 6A), in order to account for the variable number of ribosomes per fraction. This allowed calculation of an FMRP-dependent ribosome retention score (RRS), a measure of the difference between the number of ribosomes remaining on transcripts after run-off in the presence versus absence of FMRP. This analysis demonstrated a statistically significant difference between the nine FMRP target and nine non-target mRNAs in all three loss-of-function models (Figure 6B). To generalize these findings, we assessed the distribution of a total of 39 transcripts (20 FMRP targets and a set of comparable non-targets, controlling for length, abundance and neuronal expression) after run-off in the IVTEBP assay using the kcRNA decoy (Table S6). All 39 transcripts were analyzed with no prior assumptions, and with no exclusion of outliers. The overall RRS scores correlated well with FMRP target binding assessed by CLIP (chi-square score, R2=0.53, or by tags per gene, R2=0.49) for the 39 mRNAs tested (Figure 6C), and were significantly different between targets and non-targets (Figure 6D). There was a small effect on some non-target transcripts which we do not understand, perhaps from a promiscuous association of FMRP with mRNAs from reassociation of FMRP with non-targets following cell lysis and extract preparation (Baltimore and Huang, 1970; Mili and Steitz, 2004) or from exogenous FMRP present in the rabbit reticulocyte lysate (Beaulieu, 2000). Moreover, some FMRP non-targets may be bona fide targets of FXR1/2P rather than FMRP, with ribosome stalling also relieved by the kcRNA decoy.
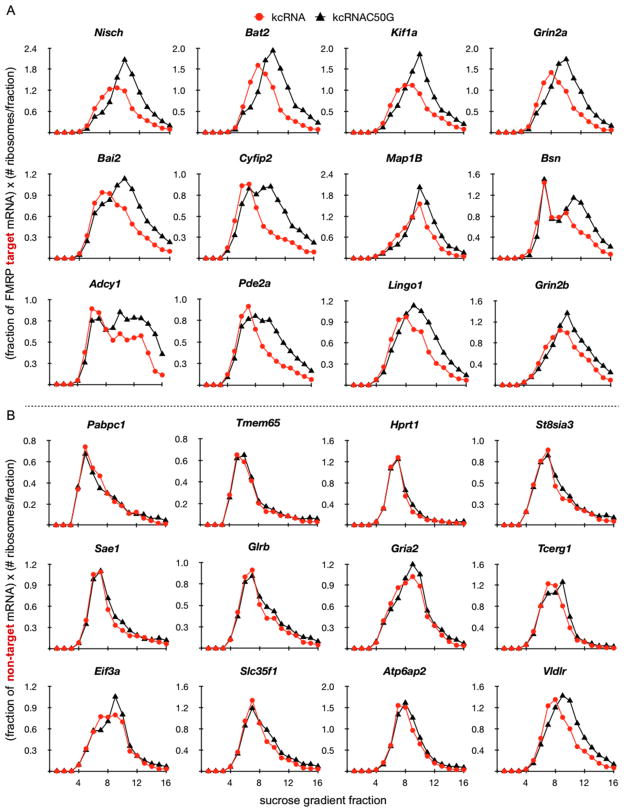
Figure 5. Ribosome-weighted mRNA profiles for FMRP targets and non-targets.
Ribosome-weighted graphs of kcRNA decoy data from Figure 4 are replotted, along with additional examples, weighting for the number of ribosomes in each fraction, as (fraction of mRNA) × (# ribosomes per fraction (from Figure 6A)). (A) Ribosome-weighted graphs of the top 12 (by RRS) of 21 FMRP target mRNA profiles. (B) Ribosome-weighted graphs of 12 FMRP non-target mRNAs. Data for all 39 transcripts is in Table S6. Error bars represent standard deviation from three technical replicates. See also Table S6 and Figure S5.
Figure 6. Quantitation of FMRP-mediated ribosome stalling.
(A) Demonstration that the sucrose gradients used in the RRS calculations were linear. Top panel: The percent sucrose (w/w) measured in each gradient fraction using a refractometer is plotted by fraction number (R2= 0.99). Fractions 1–2 correspond to lysate. Bottom panel: Determination of the approximate number of ribosomes in each sucrose gradient fraction by extrapolation from those that can be directly counted, using linear regression analysis. 9 ribosomes per mRNA (black circles as a function of gradient fraction) were counted from A254 traces, and the best-fitting equation (R2=0.999) was used to extrapolate the number of ribosomes associated with mRNAs in each fraction for the remainder of the gradient (open circles, “predicted”), based on the linearity determined in the top panel. (B) Bar graphs plotting RRS scores in three different FMRP loss of function models (I304N, KO) for 9 target (gold) and 9 non-target (red) transcripts. For kcRNA experiments (“decoy”) data represents RRS scores for 21 targets and 16 non-targets). Significant differences are evident between targets and non-targets in all three systems, as well as a significantly greater effect of kcRNA decoy than either genetic FMRP loss of function model. Error bars depict the standard error of the mean. (C) A measure of the degree of FMRP binding to target mRNAs (chi-square score) compared with a functional assay (RRS, a measure of FMRP activity in retaining mRNA on polyribosomes after puromycin run-off) showed a significant correlation for 39 tested transcripts. Target transcripts (shaded gold) showed high chi-square and RRS scores relative to non-targets (shaded red), with some outliers and two targets deemed to be in a “grey zone” (see Table S6). Two transcripts, Arc and Gria1 mRNA, are in the “grey zone”. Arc falls just below the FDR <0.1 cutoff for the robust FMRP target list, likely in part because of very low abundance in resting mice (Table S6). Neither was included in statistical analyses of FMRP targets vs. nontargets in (D). (D) 20 FMRP targets and 16 non-targets show a significant difference in RRS (RRSAvg(target)= 148.1 +/− 14.6; RRSAvg(nontarget)= 58.3 +/− 8.9; p=9.7 × 10−6). This analysis includes every mRNA we have tested to date, except Arc and Gria1 (E) RRS is highly correlated with CDS length for FMRP targets (excluding an outlier, Bsn; CDS 11,829 nts). (F) Six FMRP targets and seven non-targets matched for length (in the 1–2 kbp window) show a significant difference in RRS (RRSAvg(target)= 93.7 +/− 17.6; RRSAvg(nontarget)= 44.8 +/− 8.6; p=0.040). (G) 35S-methionine labeled protein synthesized from two FMRP mRNA targets (Camk2a and Lingo1; gold shading) or one non-target (Pabpc1; red shading) in the IVTEBP system were compared by immunoprecipitation, SDS-PAGE and PhosphorImaging, normalizing against irrelevant bands in the IP and a CHX-treated control sample. Error bars represent s.e.m. in all panels, and P values were determined using a two-tailed Student’s t test. See also Table S6 and Figure S6.
The RRS was not dependent on transcript abundance (R2=0.01), nor how many ribosomes were associated with the transcript in the steady state (R2=0.07). We observed some correlation between coding sequence (CDS) length and RRS among all transcripts (R2=0.41); this was largely due to FMRP targets (Figure 6E, R2=0.56 for targets; R2=0.17 for non-targets), consistent with FMRP stalling ribosomes across the length of the CDS of its target transcripts. There was a significant difference in the RRS of target and non-target mRNAs when matched for CDS length (Figure 6F). Taken together, these data provide quantitative support for the conclusion that FMRP stalls ribosomes on transcripts to which it is directly bound.
To assess whether the relief of FMRP-dependent ribosome stalling by kcRNA is accompanied by increased protein synthesis, we measured 35S-methionine incorporation in the IVTEBP assay into two FMRP targets (Camk2a and Lingo1) and one non-target for which we were able to quantitatively immunoprecipitate protein products. We found a small but significant increase in protein synthesis of both FMRP targets in the absence of functional FMRP, similar to results reported for Camk2a by Bear and colleagues in slice cultures (Osterweil et al., 2010), with no significant change in either overall protein synthesis or in the synthesis of the non-target PABP (Figure 6G and Figure S6). Therefore FMRP-mediated ribosomal stalling can be functional and dynamic, as acutely relieving it leads to increased synthesis of new protein.
Characterization of the FMRP-stalled complex
In some studies, what were initially believed to be translocation-blocked polyribosomes were revealed to be “pseudo-polysomes” or large mRNP complexes (Thermann and Hentze, 2007), underscoring the need for caution in making inferences about the components of heavy complexes on sucrose gradients. To further characterize the FMRP-stalled complex, we first obtained electron-microscopic images from sucrose gradient fractions after puromycin runoff in the IVT-EBP system (corresponding to fractions 7–8, Figure 3C, middle panel). These fractions harbor complexes containing multiple ribosomes (Figure 7A) identified as such by immuno-EM on the same gradient fraction prepared from a transgenic mouse expressing the tagged ribosomal protein EGFP-rpL10a (Heiman et al., 2008; Doyle et al., 2008) (Figure 7B). To investigate the association of FMRP with these complexes we expressed EGFP-tagged FMRP in cells, performed a puromycin run-off experiment in vivo, and visualized EGFP-FMRP association with these structures from the puromycin-resistant fractions (Figure 7C, S7A). The labeling of these complexes was dependent on FMRP, relative to an overexpressed negative control (EGFP alone; Figure S7B), demonstrating the physical association of FMRP with stalled polyribosomes in vivo (Figure 7C).
Figure 7. Characterization of the FMRP stalled complex.
(A) Electron microscopic images of sucrose gradient fractions after puromycin-induced run-off of translocating ribosomes (as in Figure 3C). Negative staining with uranyl acetate revealed structures confirmed to be polyribosomes in (B). Scale bar is indicated. (B) Sucrose gradients were prepared from cerebellum of the Purkinje cell-specific _Pcp2_-promoter driven EGFP-tagged rpL10a BAC transgenic mice (Doyle et al., 2008; Heiman et al., 2008), and polyribosome fractions were treated with 6 nM gold-labeled anti-EGFP monoclonal antibody and processed for electron microscopy. Specific staining on clustered structures (white arrows) demonstrates that they correspond to polyribosomes. Only ~1% of polyribosomes showed labeling, consistent with the fact that whole cerebellum was used for analysis, and indicating specificity of the immunogold label. (C) Immunoelectron microscopic images of EGFP-FMRP association with stalled polyribosomal complexes after puromycin run-off in vivo in transfected cells. EGFP-FMRP was detected using 12 nm gold-labelling and antibodies against EGFP. 13.5% of polyribosomes were labeled in the presence of EGFP-FMRP (n=500) while only 0.02% of polyribosomes from EGFP-expressing cells were associated with gold (n=500). (D–E) Western blot (WB) analysis (D) and quantitation (E) of the micrococcal nuclease (MN) resistance of FMRP and ribosome co-sedimentation. Polyribosome-containing sucrose gradient fractions of mouse brain extracts were treated with (+) or without (−) 1000 U/ml MN, centrifuged through 15–20% sucrose, separating proteins as released supernatant (S) or heavy pelleted (P) particles, which were analyzed for the indicated proteins by WB. (F) Brain extracts were subjected to puromycin runoff, treated with the indicated concentration of MN, purified on 20–50% sucrose gradients and the indicated proteins analyzed by TCA precipitation and WB; only 25% of fractions 1–3 were precipitated to compensate for their high protein concentrations. Bottom panel: quantitation of FMRP from samples treated with the indicated concentrations of MN. Signals in lanes 1–3 were multiplied by 4 to compensate for the amount precipitated. (G) Samples from (A) were treated with 1000 U/ml MN prior to sucrose gradient purification and analysis by EM. (H) Samples prepared as in (G) from WT or Fmr1 KO brain were IP’d with anti-rpP0 or control (C; anti-Nova) antibodies and probed for the co-precipitation of the indicated proteins by WB. (I) Correlation of tags/transcript pooled from two biologic replicate FMRP CLIP experiments crosslinked from steady-state (CHX treated) versus stalled (puromycin run-off) brain polyribosomes. (J) Representative results from the experiments in (I) showing CLIP tag distribution on puromycin-resistant FMRP-stalled mRNA complexes (Kif1a; representative of 10 genes assessed; comparison of steady-state tags (blue) and stalled complexes (green)). Individual tags are shown below the mRNA cartoon (CDS is gold, 5′UTR blue, 3′UTR red). Biochemical purification, including co-IP and nuclease sensitivity was done in three independent experiments, EM and FMRP immuno-EM in 2–3 independent experiments, respectively, and CLIP on the stalled complex in two biologic replicates. See also Table S7 and Figure S7.
We then used micrococcal nuclease (MN) as a means of assessing whether the FMRP stalled complex has the features of a typical polyribosome. MN degrades the mRNA exposed between ribosomes, dissociating polyribosomes (but not the ribosomes themselves). Incubation of sucrose-gradient purified polyribosomes with MN, followed by separation of ribosomes from released material on a sucrose cushion, revealed that the majority of FMRP pelleted with the ribosomes (Figure 7D–E). As controls, PABP (bound to mRNA poly(A) tails) was readily released from polyribosomes by MN, while rpP0 was MN resistant. Surprisingly, sizing of the puromycin-resistant, MN-treated complex revealed that FMRP remained associated with the large complex up to 1 U/ul MN. At very high MN concentrations (20 U/ul, used to cleave between SRP-associated stacked ribosomes; (Wolin and Walter, 1988)), FMRP is partially released to light fractions, although a substantial fraction (~60%) remains associated with the complex (Figure 7F). Taken together, these results suggest a close association of FMRP with ribosomes that are stacked or condensed in such a way as to largely prevent MN from cleaving between either FMRP and the ribosomes or between the ribosomes themselves. Consistent with this interpretation, we found that the ultrastructure of the polyribosomal complexes in the MN-treated fractions was indistinguishable from those in untreated fractions (Figure 7G vs. 7A).
To determine whether FMRP, ribosomes and repressed FMRP target mRNAs are indeed in the same macromolecular complex in the brain, antibodies against rpP0 were used to IP ribosomes from puromycin- and MN-resistant complexes purified on sucrose gradients. FMRP co-IP’d with rpP0-containing complexes, as did the ribosomal proteins rpL5 and rpS6 (Figure 7H), demonstrating a direct association of FMRP with ribosome-containing complexes, reminiscent of previous findings in cultured cells (Khandjian et al., 1996; Siomi et al., 1996).
To directly assess FMRP interaction with mRNA in the stalled complexes, we performed FMRP HITS-CLIP on polyribosomes crosslinked after puromycin- run-off in the IVTEBP system. In parallel, CLIP was done on polyribosomes taken from the steady-state (CHX) in the IVTEBP system. In two biological replicate CLIP experiments, we found that FMRP was crosslinked to the same target transcripts (Figure 7I; Table S7) and with the same CDS distribution in puromycin-insensitive (stalled) and steady-state complexes (Figure 7J). Taken together, the most judicious interpretation of these experiments is that FMRP represses translation on polyribosomes in a large complex consisting of target mRNAs and stalled ribosomes.
Discussion
Understanding the pathophysiology of Fragile X Syndrome and its extensive overlaps with autism offers the hope of gaining insights into the molecular basis of cognition and behavior. Learning and memory almost certainly involve changes in long-term synaptic strength and require new protein synthesis. This activity-dependent protein synthesis must be finely tuned, as exemplified by cases of autism caused by mutations in proteins that normally limit translation (Kelleher and Bear, 2008), and by FXS, as FMRP has been hypothesized to inhibit translation of mRNAs encoding “plasticity-related proteins” (Bear et al., 2004). Here we provide a molecular basis for the overlap between FXS, autism and defects in synaptic plasticity by first showing that many transcripts bound by FMRP encode plasticity-related proteins, and then by demonstrating that FMRP represses their translation by stalling ribosomal translocation. These results are set apart from previous studies in their overlay of unbiased, genome-wide, direct biochemical assays of endogenous interactions (Licatalosi and Darnell, 2010); while they cannot describe FMRP function on every individual candidate transcript, they provide a statistically robust model for the predominant action of FMRP in translational regulation (Figure S7C).
Our data support a model in which FMRP acts to stall ribosomal translocation during elongation as part of a complex containing target mRNAs and multiple ribosomes. This complex is relatively resistant to MN treatment, similar to mRNA-associated stacked ribosomes stalled by the action of the signal recognition particle (SRP), which represses translation as transcripts encoding secreted and transmembrane proteins are transported to the ER (Wolin and Walter, 1988). It is increasingly recognized that a large fraction of cellular mRNA may be translated within subcellular domains (Martin and Ephrussi, 2009), and our data are consistent with FMRP playing a role in controlling such processes.
Translational repression by ribosome stalling and stacking may confer several advantages to FMRP in regulating neuronal translation. Stalling of ribosomes may permit translocation to sites of axonal or dendritic protein synthesis (Krichevsky and Kosik, 2001). Pre-loading of mRNAs with ribosomes would permit very rapid protein synthesis in response to synaptic activation, as seen for example in response to local application of BDNF (Aakalu et al., 2001). The stalling of a single ribosome almost immediately slows protein production, while recycled ribosomes continue to re-initiate and restore the “loaded” state (Figure 7C) (Wolin and Walter, 1988). Stalled ribosomes may also protect mRNA from degradation during transport or storage. Although the biochemical mechanism by which FMRP stalls ribosomes remains to be determined, it is likely to be dynamic, as it can be acutely reversed by RNA decoys in run-off assays. Such reversibility could be mediated in vivo by FMRP phosphorylation, which has been hypothesized to regulate FMRP’s association with apparently stalled polyribosomes in mouse fibroblasts (Ceman et al., 2003), by FMRP degradation (Hou et al., 2006), or other means. We suggest that agents such as antibiotics that slow ribosomal translocation may restore the brake on translation that is lost in FXS, and may be of worthy of clinical consideration.
In neurons, translational inhibition by ribosomal stalling is associated with NMDAR activation, eEF2 phosphorylation, and other unknown factors (Sutton et al., 2007) which may include FMRP. Moreover, FMRP may inhibit translation in response to activation of mACh or BDNF receptors, as there are synaptic plasticity defects in these pathways in Fmr1 null mice (Lauterborn et al., 2007; Volk et al., 2007). A small fraction of FMRP is reported to inhibit translation initiation (Napoli et al., 2008), suggesting additional mechanisms for FMRP action are likely to exist. However, our data indicate that in the brain at steady-state, the majority of FMRP is associated with stalled ribosomal complexes.
While much attention has been paid to the role of FMRP in regulating Hebbian synaptic plasticity through the control of local translation, the bulk of the protein is in the cell body (Christie et al., 2009). Moreover, FMRP regulates many mRNAs that are probably not localized to the synapse: for example, Bsn and Pclo mRNAs encode proteins that are synthesized in the cell body and transported to the synapse as a complex (Shapira et al., 2003). These observations suggest that FMRP may play a role in homeostatic synaptic plasticity (Turrigiano, 2008). In this model, FMRP would act to repress the translation stimulated by neuronal activity to generate a feedback loop limiting neuronal responses to activity at the level of the neuron rather than a specific synapse.
In contrast to HITS-CLIP studies that have yielded interaction “maps” of regulatory sites involved in position-dependent alternative splicing and polyadenylation (reviewed in (Darnell, 2010)), FMRP HITS-CLIP revealed a different and unexpected mode of protein-RNA interaction. We did not find peaks of RNA tags indicating specific high affinity binding sites for FMRP, such as those suggested by previous in vitro FMRP RNA selection experiments (G-quadruplex or kissing complex motifs; (Darnell et al., 2001; Darnell et al., 2005a; Schaeffer et al., 2001)). We considered the possibility that FMRP might interact with high affinity binding sites prior to its association with polyribosomes and subsequently redistribute on the same target transcripts, such that FMRP crosslinking in polyribosomes does not reflect the initial sites of transcript interaction. However, to date we have found no biochemical evidence to support such a model, and using the RNABob motif-finding program (Darnell et al., 2001), we found that bioinformatically predicted G-quadruplex motifs are no more abundant in the set of 842 robust FMRP target mRNAs than in an equivalent set of 842 non-targets (controlled for neuronal expression and length; 1112 vs. 1068, or 43% vs. 39% of targets versus controls, respectively). Indeed, previous studies have shown that the RGG box is not necessary for polyribosome association (Darnell et al., 2005b) and G-quadruplex RNA ligands cannot compete FMRP off polyribosomes (Darnell et al., 2005a). This leaves the question of how FMRP associates with a specific set of mRNAs unanswered. In addition to the possibility that FMRP is binding to occult RNA motifs, FMRP may be recruited to its targets via a protein-protein interaction.
The proteins encoded by FMRP target mRNAs indicate a high level of control over the balance of activity-dependent translation in synaptic plasticity. First, mGluR5 and the NMDAR subunits, the only glutamate receptors known to mediate protein-synthesis dependent long-term synaptic plasticity, are themselves FMRP targets, as are many other protein components of their macromolecular complexes at the synapse. These findings are consistent with the finding that mGluR and NMDAR-dependent synaptic plasticity are altered in FXS mouse models (Harlow et al., 2010; Pilpel et al., 2009). Second, FMRP regulates the expression of components of the ERK and mTOR signal transduction pathways that convert receptor activity into translational output. Finally, FMRP controls the expression of many downstream pre- and post-synaptic structural, scaffolding, catalytic, receptor and channel proteins that are likely to be final determinants of changes in synaptic strength.
FMRP target mRNAs are a valuable dataset for considering pharmacologic therapy for FXS. mGluR5 inhibitors have shown some clinical efficacy, and NMDAR antagonists may also be worthy of clinical consideration. FMRP also targets ERK1 and the mTOR inhibitors Pten, Nf1 and Tsc2, proteins closely linked to autism, supporting the possibility that pharmacologic agents acting on the mTOR and ERK pathways may be clinically relevant for FXS and autism (Hoeffer and Klann, 2010; Kelleher and Bear, 2008). However, FMRP also targets the PI3K-enhancer PIKE (Centg1), and PIKE overexpression in FMRP null mice results in elevated PI3K signaling to the mTOR pathway (Sharma et al., 2010), indicating the need for care in translating these findings into therapy.
The molecular basis for the overlap in symptoms between Fragile X Syndrome and autism is poorly understood. The overlap between FMRP targets and the current list of autism susceptibility genes and loci is extensive and sheds light on common pathways, supporting the hypothesis that synaptic dysfunction is critical to the development of autistic features common to both disorders (Kelleher and Bear, 2008). We find that FMRP targets fall into several functional categories, as do ASD candidate genes (Sebat et al., 2007; Pinto et al., 2010) and the overlap is enriched in several inter-related functional categories, including synaptic cell adhesion molecules, the NMDAR complex, the mTOR pathway, and regulators of the small GTPases.
The FMRP target list also provides a valuable tool for focusing attention on specific gene candidates within multigenic loci for ASD. These include the three most common syndromic duplications linked to ASD, each containing between 13–28 protein-coding genes, and each locus harbors at least one FMRP target gene (Table S5). Moreover, two well-studied candidate genes present in the 17p11.2 and 15q11-13 duplications, Rai1 and Ube3a, harbored FMRP CLIP tags in 6/7 and 3/7 experiments, respectively; their extremely low expression levels in brain polyribosomes may have precluded their inclusion on the statistically robust FMRP target list. Although the FMRP target set disproportionately overlaps amplified versus deleted ASD-related CNVs, they include genes whose loss results in autism. This is consistent with the gene balance hypothesis, which posits that the same phenotype can arise from under- or overexpression of dosage-sensitive proteins because either disrupts stoichiometry of the same complex (Conrad and Antonarakis, 2007). It seems likely that FMRP tightly controls the synthesis of dosage-sensitive genes in neurons, and the overlap of FMRP targets with genes whose loss of function leads to ASD is highly significant in this regard. Taken together, the relationship between FMRP target transcripts and genes linked to the ASDs, particularly overexpressed genes, provides a new connection between loss-of-function of FMRP and the development of autistic symptoms in patients.
Experimental Procedures
These briefly described methods are supplemented with additional detail in the Extended Experimental Procedures section provided in Supplemental Information available online.
Mice used as experimental controls were littermates, and experimental results were reproduced in multiple independent experiments as noted throughout. Male wild-type and Fmr1tm1Cgr (Fmr1 KO) mouse brain (P11-P25) was used for polyribosome CLIP experiments as described in Extended Experimental Procedures. Following high throughput sequencing, unique CLIP tags were identified and mapped to the mouse mm9 genomic database or RefSeq transcripts. FMRP targets were ranked using a nonparametric method that considered both the total number of tags/gene in seven different CLIP experiments and experimental reproducibility.
GO functional category enrichment was analyzed using DAVID 6.7 software. Transcript abundance on polyribosomes was determined from RNA obtained from 6 littermate pairs of wild-type and Fmr1tm1Cgr mice (FVB background, P8 males), and data analyzed with Mouse Exon 1.0 ST Array chips using extended probe sets and the IterPLIER model using Affymetrix power tools. FMRP target pathway analysis used the core analysis module of the Ingenuity Pathways Analysis (IPA) knowledge base (www.ingenuity.com).
The IVTEBP system (in vitro translation from elongating brain polyribosomes) was generated from post-nuclear brain extracts mixed with ATP, amino acids and rabbit reticulocyte lysate, and ribosomes were run-off in the presence of puromycin or hippuristanol or allowed to continue natural elongation without additions. The effects of FMRP were measured using three loss-of-function models including the Fmr1 KO and I304N point mutant knock-in mice, and where indicated, addition of in vitro transcribed kcRNA (Darnell et al., 2005a) as a competitive RNA ligand (decoy) displacing all three FXRP family members off polyribosomes resulting in an acute, triple loss-of-function model for polyribosome-associated functions (Darnell et al., 2009). mRNA distribution on sucrose gradients was quantitated by qRT-PCR on an iQ5 Multicolor Real-Time PCR Detection System (Biorad). Experiments gave reproducible results in 4/4, 2/2 and one biologic replicate with kcRNA decoy, I304N mice, and KO mice, respectively. In sum, these results were consistent in 7/7 experiments using three loss of function models.
The microarray data have been deposited in the GEO database under accession number GSE26809.
Supplementary Material
Extended Exptl Procedures
Suppl Table S1
Suppl Table S2A-C
Suppl Table S3
Suppl Table S4
Suppl Table S5
Suppl Table S6
Suppl Table S7
Suppl Author Interview
Suppl Fig S1
Suppl Fig S2
Suppl Fig S3
Suppl Fig S4
Suppl Fig S5
Suppl Fig S6
Suppl Fig S7
Highlights.
- We identify a robust, reproducible and novel set of mRNAs FMRP binds in vivo.
- FMRP targets encode key pre- and postsynaptic proteins and autism candidate genes.
- FMRP stalls ribosomes along the coding region of its mRNA targets.
- Acute and genetic loss of FMRP relieves stalling and increases protein synthesis.
Acknowledgments
We thank J. Pelletier for providing us with hippuristanol, E. Khandjian for anti-FXR1P antibody, N. Heintz for the transgenic mice expressing tagged ribosomal protein EGFP-rpL10a, and the members of the Darnell laboratory for advice and suggestions throughout the course of this work. We are grateful to E. Sphicas and K. Uryu for help with electron microscopy. This work was supported by the NIH (J.C.D., J.D.R. and R.B.D.), the Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program (C.E.F.), an NSF grant to S.J.V., and the Rockefeller University Hospital CTSA Grant. R.B.D. is a Howard Hughes Medical Institute Investigator.
Footnotes
Author contributions: J.C.D. and R.B.D. conceived, designed and supervised the experiments, analysed the data and wrote the paper. J.C.D., S.J.V., K.Y.S.H., and C.E.F. did the experiments with assistance from A.M., E.F.S., C.C., and J.J.F. C.Z., S.W.C., A.M. and D.D.L. analyzed the high throughput sequence data and C.Z. performed statistical and comparative analyses. J.D.R. helped design the IVTEBP system and analyze the results.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Abul-Husn NS, Bushlin I, Moron JA, Jenkins SL, Dolios G, Wang R, Iyengar R, Ma’ayan A, Devi LA. Systems approach to explore components and interactions in the presynapse. Proteomics. 2009;9:3303–3315. doi: 10.1002/pmic.200800767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Huang AS. Interaction of HeLa cell proteins with RNA. J Mol Biol. 1970;47:263–273. doi: 10.1016/0022-2836(70)90301-3. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–6. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Beaulieu MA. A distinct FMRP polysomal population at an advanced stage of mammalian erythropoiesis. Biochem Biophys Res Commun. 2000;275:608–610. doi: 10.1006/bbrc.2000.3313. [DOI] [PubMed] [Google Scholar]
- Bechara EG, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braat AK, Yan N, Arn E, Harrison D, Macdonald PM. Localization-dependent oskar protein accumulation; control after the initiation of translation. Dev Cell. 2004;7:125–131. doi: 10.1016/j.devcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in Fragile X Syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Wyckoff D, Gavis ER. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr Biol. 2000;10:1311–1314. doi: 10.1016/s0960-9822(00)00754-5. [DOI] [PubMed] [Google Scholar]
- Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the Genes to Cognition database. Nucleic Acids Res. 2009;37:D846–51. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005a;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G Quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005b;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet. 2009;18:3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van Den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nature Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997a;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein--nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997b;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto CL, Broadie K. The fragile X mental retardation protein in circadian rhythmicity and memory consolidation. Mol Neurobiol. 2009;39:107–129. doi: 10.1007/s12035-009-8057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman PJ. Fragile X Syndrome: Diagnosis, Treatment and Research. Baltimore, MD: Johns Hopkins University Press; 2002. [Google Scholar]
- Harlow EG, Till SM, Russell TA, Wijetunge LS, Kind P, Contractor A. Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron. 2010;65:385–398. doi: 10.1016/j.neuron.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RN, Feinberg RL, Vaurio R, Passanante NM, Thompson RE, Kaufmann WE. Autism spectrum disorder in fragile X syndrome: a longitudinal evaluation. Am J Med Genet A. 2009;149A:1125–1137. doi: 10.1002/ajmg.a.32848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Huot ME, Tremblay S, Davidovic L, Mazroui R, Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc Natl Acad Sci U S A. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lauterborn JC, Rex CS, Kramar E, Chen LY, Pandyarajan V, Lynch G, Gall CM. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J Neurosci. 2007;27:10685–10694. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM. The state of synapses in fragile X syndrome. Neuroscientist. 2009;15:549–567. doi: 10.1177/1073858409333075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Kolleker A, Berberich S, Ginger M, Frick A, Mientjes E, Oostra BA, Seeburg PH. Synaptic ionotropic glutamate receptors and plasticity are developmentally altered in the CA1 field of Fmr1 knockout mice. J Physiol. 2009;587:787–804. doi: 10.1113/jphysiol.2008.160929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TJ, Rashid MH, Millecamps M, Sanoja R, Entrena JM, Cervero F. Decreased nociceptive sensitization in mice lacking the fragile X mental retardation protein: role of mGluR1/5 and mTOR. J Neurosci. 2007;27:13958–13967. doi: 10.1523/JNEUROSCI.4383-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Zhai RG, Dresbach T, Bresler T, Torres VI, Gundelfinger ED, Ziv NE, Garner CC. Unitary assembly of presynaptic active zones from Piccolo-Bassoon transport vesicles. Neuron. 2003;38:237–252. doi: 10.1016/s0896-6273(03)00207-1. [DOI] [PubMed] [Google Scholar]
- Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. Journal of Neuroscience. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Symons FJ, Clark RD, Hatton DD, Skinner M, Bailey DB., Jr Self-injurious behavior in young boys with fragile X syndrome. Am J Med Genet A. 2003;118A:115–121. doi: 10.1002/ajmg.a.10078. [DOI] [PubMed] [Google Scholar]
- Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Volk LJ, Pfeiffer BE, Gibson JR, Huber KM. Multiple Gq-coupled receptors converge on a common protein synthesis-dependent long-term depression that is affected in fragile X syndrome mental retardation. J Neurosci. 2007;27:11624–11634. doi: 10.1523/JNEUROSCI.2266-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zang JB, et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukin RS, Richter JD, Bagni C. Signals, synapses, and synthesis: how new proteins control plasticity. Front Neural Circuits. 2009;3:14. doi: 10.3389/neuro.04.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Exptl Procedures
Suppl Table S1
Suppl Table S2A-C
Suppl Table S3
Suppl Table S4
Suppl Table S5
Suppl Table S6
Suppl Table S7
Suppl Author Interview
Suppl Fig S1
Suppl Fig S2
Suppl Fig S3
Suppl Fig S4
Suppl Fig S5
Suppl Fig S6
Suppl Fig S7