Rare copy number variants are an important cause of epileptic encephalopathies (original) (raw)
. Author manuscript; available in PMC: 2012 Dec 1.
Published in final edited form as: Ann Neurol. 2011 Dec;70(6):974–985. doi: 10.1002/ana.22645
Abstract
Objective
Rare copy number variants (CNVs) – deletions and duplications – have recently been established as important risk factors for both generalized and focal epilepsies. A systematic assessment of the role of CNVs in epileptic encephalopathies, the most devastating and often etiologically obscure, group of epilepsies, has not been performed.
Methods
We evaluated 315 patients with epileptic encephalopathies characterized by epilepsy and progressive cognitive impairment for rare CNVs using a high-density, exon-focused whole-genome oligonucleotide array.
Results
We found that 25/315 (7.9%) of our patients carried rare CNVs that may contribute to their phenotype, with at least half being clearly or likely pathogenic. We identified two patients with overlapping deletions at 7q21 and two patients with identical duplications of 16p11.2. In our cohort, large deletions were enriched in affected individuals compared to controls, and four patients harbored two rare CNVs. We screened two novel candidate genes found within the rare CNVs in our cohort but found no mutations in our patients with epileptic encephalopathies. We highlight several additional novel candidate genes located in CNV regions.
Interpretation
Our data highlight the significance of rare copy number variants in the epileptic encephalopathies, and we suggest that CNV analysis should be considered in the genetic evaluation of these patients. Our findings also highlight novel candidate genes for further study.
INTRODUCTION
Epilepsy is often associated with major co-morbidities, most frequently cognitive difficulties, which can be static or progressive. In the former, a variety of disorders are believed to cause both static intellectual disability and epilepsy; in the latter, the epileptic process contributes to cognitive impairment. Epileptic encephalopathies (EE), recently defined as severe epilepsies where the epileptic activity, in addition to the seizures, contributes to cognitive impairment or regression, account for a significant proportion of the refractory epilepsies usually associated with poor outcome.1 Most of the EE begin in infancy or childhood, often in the setting of normal development with subsequent cognitive decline. In this way, the EE differ considerably from disorders with static intellectual disability.
Copy number variants (CNVs) have been established as an important source of mutation in many neurocognitive and neuropsychiatric conditions, including intellectual disability (ID), autism spectrum disorders, and schizophrenia.2 CNVs are regarded as causative in >10% of cases of ID.3 More recently, both targeted4–6 and genome-wide7, 8 discovery of CNVs in individuals with epilepsy have established the importance of CNVs in the epilepsies as well. We previously evaluated >500 individuals with various types of pharmacoresponsive epilepsy for rare, potentially pathogenic CNVs, which were present in nearly 10% of patients.7 The majority of patients in that series had genetic (idiopathic) generalized or focal epilepsies. However, a small number had more severe forms of epilepsy, and a greater percentage of these patients carried rare, potentially pathogenic CNVs. Although there have been reports of CNVs in some cases9–11, the EE have not been systematically interrogated for CNVs as a group.
In this study, we hypothesized that some EE could be caused by CNVs and that genes within those CNVs would be novel candidate genes for EE. We selected a cohort of 315 patients with EE. We performed high-density whole-genome array comparative genomic hybridization (CGH) to determine what proportion of severe epilepsies may be caused by rare CNVs and to identify novel candidate genes within those regions.
METHODS
Patient samples
Patients with EE for whom no cause was known were ascertained by referral and from the investigators’ clinical practices. Detailed epilepsy, developmental and general medical history was obtained with EEG and neuroimaging results. A seizure questionnaire was completed with the parents or caregivers where possible.12 Strenuous efforts were made to obtain all previous medical records. Data were analysed to determine each patient’s phenotype and epilepsy syndrome according to the ILAE classifications.1, 13–15 Where patients did not have a phenotype consistent with a well known epilepsy syndrome, but they did have an encephalopathic EEG with high voltage diffuse background slowing and frequent epileptiform activity, and developmental slowing or regression, they were classified according to the epileptiform pattern. For example, those with generalized spike and slow wave or sharp and slow wave activity were called symptomatic generalized epilepsies (SGEs) and those with unifocal or multifocal epileptiform abnormalities were classified as focal epilepsies with regression.
This study was carried out with approval from the Human Research Ethics Committees of Austin Health and the Royal Children’s Hospital (Victoria, Australia) and the human subjects review board at the University of Washington (Seattle, WA). All subjects or, in the case of minors or individuals with intellectual disability, parents or legal guardians gave informed consent to participate.
Array CGH and analysis
We performed oligonucleotide array CGH using commercially available whole-genome exon-focused arrays with 720,000 isothermal probes (Human CGH 3×720K Whole-Genome Exon-Focused Array, Roche NimbleGen, Madison, WI). Probes were preferentially placed in exonic sequences but also distributed throughout non-exonic regions. Probe spacing was variable with a mean spacing of ~4.2 kb. Data were analyzed using NimbleScan software followed by a three-state Hidden Markov Model as previously described.7 CNV calls were then filtered to eliminate (i) events comprising <5 probes; (ii) events that did not overlap any RefSeq genes; (iii) events entirely within segmental duplications; and (iv) events with >50% overlap with a CNV detected in 4519 published controls, taking into account probe coverage and ability to detect a given CNV in those controls.16, 17 For a subset of genomic regions that are prone to recurrent rearrangement and known to be associated with a range of neurocognitive disorders, we did not require absence of CNVs in controls, as it is well established that there is incomplete penetrance and a small number of controls carry CNVs in some of these regions (e.g. 15q13, 16p11.2, 16p13, 15q11.2; see below). All filtered events were also visually inspected in a genome browser. Candidate rare CNVs not seen in controls (all CNVs listed in Table 2) were validated using custom high-density arrays (Agilent Technologies, Santa Clara, CA). Parents were analyzed, where available, to determine if the CNV had arisen de novo or was inherited; parental phenotypes were taken into account to help to determine if the CNV was significant. We also considered CNV size and gene content when evaluating the likely pathogenicity of a CNV, following guidelines that have been published for the interpretation of clinical array CGH results in patients with ID, autism or multiple anomalies.3 Deletions involving known epilepsy genes (CDKL5, UBE3A, CNTNAP2) were considered pathogenic. De novo deletions were considered pathogenic; de novo duplications were considered likely pathogenic; CNVs >1 Mb that were inherited or of unknown inheritance were deemed likely pathogenic. In two cases, an inherited 500-kb duplication of 16p11.2 was considered likely pathogenic because of the known disease associations with duplications at this locus as it may be contributing to the epilepsy phenotype. All other CNVs were considered of unknown significance due to size (<500 kb), inheritance (inherited or unknown), or gene content (no known epilepsy genes).
TABLE 2.
Rare copy number variants identified in a cohort of 315 affected individuals
| Proband | Chromosome | Build 36 coord (Mb) | Size | CNV | Inheritance | Syndrome | Seizure type(s) and intellect | Causal? | Candidate Gene(s) |
|---|---|---|---|---|---|---|---|---|---|
| EPILEPTIC ENCEPHALOPATHY (n=315) | |||||||||
| T2363 | 1q44 | chr1:242.62–244.06 | 1.44 Mb | del | Not in mother | SGE | FS, GTCS, M, CSE, T; severe ID | L | HNRNPU, EFCAB2 |
| T2761 | 7q35 | chr7: 146.09–146.15 | 60 kb | delh | Parents heterozygous | FE with Regression | FDS, CSE; moderate ID | P | CNTNAP2^ |
| T2959 | Xp22 | chrX:18.21–18.50 | 290 kb | del | de novo | IS | T, Sp, M, severe ID | P | CDKL5 |
| T3334 | 15q11 | chr15:23.21–23.48 | 270 kb | del | de novom | MAE | At, M, MSE; severe ID | P | UBE3A |
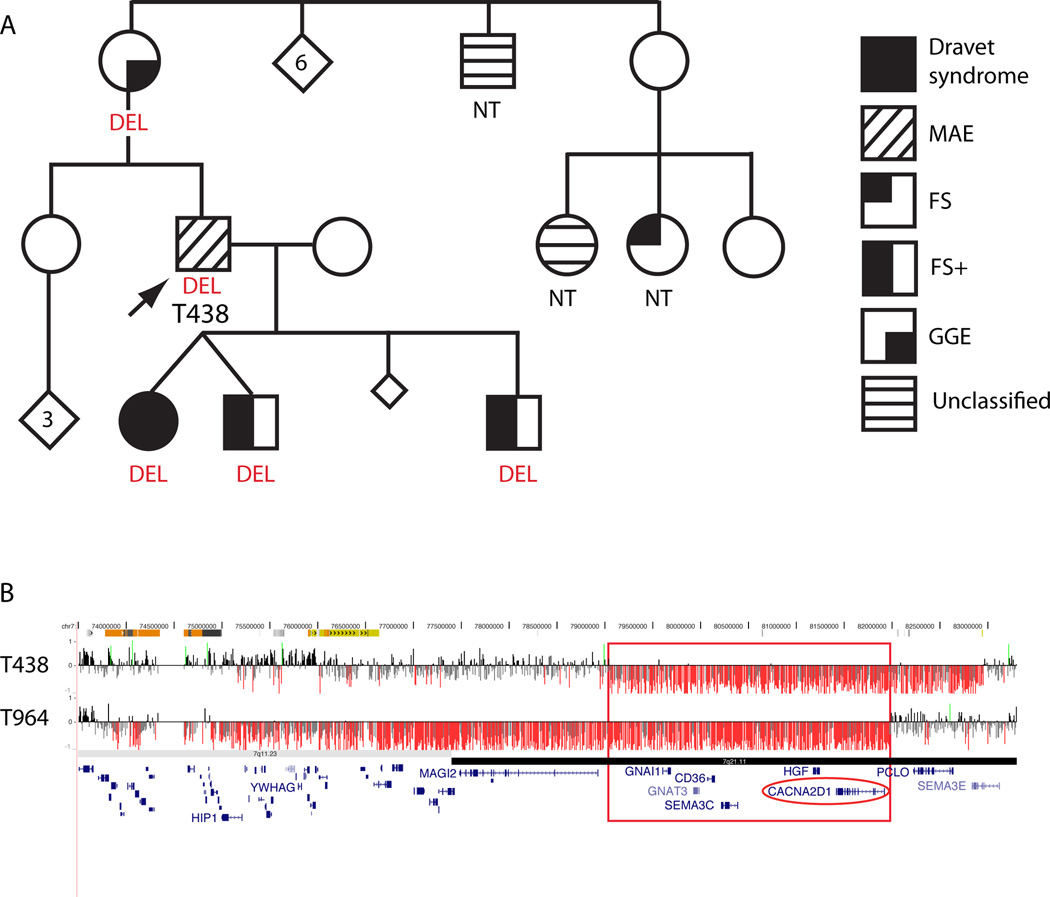
| T438 | 7q21 | chr7:79.06–82.95 | 3.9 Mb | del | Inherited (M)* | MAE | FS, At DA, Abs, GTCS; borderline intellect | L | CACNA2D1, PCLO |
| T964 | 7q11-q21 | chr7:73.78–81.98 | 8.2 Mb | del | de novo | SGE | FS, GTCS, M, Atyp Abs; severe ID | P | MAGI2, CACNA2D1 |
| 7257 | 10p13 | chr10:12.25–17.5 | 5.25 Mb | del | de novo | MAE | GTCS, Abs, M, At DA, T; mild ID | P | |
| 8893 | 5q33-q34 | chr5:156.18–162.63 | 6.45 Mb | del | de novo | MAE | FS, GTCS, M, Abs; mild ID | P | GABRA1, GABRG2 |
| T1962 | 4p16 | chr4: pter-1.50 | 1.5 Mb | del | de novo | EAS | FS, GTCS, FDS; moderate ID | P | SLC26A1 |
| T3729 | 9p24-p21 | chr9: 1–9.22chr9:9.22 –27.70 | 9.22 Mb18.5 Mb | deldup | de novode novo | SGE | T, GTCS, CSE, FDS; moderate ID | P | |
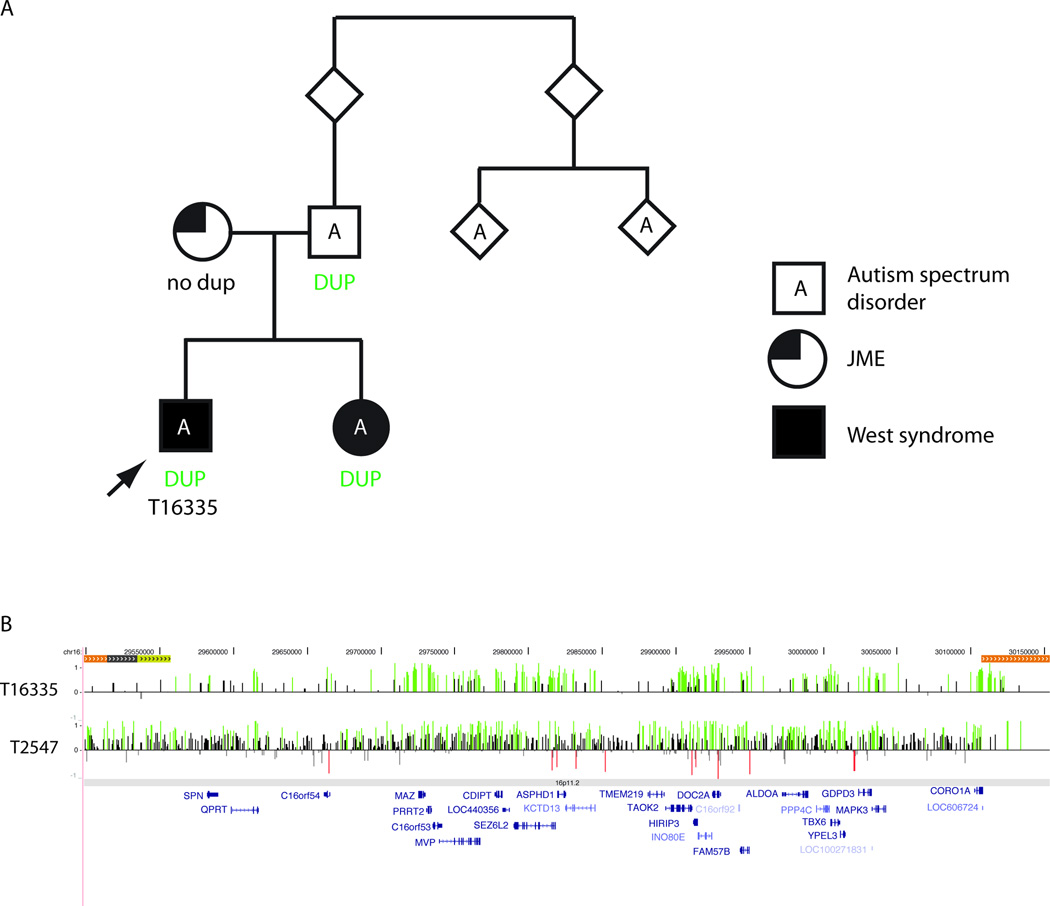
| T16335 | 16p11.2 | chr16: 29.5–30.0 | 500 kb | dup | Inherited (P) | IS | Sp; mild ID | L | |
| T2547 | 16p11.2 | chr16: 29.5–30.0 | 500 kb | dup | Inherited (P) | MFEE | GTCS, HC, SGS, FDS, T; mild ID | L | |
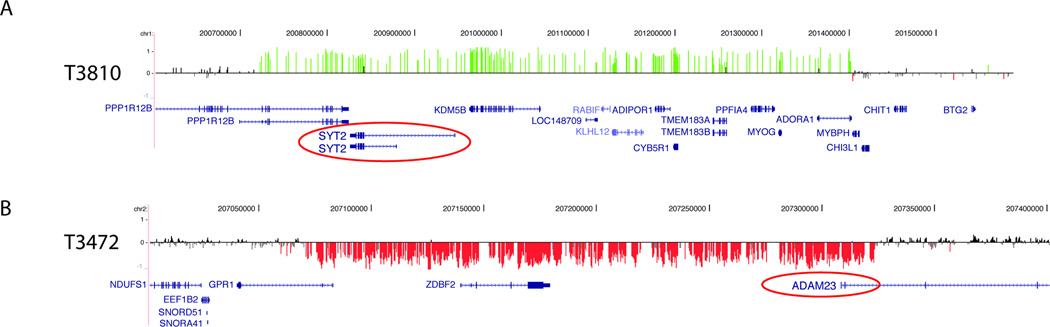
| T3810 | 1q32 | chr1:200.72–201.40 | 700 kb | dup | de novo | EAS | EEG only; mild ID | L | SYT2 |
| T2709 | 12q124q28 | chr12:38.86–39.05chr4:130.14–130.21 | 190 kb70 kb | deldel | Inherited (M)Inherited (P) | MPSI | FDS, MFS, Sp, severe ID | U | LRRK2^SCLT1^ |
| T3472 | 2q33 | chr2:206.79–207.03 | 510 kb | del | Not in mother | DS | Febrile hemiclonic status, GTCS, HC, M, At DA, CSE; moderate ID | U | GPR1, ZDBF2, ADAM23 |
| 8245 | 15q13 | chr15: 26.80–28.50 | 1.7 Mb | del | Inherited (P) | MAE | FS, GTCS, At DA, Abs; normal intellect | U | |
| T1466 | 5p13 | chr5: 37.4–38.0 | 600 kb | del | Inherited (M) | SGE | GTCS, FS, T DA, Abs, Atyp Abs, T, At DA, M, NSE; severe ID | U | GDNF |
| T1456 | 6q23 | chr6: 137.32–137.40 | 80 kb | del | Inherited (P) | SGE | Cyanotic, Abs, M, T DA, AT DA, MSE ; severe ID | U | NHEG |
| 6184 | Xq28 | chrX: 148.49–148.55 | 60 kb | del | Inherited (M) | SGE | GTCS, T DA, Abs, FDS, Atyp Abs, At, M, severe ID, | U | TMEM185A^ |
| T3467 | 20q1322q11 | chr20: 52.67–53.09chr22: 18.62–20.29 | 420 kb1.67 Mb | dupdup | Inherited (M)Inherited (M) | CSWS | FDS, SGS; mild ID | U | DOK5^ |
| T892 | 3q11 | chr3: 98.05–99.69 | 1.64 Mb | dup | Inherited (M) | IS | GTCS, Sp, CSE, T, FDS; severe ID | U | EPHA6 GABRR3 |
| T18349 | 16p11.25p13 | chr16: 28.7–29.0chr5: 36.55–36.85 | 300 kb300 kb | deldel | Neither in mother | SGE | FS, GTCS, Abs, Atyp Abs, T, CSE, M; moderate ID | U | SLC1A3^ |
| T19083 | 14q22 | chr14:50.23–50.37 | 135 kb | del | Inherited (M) | MFEE | FDS, T DA, M; mild ID | U | NIN^ |
| T16681 | 22q11.21 | chr22:16.44–16.52 | 80 kb | del | Inherited (P) | MFEE | FSD, aura, SGS, Ge, GTCS; mild ID | U | SLC25A18 |
| T18721 | 4q35 | chr4:184.18–185.23 | 1.05 Mb | dup | Inherited (P) | MAE | FS, GTCS, Abs, Abs status, M, At DA; mild ID | U |
Sequence analysis
For each candidate gene (CACNA2D1, LRRK2), targeted sequencing was performed for all exons, exon-intron boundaries and 2 kb upstream of the transcription start site using standard Sanger sequencing. Primer sequences are available upon request. CACNA2D1 was resequenced in 94 probands including 80 cases with epilepsy with myoclonic atonic seizures (MAE), 13 with Dravet syndrome and 1 with genetic epilepsy with febrile seizures plus (GEFS+) phenotype. Of these, 92 were from our cohort of 315 cases; two additional cases were selected for resequencing only. LRRK2 was resequenced in all eleven cases from our cohort with a diagnosis of migrating partial seizures of infancy (MPSI).
RESULTS
We studied a cohort of 315 unrelated patients with epileptic encephalopathy (EE; Table 1). We identified 25/315 (7.9%) patients with one or more rare CNVs not seen in controls (Table 2). The mean CNV size was 2.26 Mb and median size was 510 kb. Of these, 13 patients (4.1%) had CNVs that were clearly pathogenic (n=8) or likely pathogenic (n=5), and 12 had one or more rare CNVs that were not seen in controls but were of unclear clinical significance. Four individuals had two rare CNVs.
TABLE 1.
Phenotypes of patients included in this study
| DIAGNOSIS | N | N with rare CNV(s) | |
|---|---|---|---|
| MAE | 77 | 6 | (7.8%) |
| SGE | 65 | 7 | (10.8%) |
| Infantile spasms | 44 | 3 | (6.8%) |
| Epilepsy-Aphasia Syndrome / CSWS | 29 | 3 | (10.3%) |
| Lennox-Gastaut Syndrome | 20 | 0 | (0.0%) |
| Dravet syndrome* | 19 | 1 | (5.3%) |
| MPSI | 11 | 1 | (9.1%) |
| DESC / FIRES | 10 | 0 | (0.0%) |
| FE with regression^ | 17 | 4 | (23.5%) |
| Other# | 23 | 0 | (0.0%) |
| Total | 315 | 25 | (7.9%) |
Inheritance
We evaluated one or both parents in all 25 cases to determine the inheritance pattern of all 29 rare CNVs (Table 2). We identified nine de novo, eight maternally inherited and seven paternally inherited CNVs. In one case (T2761), the proband had a homozygous deletion and both parents were confirmed to be heterozygous carriers. In three cases (four CNVs total) the CNVs were not present in the mother, but the father was unavailable for analysis.
Pathogenic CNVs
In thirteen patients we considered the CNV(s) to be pathogenic or likely pathogenic based on size, gene content, de novo inheritance or the previous literature (Table 2). Clearly pathogenic CNVs include heterozygous deletions that disrupt a single, known gene in two cases: UBE3A in case T3334 and CDKL5 in case T2959. We also identified a homozygous deletion removing exon 2 of CNTNAP2 in case T2761 and his affected sibling. Ten other cases have CNVs encompassing multiple genes. Two probands have overlapping deletions of 7q21: T964 with a de novo 8-Mb deletion of 7q11-q21 and T438 with a 4-Mb deletion of 7q21 (Figure 1). The 7q21 deletions have a 3-Mb region of overlap that includes six genes: GNAI1, GNAT3, CD36, SEMA3C, HGF, CACNA2D1. Other pathogenic CNVs include deletions of 10p13 (7257), 5q33-q34 (8893) and 4p16 (T1962) as well as one case with deletion of 9p24-p23 and adjacent duplication of 9p23-p21 (T3729). Likely pathogenic CNVs include a duplication of 16p11.2 in two cases (Figure 2) and a deletion of 1q44 (T2363). In addition, we identified a likely pathogenic, novel, de novo 700-kb duplication of 1q32 in case T3810 (Figure 3). Additional rare CNVs of uncertain significance were present in 12 patients (Table 2, Figure 3).
Figure 1. Overlapping deletions of 7q21 in two probands.
A) Pedigree for proband T438, who has a ~4 Mb deletion of 7q21. The proband’s mother and three children, who have all had one or more seizures, also have the same deletion. B) Array CGH data for T438 and T964. The red box highlights the region on 7q21 that is deleted in both patients. X-axis represents genomic coordinates (chr7: 73.5–83.5 Mb, NCBI Build 36). For each individual, deviations of probe log2 ratios from zero are depicted by vertical grey/black lines, with those exceeding a threshold of 1.5 standard deviations from the mean probe ratio colored green and red to represent relative gains and losses, respectively. Genes are represented by blue lines at the bottom.
MAE = Epilepsy with myoclonic-atonic seizures; FS = febrile seizures; FS+ = febrile seizures plus; GGE = genetic generalized epilepsies; DEL = deletion; NT = not tested
Figure 2. Recurrent duplications of 16p11.2 in two probands.
A) Pedigree for proband T16335, who has a duplication of 16p11.2. The duplication in inherited from his father and also present in his sister with West syndrome. B) Array CGH data for T16335 and T2547, with log2 ratios displayed as in Figure 1.
JME = juvenile myoclonic epilepsy; DUP = duplication
Figure 3. Copy number changes affecting neuronal genes of interest.
A) 700-kb de novo duplication of 1q32 in patient T3810. B) 300-kb deletion of 5p13 involving the ADAM23 gene in T18349. Array CGH data displayed as in Figures 1 and 2.
Hotspot CNVs
Certain regions of the genome are “hotspots” for recurrent CNVs due to the presence of large blocks of duplicated DNA that facilitate non-allelic homologous recombination at meiosis.18 We identified five individuals with CNVs at hotspot regions. Two individuals (T16335 with West syndrome and T2547 with a multifocal epileptic encephalopathy) have duplications of proximal 16p11.2 (chr16:29.5–30.0 Mb; Figure 2). We also identified rearrangements of uncertain significance at hotspot regions in case T18349 with SGE (distal 16p11.2 deletion), case T3467 with continuous spike and wave during slow sleep (CSWS; duplication of distal 22q11) and case 8245 with MAE (deletion of proximal 15q13.3, BP3–BP4) (Table 2). We did not detect CNVs at distal 15q13.3 (BP4–BP5), 16p13.11 or 15q11.2, loci known to be important for GGE and some focal epilepsies.4, 6, 8
Sequence analysis of candidate genes
We selected two candidate genes for sequence analysis: CACNA2D1 and LRRK2 (Table 3). CACNA2D1, which is deleted in two patients (T438 with MAE and T964 with SGE), was resequenced in 94 probands. No convincing mutations were identified (Table 3). LRRK2, deleted in patient T2709 with MPSI, was resequenced in ten additional patients from our cohort with MPSI. In addition, the non-deleted allele was sequenced in the original proband. No deleterious changes were identified.
Table 3.
CACNA2D1 resequencing results
| Gene | Patient | Sequencechange | AA change | Phenotype | Inheritance |
|---|---|---|---|---|---|
| CACNA2D1 | T1570 | Exon 1, c.2382G>A# | S709N | MAE | Inherited from father |
| 9402 | Exon 38, c.3390A>C* | D1045A | Mild MAE | Unaffected mother and dizygotic twin have same change | |
| T17741 | Exon 38, c.3390A>C | D1045A | MAE | Unaffected father and affected monozygotic twin have same change |
DISCUSSION
We performed genome-wide exon-focused array CGH in a series of 315 patients with epileptic encephalopathies in order to identify novel genomic regions and candidate genes and to investigate how frequently CNVs were responsible for these phenotypes. Overall, we found that 7.9% of affected individuals carried at least one rare CNV. In 4.1%, we identified clearly pathogenic CNVs. These findings suggest that array CGH should be considered in the genetic evaluation of individuals with EE. Rare CNVs were most commonly identified in patients with focal epilepsy with regression, epilepsy-aphasia syndrome and symptomatic generalized epilepsy (Table 1) but were not identified in any of the patients with Lennox Gastaut syndrome or devastating epileptic encephalopathy in school age children. Investigation of larger cohorts of specific EE should be carried out to further evaluate whether this apparent difference is significant.
Within our cohort, we identified both de novo and inherited CNVs. While we considered those that are de novo most likely to be pathogenic, many of the rare inherited CNVs contain brain-expressed genes involved in synaptic transmission and axonal guidance and are likely to contribute to the patient’s phenotype. Indeed, even for the recurrent deletions at 15q13.3 and 16p13.11, which are clearly associated with epilepsy risk, there are many examples of unaffected carrier parents, incomplete penetrance and variable expressivity. The same may be true for some of the rare inherited CNVs identified in this study, and in these cases there may be additional genetic or non-genetic factors that also play a role in the clinical presentation.
Pathogenic events and novel candidate genes
In several cases we found clearly pathogenic deletions that involved known epilepsy genes and had clear genotype-phenotype correlation, albeit atypical features in some instances. These include deletion of Xp22 resulting in the disruption of CDKL5 in a boy who had an EE with the typical picture of a three stage evolution of seizures characteristically seen in girls with CDKL5 encephalopathy19 and also reported in rare male cases.10 We also found an atypical deletion of 15q11 that disrupted UBE3A in a man with MAE who had features reminiscent of Angelman syndrome. Another man with MAE had a 6.5-Mb deletion of 5q33-q34 encompassing a cluster of genes encoding subunits of the GABA-A receptor. Mutations in two of the genes in the deleted region, GABRA1 and GABRG2, have been associated with JME20 and GEFS+ and CAE,21, 22 respectively. We identified a homozygous deletion of exon 2 of the CNTNAP2 gene in a pair of siblings of which the proband had focal epilepsy with regression; they shared some features in common with the Amish families reported by Strauss and colleagues.23
We also identified several CNVs that provide insight into potentially novel candidate genes for epilepsy. Two probands had overlapping deletions of 7q21. Deletions of 7q11-q21 distal to the Williams-Beuren syndrome locus have been associated with infantile spasms, with considerable interest in the gene MAGI2.24 YWHAG and HIP1 have also been proposed as candidate genes for the infantile spasm phenotype.25 While patient T964 with SGE carried a large de novo deletion that included MAGI2, YWHAG and HIP1, patient T438 had a ~4-Mb deletion that did not include these genes. Furthermore, neither of our patients had spasms. Given the similarity in phenotypes of the two probands, we focused on the six genes that were deleted in both patients as candidates for sequencing. One of the genes in the region of overlap is CACNA2D1, which encodes the alpha-2/delta subunit of brain voltage-dependent calcium channels that bind the anti-epileptic drug, gabapentin.26 Therefore, we considered CACNA2D1 an excellent candidate gene for MAE and related phenotypes. We performed sequence analysis in 94 probands with MAE or Dravet syndrome phenotypes but did not detect any clearly deleterious mutations.
Interestingly, the smaller 4-Mb deletion in the patient with MAE segregated with a range of epilepsy and cognitive phenotypes in the proband’s family (Figure 1). The deletion was found in family members with markedly different phenotypic severity ranging from the EEs of MAE and Dravet syndrome to febrile seizures plus, consistent with the GEFS+ spectrum, and even GGE.27 All affected family members had learning difficulties; affected children all had marked behavioural problems. Thus the 7q21 deletion may be acting as one of several genetic factors that contribute to the more severe phenotype of MAE and Dravet syndrome in the setting of complex inheritance rather than acting as a monogenic cause. We have not screened CACNA2D1 in other phenotypes, but given the presence of GGE in the proband’s mother, a cohort of patients with GGE would be worth testing. Alternatively, another gene in the deleted region may be responsible for the phenotypes in this family.
We identified a 510-kb deletion in patient T3472 with Dravet syndrome that disrupts the ADAM23 gene (Figure 3). The deletion is not found in the mother’s genome, but we were unable to evaluate the father. ADAM23 knockout mice have aberrant dendrite morphology and exhibit seizures in the neonatal period, and heterozygous mice are susceptible to PTZ-induced seizures.28 Furthermore, ADAM23 is part of a complex that contains LGI1, ADAM22 and Kv1.129 and has been shown to directly bind LGI1,28 mutations of which cause autosomal dominant partial epilepsy with auditory features in humans.30, 31 It is possible that disruption of ADAM23 in our patient leads to Dravet syndrome.
Case T3810 with an epilepsy-aphasia syndrome carries a de novo 700-kb duplication of 1q32 that involves 13 genes (Figure 3). One of these is SYT2, a member of the synaptotagmin family of genes that encode membrane proteins expressed at the synapse that are thought to act as calcium sensors.32, 33 It is possible that excess SYT2 disrupts normal synaptic transmission in our patient.
Hotspot CNVs
Recurrent deletions at three rearrangement “hotspots” – 15q13.3 (BP4–BP5), 16p13.11 and 15q11.2 – have recently been identified as important risk factors for epilepsy.4–6, 8 Each of these deletions is found in up to 1% of individuals with epilepsy, and each has also been associated with ID, autism and/or schizophrenia.34–41 While heterozygous 15q13.3 deletions have been found almost exclusively in patients with GGE, deletions at the other loci have been found in patients with a broader range of epilepsies. We identified one patient with an inherited deletion BP3–BP4 on 15q13. This deletion is proximal to the BP4–BP5 deletion that has been associated with epilepsy, and the clinical significance is not clear.42 Interestingly, we did not identify any CNVs at 15q13.3 (BP4–BP5), 16p13.11 or 15q11.2, suggesting that heterozygous deletions of these regions may be primarily associated with the more common and milder forms of epilepsy, although homozygous 15q13.3 deletions have been associated with a severe phenotype.43
We did identify CNVs at several other hotspot regions that have been associated with neurocognitive disorders. Interestingly, two individuals in the EE cohort have duplications of proximal 16p11.2 (chr16: 29.5–30.0 Mb), which we considered to be likely pathogenic. T16335 had West syndrome and autism spectrum disorder and T2547 had multifocal epileptic encephalopathy. Duplications of this region have been associated with ID, schizophrenia and autism44–46. We previously reported one individual with JME and the same duplication7, and Bedoyan and colleagues47 reported a de novo duplication of this region in a child with EE that the authors felt was consistent with MPSI. We did not find this duplication in 11 cases with MPSI. We were able to evaluate relatives of case T16335 and found that the duplication was also present in the proband’s sister who also had West syndrome, and father, both of whom had autism spectrum disorder. Similar to the deletions of 15q13.3, 15q11.2 and 16p13.11, duplications at 16p11.2 appear to be a risk factor for a wide range of neurocognitive and neuropsychiatric disorders including different types of epilepsy, autism, ID and schizophrenia, though the mechanism underlying such highly variable expression is not yet known.48, 49 As discussed below, we also identified a deletion of distal 16p11.2 in a patient with a second deletion.
Two hits
Four patients (1.3% of cohort) in our series carry two rare CNVs (16% of patients with CNVs, similar to controls in which 29% of patients with rare CNVs have carry at least two; p=0.18). Patient T18349 carries two deletions: a deletion of 16p11.2 (chr16:28.8–29.0 Mb) and a deletion of the SLC1A3 gene. The 16p11.2 deletion has been associated with variable phenotypes including early-onset obesity50 and variable developmental delay,51 with a minority of patients reported to have seizures. Interestingly our patient was also obese. A de novo missense mutation in SLC1A3 has been reported in one patient with episodic ataxia, seizures and hemiplegia,52 and a different missense mutation segregates with a mild form of episodic ataxia in another family.53 Given that both the 16p11.2 deletion and the SLC1A3 gene have been associated with epilepsy, it is possible that the two hits together in this patient result in a more severe phenotype of SGE with abnormal early development and moderate ID.54
Patient T2709 with MPSI has a maternally inherited deletion encompassing the entire LRRK2 gene and a paternally inherited deletion involving the SCLT1 gene. SLCT1 acts as a linker protein between the voltage-gated sodium channel Nav1.8 and clathrin.55 LRRK2 is a brain-expressed serine/threonine protein kinase in which gain-of-function mutations cause autosomal dominant early-onset Parkinson disease.56 We hypothesized that complete loss of function of LRRK2 might lead to the MPSI phenotype in our patient, but we were unable to detect a mutation in the non-deleted allele (see below; Table 3). Although each deletion in T2709 is inherited, the combination of both may contribute to the phenotype.
Case T3729 has a 9-Mb terminal deletion of chromosome 9p and an adjacent 18.5-Mb duplication. The terminal deletion in our patient is similar to one described by Heinzen and colleagues8 in a patient with unclassified epilepsy, developmental delay, dysmorphic features and spastic quadriplegia. Finally, patient T3467 with CSWS and her affected brother both have two duplications. One involves the distal part of the common 22q11 deletion syndrome region. The second duplication involves the 3’ end of the DOK5 gene. However, both duplications are inherited from their unaffected mother, making it less likely that either is a highly penetrant pathogenic CNV.
Excess of large deletions
In our cohort, we find 12 CNVs that are >1Mb in 11 (3.5%) individuals (Table 2). Of these, 7 (2.2%) have a large deletion, 3 (1.0%) have a large duplication, and one patient has one of each. In contrast, in a set of 2493 control individuals, Itsara and colleagues16 found that 1.6% of controls had a CNV >1 Mb; 0.3% had a large deletion and 1.3% had a large duplication. Therefore, we find a clear excess of large deletions in our cohort (8/315 patients vs. 8/2493 controls, p=0.00013, Fisher’s exact test). Our findings are similar to those of Heinzen and colleagues8 who also noted an excess of large (>1 Mb) deletions in their cohort of patients with primarily focal epilepsies, compared to controls without ID.
Comparison with previous studies of epilepsies
We compared our CNVs in the EE to those of two recently published studies of epilepsy in which whole-genome CNV analysis was performed.7, 8 Heinzen et al8 reported 36 patients with rare deletions >1 Mb in a series of 3812 patients with focal (>90%) or generalized (<10%) epilepsy. At least three have significant overlap with CNVs in our study: 5q34 deletion, 9p24.3-p23 deletion and 7q21.1-q21.13 deletion. We also find some overlapping CNVs with those from our recent report7 of CNVs in 517 patients with generalized (n=399), focal (n=63) or other (n=55) epilepsies: 16p11.2 duplication, 5q33.2 deletion, and intragenic deletion of CNTNAP2. Together, these reports confirm the importance of rare CNVs in the genetic etiology of the epilepsies and emphasize the need for the evaluation of larger series of patients to better understand the pathogenic significance, especially for rare inherited CNVs.
Conclusions
In summary, this is the first report of genome-wide CNV discovery in a large series of patients with epileptic encephalopathies. We identified rare CNVs in 7.9% of 315 patients, at least half of which are likely to be pathogenic and which largely differ from CNVs found in milder epilepsies. Our results also illuminate several novel candidate genes for epilepsy in humans that deserve further study, including CACNA2D1, SLC1A3 and ADAM23. Comprehensive sequence analysis of these and other candidate genes in extended cohorts will help determine whether point mutations contribute to the genetic etiology of each EE.
We suggest that array CGH should be considered in the genetic evaluation of this patient population, where the occurrence of severe epilepsy and regression often prompts extensive and frequently negative investigations. While the interpretation of array CGH results in the clinical setting can be complicated by results of unclear significance, the application of established guidelines can facilitate interpretation and counseling3. Furthermore, as this tool is applied more widely to epilepsy cohorts in both the clinic and the research lab, comparison of CNVs across studies will clarify which specific CNVs are clearly pathogenic and begin to illuminate novel genes, pathways and syndromes in the epileptic encephalopathies.
Supplementary Material
Supp Table S1
Acknowledgments
We thank the patients and their families for participating in our research studies and referring clinicians. We thank Drs Lindsay Smith, Simon Harvey, Sophie Calvert, Kent Kelley, Frederick Andermann and Daniel Keene for referral of patients. HCM is funded by NIH (NINDS 1R01NS069605) and is a recipient of a Career Award for Medical Scientists from the Burroughs Wellcome Fund. EEE is an investigator of the Howard Hughes Medical Institute. Program grant funding was received from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Berg AT, Berkovic SF, Brodie MJ, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010 Apr;51(4):676–685. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- 2.Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009 Jun;19(3):196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010 May 14;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Kovel CG, Trucks H, Helbig I, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2009 Oct 20; doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dibbens LM, Mullen S, Helbig I, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet. 2009 Oct 1;18(19):3626–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009 Feb;41(2):160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genetics. 2010;6 doi: 10.1371/journal.pgen.1000962. e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinzen EL, Radtke RA, Urban TJ, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010 May 14;86(5):707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saitsu H, Kato M, Mizuguchi T, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008 Jun;40(6):782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 10.Castren M, Gaily E, Tengstrom C, Lahdetie J, Archer H, Ala-Mello S. Epilepsy caused by CDKL5 mutations. Eur J Paediatr Neurol. 2010 May 19; doi: 10.1016/j.ejpn.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Paciorkowski AR, Thio LL, Rosenfeld JA, et al. Copy number variants and infantile spasms: evidence for abnormalities in ventral forebrain development and pathways of synaptic function. Europeanjournal of human genetics : EJHG. 2011 Jun 22; doi: 10.1038/ejhg.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reutens DC, Howell RA, Gebert KE, Berkovic SF. Validation of a questionnaire for clinical seizure diagnosis. Epilepsia. 1992 Nov–Dec;33(6):1065–1071. doi: 10.1111/j.1528-1157.1992.tb01760.x. [DOI] [PubMed] [Google Scholar]
- 13.Proposal for revised clinical and electroencephalographic classification of epileptic seizures. From the Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia. 1981 Aug;22(4):489–501. doi: 10.1111/j.1528-1157.1981.tb06159.x. [DOI] [PubMed] [Google Scholar]
- 14.Proposal for classification of epilepsies and epileptic syndromes. Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia. 1985 May–Jun;26(3):268–278. [PubMed] [Google Scholar]
- 15.Proposal for revised classification of epilepsies and epileptic syndromes. Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia. 1989 Jul–Aug;30(4):389–399. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- 16.Itsara A, Cooper GM, Baker C, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009 Feb;84(2):148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaikh TH, Gai X, Perin JC, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009 Sep;19(9):1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey JA, Gu Z, Clark RA, et al. Recent segmental duplications in the human genome. Science. 2002 Aug 9;297(5583):1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 19.Bahi-Buisson N, Kaminska A, Boddaert N, et al. The three stages of epilepsy in patients with CDKL5 mutations. Epilepsia. 2008 Jun;49(6):1027–1037. doi: 10.1111/j.1528-1167.2007.01520.x. [DOI] [PubMed] [Google Scholar]
- 20.Cossette P, Liu L, Brisebois K, et al. Mutation of GABRA1 in an autosomal dominant form of juvenile myoclonic epilepsy. Nat Genet. 2002 Jun;31(2):184–189. doi: 10.1038/ng885. [DOI] [PubMed] [Google Scholar]
- 21.Baulac S, Huberfeld G, Gourfinkel-An I, et al. First genetic evidence of GABA(A) receptor dysfunction in epilepsy: a mutation inthe gamma2-subunit gene. Nat Genet. 2001 May;28(1):46–48. doi: 10.1038/ng0501-46. [DOI] [PubMed] [Google Scholar]
- 22.Wallace RH, Marini C, Petrou S, et al. Mutant GABA(A) receptor gamma2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet. 2001 May;28(1):49–52. doi: 10.1038/ng0501-49. [DOI] [PubMed] [Google Scholar]
- 23.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006 Mar 30;354(13):1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 24.Marshall CR, Young EJ, Pani AM, et al. Infantile spasms is associated with deletion of the MAGI2 geneon chromosome 7q11.23-q21.11. Am J Hum Genet. 2008 Jul;83(1):106–111. doi: 10.1016/j.ajhg.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komoike Y, Fujii K, Nishimura A, et al. Zebrafish gene knockdowns imply roles for human YWHAG in infantile spasms and cardiomegaly. Genesis. 2010 Apr;48(4):233–243. doi: 10.1002/dvg.20607. [DOI] [PubMed] [Google Scholar]
- 26.Marais E, Klugbauer N, Hofmann F. Calcium channel alpha(2)delta subunits-structure and Gabapentin binding. Mol Pharmacol. 2001 May;59(5):1243–1248. doi: 10.1124/mol.59.5.1243. [DOI] [PubMed] [Google Scholar]
- 27.Scheffer IE, Zhang YH, Jansen FE, Dibbens L. Dravet syndrome or genetic (generalized) epilepsy with febrile seizures plus? Brain Dev. 2009 May;31(5):394–400. doi: 10.1016/j.braindev.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Owuor K, Harel NY, Englot DJ, Hisama F, Blumenfeld H, Strittmatter SM. LGI1-associated epilepsy through altered ADAM23-dependent neuronal morphology. Mol Cell Neurosci. 2009 Dec;42(4):448–457. doi: 10.1016/j.mcn.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukata Y, Lovero KL, Iwanaga T, et al. Disruption of LGI1-linked synaptic complex causes abnormal synaptic transmission and epilepsy. Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3799–3804. doi: 10.1073/pnas.0914537107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalachikov S, Evgrafov O, Ross B, et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet. 2002 Mar;30(3):335–341. doi: 10.1038/ng832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou YD, Lee S, Jin Z, Wright M, Smith SE, Anderson MP. Arrested maturation of excitatory synapses in autosomal dominant lateral temporal lobe epilepsy. Nat Med. 2009 Oct;15(10):1208–1214. doi: 10.1038/nm.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilbush BS, Morgan JI. A third synaptotagmin gene, Syt3, in the mouse. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8195–8199. doi: 10.1073/pnas.91.17.8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang ZP, Xu W, Cao P, Sudhof TC. Calmodulin suppresses synaptotagmin-2 transcription in cortical neurons. J BiolChem. 2010 Oct 29;285(44):33930–33939. doi: 10.1074/jbc.M110.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannes FD, Sharp AJ, Mefford HC, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009 Apr;46(4):223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 Sep 11;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mefford HC, Cooper GM, Zerr T, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009 Sep;19(9):1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller DT, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J Med Genet. 2008 Nov 26;46(4):242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagnamenta AT, Wing K, Akha ES, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009 Dec 3;17(5):687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008 Mar;40(3):322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008 Sep 11;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullmann R, Turner G, Kirchhoff M, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007 Jul;28(7):674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 42.van Bon BW, Mefford HC, Menten B, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009 Aug;46(8):511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endris V, Hackmann K, Neuhann TM, et al. Homozygous loss of CHRNA7 on chromosome 15q13.3 causes severe encephalopathy with seizures and hypotonia. American journal of medical genetics Part A. 2010 Nov;152A(11):2908–2911. doi: 10.1002/ajmg.a.33692. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009 Nov;41(11):1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010 May;47(5):332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008 Feb 14;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 47.Bedoyan JK, Kumar RA, Sudi J, et al. Duplication 16p11.2 in a child with infantile seizure disorder. Am J Med Genet A. 2010 Jun;152A(6):1567–1574. doi: 10.1002/ajmg.a.33415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010 Oct 15;19(R2):R176–R187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mefford HC. Genotype to phenotype-discovery and characterization of novel genomic disorders in a "genotype-first" era. Genet Med. 2009 Dec;11(12):836–842. doi: 10.1097/GIM.0b013e3181c175d2. [DOI] [PubMed] [Google Scholar]
- 50.Bochukova EG, Huang N, Keogh J, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010 Feb 4;463(7281):666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachmann-Gagescu R, Mefford HC, Cowan C, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet Med. 2010 Aug 30; doi: 10.1097/GIM.0b013e3181ef4286. [DOI] [PubMed] [Google Scholar]
- 52.Jen JC, Wan J, Palos TP, Howard BD, Baloh RW. Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology. 2005 Aug 23;65(4):529–534. doi: 10.1212/01.wnl.0000172638.58172.5a. [DOI] [PubMed] [Google Scholar]
- 53.de Vries B, Mamsa H, Stam AH, et al. Episodic ataxia associated with EAAT1 mutation C186S affecting glutamate reuptake. Arch Neurol. 2009 Jan;66(1):97–101. doi: 10.1001/archneurol.2008.535. [DOI] [PubMed] [Google Scholar]
- 54.Girirajan S, Rosenfeld JA, Cooper GM, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010 Mar;42(3):203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu C, Cummins TR, Tyrrell L, Black JA, Waxman SG, Dib-Hajj SD. CAP-1A is a novel linker that binds clathrin and the voltage-gated sodium channel Na(v)1.8. Mol Cell Neurosci. 2005 Apr;28(4):636–649. doi: 10.1016/j.mcn.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Dachsel JC, Farrer MJ. LRRK2 and Parkinson disease. Arch Neurol. 2010 May;67(5):542–547. doi: 10.1001/archneurol.2010.79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Table S1