Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation (original) (raw)
Long noncoding RNAs (lncRNAs) are currently generating much interest as potential regulators of diverse cellular processes. Lodish and colleagues for the first time identify a lncRNA with a critical role in erythropoiesis. This erythroid-specific lncRNA-EPS exhibits anti-apoptotic activity and is required for terminal differentiation of erythroid progenitors. The authors identify multiple apoptotic genes that are regulated by lncRNA-EPS; in particular, the proapoptotic gene Pycard. Significantly, this lncRNA-EPS function is observed in both mice and humans.
Keywords: long noncoding RNA, erythroid terminal differentiation, apoptosis
Abstract
Long noncoding RNAs (lncRNAs) are differentially expressed under both normal and pathological conditions, implying that they may play important biological functions. Here we examined the expression of lncRNAs during erythropoiesis and identified an erythroid-specific lncRNA with anti-apoptotic activity. Inhibition of this lncRNA blocks erythroid differentiation and promotes apoptosis. Conversely, ectopic expression of this lncRNA can inhibit apoptosis in mouse erythroid cells. This lncRNA represses expression of Pycard, a proapoptotic gene, explaining in part the inhibition of programmed cell death. These findings reveal a novel layer of regulation of cell differentiation and apoptosis by a lncRNA.
Erythropoiesis is highly regulated to ensure the proper generation of mature red blood cells needed under multiple physiological conditions. Although the regulation of erythropoiesis by transcription factors and microRNAs is becoming well understood (Kerenyi and Orkin 2010; Zhao et al. 2010), the modulation of red blood cell development by long noncoding RNAs (lncRNAs) is still unknown. lncRNAs are RNA transcripts that are longer than 200 nucleotides (nt) and do not have functional protein-coding capacity. Transcriptomic studies indicated that lncRNAs constitute a significant fraction of the mammalian transcriptome (Carninci et al. 2005; Harrow et al. 2006; Amaral et al. 2008). Interestingly, many lncRNAs are differentially expressed in different tissues and under various developmental and pathological conditions (Guttman et al. 2009; Ponting et al. 2009; Cabili et al. 2011; Wapinski and Chang 2011), suggesting that these RNA transcripts may play important biological roles. Indeed, lncRNAs are involved in various biological processes, such as X-chromosome inactivation (Payer and Lee 2008), genomic imprinting (Koerner et al. 2009), p53 signal pathway (Huarte et al. 2010), cancer metastasis (Gupta et al. 2010), development (Rinn et al. 2007), maintenance of pluripotency of stem cells (Guttman et al. 2011), reprogramming of somatic cells (Loewer et al. 2010), etc. Interestingly, mechanistic studies revealed that unlike small RNAs, such as microRNAs, lncRNAs can regulate gene expression via diverse mechanisms (for review, see Hung and Chang 2010), such as chromatin modification, enhancing transcription, and promoting mRNA degradation. Collectively, these observations revealed that lncRNAs are a novel class of regulatory factors in modulating gene expression under various biological processes.
From embryonic day 12 (E12) to E16, mouse fetal liver serves as the primary erythropoietic site for the embryo: Cells of the erythroid lineage comprise >90% of total fetal liver cells. Using murine fetal liver cells, our laboratory previously developed a series of methods to monitor erythroid differentiation both in vivo and in vitro and purify large quantities of primary erythroid progenitors (burst-forming unit erythroids [BFU-Es] and colony-forming unit erythroids [CFU-Es]) to high purity (Zhang et al. 2003; Ji et al. 2008; Flygare et al. 2011). Furthermore, our culture system for primary mouse fetal liver cells recapitulates erythroid development in vivo (Zhang et al. 2003; Ji et al. 2008). Thus, we have an excellent system to identify and functionally characterize lncRNAs in erythropoiesis.
Here we examined the expression of lncRNAs during erythropoiesis. We observed that >400 putative lncRNAs are differentially expressed during this important developmental process. We functionally characterized an erythroid-specific long intergenic noncoding RNA (lincRNA), which we named LincRNA erythroid prosurvival (LincRNA-EPS). LincRNA-EPS is highly induced during the terminal differentiation of erythroid cells. Knocking down this lincRNA inhibits differentiation and promotes apoptosis. Conversely, ectopic expression of this lincRNA can prevent apoptosis in mouse erythroid cells. We observed that this lincRNA can repress many apoptotic genes, and among these genes, Pycard, a proapoptotic gene, has a functional link to LincRNA-EPS. Altogether, these results revealed a novel layer of regulation of erythroid cell differentiation and apoptosis by a lincRNA.
Results and Discussion
Identification of lncRNAs expressed during erythropoiesis
We identified >400 putative lncRNAs that are expressed during mouse erythropoiesis through an RNA-seq analysis (Supplemental Fig. S1). Specifically, a list of lncRNAs was obtained from the NCode noncoding RNA arrays (Invitrogen) that contain 10,802 putative lncRNAs (Supplemental Table S1). To determine the expression of each of these putative lncRNAs during erythropoiesis, we examined the transcriptomic data from next-generation sequencing on polyadenylated RNAs from purified mouse fetal liver BFU-Es, CFU-Es, and Ter119+ cells; these represent three key developmental stages of erythropoiesis (Lodish et al. 2010). lncRNAs with similar expression patterns in these three types of cells were clustered together (Supplemental Fig. S1). During erythropoiesis, there are 163 putative lncRNAs that have a gradual increase in expression and 42 putative lncRNAs that have a gradual decrease in expression. The other lincRNAs had a fluctuating expression pattern. In total, we observed that 427 putative lncRNAs are expressed during this developmental process (Supplemental Table S2).
Here we focused on the group of lncRNAs that undergo a large increase in expression from CFU-Es to Ter119+ differentiated erythroblasts, since this expression pattern suggests that they may regulate the terminal differentiation of erythroid cells. We used several criteria to choose the lncRNAs for detailed characterization. First, we focused on lncRNAs that have high absolute expression levels in Ter119+ cells and that show the greatest increase in expression during terminal erythropoiesis. Second, we avoided lncRNAs that overlap with known genes (including introns and untranslated regions [UTRs]) in the mouse genome. Eventually, we focused on one top candidate intergenic lncRNA, named LincRNA erythroid prosurvival (LincRNA-EPS), because it is highly specific to terminal differentiating erythroid cells and has potent anti-apoptotic activity (see below).
Molecular and cellular characterization of LincRNA-EPS
To determine the complete molecular structure of LincRNA-EPS, we performed 5′ and 3′ rapid amplification of cDNA ends (RACE) to characterize the very 5′ and 3′ ends of this RNA transcript (Supplemental Fig. S2A,B). Using the sequence information from these two ends, we cloned the processed full-length LincRNA-EPS transcript. Sequence analysis indicated that LincRNA-EPS is a 2531-nt transcript with four exons and three introns (Supplemental Fig. S2C). The 5′ end cap structure, as indicated by the 5′RLM-RACE, and the 3′ end poly(A) tail, as implied by the 3′RACE, indicate that LincRNA-EPS is a PolII transcript. To test that the full-length transcript we obtained is truly a single transcript, we performed RT–PCR using primer sets targeting its 5′ end and 3′ end (Supplemental Fig. S2D). Both PCR products were strongly detected in terminal differentiating erythroid cells (Ter119+ cells) and were barely detectable in erythroid progenitors (Ter119− cells) (Supplemental Fig. S2D). This result is consistent with the notion that the cloned full-length lincRNA-EPS is truly a single transcript.
To determine the cellular localization of LincRNA-EPS, we fractionated mouse fetal liver Ter119+ cells into nuclear and cytoplasmic fractions. We can cleanly separate nucleus from cytoplasm, as indicated by the results showing that GAPDH protein was exclusively detected in the cytoplasmic fraction, while the nucleus-retained 47S pre-rRNA was predominantly found in the nuclear fraction (Supplemental Fig. S3A). RT–PCR analysis revealed that LincRNA-EPS was predominantly detected in the nuclear fraction (Supplemental Fig. S3A,B).
Expression pattern of LincRNA-EPS
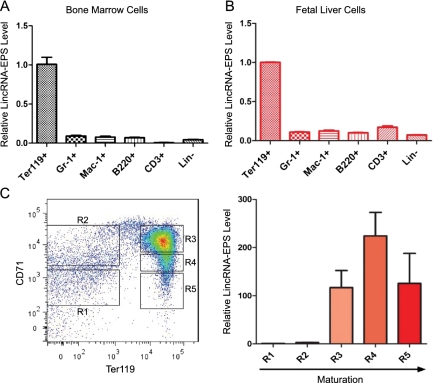
RNA-seq results indicated that LincRNA-EPS is highly induced in terminally differentiating erythroblasts (Supplemental Fig. S6A). Interestingly, real-time RT–PCR analysis revealed that LincRNA-EPS is enriched in hematopoietic organs, such as spleen, bone marrow, and fetal liver cells, among all mouse organs and cells tested (Supplemental Fig. S6B). In addition, fractionation of total bone marrow cells or fetal liver cells using different cell lineage surface markers followed by real-time RT–PCR revealed that LincRNA-EPS is highly specific to terminal differentiating erythroid cells compared with terminally differentiated cells of other hematopoietic lineages in both bone marrow and fetal liver cells (Fig. 1A,B). To further determine when LincRNA-EPS is induced during erythropoiesis, we fractionated total fetal liver cells into five different fractions (R1–R5) based on expression of CD71 and Ter119 (Fig. 1C). CFU-E progenitors are predominantly in the R1 and R2 fractions, and differentiating erythroblasts fall in the R3–R5 windows (Zhang et al. 2003). Real-time RT–PCR revealed that LincRNA-EPS is dramatically induced during the transition from R2 to R3 (Fig. 1C), which corresponds to the transition from CFU-Es to Ter119+ cells (Zhang et al. 2003; Flygare et al. 2011); during this transition, hemoglobins and many other erythroid-important genes are induced (Hattangadi et al. 2010). This expression pattern strongly suggests that LincRNA-EPS plays an important role during the terminal differentiation of erythroid cells.
Figure 1.
LincRNA-EPS is highly specific to the terminal differentiating erythroid cells. Total nucleated mouse bone marrow cells (A) or fetal liver cells (B) were fractionated into erythroid cells (Ter119+), granulocytes (Gr-1+), monocytes (Mac-1+), B cells (B220+), T cells (CD3+), and lineage-depleted cells (Lin−) using antibodies to cell surface antigens and magnetic-assisted cell sorting (MACS), followed by total RNA extraction, reverse transcription, and quantitative PCR. The level of LincRNA-EPS in each fraction was normalized to that of 18S rRNA, and the normalized LincRNA-EPS level in Ter119+ erythroid cells was set as 1. (C) Total mouse fetal (E14.5) liver cells were fractionated into R1–R5 based on the expression levels of Ter119 and CD71; cells in each fraction were collected by fluorescence-activated cell sorting (FACS). Then, LincRNA-EPS levels were quantified by real-time RT–PCR and normalized to 18S rRNA; the normalized LincRNA-EPS level in R1 was set as 1. This quantification was based on three independent experiments, with the mean and the standard error of the mean indicated on the panels.
Inhibition of LincRNA-EPS blocks differentiation and promotes apoptosis
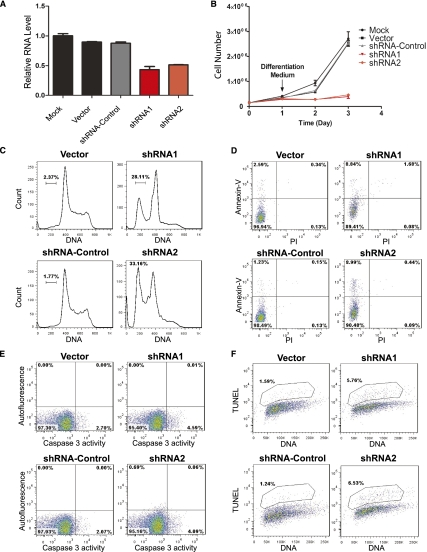
Loss-of-function studies revealed that inhibition of LincRNA-EPS induction results in apoptosis and blockage of proliferation of erythroid progenitors. We employed lineage-negative (Lin−) fetal liver cells, highly enriched for erythroid progenitors (Flygare et al. 2011), and inhibited LincRNA-EPS induction by retroviral transduction of shRNA-expressing vectors. Progenitors were first cultured for 24 h in maintenance medium to allow the expression of shRNAs and then switched to medium containing erythropoietin (Epo), which induces their differentiation into mature erythrocytes (Lodish et al. 2010). The 24-h incubation in maintenance medium does not result in changes in the apoptotic states of the transduced cells (Supplemental Fig. S7). These shRNAs inhibit the induction of LincRNA-EPS by 40%–50% (Fig. 2A). Knocking down LincRNA-EPS strongly inhibited the proliferation of erythroid cells during their terminal differentiation (Fig. 2B). To determine how LincRNA-EPS regulates erythroid cell proliferation, we performed cell cycle analysis. Compared with the control cells, LincRNA-EPS knockdown cells are enriched in the G1 phase and, importantly, a significant fraction of these cells are in the sub-G1 population (Fig. 2C), suggesting that they are undergoing apoptosis and/or necrosis.
Figure 2.
Inhibition of LincRNA-EPS leads to apoptosis during terminal differentiation of erythroid cells. (A) LincRNA-EPS was knocked down by shRNAs encoded by retroviral vectors. Mouse fetal liver Lin− cells were transduced by retroviral vectors and cultured in maintenance medium for 24 h to allow shRNA expression and then switched to differentiation medium. The LincRNA-EPS level was determined on transduced cells (GFP+) at 24 h in differentiation medium by real-time RT–PCR and normalized to the level of 18S rRNA; the normalized LincRNA-EPS level in mock-treated cells was set as 1. shRNA1 and shRNA2 target different regions on LincRNA-EPS, and shRNA targeted to firefly luciferase was used as an shRNA control. (B) Growth curves of Lin− cells mock-treated or transduced by vector control, shRNA control, LincRNA-EPS shRNA1, or LincRNA-EPS shRNA2. The cell number at each time point is the average of three measurements. The cell cycle status (C) and apoptotic status determined by Annexin-V staining (D), caspase3 activity measurement (E), and TUNEL assay (F) were measured by flow cytometry at 24 h in differentiation medium. The DNA content in the cell cycle analysis (C) and TUNEL assay (F) was determined by Hoechst33342 and propidium iodide staining, respectively. The apoptosis analyses shown here are representative of three independent experiments.
We used three independent approaches to confirm the increased apoptotic state in LincRNA-EPS knockdown cells (Supplemental Fig. S8). First, LincRNA-EPS knockdown cells have an increased fraction of Annexin-V-positive cells (Fig. 2D). Second, caspase 3 activity increased when LincRNA-EPS induction was inhibited (Fig. 2E). Third, the level of an important apoptotic marker, 3′-hydroxyl ends of DNA, was significantly higher in LincRNA-EPS knockdown cells compared with the controls, as determined by the terminal transferase dUTP nick end-labeling (TUNEL) assay (Fig. 2F). Thus, inhibition of LincRNA-EPS results in apoptosis of erythroid progenitors (Supplemental Fig. S8B). Consequently, subsequent erythroid terminal differentiation and enucleation were severely blocked in LincRNA-EPS knockdown cells (Supplemental Fig. S9). Collectively, these results indicate that LincRNA-EPS plays an anti-apoptotic role during erythroid terminal differentiation.
Ectopic expression of LincRNA-EPS in erythroid progenitor cells can protect them from apoptosis caused by Epo deprivation
Epo is an essential cytokine for the terminal differentiation of erythroid cells; it prevents erythroid progenitors from undergoing apoptosis and promotes their proliferation and terminal differentiation (Wojchowski et al. 2010). The time frame of LincRNA-EPS induction—at the transition from Ter119− to Ter119+ cells—correlates well with the time window at which Epo exerts its biological function, suggesting that LincRNA-EPS's anti-apoptotic ability could contribute to cell survival mediated by Epo.
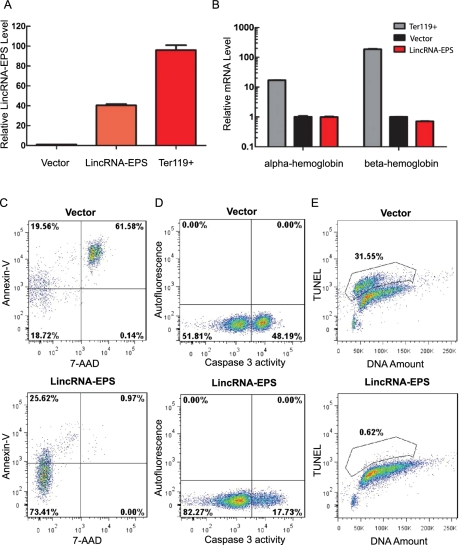
To determine whether LincRNA-EPS is involved in the cell survival pathway mediated by Epo, we expressed LincRNA-EPS in fetal liver Lin− cells by retroviral transduction (Fig. 3A), and then cultured these transduced cells in maintenance medium that does not contain Epo. The level of the expressed LincRNA-EPS was less than that in normal Ter-119+ erythroblasts and thus is within a physiological range (Fig. 3A). As expected, most erythroid progenitors undergo apoptosis after 40–48 h of Epo starvation (Fig. 3C–E, top panels), as measured by Annexin-V binding, caspase 3 activation, and TUNEL assay, respectively. Dramatically, ectopic expression of LincRNA-EPS protects erythroid progenitor cells from apoptosis (Supplemental Fig. S10), as evidenced by the decreased percentage of Annexin-V-positive cells (Fig. 3C), lower caspase 3 activity (Fig. 3D), and a dramatically reduced TUNEL signal (Fig. 3E) compared with cells transduced by an empty retroviral vector (Supplemental Fig. S10B). Annexin-V and 7-aminoactinomycin D (7-AAD) double-positive cells represent those in a late apoptotic state, and their level was significantly reduced following LincRNA-EPS ectopic expression (Supplemental Fig. S10B). Consistent with the anti-apoptosis phenotype conferred by LincRNA-EPS, following LincRNA-EPS ectopic expression, more cells become localized in the S and G2/M phases of the cell cycle (Supplemental Fig. S11A) and undergo some proliferation (Supplemental Fig. S11B). Ectopic expression of LincRNA-EPS did not induce terminal differentiation of erythroid cells, as determined by the low hemoglobin mRNA levels in the transduced cells compared with terminal differentiating Ter119+ erythroblast cells (Fig. 3B). Taken together, these data demonstrate that LincRNA-EPS possesses a potent anti-apoptotic activity.
Figure 3.
Ectopic expression of LincRNA-EPS protects mouse erythroid progenitors from apoptosis induced by Epo deprivation. Lin− mouse fetal liver cells were transduced by retroviral vectors and maintained in maintenance medium for 40–48 h. The LincRNA-EPS levels (A) and hemoglobin mRNAs level (B) in transduced (GFP+) cells were quantified by real-time RT–PCR and normalized to 18S rRNA; the RNA level in the empty vector transduced cells was set as 1. Ter119+ cells were isolated by MACS from total nucleated fetal liver cells. Cell apoptosis was assayed by flow cytometry using Annexin-V staining (C), caspase 3 activity staining (D), and TUNEL assay (E). The top panels indicate the apoptotic state in the empty vector transduced Lin− cells, and the bottom panels indicate the apoptotic state in Lin− cells ectopically expressing LincRNA-EPS. The apoptosis analyses shown here are representative of three independent experiments.
To determine which region of LincRNA-EPS mediates the anti-apoptotic activity, we performed a structure–function analysis. Specifically, we made a series of LincRNA-EPS truncations from either its 5′ end or 3′ end (Supplemental Fig. S12A) and then transduced each of them into Lin− fetal liver cells. The apoptotic state of the transduced cells was determined by Annexin-V and 7-AAD staining after Epo starvation for 40 h. All of the LincRNA-EPS 5′ end truncations retained anti-apoptotic activity at the wild-type level (Supplemental Fig. S12B–E,J–L), while truncations from the 3′ end of LincRNA-EPS severely abolished this activity (Supplementl Fig. S12F–I,J–L). Thus, the 500 nt at the 3′ end of LincRNA-EPS, which lacks any ORF, is sufficient for the anti-apoptotic activity.
There is a functional link between LincRNA-EPS and Pycard
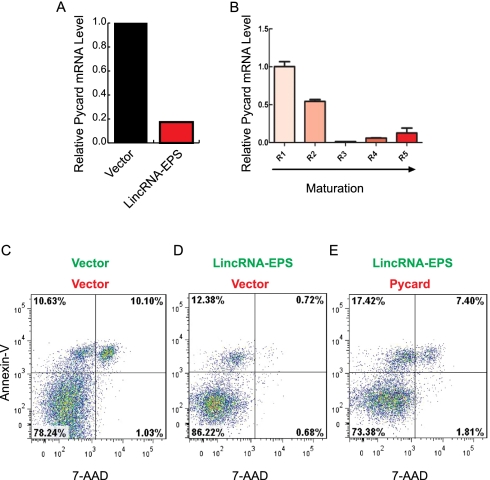
To obtain mechanistic insights on how LincRNA-EPS inhibits apoptosis, we used microarray analysis to profile gene expression in Lin− cells ectopically expressing LincRNA-EPS. Many genes involved in apoptosis, as defined by gene ontology (see the Supplemental Material), were repressed when LincRNA-EPS was ectopically expressed (Supplemental Fig. S13). This observation was further verified by real-time RT–PCR on 27 selected apoptotic genes that are expressed during erythropoiesis (Supplemental Fig. S14). Among these genes, Cideb and Pycard have the most dramatic repression upon LincRNA-EPS expression. We focused on Pycard because it has much higher absolute expression level than does Cideb (Supplemental Fig. S14B). Pycard is a proapoptotic gene encoding an adaptor protein that can activate caspase in apoptosis (Ohtsuka et al. 2004). Several lines of evidence suggest that Pycard is one target of LincRNA-EPS. First, Pycard was dramatically repressed following LincRNA-EPS ectopic expression (Fig. 4A; Supplemental Fig. S14). Second, the expression of Pycard during normal erythropoiesis is inversely correlated with that of LincRNA-EPS (Fig. 1C vs. Fig. 4B). Third, Pycard overexpression results in similar phenotypes on erythroid terminal differentiation, as does inhibition of LincRNA-EPS induction (Supplemental Fig. S15). Specially, overexpression of Pycard inhibits the proliferation of erythroid cells (Supplemental Fig. S15B), promotes their apoptosis (Supplemental Fig. S15C), and interferes with their terminal differentiation and enucleation (Supplemental Fig. S15D,E). Moreover, knocking down Pycard in Epo-deprived erythroid progenitors by shRNA mimics the anti-apoptotic phenotype conferred by LincRNA-EPS ectopic expression, as indicated by the reduction of late apoptotic cells (Supplemental Fig. S16). Conversely, overexpression of Pycard suppresses the anti-apoptotic phenotype mediated by ectopic expression of LincRNA-EPS (Fig. 4C–E; Supplemental Fig. S17). In this experiment, we transduced erythroid progenitors with two retroviral vectors, which express LincRNA-EPS and Pycard, respectively, and then cultured them in maintenance medium without Epo. The double-transduced cells could be identified by the GFP and the DsRed marker proteins expressed by these vectors, and their apoptotic state was monitored by flow cytometry using Annexin-V and 7-AAD staining. The GFP and DsRed double-positive cells are biased toward live cells, as indicated by the different percentage of apoptotic cells in the two empty vector controls compared with the single GFP transduced cells (Fig. 4, D vs. C). Nonetheless, ectopic expression of LincRNA-EPS along with an empty DsRed vector clearly reduced the number of Annexin-V+/7-AAD+ apoptotic cells. In contrast, ectopic expression of LincRNA-EPS along with Pycard restored the apoptotic state to the level of that of the control cells (two empty vectors) (Fig. 4, E vs. C). Thus, there is a functional link between LincRNA-EPS and Pycard in regulating apoptosis of Epo-deprived progenitor cells. Collectively, these results indicate that LincRNA-EPS modulates apoptosis at least in part through repressing Pycard expression.
Figure 4.
Pycard is one functional target of LincRNA-EPS. (A) Relative Pycard mRNA level in Lin− cells transduced by the empty vector or the vector with LincRNA-EPS and cultured in maintenance medium. (B) Expression pattern of Pycard mRNA in R1–R5 fetal liver cells. The mRNA level was normalized to 18S rRNA, and the normalized mRNA level in R1 was set as 1. (C–E) Apoptotic state of 40-h Epo-starved Lin− cells doubly transduced with LincRNA-EPS ectopically expressed from a GFP-containing vector and Pycard overexpressed from a DsRed-expressing vector. The apoptosis analyses shown here are representative of three independent experiments.
In sum, we identified a bona fide long noncoding transcript highly specific to terminal differentiating erythroid cells. Functional characterization indicates that the LincRNA-EPS is required for the terminal differentiation of erythroid cells by inhibiting apoptosis. Mechanistic studies suggest that the anti-death ability of LincRNA-EPS is mediated in part through repressing Pycard.
Regulation of apoptosis is essential for erythropoiesis and many other developmental processes (Testa 2004). Our finding of an anti-apoptotic lncRNA reveals a novel layer of modulation of the tightly controlled cell death program that is required for the proper generation of mature red blood cells in response to various physiological and pathological stimuli. It will be of great interest to explore how this lncRNA-mediated anti-apoptotic activity is integrated into other regulatory networks that are essential for erythropoiesis. Unlike small noncoding RNAs, lncRNAs regulate gene expression via diverse mechanisms (Hung and Chang 2010). The nuclear localization of LincRNA-EPS suggests that it regulates gene expression via modulating certain nuclear events, such as epigenetic modifications, transcription, or mRNA splicing. One hypothesis is that, like other lncRNAs (Guttman et al. 2011), LincRNA-EPS associates with chromatin modifiers to repress the transcription of Pycard and perhaps other genes whose encoded proteins promote cell apoptosis. Future identification of protein factors associated with LincRNA-EPS will test this hypothesis. In addition, it will be of great interest to identify the LincRNA-EPS homolog in humans. Although some lncRNAs are evolutionarily conserved, unlike most protein-coding genes, many lncRNA homologs, such as Xist, are not conserved in primary sequences in different species (Pang et al. 2006). Thus, poorly conserved sequence information cannot guarantee the absence of homologs in different species. Although it may not be surprising that humans may have a LincRNA-EPS homolog, improved computational methods and/or experimental approaches are needed to identify it. Finally, our studies revealed that in addition to transcriptional factors and microRNAs, lncRNAs also play an important role during erythropoiesis.
Materials and methods
Plasmids and oligonucleotides
The plasmids and oligonucleotides used in this study are listed in Supplemental Table S3.
Cell culture
293T cells for retroviral packing were maintained in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS) (Invitrogen). The maintenance medium for primary mouse fetal liver erythroid progenitors was StemSpan-SFEM (StemCell Technologies) medium containing 100ng/mL recombinant mouse stem cell factor (Amgen), and the maintenance medium for 40 ng/mL recombinant liver erythroid progenitors was Iscove modified Dulbecco's medium containing 15% FBS (StemCell Technologies), 1% detoxified bovine serum albumin (BSA) (StemCell Technologies), 250 μg/mL holo-transferrin (Sigma-Aldrich), 2 U/mL Epo (Amgen), 10 μg/mL recombinant human insulin (Sigma-Aldrich), and 2 mM L-glutamine (Invitrogen). All of the cells used in this study were cultured at 37°C with 5% CO2. Jurkat cells were maintained in RPMI-1640 medium supplemented with 10% FBS.
Isolation of erythroid progenitors from primary mouse fetal liver cells
Total fetal liver cells were obtained from E14.5 C57BL/6 mouse embryos. The procedure for isolating mouse erythroid progenitor cells was described previously (Zhang et al. 2003; Hattangadi et al. 2010).
Retroviral transduction
Isolated erythroid progenitors were transduced by MSCV-based retroviruses using the protocols described in Hattangadi et al. (2010).
RNA analysis
Total RNA was extracted from cells using the miRNeasy kit (Qiagen), including the DNase I (Qiagen) on-column digestion, and then cDNAs were synthesized using the SuperScript II reverse transcriptase (Invitrogen) with random primers. Real-time PCR was performed on an ABI Prism 7900 sequence detection system using SYBR Green PCR Master Mix (Applied Biosystems). The 18S rRNA level was used for normalization. 5′RLM-RACE and 3′RACE were performed using the protocols provided in the FirstChoice RLM-RACE kit (Ambion). The nucleus and cytoplasm fractionation was performed using the protocols detailed in the PARIS kit from Ambion.
Codon usage frequency analysis
The codon usage frequencies of the four putative ORFs on LincRNA-EPS were calculated and compared with those of all known mouse protein-coding genes by the Graphical Codon Usage Analyzer (http://gcua.schoedl.de).
Flow cytometry analysis of erythroid differentiation and enucleation
In all of the flow cytometry experiments, we gated on transduced cells, defined as GFP+ and/or RFP+ population, for phenotypic analysis. Procedures for immunostaining and flow cytometry analysis of erythroid differentiation and enucleation were described previously (Ji et al. 2008; Hattangadi et al. 2010).
Apoptosis assays
All apoptosis assays were performed using flow cytometry on a BD LSRII flow cytometer. Annexin-V and 7-AAD/PI staining was performed using the Annexin-V Apoptosis Detection kit I (BD Pharmingen). Annexin-V was conjugated with either phycoerythrin (PE) or Pacific Blue. Caspase 3 activity was detected using the Caspase-3 Detection kit (Red-DEVD-FMK) (EMD4Biosciences). TUNEL assay was performed using the FlowTACS Flow Cytometry Apoptosis Detection kit (R&D Systems).
Cell cycle analysis
Cell cycle analysis was performed following Hoechest 33342 staining in living cells. Specifically, Hoechst 33342 was added to the cell culture medium at a final concentration of 10 ug/mL. Cells were incubated for 30 min at 37°C, and then the cellular DNA amount was determined by flow cytometry (Belloc et al. 1994).
Microarray analysis
Total RNA was extracted from vector and LincRNA-EPS transduced erythroid progenitor cells (sorted as GFP-positive cells), respectively. The quality of the RNA sample was determined by a 2100 Bioanalyzer (Agilent Technologies). The Aligent 8X66K multiplex array was used for the analysis. Reverse transcription, labeling, and hybridization were performed by the Genome Technology Core at the Whitehead Institute using the protocols as detailed in Hattangadi et al. (2010).
Microarray data were analyzed using the limma package from Bioconductor. A loess method was used to make intensities consistent within each array, and a quantile method was applied to achieve consistency between arrays. Differential expression was determined by a moderated _t_-test with limma and corrected for false discovery rate. The microarray analysis data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (repository number GSE30279).
Acknowledgments
We thank Drs. Jeff Coller, John Rinn, and Shilpa Hattangadi for critical comments on this manuscript, and all of the members of the Lodish laboratory, the flow cytometry cores at the Whitehead Institute and MIT, and the genome technology core at the Whitehead Institute for technical support. W.H. is a Merck Fellow of the Life Sciences Research Foundation. This research was supported by National Institute of Health (NIH) grants DK068348 and 5P01 HL066105 to HFL.
Footnotes
Supplemental material is available for this article.
References
- Amaral PP, Dinger ME, Mercer TR, Mattick JS 2008. The eukaryotic genome as an RNA machine. Science 319: 1787–1789 [DOI] [PubMed] [Google Scholar]
- Belloc F, Dumain P, Boisseau MR, Jalloustre C, Reiffers J, Bernard P, Lacombe F 1994. A flow cytometric method using Hoechst 33342 and propidium iodide for simultaneous cell cycle analysis and apoptosis determination in unfixed cells. Cytometry 17: 59–65 [DOI] [PubMed] [Google Scholar]
- Cabili TC, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Flygare J, Rayon Estrada V, Shin C, Gupta S, Lodish HF 2011. HIF1α synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood 117: 3435–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D et al. 2006. GENCODE: Producing a reference annotation for ENCODE. Genome Bio 7: S4 doi: 10.1186/gb-2006-7-s1-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattangadi SM, Burke KA, Lodish HF 2010. Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood 115: 4853–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Chang HY 2010. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol 7: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Jayapal SR, Lodish HF 2008. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol 10: 314–321 [DOI] [PubMed] [Google Scholar]
- Kerenyi MA, Orkin SH 2010. Networking erythropoiesis. J Exp Med 207: 2537–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner MV, Pauler FM, Huang R, Barlow DP 2009. The function of non-coding RNAs in genomic imprinting. Development 136: 1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Flygare J, Chou S 2010. From stem cell to erythroblast: Regulation of red cell production at multiple levels by multiple hormones. IUBMB Life 62: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. 2010. Large intergenic noncoding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI, Aaronson SA, Lee SW 2004. ASC is a Bax adaptor and regulates the p53–Bax mitochondrial apoptosis pathway. Nat Cell Biol 6: 121–128 [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS 2006. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet 22: 1–5 [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa U 2004. Apoptotic mechanisms in the control of erythropoiesis. Leukemia 18: 1176–1199 [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY 2011. Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361 [DOI] [PubMed] [Google Scholar]
- Wojchowski DM, Sathyanarayana P, Dev A 2010. Erythropoietin receptor response circuits. Curr Opin Hematol 17: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: Functional analysis by a flow cytometry-based novel culture system. Blood 102: 3938–3946 [DOI] [PubMed] [Google Scholar]
- Zhao G, Yu D, Weiss MJ 2010. MicroRNAs in erythropoiesis. Curr Opin Hematol 17: 155–162 [DOI] [PubMed] [Google Scholar]