Mapping a Dynamic Innate Immunity Protein Interaction Network Regulating Type I Interferon Production (original) (raw)
. Author manuscript; available in PMC: 2012 Sep 23.
SUMMARY
To systematically investigate innate immune signaling networks regulating production of type I interferon, we analyzed protein complexes formed after microbial recognition. Fifty-eight baits were associated with 260 interacting proteins forming a human innate immunity interactome for type I interferon (HI5) of 401 unique interactions; 21% of interactions were modulated by RNA, DNA, or LPS. Overexpression and depletion analyses identified 22 unique genes that regulated NF-κB and ISRE reporter activity, viral replication, or virus-induced interferon production. Detailed mechanistic analysis defined a role for mind bomb (MIB) E3 ligases in K63-linked ubiquitination of TBK1, a kinase that phosphorylates IRF transcription factors controlling interferon production. Mib genes selectively controlled responses to cytosolic RNA. MIB deficiency reduced antiviral activity, establishing the role of MIB proteins as positive regulators of antiviral responses. The HI5 provides a dynamic physical and regulatory network that serves as a resource for mechanistic analysis of innate immune signaling.
INTRODUCTION
The innate immune system is genetically programmed for detection of invading pathogens. Host recognition of conserved microbial products depends on germline-encoded receptors, collectively termed pattern recognition receptors (PRRs). The tailoring of innate responses is mediated through different sets of PRRs including the transmembrane Toll-like receptors (TLRs), which recognize extracellular microbial by-products, and the RIG-I-like receptors (RLRs), which sense infection in the cytosolic compartment (Takeuchi and Akira, 2010; Wilkins and Gale, 2010). Detection of microbial components by TLRs and RLRs activates signaling cascades, leading to production of antimicrobial cytokines.
Different TLRs recognize distinct ligands. For example, TLR3 senses microbial nucleic acids whereas TLR4 can recognize bacterial lipopolysaccharides (LPS) and viral coat proteins. Signaling through TLR3 and TLR4 activate TBK1 kinase activity and induce production of potent antimicrobial cytokines including type I interferons (IFN). However, TLRs have a restricted tissue distribution with expression generally limited to immunocytes. RLRs sense viral RNAs in the cytoplasm of nearly all cell types (Gitlin et al., 2006; Kato et al., 2005, 2006; Yoneyama et al., 2004). Activation of RLRs engages the mitochondrial adaptor MAVS (also termed IPS-1, VISA, and Cardif), which recruits TBK1, leading to phosphorylation of the IRF3 transcription factor (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). Once phosphorylated, IRF3 helps drive IFN expression (Sharma et al., 2003). Infection with DNA viruses or transfection with double-stranded DNA (dsDNA) also leads to TBK1 activation and IFN production through a series of still poorly defined DNA sensors and adaptors. Thus, the signaling pathways for TLR3, TLR4, RIG-I, MDA5, and DNA sensors converge at the level of TBK1 activation (Fitzgerald et al., 2003; Stetson and Medzhitov, 2006; Takeuchi and Akira, 2010). Because excessive or prolonged cytokine production leads to inflammation and tissue damage, IFN responses are strictly regulated to avoid pathologic consequences including autoimmunity.
To systematically explore the signal transduction pathways responsible for regulating cellular antiviral defense and IFN production, we initiated a global proteomic analysis of the human innate immunity interactome for type I interferon (HI5). We followed the dynamic changes in protein-protein interactions resulting from encounter with ligand. Analysis of 58 HI5-associated baits revealed connections with 260 proteins forming a framework of 401 protein interactions. Some of these interactions represent signaling modules that may participate in assembly, recruitment, and activation or disruption of the IFN signaling circuit. Functional studies demonstrated the biologic activity of 22 proteins in the HI5. Detailed mechanistic analysis defined the role of the E3 ligases mind bomb 1 and 2 (MIB1 and MIB2) in response to RNA viruses. MIBs are responsible for TBK1 K63-linked ubiquitination, promoting IFN production and controlling antiviral immunity.
All cells have basic defenses against infection. We performed a systematic proteomic analysis to discover unique molecules capable of regulating innate antiviral responses. More than 20% of the protein interactions were up- or downregulated after stimulation with microbial byproducts, demonstrating dynamic remodeling of the interactome. Functional analyses identified 22 molecules that modulated IFN expression and antiviral activity. The HI5 network integrates candidate genes into a dynamic antimicrobial network and serves as a resource for mechanistic analysis of innate immune signaling.
RESULTS
Proteomic Analysis of the Human Innate Immunity Interactome for Type I Interferon
Fifty-eight genes with known or suspected involvement in transcriptional regulation of type I IFN production were tagged with the FLAG epitope (see Figure S1A and Table S1A available online). Each stable cell line was also stimulated with poly(rI:rC), poly(dA:dT), LPS, and/or CpG (Figure S1B and Table S1A). Anti-FLAG affinity purifications were repeated in at least one independent experiment (Figure S1C). A total of 264 complexes were purified and analyzed by mass spectrometry.
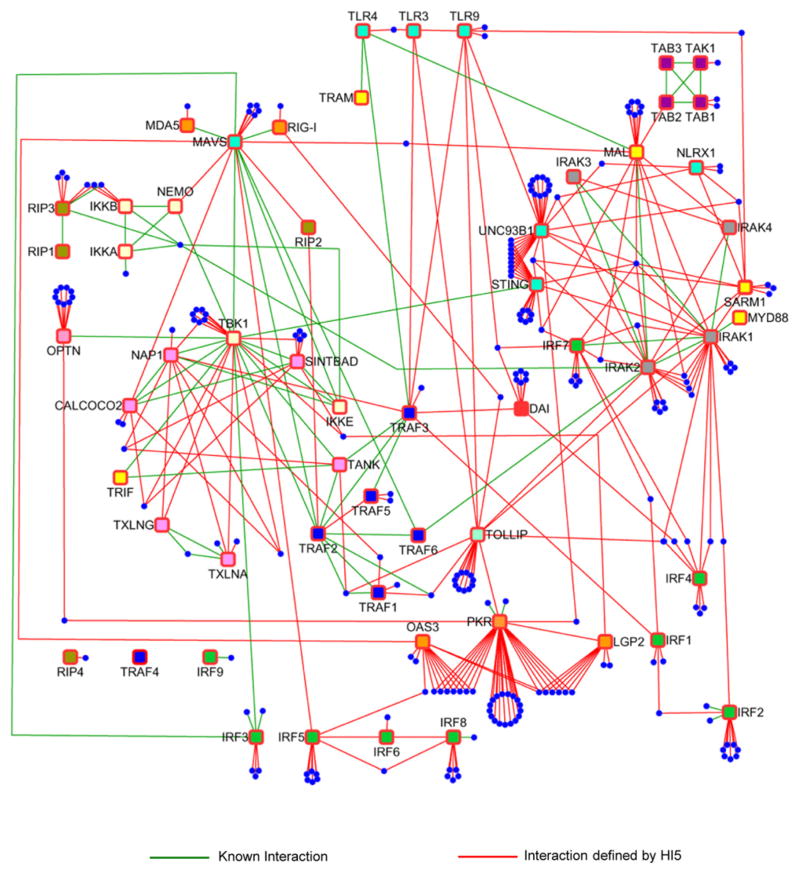
Total spectral counts (TSC) of 1218 unique proteins were detected in complexes of 58 genes involved in innate immunity plus FLAG-GFP and 4 other controls (Table S1B). Statistical methods including ANOVA and Z score plus criteria for reproducibility were integrated to filter out nonspecific binding proteins (NSBP) (Figures S1D–S1H, Supplemental Experimental Procedures). We identified 260 nonredundant high-confidence candidate interacting proteins (HCIP) making a HI5 network of 401 unique interactions (Figures 1 and S1F and Table S1B). Cytoscape software was used to visualize the interconnectivity of the HI5 and combine the interactions into one map (Figure 1). By using the BIOGRID and STRING protein interaction databases and curated literature, we identified 71 known interactions although 330 interactions were not present in these databases (Figure S1I and Table S1B). Thus, our analysis greatly expands the number of candidate proteins in the innate immunity network and connects the network to other cellular processes.
Figure 1. Overview of Human Innate Immunity Interactome for Type I Interferon.
Baits and HCIP are shown as squares and circles, respectively. Green lines represent known interactions and red lines reflect interactions defined by HI5.
Dynamics of the HI5
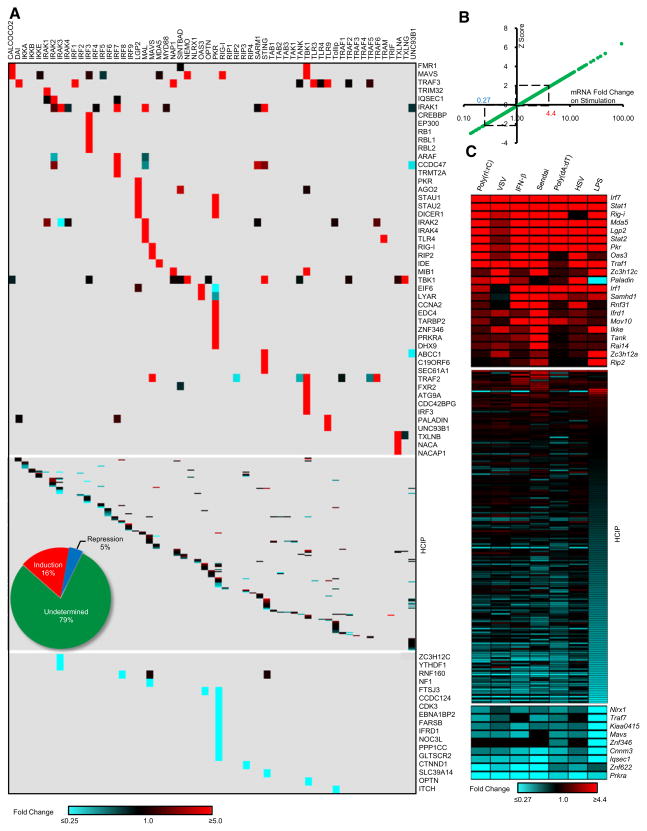
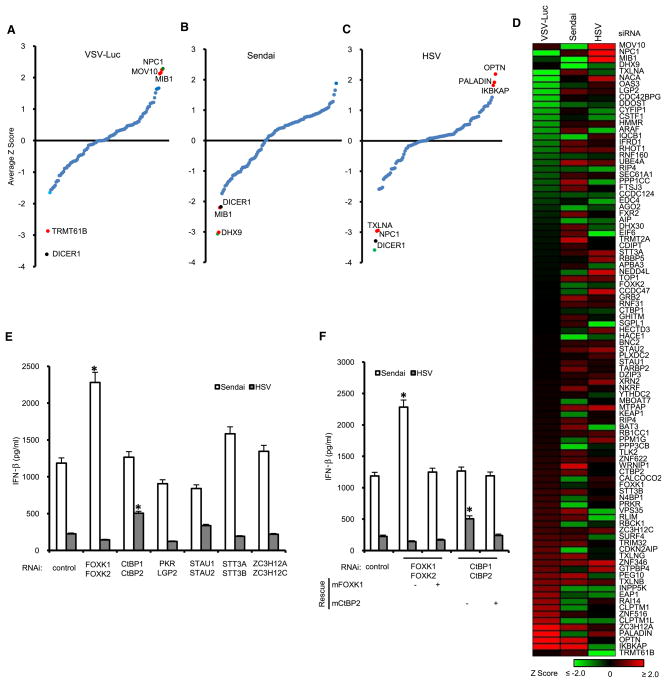
To determine ligand-dependent interactions in the HI5, we first compared the changes in bait normalized TSC (BNT) for each HCIP with versus without stimulation. Figure 2A provides a global view of the dynamic profile of each HCIP with its bait as a heat map. To determine the statistical significance of ligand-induced perturbations in the network, Z scores were calculated based on BNT ratios. An increase >5-fold was statistically significant as was a ligand-dependent reduction of >4-fold. To insure reproducibility, the BNT ratios in each replicate experiment had to meet the >5 or <0.25 standard to be classified as a dynamic interaction. In total, 65 inducible (16%) and 19 repressive (5%) interactions were identified (Figures 2A and S2). For example, treatment of TLR9-expressing cells with unmethylated CpG DNA induced association with PALADIN, revealing mechanistic leads for TLR9 signaling pathways.
Figure 2. Dynamics of the HI5.
(A) Heat map profile showing spectral peptide fold change for each HCIP upon stimulation. The ratio of bait normalized TSC (BNT) was used to calculate the fold change. For bait k BNT (k) ratio: [(BNT)ks1+(BNT)ks2]/[(BNT)ku1+(BNT)ku2], s stands for stimulation, u for unstimulated, 1 and 2 for replicate experiments. Inserted pie chart summarizes data.
(B) Z score analysis of HCIP mRNA fold change upon viral infection and ligand stimulation.
(C) HCIP gene expression profile. Mouse macrophage RAW264.7 cells were treated with poly(rI:rC), poly(dA:dT), LPS, IFN-β, VSV, Sendai virus, or HSV.
In addition to altering protein-protein interactions, viral infection or ligand stimulation can induce changes in gene transcription. To characterize the early transcriptional responses of HCIP, mouse macrophages were infected with virus or stimulated with relevant ligands. In total, 263 HCIP (including 44 baits) plus 24 reference genes were examined by real-time PCR (Table S2). Cells were infected with vesicular stomatitis virus (VSV), Sendai virus, and Herpes simplex virus (HSV), or treated with viral RNA and DNA mimics [poly(rI:rC) and poly(dA:dT)], IFN-β, or LPS. An increase >4.4-fold was statistically significant as was a ligand-dependent decrease of >3.7-fold (Figure 2B). In total, 21 HCIP mRNA levels were upregulated whereas 9 were down-regulated upon viral infection or ligand stimulation. Overall, 11% of HCIP genes are transcriptionally regulated subsequent to recognition of microbial ligands (Figure 2C). Among the most highly regulated genes are several that encode nucleic acid sensing domains (RIG-I, MDA5, LGP2, PKR, and OAS3). The reduced transcriptional response to VSV infection may be attributed to its established ability to inhibit IFN production (Ferran and Lucas-Lenard, 1997). Both Sendai and HSV displayed a broader spectrum of mRNA regulation than viral RNA and DNA analogs or IFN-β, suggesting that host cells responded to additional ligands.
Taken together, these results illustrate the dynamic landscape of the interactome and transcriptome triggered by recognition of microbial byproducts. Defining these dynamics on a global scale allows insights into the internal connections of the innate immune system and offers clues into communication with other signaling pathways and cellular processes.
Validation of the HI5 Interactions
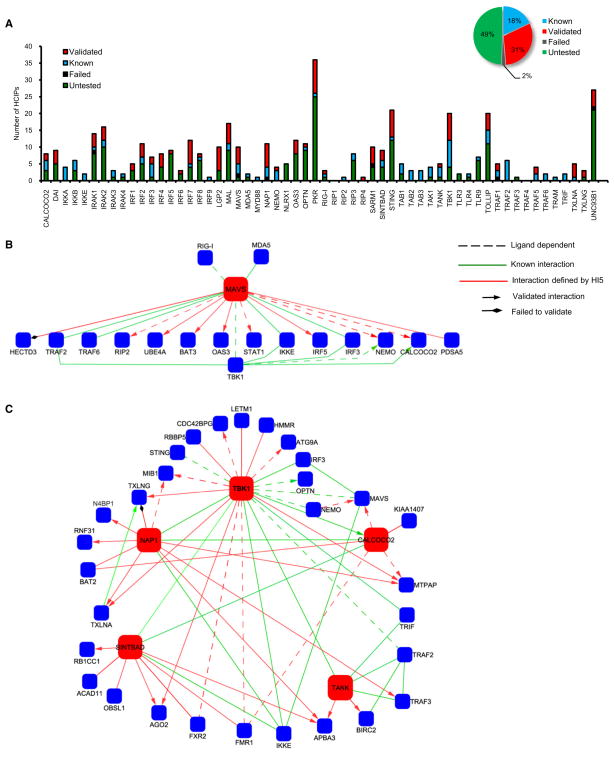
In vitro immunoprecipitation was performed to validate the interactions not found in the BIOGRID or STRING databases. In total 124 of 131 (95%) interactions were confirmed by coimmunoprecipitation in HEK293 cells or by endogenous coimmunoprecipitation in THP-1 cells (Figures 3A and S3). Combined with the 71 previously established protein-protein associations, 195 interactions have been validated in the HI5 (Figure 3A; Table S3).
Figure 3. In Vitro Validation of Protein Interactions in the HI5.
(A) Validation of physical associations for bait-prey interactions. Summary of total interactions is shown in the inset.
(B and C) Validation of MAVS (B) and TBK1 (C) modules.
Dashed line indicates ligand-dependent interaction; green line indicates known interaction; red line indicates interactions defined by HI5; diamond head indicates failed to confirm; arrowhead indicates interaction confirmed by in vitro coimmunoprecipitation.
Because innate immune responses to RNA viruses are dependent upon MAVS and TBK1, we concentrated on HCIP associated with these baits. Proteomic analysis identified 17 preys that interact with MAVS including 8 of 9 interactions defined by the HI5 that were confirmed by coimmunoprecipitation in HEK293 and THP-1 cells (Figure S3A). The interaction of MAVS with four proteins (RIP2, STAT1, NEMO, and CALCOCO2) was poly(rI:rC) dependent (Figures 3B and S3A). We established that the E4 ubiquitin ligase UBE4A and two ubiquitin binding proteins (NEMO and CALCOCO2) interacted with MAVS (Figures 3B and S3A). The latter findings are consistent with reports that the antiviral activity of MAVS is dependent upon ubiquitination (Zeng et al., 2009; Zhao et al., 2007).
The TBK1 kinase complex is located in the hub of the HI5 (Figure 3C) linking upstream intermediates (MAVS and STING) with downstream IRF substrates. TBK1 was associated with 24 proteins including 12 HCIP defined by the HI5. Mass spectrometric analysis of TBK1 interacting proteins found 13 proteins associated with two or more common partners, demonstrating the high interconnectivity in the TBK1 module (Figures 3C and S3B–S3I). For example, APBA3 (also termed MINT3) is associated with NAP1, SINTBAD, and TANK (Figures S3F–S3I). Among the 24 novel HCIP interactions tested within the TBK1 subnetwork, 23 were validated by coimmunoprecipitation (Figure 3C). The NAP1, SINTBAD, and TANK adaptors share similar TBK1 binding domains and contribute to assembly of TBK1 complexes (Chau et al., 2008; Ryzhakov and Randow, 2007). However, the data reveal that these structurally related molecules each have several unique binding partners (e.g., NAP1 interacts with MIB1), suggesting distinct functional roles for these scaffolds.
Other nodes of high interconnectivity include the IRAK complex, which is wired to the UNC93B1-STING module, and PKR, which is linked with LPG2 or OAS3 (Figure 1). Again >90% of the interactions were confirmed by coimmunoprecipitation (Figures S3J–S3M and Table S3). In the HI5, 19 baits were connected to >3 unique HCIP, establishing distinct subnetworks. We validated 10 subnetworks in vitro including MAL, IRAK2, IRF3, IRF4, IRF7, and DAI (Figures S3N–S3S and Table S3).
In summary, 124 of 131 protein interactions were validated, yielding an expanded outlook of the contacts linked to the MAVS, TBK1, IRAK, and IRF complexes. These connections include several adaptors that may regulate signalosome assembly.
Gain-of-Function Analysis of the HI5
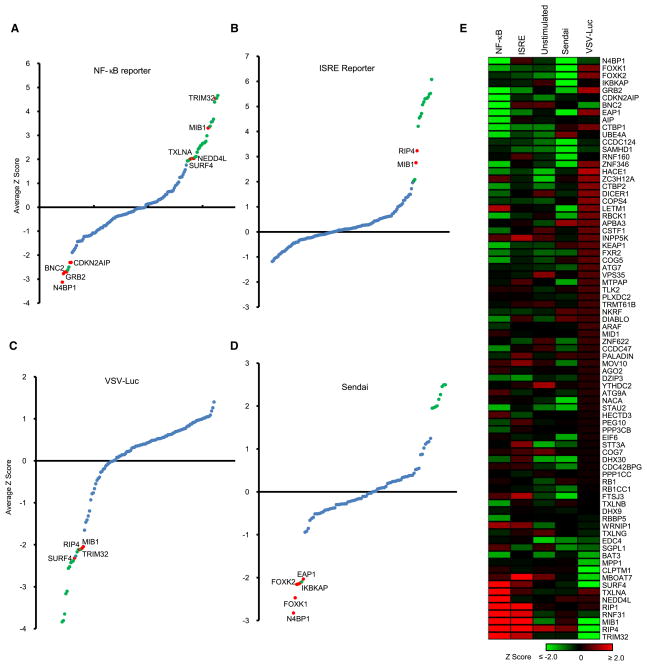
To establish the activity of HI5 genes, 132 HCIP and bait proteins were tagged into expression vectors. The effect of transient transfection was first examined for induction of NF-κB reporter activity in HEK293 cells. For statistical analysis, the data were combined with our laboratory database containing results from 446 other genes tested for NF-κB reporter activity in HEK293 cells. Genes that either stimulated (>6.6-fold) or suppressed (>5.3-fold) NF-κB activity were considered significant (Z-score ≥2 or ≤ −2). Four genes (N4BP1, BNC2, GRB2, and CDKN2AIP) were found to suppress basal NF-κB activity. In addition, three known negative regulators of the NF-κB pathway or IFN-β transcription were identified. Overexpression of 28 genes induced NF-κB activity and we report here that 5 of them (TRIM32, MIB1, NEDD4L, TXLNA, SURF4) are positive regulators of NF-κB (Figure 4A) and all are linked to the IRAK1 or TBK1 kinase complexes (Figure S2).
Figure 4. Gain-of-Function Analysis.
(A and B) NF-κB (A) and ISRE (B) reporter assay in HEK293 cells. Reporter constructs and 132 HCIP and baits were transfected into HEK293 cells. Each assay was repeated three times. Z scores were calculated based on average normalized reporter activity.
(C) VSV replication assay in HEK293 cells. 48 hr after transfection with HCIP, cells were infected with 0.1 M.O.I. VSV-Luc. VSV replication was determined by luciferase activity. The average value of triplicates was used for calculation of Z scores.
(D) ELISA assay for Sendai virus-induced IFN-β production. 48 hr after transfection with HCIP, HEK293 cells were infected with 5 HA Sendai virus. Blue dots indicate −2 < Z < 2; red dots indicate regulator defined by HI5; green dots indicate known function.
(E) Heat map summary comparing effects of ectopic expression of interactors defined by HI5 on transcription factors, VSV replication, and Sendai virus-induced IFN production. Genes with established functions in innate immunity are not shown.
We next examined the effect of ectopic HCIP expression on ISRE reporter activity. For statistical analysis, the data were combined with our ISRE database of 437 genes tested in HEK293 cells. Fifteen HCIP were found to induce ISRE activity including two positive regulators (RIP4 and MIB1) that we report here. None of the HCIP suppressed basal ISRE activity (Figure 4B).
The effect of HCIP expression on VSV replication was examined by infection of HEK293 cells with the VSV-Luc reporter virus. The results were combined with similar data from 124 other genes to calculate Z-scores. Seventeen genes including four defined by the HI5 (MIB1, RIP4, TRIM32, and SURF4) significantly inhibited VSV infection or replication (Figure 4C).
Finally, the effect of HCIP expression on IFN production was examined by ELISA in control and Sendai virus-infected HEK293 cells. Twelve genes previously reported to modulate IFN production were confirmed in this study (Table S4). No genes were found to significantly enhance Sendai virus-induced IFN production (Table S4), whereas five genes (FOXK1, FOXK2, EAP1, N4BP1, and IKBKAP) showed significant (>3.5-fold) inhibition of Sendai virus-induced IFN-β production (Figure 4D). Among these, FOXK1, FOXK2, EAP1, and IKBKAP were found in complexes with the known IFN transcriptional repressors IRF2 and IRF4.
In a side-by-side comparison, 14 HCIP were expressed in HEK293 and THP-1 cells and examined for regulation of reporter activity, Sendai virus-induced IFN production, and viral replication (Figures S4A and S4B). NF-κB or ISRE reporter activity, virus-induced IFN production, and VSV replication were regulated in similar fashion in both cell types. To exclude the effect of transient gene expression on cell proliferation, we also monitored cell viability with MTT (Figure S4C). The effects of 80 new HCIP on NF-κB and ISRE reporter activity, VSV replication, and IFN production were summarized as a heat map (Figure 4E). These data underscore the functional capacity of the latest components in the HI5.
RNAi Analysis of the HI5
RNA interference was used to systematically investigate the function of 97 HCIP, including 73 genes also studied by gain of function (Figure S5A). VSV-Luc virus was used to assess the effect of RNAi on viral growth. Targeting of two genes (DICER1, TRMT61B) inhibited VSV luciferase activity in HEK293 cells (Figure 5A). However, DICER1 is known to regulate cell cycle and silencing DICER1 with RNAi-inhibited cell proliferation (Figure S5B). RNAi of three genes (MOV10, NPC1, and MIB1) significantly increased VSV replication (Figure 5A). A RNA virus (Sendai) and a DNA virus (HSV) were used to probe the effect of HCIP gene silencing on IFN production. Silencing MIB1 or DHX9 with RNAi specifically inhibited Sendai virus-induced IFN production. None of the genes tested displayed significant synergy with Sendai virus for IFN production (Figure 5B). Knockdown of the TBK1- and STING-associated genes TXLNA and NPC1 showed significant inhibitory effects on HSV-induced IFN-β production whereas OPTN, IKBKAP, and PALADIN enhanced HSV-induced IFN-β production (Figure 5C).
Figure 5. RNAi Analysis.
(A–C) HEK293 cells were transfected with 98 HCIP including TBK1 siRNA SMARTpools. After 48 hr, cells were infected with 0.1 M.O.I. VSV-Luc (A), 5 HA Sendai virus (B), and 1 M.O.I. HSV (C). VSV luciferase assays were repeated three times. Sendai virus- and HSV-induced IFN production was assayed in duplicate by ELISA. The average values were used for calculation of Z scores. Blue dots indicate −2 < Z < 2; red dots indicate regulator defined by HI5; green dot indicates positive control, TBK1; black dot indicates failed MTT assay.
(D) Heat map summary of the effect of siRNAs on VSV replication and Sendai virus- or HSV-induced IFN production.
(E) HEK293 cells were treated with a combination of two pools of RNAi. ELISA assays for IFN were performed after Sendai virus and HSV infection. An asterisk indicates p < 0.05. Data depict the mean ± SD of triplicate samples.
(F) RNAi rescue for FOXK1, FOXK2 and CtBP1, CtPB2. Mouse FOXK1 and CtBP2 were transfected with siRNA against human FOXK1, FOXK2 and CtBP1, CtBP2 into HEK293 cells. ELISA assay was performed with 24 hr supernatants from Sendai virus- or HSV-infected cells.
*p < 0.05, data depict mean ± SD of triplicate samples.
To compare the roles of these HCIP in different cell types, siRNA pools for nine genes were transfected into HEK293 or THP-1 monocytic cells. RNAi regulated VSV replication in similar fashion in both cell lines (Figures S5C and S5D). Sendai virus-and HSV-induced IFN responses were generally similar in both cell types except OPTN failed to enhance HSV-induced IFN responses in THP-1 cells (Figures S5C and S5D). The consolidated profile of siRNA effects on VSV, Sendai virus, and HSV illustrates the virus-specific effects of HI5 genes (Figure 5D). To exclude off-target effects of siRNA pools, we rescreened the four individual siRNAs from each of the nine target-specific pools separately. At least two individual siRNA oligo pairs from each pool produced significant effects on viral replication or virus-induced IFN production (Figure S5E). The knockdown efficiency of the siRNA oligos was validated at the protein or mRNA level (Figures S5F–S5G). Taken together, the data demonstrate RNAi to nine HCIP defined by the HI5 specifically regulate antiviral responses.
Based on the association of structurally related HCIP with the same bait, we next examined the potential redundancy of selected gene pairs. Knockdown of six pairs of genes had no significant effect on VSV-Luc replication (Figure S5H). However, FOXK1 and FOXK2 were redundant for regulation of Sendai-induced IFN production (Figure 5E), which is consistent with the effects observed with FOXK1 and FOXK2 overexpression (Figure 4D). The combination of CtBP1 and CtBP2 RNAi enhanced HSV-induced IFN production (Figure 5E). CtBPs are corepressors of transcription; they associate with the DNA sensor DAI, suggesting a role for these complexes in regulating antiviral activity. The virus-specific effects of FOXK1 and FOXK2 or CtBP1 and CtBP2 were rescued by overexpression of mouse FOXK1 and CtBP2, thereby excluding off-target effects (Figure 5F). Together with the gain-of-function assays, we discovered 22 genes that regulated NF-κB and ISRE reporter activity, VSV replication, or Sendai virus- and HSV-induced interferon production (Table S5).
Role of Mind Bomb Genes in TBK1 Ubiquitination
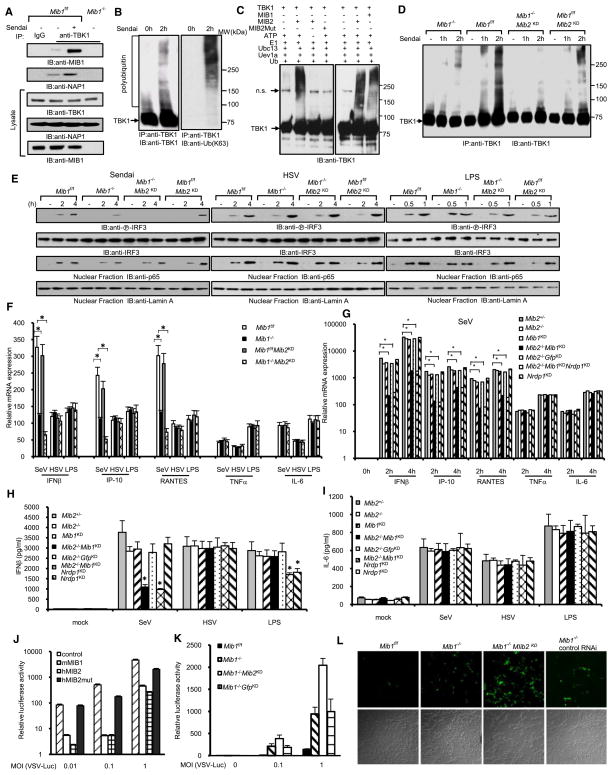
MIB1 activated NF-κB and ISRE reporter activity, inhibited viral replication, and was required for IFN production (Table S5). Overexpression of MIB2, a MIB1 homolog, also activated NF-κB and ISRE reporter activity (Figure S6A). Sendai virus infection enhanced the binding of endogenous TBK1 to endogenous MIB1 and endogenous NAP1 (Figure 6A). These data are consistent with the poly(rI:rC)-inducible interaction of TBK1 and NAP1 with MIB1 (Table S1). Similarly, MIB2 associations with TBK1 and NAP1 were also dependent upon viral infection (Figure S6B).
Figure 6. Mind Bomb E3 Ligases Regulate Innate Antiviral Immunity through TBK1 Ubiquitination.
(A) Lysates from _Mib1_f/f and _Mib1_−/− cells (_Mib1_f/f MEFs infected with Adeno-Cre for removal of the floxed allele) were infected with Sendai virus, and after 2 hr, cell lysates were immunoprecipitated with control IgG or TBK1 antibody and immunoblotted with MIB1 or NAP1 antibody to examine endogenous interactions induced by Sendai virus infection.
(B) Sendai virus induces TBK1 ubiquitination. HEK293 cells were treated with 50 HA Sendai virus for the designated times. The immune complexes were immunoblotted with TBK1 or antibody specific for K63-linked ubiquitin.
(C) In vitro ubiquitination reaction of TBK1 in the presence of MIB1, MIB2, or MIB2 mutant.
(D) _Mib1_f/f and _Mib1_−/− cells were transfected with MIB2 siRNA (_Mib2_KD). 48 hr later, cells were treated with 50 HA Sendai virus for the designated times. Cell lysates were immunoprecipitated and immunoblotted with TBK1 antibody.
(E) Mouse embryonic fibroblasts (MEFs) from _Mib1_f/f and _Mib1_−/− mice were transfected with control or MIB2 siRNA. After 48 hr, cells were treated with indicated stimulus for the designated times. Cell lysates or nuclear extracts were collected for immunoblotting with the indicated antibodies.
(F) MEFs from _Mib1_f/f and _Mib1_−/− mice were transfected with control or MIB2 siRNA. After 48 hr, cells were treated with indicated stimulus. Cytokine mRNA levels were determined by real-time PCR. Data represent the mean ± SD of triplicate samples. *p < 0.05.
(G) Bone marrow-derived macrophages (BMDM) from Mib2+/− and _Mib2_−/− mice were transfected with IKKε siRNA together with the indicated siRNA. After 48 hr, cells were infected with Sendai virus for the designated times. Cytokine mRNA expression was determined by real-time PCR. *p < 0.05. Data depict the mean ± SD of triplicate samples.
(H and I) BMDM from Mib2+/− and _Mib2_−/− mice were transfected with IKKε siRNA together with the indicated siRNA. After 48 hr, cells were treated with Sendai virus, HSV, and LPS. IFN-β and IL-6 protein levels were determined by ELISA. Data represent the mean ± SD of triplicate samples. *p < 0.05.
(J) VSV-Luc replication in HEK293 cells expressing control (cross-hatched), MIB1 (open), MIB2 (checkered), or MIB2 mutant (solid) constructs. Cells were infected with the indicated amount of virus for 24 hr. Data represent the mean ± SD of triplicate samples.
(K) VSV-Luc replication in _Mib1_f/f (solid), _Mib1_−/− (cross-hatched), and _Mib1_−/− MEFs transfected with MIB2 siRNA (open) or control (checkered). Cells were infected with the indicated dose of VSV. Data represent the mean ± SD of triplicate samples.
(L) VSV-eGFP replication in _Mib1_f/f, _Mib1_−/−, or _Mib1_−/− MEFs treated with control or MIB2 siRNA. Upper panel, green fluorescence; lower panel, bright field.
MIB proteins encode multiple RING finger motifs that have the highest similarity to those present in several E3 ligases associated with NF-κB signaling, including IAP-1, IAP-2, and XIAP (Jin et al., 2002). These domains suggest that MIBs may catalyze ubiquitination of TBK1 and/or NAP1. Previous reports described TBK1 ubiquitination after poly(rI:rC) stimulation (Friedman et al., 2008; Wang et al., 2009). We confirmed that poly(rI:rC) induced TBK1 ubiquitination but this was not observed after poly(dA:dT) treatment (Figure S6C). Although MIB1 also binds NAP1, we could not detect evidence of NAP1 ubiquitination.
To determine the type of ubiquitin-linked chains conjugated to TBK1, we used a K63-linkage-specific polyubiquitin antibody. K63-linked ubiquitin was detected on endogenous TBK1 after infection with Sendai virus (Figure 6B). We also transfected HA-tagged ubiquitin point mutants (K63R and K48R) and mutants containing only one lysine at either position 48 (K48) or 63 (K63) into macrophages. After Sendai virus infection, TBK1 robustly coupled with K63 and K48R ubiquitin whereas conjugation with K48 and K63R ubiquitin were not detected (Figure S6D). In vitro ubiquitination assays demonstrated that both MIB1 and MIB2 effectively delivered K63 and K48R ubiquitin to recombinant TBK1 proteins (Figures 6C and S6E). In contrast, K48 and K63R ubiquitin could not be conjugated with TBK1 (Figure S6E). A MIB2 ligase mutant with disrupted RING domains (MIB2mut) also failed to induce in vitro ubiquitination of TBK1 (Figures 6C and S6E).
To corroborate the role of MIB proteins in TBK1 ubiquitination, we expressed TBK1 with MIB1, MIB2, or a MIB2 mutant. MIB1 or MIB2 expression augmented TBK1-driven expression of IFN-β and RANTES mRNA (Figure S6F). Mutation of the MIB2 catalytic domain failed to augment IFN and RANTES transcription (Figure S6F). MEFs deficient in MIB1 and MEFs transfected with MIB2 siRNA (_Mib2_KD) were used to provide genetic evidence that both MIB1 and MIB2 contribute to TBK1 polyubiquitination in response to viral infection. As shown in Figure 6D, Sendai virus-induced TBK1 ubiquitination was dramatically reduced in MIB1, MIB2 double-deficient (_Mib1_−/−_Mib2_KD) MEFs compared to control _Mib1_f/f MEFs. Mouse fibroblasts deficient in either MIB1 (_Mib1_−/−) or MIB2 (_Mib1_f/f MEFs transfected with MIB2 siRNA) alone displayed a partial reduction of TBK1 ubiquitination (Figure 6D).
The mechanisms regulating TBK1 catalytic activity in response to viral infection are incompletely understood. The present data suggest that MIB proteins play a critical role in TBK1 K63-linked ubiquitination and activation.
MIBs Are Essential for Anti-RNA Viral Activity
To examine the effect of MIB deficiency on TBK1 activation, MIB siRNA oligos were transfected along with ISRE reporters and TBK1, MAVS, IKKε, or the constitutively active form of IRF3 [IRF3(5D)] into HEK293 cells. Gene targeting of MIB1 or MIB1 and MIB2 blocked TBK1- and MAVS-induced ISRE activation but had no effect on IKKε- and IRF3(5D)-induced ISRE activity (Figure S6G). Rescue with MIB1 restored TBK1- and MAVS-induced reporter activity (Figure S6H). The data tentatively position MIBs between MAVS and IRF3 and imply that IKKε utilizes different mechanisms to activate IRFs.
Silencing MIB1 or MIB1 plus MIB2 selectively inhibited Sendai-mediated IRF3 phosphorylation but had little effect on HSV- and LPS-driven IRF3 activation in MEFs (Figure 6E). MIB2 knockdown has marginal effects on Sendai virus-induced IRF3 phosphorylation (Figure 6E), which is consistent with the low amount of MIB2 expression in MEFs. We next investigated the role of MIBs in NF-κB activation. Depletion of MIBs has no apparent effect on Sendai virus-induced p65 nuclear translocation (Figure 6E). Furthermore, MIB1, MIB2 double-deficient and _Mib1_−/− MEFs exhibited reduced IFN-β, IP-10, and RANTES mRNA expression upon Sendai virus infection whereas little effect was found after infection with HSV and LPS (Figure 6F). Moreover, NF-κB-regulated genes TNF and IL-6 were expressed at comparable levels after Sendai virus infection (Figure 6F). The combined data suggest that Mib genes selectively regulate IRF3 activation in response to RNA and RNA viruses.
We also examined the role of MIBs in bone marrow-derived macrophages (BMDM). Because the TBK1-related kinase IKKε is constitutively expressed in macrophages, we depleted IKKε from _Mib2_−/− or heterozygous control macrophages to focus studies on the sole role of TBK1. After MIB1 was silenced with siRNA, BMDM were treated with Sendai virus, HSV, and LPS. IFN-β, IP-10, and RANTES mRNA levels were significantly reduced in MIB1, MIB2 double-deficient macrophages upon Sendai virus infection, whereas neither TNF nor IL-6 mRNA displayed significant change (Figure 6G). As noted with MEFs, MIB1, MIB2 double deficiency in BMDM had little effect on HSV- and LPS-induced cytokine responses (Figures S6I and S6J), whereas Nrdp1-deficient BMDM display defective IFN-β and IP-10 mRNA responses to LPS (Figure S6I). MIB1, MIB2 double-deficient BMDM also demonstrated defective IFN-β and RANTES protein production after Sendai virus infection (Figures 6H and S6K) although IL-6 synthesis was not affected (Figure 6I). The specificity and efficiency of MIB1, MIB2, IKKε, and Nrdp1 silencing were validated (Figures S6L–S6O). We also found that macrophages express relatively high amounts of MIB2 mRNA compared with MEFs (Figure S6P).
Finally, we examined the roles of MIBs on viral replication. Ectopic expression of MIB1 and MIB2 significantly suppressed VSV-Luc viral replication in HEK293 cells, whereas expression of the MIB2 mutant had little effect (Figure 6J). Conversely, upon VSV-Luc infection, _Mib1_−/− and MIB1, MIB2 double-deficient MEFs showed remarkably elevated levels of viral replication as measured by luciferase activity compared to control _Mib1_f/f fibroblasts (Figure 6K). MIB-deficient cells were protected against viral replication by reconstitution with MIB1 or MIB2 but not the catalytically inactive MIB2 mutant (Figure S6Q). Similarly, after infection with eGFP-tagged VSV, _Mib1_−/− MEFs showed an increased number of VSV-eGFP-positive cells (Figure 6L). Collectively, these results indicate that MIB E3 ligases are critical for production of type I interferon and regulate innate immune responses to RNA viruses.
DISCUSSION
Here we have constructed a comprehensive map describing the dynamic physical and functional protein interactions regulating innate immunity. This architectural representation of the HI5 may help unravel the multitiered signaling mechanisms underlying the innate responses and connect innate immunity to other cellular pathways. We assembled a map of 401 unique interactions between 58 baits and 260 HCIP. As with any screening approach, the database does not represent a complete interaction network and the absence of some known interactors illustrates the limitations of this technology (see Supplemental Experimental Procedures). Many of the hits identified in this screen require additional validation by more directed experiments. The regulatory responses of genes in the HI5 were also detailed by transcriptional profiling of macrophages exposed to virus, viral RNA and DNA mimics, LPS, or IFN. Furthermore, to examine HCIP activity, we assessed the gain- and loss-of function effects in eight independent assays in HEK293 and THP-1 cells and identified 22 genes that functionally regulate NF-κB and ISRE reporter activity, VSV replication, or Sendai virus- and HSV-induced IFN production. Finally, detailed mechanistic analysis of MIB proteins established their roles in regulation of innate immunity to RNA viruses.
MIB proteins were shown here to physically interact with TBK1. They selectively participate in ubiquitination of TBK1 after stimulation with dsRNA or RNA viruses. MIBs are best known for their role in the Notch pathway, an evolutionarily conserved signal transduction cascade that plays an essential role in organ formation and other developmental processes in flies, fish, and mammals (Itoh et al., 2003). Mice lacking MIB1 die during early embryogenesis with defects in neurogenesis and vasculogenesis resulting from the failure to activate Notch signaling (Koo et al., 2005a). MIB1 is constitutively and ubiquitously expressed in both embryo and adult tissues, whereas MIB2 is primarily found in some adult tissues (Koo et al., 2005b). Consequently, MIB2-deficient mice do not share the same developmental defects noted for MIB1-deficient animals (Koo et al., 2007; Wu et al., 2007).
A recent report identified Nrdp1 as an E3 ligase involved in LPS-induced TBK1 ubiquitination (Wang et al., 2009). Nrdp1 is selectively expressed in mouse hematopoietic cells and was not detectable in other cells (Abdullah et al., 2001). Cell-autonomous defense against infection with viruses is a fundamental process affecting all cell types. MIB1 is ubiquitously expressed (Song et al., 2008) and may play a central role in innate defense against RNA viruses in all tissues. However, there is no evidence that MIBs participate in responses to LPS, dsDNA, or DNA virus, suggesting that these stimuli trigger alternate pathways to TBK1 activation. We provide genetic evidence to underscore the role of MIB proteins in TBK1 ubiquitination in response to dsRNA and RNA viruses, in both MEFs and macrophages. The data indicate that MIBs selectively control responses to cytosolic RNA and regulate type I interferon transcription.
IFN transcriptional activation is regulated by the cooperative assembly of the IRF3 and NF-κB transcription factors (Panne et al., 2007). Although we demonstrate that overexpression of MIB1 and MIB2 activates both NF-κB and IRF3 signaling, with RNAi we noted that MIBs were not required for NF-κB activation or IL-6 production in response to RNA virus. Similarly, it has been shown that TBK1 is not essential for NF-κB activation induced by virus although overexpression of TBK1 strongly activates NF-κB signaling (Perry et al., 2004). Thus, it is possible that over-expression of MIBs activates TBK1, which in turn elicits NF-κB signaling.
Through proteomic analysis of TBK1 and IKKε complexes, we found that IKKε but not TBK1 can directly associate with MAVS (Figure S2), suggesting that MAVS may use different mechanisms to activate IKKε. Silencing MIB1 in HEK293 cells inhibits TBK1-induced ISRE reporter activity but not IKKε-induced ISRE reporter activity. Further studies are required to define the molecular requirements for IKKε-mediated signal transduction, but these data are consistent with the hypothesis that TBK1 and IKKε utilize different signaling cascades (Paz et al., 2009).
K63-linked ubiquitination plays an essential role in transmitting signals required for activation of innate antiviral responses (Bhoj and Chen, 2009). The TRIM25 and TRIM56 E3 ligases target upstream elements in the IFN pathway (Gack et al., 2007; Tsuchida et al., 2010). TBK1 K63-linked ubiquitination may link NEMO to TBK1. NEMO was shown to be critical for IFN production in response to dsRNA (Zeng et al., 2009; Zhao et al., 2007). Proteomic analysis of NEMO complexes demonstrated inducible interactions with both TBK1 and MAVS after poly(rI:rC) stimulation (Figures S2 and S3B). NEMO has two ubiquitin binding domains (UBD) and both are required for IRF3 activation (Zeng et al., 2009). These two independent UBD potentially allow NEMO to bridge TBK1 with other ubiquitinated components of the signalosome.
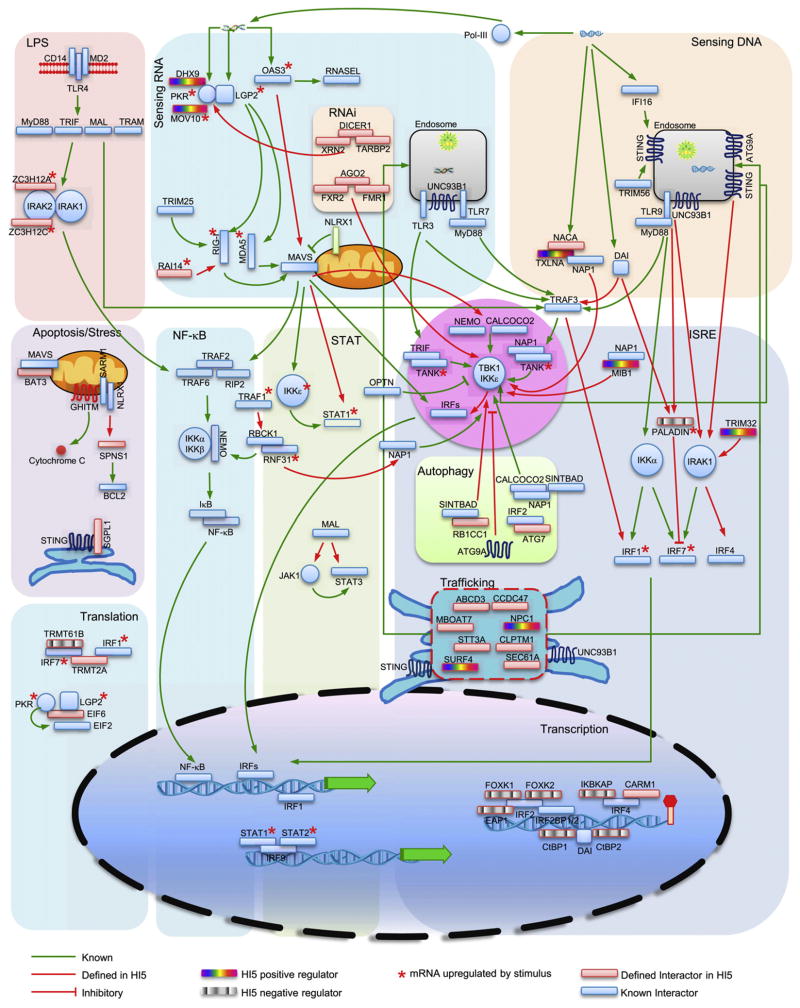
Computational analyses allowed us to integrate several candidate genes into a dynamic antiviral innate immune network. This network covers several processes including RNA and DNA sensing, TLR4 engagement, activation of NF-κB, IRF and STAT transcription factors, translation, transcription, apoptosis and ER stress, RNA interference, autophagy, and trafficking (Figure 7). At least four proteins (CALCOCO2, ATG7, ATG9A, and RB1CC1) appear to connect the HI5 to autophagy. SARM1 associates with NLRX1 on mitochondria where they may regulate IFN production (Carty et al., 2006) and oxidative stress (Tattoli et al., 2008). Their binding partners, GHITM and SPNS1, have been reported to regulate apoptosis (Oka et al., 2008; Yanagisawa et al., 2003). Similarly, BAT3 and SGPL1 also regulate cell apoptosis (Colié et al., 2010; Tsukahara et al., 2009). These protein interactions suggest links between innate antiviral responses and autophagy, cell stress, and apoptosis.
Figure 7. Functional Integration of Selected Components of the HI5.
Baits and HCIP are shown within selected pathways, suggesting functional roles and modes of regulation. Line style and color fill are indicated.
Recently, LGP2 was shown to support both RIG-I- and MDA5-mediated antiviral responses (Satoh et al., 2010). We found that LGP2 associated with PKR and this combination interacted with six common proteins including DICER1. However, _Dicer1_-deficient mouse cells do not have a generalized defect in IFN transcription (Otsuka et al., 2007). Crosstalk between LGP2 and PKR with the RNAi pathway needs further study because it represents a potential mechanism for processing viral RNA prior to RLR recognition. In addition, two PKR binding proteins, MOV10 and DHX9, were found to regulate VSV replication and Sendai-mediated IFN production, respectively, but the mechanisms underlying this activity are unknown.
Several cytosolic DNA sensors have been identified but little is known about how DNA recognition signals IFN transcription. Genetic deficiency of a DNA sensor termed DAI suggests that it is dispensable for DNA-induced IFN production and implies that other DNA sensors exist (Ishii et al., 2008; Takaoka et al., 2007). Additional DNA receptors including DHX9 and IFI16 have recently been identified but both are cell type specific (Kim et al., 2010; Unterholzner et al., 2010). Recent studies also showed that poly(dA:dT) induces RNA polymerase III-dependent innate immune responses that activate the RIG-I/MAVS signaling axis. However, in primary cells, dsDNA induces innate immune responses independently of MAVS (Ablasser et al., 2009; Chiu et al., 2009). Our analysis of DAI complexes indicated that DAI associates with a transcriptional repressor complex containing CtBP1 and CtBP2. RNAi of CtBP1 and CtBP2 compensated for HSV-induced IFN production (Figure 5E). Whether these corepressors inhibit viral or host DNA transcription needs further investigation. Another DAI binding protein, PALADIN, was found to interact with TLR9 and IRF7. PALADIN contains a protein tyrosine phosphatase domain and negatively regulates insulin signaling (Huang et al., 2009). PALADIN mRNA levels are upregulated after HSV infection (Figure 2C). Silencing of PALADIN enhanced HSV-induced IFN-β production (Figure 5C), suggesting that PALADIN negatively regulates a pathway controlling responses to DNA viruses.
Recently, STING (also known as MITA or MPYS) has been shown to regulate DNA-mediated IFN transcription (Ishikawa and Barber, 2008). SEC61A, a central component of the protein translocation apparatus in the ER, is associated with STING, suggesting potential crosstalk with translocation functions. It was reported that ATG9A interacted with STING and inhibited its association with TBK1 (Saitoh et al., 2009). We found that poly(dA:dT) induced an association between TBK1 and ATG9A. Furthermore, we found that STING and UNC93B1 interacted with 10 common binding partners and most of them are ER membrane proteins involved in protein translocation. Among them, SURF4 overexpression activated NF-κB reporter activity and inhibited VSV replication whereas RNAi of NPC1 inhibited HSV-mediated IFN production.
We also found that the TBK1 binding protein α-taxilin (TXLNA) was required for HSV-induced IFN production. The taxilin family consists of three members that share an extraordinarily long coiled-coil region (Nogami et al., 2004). TXLNA interacts with the nascent polypeptide-associated complex and coactivator alpha (NACA) transcriptional coactivator (Yoshida et al., 2005). Because NACA has DNA-binding ability (Akhouayri et al., 2005), it may sense DNA in the cytoplasm and transduce signals to TXLNA. This hypothesis is currently under investigation in our laboratory.
Several proteins in the HI5 were found to associate with IRF transcriptional regulators. EAP1, FOXK1, and FOXK2 bind IRF2 and negatively regulate Sendai virus-induced IFN production. IKBKAP interacts with IRF4 and negatively regulates HSV-induced IFN production. In addition to transcriptional regulation, several HCIP may regulate translation of viral and/or host proteins. TRMT61B is a tRNA methyltransferase regulating RNA translation. Gene targeting of TRMT61B severely impaired VSV replication ability. Thus, mapping the molecular circuits of the HI5 can be a valuable tool for understanding the connections between innate immunity and other cell processes.
The HI5 is a resource for the study of molecular mechanisms controlling innate immune responses. Dynamic remodeling of the HI5 occurs after perturbation with ligand. The genes identified by these proteomic approaches offer potential for therapeutic intervention of infectious and autoimmune diseases.
EXPERIMENTAL PROCEDURES
Cell Lines and Viruses
HEK293, THP-1, and RAW264.7 cells were purchased from ATCC. HEK293/TLR4/MD2/CD14 cell line was purchased from Invivogen (San Diego, CA). HEK293/TLR3 stable cell line was provided by V. Dixit (Genentech). Embryonic fibroblasts derived from _Mib1_f/f mice (Koo et al., 2007) were immortalized by transfection with SV40 LT. _Mib2_−/− mice were derived from founders purchased from MMRRC (Chapel Hill, NC). Mice were bred and maintained in animal facilities at Harvard Medical School in accordance with the Institutional Animal Care and Use Committee. To generate macrophage cultures, bone marrow cells were cultured for 7 days in DMEM supplemented with 30% L929 cell supernatant as a source of GM-CSF. Sendai virus (ΔM) and HSV (KOS strain) were purchased from Charles River (Wilmington, MA) and ATCC (Manassas, VA), respectively. VSV (Indiana) viruses were donated by S. Whelan (Harvard Medical School).
Protein Purification
For purification, 1 × 107 cells were lysed in 10 ml lysis buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM NaCl, 0.5% Nonidet P40, 10% glycerol, phosphatase inhibitors, and protease inhibitors). Detailed methods are provided in Supplemental Experimental Procedures.
Mass Spectrometry
Excised gel bands were cut into approximately 1 mm3 pieces and subjected to in-gel trypsin digestion as detailed in Supplemental Experimental Procedures. Samples were separated on a nano-scale C18 spherical silica bead reverse-phase HPLC capillary column. As peptides eluted, they were subjected to electrospray ionization and entered into an LTQ Velos ion-trap mass spectrometer (ThermoFisher, San Jose, CA). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein databases with the acquired fragmentation pattern by the software program Sequest (ThermoFisher, San Jose, CA).
Functional Analyses
Unless indicated otherwise, 1 μg/ml poly(rI:rC), 1 μg/ml poly(dA:dT), 10 μg/ml LPS, 1.5 U/ml IFN-β, 5 HA Sendai virus, 1 M.O.I. VSV-Luc, and 1 M.O.I. HSV were used to treat cells. Detailed methods, including HCIP mRNA profiling, validation of interactions, reporter assays, and Sendai virus- and HSV-induced IFN production are provided as Supplemental Experimental Procedures.
Human MIB1 and MIB2 cDNA were ordered from ATCC. Murine Mib1 and Mib2 were cloned from a mouse astrocyte line (Li et al., 2006). The first and third Cys in each MIB2 RING domain were mutated to Ala to destroy E3 ligase activity.
Supplementary Material
01
02
03
04
05
06
07
Acknowledgments
This work was supported by NIH grants NS057162 and AI089829. We thank J. Peng (Emory) for MIB1 antibody, J. Hiscott (McGill) for IRF3(5D), and the services of the Taplin Mass Spectrometry Facility (Harvard). Because of space limitations we list the donors of plasmids in the Supplemental Tables.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and six tables and can be found with this article online at doi:10. 1016/j.immuni.2011.06.014.
References
- Abdullah JM, Li X, Nachtman RG, Jurecic R. FLRF, a novel evolutionarily conserved RING finger gene, is differentially expressed in mouse fetal and adult hematopoietic stem cells and progenitors. Blood Cells Mol Dis. 2001;27:320–333. doi: 10.1006/bcmd.2001.0390. [DOI] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhouayri O, Quélo I, St-Arnaud R. Sequence-specific DNA binding by the alphaNAC coactivator is required for potentiation of c-Jun-dependent transcription of the osteocalcin gene. Mol Cell Biol. 2005;25:3452–3460. doi: 10.1128/MCB.25.9.3452-3460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Chau TL, Gioia R, Gatot JS, Patrascu F, Carpentier I, Chapelle JP, O’Neill L, Beyaert R, Piette J, Chariot A. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem Sci. 2008;33:171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colié S, Codogno P, Levade T, Andrieu-Abadie N. Regulation of cell death by sphingosine 1-phosphate lyase. Autophagy. 2010;6:426–427. doi: 10.4161/auto.6.3.11529. [DOI] [PubMed] [Google Scholar]
- Ferran MC, Lucas-Lenard JM. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Friedman CS, O’Donnell MA, Legarda-Addison D, Ng A, Cárdenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picorna-virus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Hancock MK, Pitman JL, Orth AP, Gekakis N. Negative regulators of insulin signaling revealed in a genome-wide functional screen. PLoS ONE. 2009;4:e6871. doi: 10.1371/journal.pone.0006871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jin Y, Blue EK, Dixon S, Shao Z, Gallagher PJ. A death-associated protein kinase (DAPK)-interacting protein, DIP-1, is an E3 ubiquitin ligase that promotes tumor necrosis factor-induced apoptosis and regulates the cellular levels of DAPK. J Biol Chem. 2002;277:46980–46986. doi: 10.1074/jbc.M208585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. Aspartate-glutamatealanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005a;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Koo BK, Yoon KJ, Yoo KW, Lim HS, Song R, So JH, Kim CH, Kong YY. Mind bomb-2 is an E3 ligase for Notch ligand. J Biol Chem. 2005b;280:22335–22342. doi: 10.1074/jbc.M501631200. [DOI] [PubMed] [Google Scholar]
- Koo BK, Yoon MJ, Yoon KJ, Im SK, Kim YY, Kim CH, Suh PG, Jan YN, Kong YY. An obligatory role of mind bomb-1 in notch signaling of mammalian development. PLoS ONE. 2007;2:e1221. doi: 10.1371/journal.pone.0001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Berman MA, Zhang Y, Dorf ME. RNAi screen in mouse astrocytes identifies phosphatases that regulate NF-kappaB signaling. Mol Cell. 2006;24:497–509. doi: 10.1016/j.molcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Nogami S, Satoh S, Tanaka-Nakadate S, Yoshida K, Nakano M, Terano A, Shirataki H. Identification and characterization of taxilin isoforms. Biochem Biophys Res Commun. 2004;319:936–943. doi: 10.1016/j.bbrc.2004.05.073. [DOI] [PubMed] [Google Scholar]
- Oka T, Sayano T, Tamai S, Yokota S, Kato H, Fujii G, Mihara K. Identification of a novel protein MICS1 that is involved in maintenance of mitochondrial morphology and apoptotic release of cytochrome c. Mol Biol Cell. 2008;19:2597–2608. doi: 10.1091/mbc.E07-12-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, et al. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol. 2009;29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J Exp Med. 2004;199:1651–1658. doi: 10.1084/jem.20040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryzhakov G, Randow F. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 2007;26:3180–3190. doi: 10.1038/sj.emboj.7601743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Song R, Kim YW, Koo BK, Jeong HW, Yoon MJ, Yoon KJ, Jun DJ, Im SK, Shin J, Kong MP, et al. Mind bomb 1 in the lymphopoietic niches is essential for T and marginal zone B cell development. J Exp Med. 2008;205:2525–2536. doi: 10.1084/jem.20081344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Carneiro LA, Jéhanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Tsukahara T, Kimura S, Ichimiya S, Torigoe T, Kawaguchi S, Wada T, Yamashita T, Sato N. Scythe/BAT3 regulates apoptotic cell death induced by papillomavirus binding factor in human osteosarcoma. Cancer Sci. 2009;100:47–53. doi: 10.1111/j.1349-7006.2008.00991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Rajendra R, Barsi JC, Durfee L, Benito E, Gao G, Kuruvilla M, Hrdlicková R, Liss AS, Artzt K. Targeted disruption of Mib2 causes exencephaly with a variable penetrance. Genesis. 2007;45:722–727. doi: 10.1002/dvg.20349. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Miyashita T, Nakano Y, Yamamoto D. HSpin1, a transmembrane protein interacting with Bcl-2/Bcl-xL, induces a caspase-independent autophagic cell death. Cell Death Differ. 2003;10:798–807. doi: 10.1038/sj.cdd.4401246. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Nogami S, Satoh S, Tanaka-Nakadate S, Hiraishi H, Terano A, Shirataki H. Interaction of the taxilin family with the nascent polypeptide-associated complex that is involved in the transcriptional and translational processes. Genes Cells. 2005;10:465–476. doi: 10.1111/j.1365-2443.2005.00848.x. [DOI] [PubMed] [Google Scholar]
- Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06
07