MicroRNA-210 Modulates Endothelial Cell Response to Hypoxia and Inhibits the Receptor Tyrosine Kinase Ligand Ephrin-A3 (original) (raw)
Abstract
MicroRNAs (miRNAs) are small non-protein-coding RNAs that function as negative gene expression regulators. In the present study, we investigated miRNAs role in endothelial cell response to hypoxia. We found that the expression of miR-210 progressively increased upon exposure to hypoxia. miR-210 overexpression in normoxic endothelial cells stimulated the formation of capillary-like structures on Matrigel and vascular endothelial growth factor-driven cell migration. Conversely, miR-210 blockade via anti-miRNA transfection inhibited the formation of capillary-like structures stimulated by hypoxia and decreased cell migration in response to vascular endothelial growth factor. miR-210 overexpression did not affect endothelial cell growth in both normoxia and hypoxia. However, anti-miR-210 transfection inhibited cell growth and induced apoptosis, in both normoxia and hypoxia. We determined that one relevant target of miR-210 in hypoxia was Ephrin-A3 since miR-210 was necessary and sufficient to down-modulate its expression. Moreover, luciferase reporter assays showed that Ephrin-A3 was a direct target of miR-210. Ephrin-A3 modulation by miR-210 had significant functional consequences; indeed, the expression of an Ephrin-A3 allele that is not targeted by miR-210 prevented miR-210-mediated stimulation of both tubulogenesis and chemotaxis. We conclude that miR-210 up-regulation is a crucial element of endothelial cell response to hypoxia, affecting cell survival, migration, and differentiation.
Hypoxia occurs during several physio-pathological circumstances such as rapid tissue growth, acute and chronic ischemia, organ and tumor development, and high altitude (1). Diminished oxygen concentration induces an articulate program of responses aimed at relieving tissue hypoxia and removing irreversibly damaged cells (2,3). These responses include endothelial cell (EC)2 proliferation, migration, and angiogenesis, but also growth arrest and apoptotic cell death; the nature of the outcome depends on numerous parameters, including cell histological origin and genotype, as well as the severity and the duration of the hypoxic stress. Thus, it is of pivotal importance in understanding the molecular mechanisms triggered by cell exposure to low oxygen tension.
Little is known about the role played by microRNAs (miRNAs) in EC response to hypoxia (4). miRNAs are 21–23-nucleotide RNA molecules that regulate the stability or the translational efficiency of target messenger RNAs (5,6). The biogenesis of miRNAs begins with a primary transcript, termed the pri-miRNA, and the combined action of Drosha and Dicer ribonucleases generates the mature miRNA species. This product is loaded into the RNA-induced silencing complex, where it mediates the translational inhibition of target mRNA, albeit the opposite effect has been described as well (7). miRNAs have diverse functions, including the regulation of cellular differentiation, proliferation, and apoptosis (5). Although it is clear that miRNAs have essential functions in mammalian biology, few miRNA genes have been functionally linked to specific cellular pathways. In this study, we show that miR-210 is a key player of EC response to low oxygen tension and identify Ephrin-A3 as a relevant target of miR-210 in hypoxic conditions.
EXPERIMENTAL PROCEDURES
_Cell Cultures_—Human umbilical vein EC (HUVEC; Clonetics) were grown in EGM-2 (BioWhittaker) containing 2% fetal bovine serum. U2OS osteosarcoma cell line and Phoenix-ampho cells (American Type Culture Collection) were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The induction of acidosis and hypoxia is described in supplemental materials.
_miRNA and mRNA Quantification_—Total RNA was extracted using TRIzol (Invitrogen). Small RNA and mRNA fractions were enriched using the PureLink miRNA isolation kit and Micro-to-Midi isolation kit (Invitrogen), respectively, according to the manufacturer's instructions. miRNA levels were analyzed using the TaqMan real-time PCR (qPCR) method (1 ng/assay) and quantified with the ABI Prism 7000 SDS (Applied Biosystems). Primers for 157 miRNA, 28 positive and negative controls, and the reagents for reverse transcriptase and qPCR reactions were all obtained from Applied Biosystems. Relative expression was calculated using the comparative Ct method (2-[Δ][Δ]Ct) (8). Different samples were normalized to miR-16 expression. For miRNA profiling, miRNAs were assayed in a 96-well format, and samples were also normalized to the median Ct value, obtaining almost identical results. mRNAs levels were analyzed using the SYBR-GREEN qPCR method (1–3 μg/assay, Ambion, see supplemental materials for details). Northern blottings are described in the supplemental materials.
_Small Interfering RNA-mediated Gene Silencing_—Small interfering RNAs targeting HIF1α, HIF2α, or a scramble sequence (Santa Cruz Biotechnology) were transfected into HUVEC, according to the manufacturer's instruction (see the supplemental materials for details).
_miRNA Down-modulation and Overexpression_—Locked Nucleic Acid oligonucleotides against miR-210 or a control scramble sequence (Exiqon) were transfected by small interfering RNA transfection reagent (Santa Cruz Biotechnology) in 40% confluent HUVEC (4 × 103/cm2) at the final concentration of 40 nm. After 16 h, cells were re-fed with fresh medium, and experiments were performed 24 h later. Alternatively, cells were incubated with fresh medium for 2 h and then shifted to hypoxic conditions. miRIDIAN miR-210 Mimic or a control scramble sequence (Dharmacon) were transfected using the same protocol. To obtain stable miR-210-overexpressing cells, HUVEC were infected by retroviral vectors bearing pre-miR210 sequence (see the supplemental materials).
_miRNAs Target Prediction_—Bioinformatic prediction of target genes and miRNA binding sites was performed using four different programs: MiRanda (version 2005), TargetScan (version 3.1), Sanger MirBase (version 4), and PicTar (version 2006) (9–11). Only common targets were considered for experimental analysis.
_Additional Information_—See the supplemental materials for information on apoptosis, cell cycle analysis, adenoviral infection, capillary-like formation assay, chemotaxis, immunofluorescence, reporter construct generation and luciferase assay.
_Statistical Analysis_—Variables were analyzed by both Student's t test and one-way analysis of variance and a of p ≤ 0.05 was deemed statistically significant. Values are expressed as ±standard error (S.E.).
RESULTS
miR-210 Induction by Hypoxia_—To assess whether hypoxia regulates miRNA expression in EC, HUVEC were exposed to 1% oxygen for different time periods. To control that cells responded to low oxygen tension, cell proliferation was measured. As expected, hypoxia induced cell death and growth arrest, as assessed by growth curves (Fig. S1_A) and by the rate of DNA synthesis with bromodeoxyuridine incorporation (Fig. S1,B and C) (1–3). Low molecular weight RNA was extracted, and miRNA expression profile at each time point was determined (Tables S1 and S2). To normalize different samples, we used miR-16 expression, which previous experiments showed was not regulated by hypoxia (Fig. S2_A_).
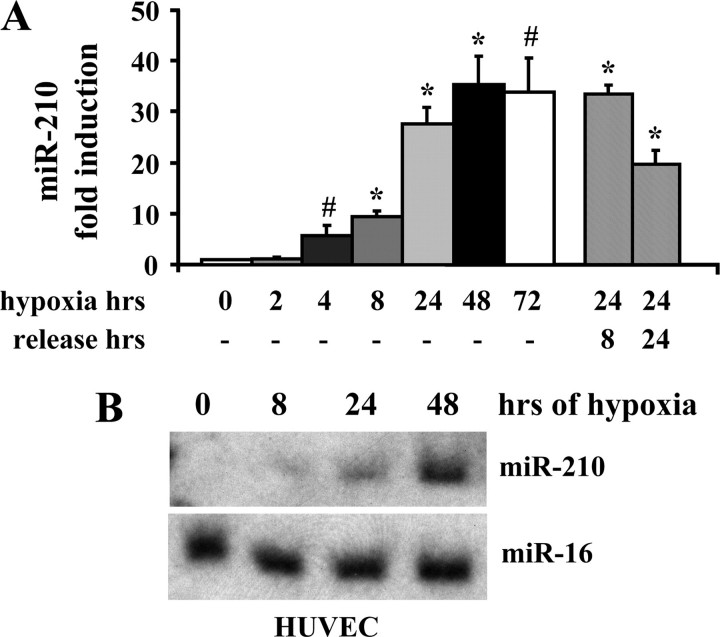
As found in different experimental systems (4), miR-210 levels were strongly induced by hypoxia. miR-210 increased as early as 4 h after hypoxia induction, it was more than 35-fold higher than normoxic control at 48 h, and miRNA up-regulation was maintained for the next 72 h (Fig. 1_A_). miR-150 and 328 were up-regulated as well, albeit to a lower extent and with slower kinetics (Fig. S2, B and C).
FIGURE 1.
miR-210 induction by hypoxia. A, activation of miR-210 by hypoxia is fast and long lasting. HUVEC were exposed to hypoxia for the indicated time. Separated bars indicate miR-210 expression of HUVEC exposed to 1% oxygen for 24 h and then exposed to atmosphere oxygen for 8 and 24 h (*, p < 0.001; #, p < 0.02; n = 4–11). B, Northern blotting showing time-dependent induction of miR-210 by hypoxia in HUVEC.
When miR-210 regulation by hypoxia was further investigated, it was found that its activation was inversely proportional to O2 tension (Fig. S3_A_). Moreover, upon HUVEC reoxygenation after 24 h of hypoxia, miR-210 levels remained high for the next 8 h and slowly declined thereafter, indicating the maintenance of this adaptive response (Fig. 1_A_). Finally, miR-210 induction by hypoxia was confirmed by northern blotting (Fig. 1_B_).
Hypoxia is associated to increased oxidative stress and acidosis (12–14). Thus, it was assessed whether either oxidative stress or acidification in the absence of hypoxia were sufficient to induce miR-210 positive modulation (Fig. S3_B_). We found that HUVEC exposure to 400 μm H2O2 for 8 and 24 h did not induce miR-210 modulation; similarly, cell culture at both pH 6.6 and pH 7.0 for 24 and 48 h did not increase miR-210. In keeping with these data, a buffered medium that prevented hypoxia-induced acidosis (15) did not inhibit miR-210 activation by hypoxia. Finally, we found that glucose supplementation did not prevent miR-210 induction by hypoxia (Fig. S3_C_) and that this event was not endothelium-specific (Fig. S4).
Many hypoxia effects are mediated by HIF transcription factor (1–3). To analyze the role of HIF in the induction of miR-210 by hypoxia in EC, the expression of HIF1α and HIF2α was knocked down by the transfection of specific small interfering RNAs. Although the knock down of both HIF isoforms was highly effective (Fig. S5, A and B), only HIF1α RNA interference decreased the induction of miR-210 by hypoxia significantly (Fig. S5_C_). In keeping with this observation, HIF1α overexpression was sufficient to induce miR-210 expression in the absence of hypoxia (Fig. S5, D and E).
We conclude that miR-210 displays a rapid and dose-dependent induction in response to hypoxia and that its up-regulation is maintained over time. Neither oxidative stress nor acidosis appears to play a role in this event and, in keeping with previous observations (4), HIF1α is necessary and sufficient for miR-210 activation.
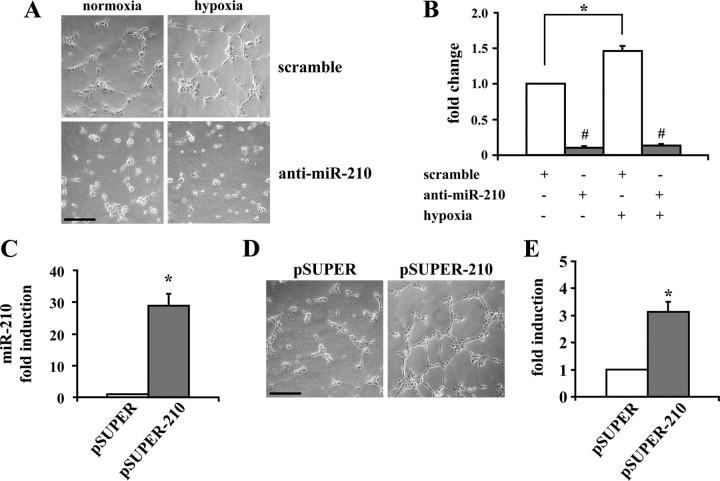
_miR-210 Expression Stimulates Capillary-like Formation and Cell Migration_—One main feature of hypoxia is the stimulation of angiogenesis (3). Several aspects of angiogenesis can be studied in vitro, taking advantage of the ability of EC to form capillary-like structures once plated on Matrigel or other extracellular components (16). To this aim, HUVEC were first exposed to 24 h of hypoxia and, afterward, their ability to form capillary-like structures in low growth factor-containing medium was assessed. In keeping with previous reports (17), hypoxia preconditioning significantly increased tubulogenesis (Fig. 2, A and B). To determine whether this event was, at least in part, dependent on miR-210 activation, HUVEC were transfected with a miR-210 complementary Locked Nucleic Acid (anti-miR-210) that binds with high affinity and specificity to the complementary miRNA, inactivating its function. Fig. 2, A and_B_, show that 24 h of miR-210 blocking greatly decreased the ability of HUVEC to form capillary-like structures, both in cells exposed to hypoxia and in control cells.
FIGURE 2.
miR-210 enhances the development of capillary-like structures. A and B, effect of hypoxia and miR-210 blockade on HUVEC tubulogenesis. HUVEC (4 × 103/cm2) were transfected with anti-miR-210. Eighteen hours later, cells were exposed to hypoxia for further 24 h, and then their organization into capillary-like structures was assayed. A shows representative phase-contrast images.Size bar = 200 μm. B shows the quantitative assessment of capillary-like structures (hypoxic control versus normoxic control:*, p < 0.01; anti-miR-210 versus control: #,p < 0.005; n = 3). C, HUVEC were transduced with a retroviral vector bearing the miR-210 pre-miRNA sequence under the control of a constitutive promoter. These cells expressed almost 30-fold more mature miR-210 than control (*, p < 0.004; n = 4).D and E, miR-210 stimulated HUVEC tubulogenesis. The differentiation into capillary-like structures of HUVEC overexpressing miR-210 (pSUPER-210) and control (pSUPER) was assayed. D shows representative phase-contrast images. Size bar = 200 μm. E shows the quantitative assessment of capillary-like structures (*, p < 0.003; n = 3).
To assess whether miR-210 induction affected capillary-like structure formation in the absence of hypoxia, HUVEC overexpressing pre-miR-210 were generated, yielding a population that expressed mature miR-210 levels comparable with these observed in hypoxic cells (Fig. 2_C_). We found that miR-210 overexpression significantly increased capillary-like formation when HUVEC were plated in low growth factor medium (Fig. 2, D and_E_).
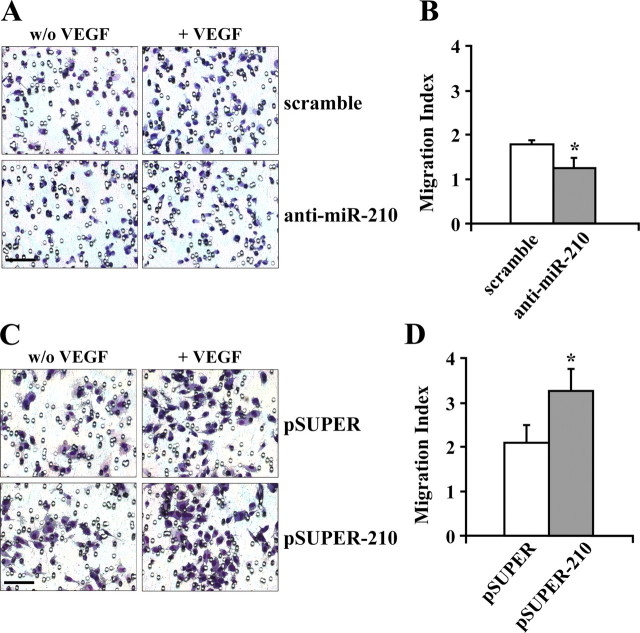
EC migration is a crucial event in the angiogenic process (3). Thus, we assessed whether the modulation of miR-210 expression affected VEGF-induced chemotaxis. To this aim, HUVEC were transfected with anti-miR-210, and then their ability to migrate in response to VEGF was assayed.Fig. 3, A and_B_, show that miR-210 blockade significantly decreased VEGF-driven chemotaxis. Conversely, miR-210 up-regulation to levels similar to these induced by hypoxia significantly increased HUVEC ability to migrate in response to VEGF (Fig. 3, C and_D_).
FIGURE 3.
miR-210 expression enhances VEGF-induced chemotaxis. A, representative experiment of the chemotactic responses of HUVECs transfected with either anti-miR-210 or control in response to 20 ng/ml VEGF. Size bar = 50 μm. B, miR-210 blockade decreases HUVEC migration index (*, p < 0.015; n = 3). C, HUVECs were transduced with either a retroviral vector encoding miR-210 (pSUPER-210) or vector alone (pSUPER), and their chemotactic responses in response to 20 ng/ml VEGF was assayed. Size bar = 50 μm.D, miR-210 expression increases HUVEC migration index (*,p < 0.001; n = 7).
Finally, to investigate the relevance of miR-210 action in the hypoxia-activated gene expression pattern, the mRNA levels of 11 hypoxia target genes were measured both in HUVEC exposed to hypoxia and in cells overexpressing miR-210 in normoxic conditions (Fig. S6). Although hypoxia up-regulated the expression of all 11 genes, miR-210 did not affect their levels. We concluded that miR-210 expression does not seem to affect the global gene expression response to hypoxia by HUVEC.
miR-210 Levels Modulate EC Survival_—It was examined whether miR-210 induction affected EC proliferation. To address this issue, we compared growth curves of HUVEC transduced with retroviral vectors expressing miR-210 or with backbone vector alone, in the presence or absence of hypoxia. It was found that miR-210 expression did not affect cell number significantly both in normoxic and in hypoxic conditions (Fig. S7_A). Using a reciprocal approach, HUVEC were transfected with anti-miR-210, and cells were counted. We found that 24 h of miR-210 blocking decreased HUVEC proliferation even in normoxic cells and that exposure to hypoxia further decreased EC number (Fig. S7_B_). This demise was, at least in part, due to apoptotic cell death since apoptotic DNA fragmentation was increased by anti-miR-210 transfection (Fig. S7_C_). Similar data were obtained by a terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay (TUNEL, not shown). Conversely, when DNA synthesis was measured, we found that anti-miR-210 only minimally affected bromodeoxyuridine incorporation rate (Fig. S7_D_).
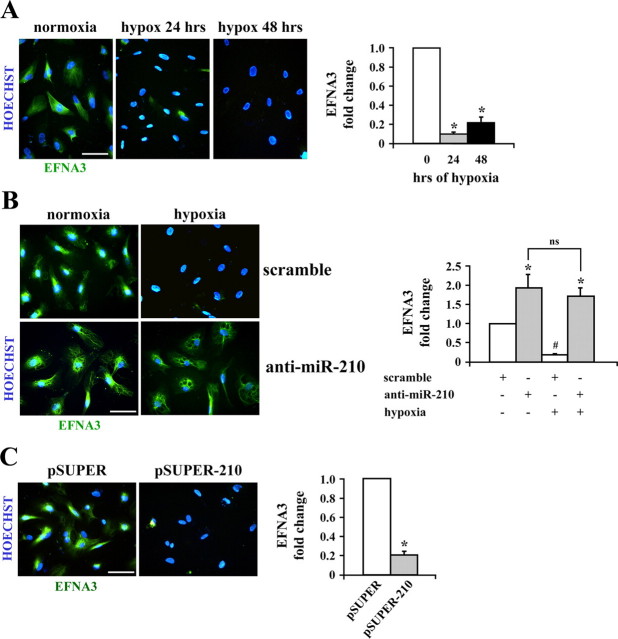
Hypoxia Down-modulates Ephrin-A3 via miR-210_—To further corroborate the biological relevance of miR-210 regulation in cell response to hypoxia, an in silico search of potential targets was performed using PicTar, miRANDA, Sanger MirBase, and Targetscan algorithms (Table S3) (9–11). One of the common targets of these software programs was Ephrin-A3 (EFNA3). Given the crucial role of Ephrins in the development of the cardiovascular system and in vascular remodeling (18), we decided to validate this target prediction. To this aim, HUVEC were exposed to hypoxia for 24 and 48 h, and EFNA3 was detected by semiquantitative immunofluorescence.Fig. 4_A_ and Fig. S8_A show that although EFNA3 was readily detectable in normoxic cells, its expression decreased to almost undetectable levels following hypoxia exposure. This down-modulation was not due to decreased mRNA levels since measurements by qPCR displayed an unexpected increase of EFNA3 mRNA (Fig. S8_B_). Although EFNA3 regulation clearly needs further investigation, we can conclude that EFNA3 is post-transcriptionally down-modulated by hypoxia.
FIGURE 4.
EFNA3 expression is inhibited by hypoxia and miR-210 expression. A, EFNA3 inhibition in hypoxic cells. HUVEC were exposed to hypoxia for 24 and 48 h. Then, cells were fixed and stained with both an antibody to EFNA3 (K-19, Santa Cruz Biotechnology, green) and the DNA intercalating agent Hoechst 33342 (blue). Size bar = 50 μm. EFNA3 signal was quantified using Scion Image software, and after 24 and 48 h of hypoxia, the decrease of EFNA3 fluorescence was about 80% when compared with the normoxic control (*, p < 0.0001;n = 3). B, HUVEC were transfected with anti-miR-210 and, 18 h later, were exposed to hypoxia for 24 h. Then, cells were fixed and stained with both α-EFNA3 (green) and the DNA intercalating agent Hoechst 33342 (blue). Size bar = 50 μm. EFNA3 signal was quantified using Scion Image software (anti-miR-210 versus control:*, p < 0.02; hypoxia versus control: #,p < 0.001; ns, not significant; n = 3).C, miR-210 expression levels comparable with these observed during hypoxia induce EFNA3 inhibition. HUVEC infected either with a retroviral vector encoding miR-210 (pSUPER-210) or vector alone (pSUPER) were stained with both an antibody to EFNA3 (green) and the DNA intercalating agent Hoechst 33342 (Blue). Size bar = 50 μm. EFNA3 signal was quantified using Scion Image software (*, p < 0.005;n = 3).
Then, it was assessed whether EFNA3 down-modulation was indeed due to increased miR-210 levels. To address this issue, HUVEC were transfected with anti-miR-210 and exposed to hypoxia. Fig. 4_B_ shows that miR-210 blocking completely prevented EFNA3 decrease.
Afterward, it was investigated whether miR-210 up-regulation in the absence of hypoxia regulated EFNA3 levels. We found that EFNA3 protein staining was strongly diminished in miR-210-overexpressing cells (Fig. 4_C_) in the absence of EFNA3 mRNA regulation (Fig. S8_C_). Taken together, these data establish a cause-and-effect relationship between mRNA-210 up-regulation and EFNA3 down-modulation by hypoxia.
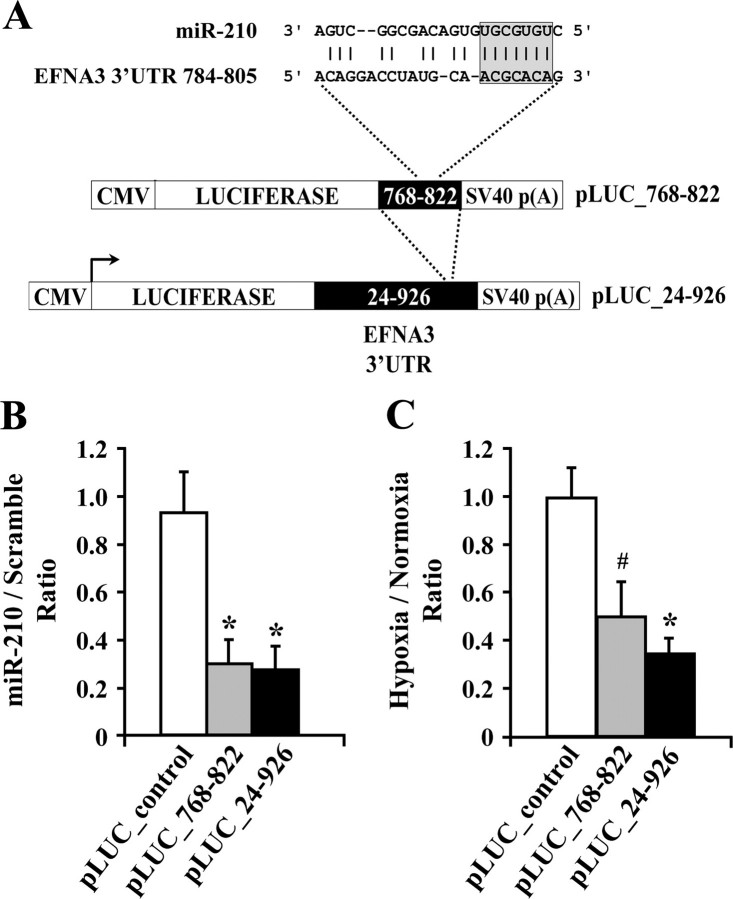
To investigate whether miR-210 directly regulated EFNA3 expression, an assay was set up in which the 3′-UTR of the EFNA3 gene was inserted downstream of a luciferase open reading frame (pLUC_24–926,Fig. 5_A_). Alternatively, to eliminate the potential interference of other miRNA binding sites, only the putative miR-210 target site and the immediately surrounding sequences were cloned (pLUC_768–822). As control, we deleted the sequence complementary to miR-210 seed sequence in this latter construct (pLUC_control). The different luciferase constructs were transfected into U2OS cells with a plasmid carrying a constitutive expression cassette for miR-210 or for a control sequence. U2OS were chosen for their high efficiency of transfection. A significant negative effect on luciferase activity was observed in both constructs bearing an intact miR-210 binding site when compared with the control (Fig. 5_B_). Likewise, endogenous levels of miR-210 induced by hypoxia were sufficient to repress pLUC_24–926 and pLUC_768–822 reporter constructs, whereas pLUC-control was unaffected (Fig. 5_C_). We concluded that EFNA3 is a direct target of miR-210.
FIGURE 5.
miR-210 inhibits EFNA3 expression directly. A, structure of pLUC firefly luciferase reporter plasmids. miR-210 seed sequence and its complementary binding site in EFNA3 3′-UTR are highlighted. CMV, cytomegalovirus. B, U2OS were transfected with pLUC derivatives along with a plasmid encoding either miR-210 or a control sequence. Luciferase values were normalized for their mRNA levels, and the ratio of luciferase activity of each construct was calculated either in the presence or in the absence of exogenous miR-210 (B;*,p < 0.003; n = 6) or in the presence or absence of hypoxia (C;#, p < 0.03; *, p < 0.005; n = 3).
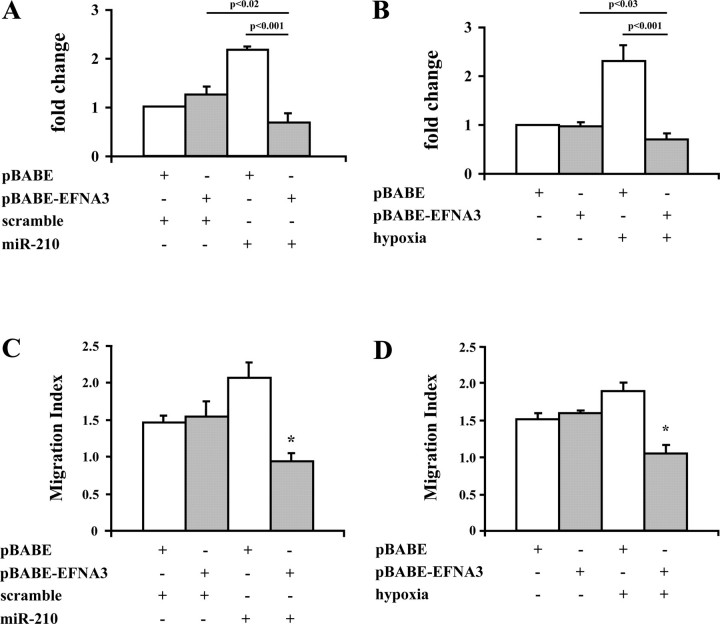
_EFNA3 Down-modulation Is Necessary for miR-210-mediated Stimulation of Both Tubulogenesis and Chemotaxis_—We investigated whether EFNA3 down-modulation is an integral part of miR-210-induced increase of EC migration and tubulogenesis. To this aim, miR-210 was overexpressed in the presence of an EFNA3 allele that is devoid of most 3′-UTR sequence (EFNA3Δ) and therefore cannot be targeted by miR-210. EFNA3Δ was expressed using a retroviral vector, whereas miR-210 was transfected as mature sequence. Fig. 6_A_ shows that EFNA3Δ expression completely prevented miR-210-induced increase of capillary-like structures. Indeed, the simultaneous expression of miR-210 and EFNA3Δ decreased tubulogenesis below control levels, whereas EFNA3Δ expression had no effect per se. We found that the same was true also in hypoxia-stimulated cells, therefore, in the absence of miR-210 overexpression.Fig. 6_B_ shows that the expression of EFNA3Δ prevented hypoxia-induced increase of EC tubulogenesis and that the level of capillary-like structures was below control. Then, we assayed whether EFNA3 inhibition played a role in miR-210 stimulation of EC chemotaxis as well. Fig. 6_C_ shows that although EFNA3Δ expression did not affect EC chemotaxis in response to VEGF, miR-210 and EFNA3Δ co-expression inhibited it completely. Likewise, the expression of EFNA3Δ prevented the increase in VEGF-driven chemotaxis induced by hypoxia (Fig. 6_D_).
FIGURE 6.
miR-210 repression of EFNA3 is necessary to stimulate capillary-like formation and cell chemotaxis. HUVEC expressing either EFNA3Δ or vector alone (4 × 103/cm2) were transfected with mature miR-210 RNA or a scramble sequence and further cultivated for 36 h (A and C). Alternatively, HUVEC expressing either EFNA3Δ or vector alone were exposed to hypoxia or normoxia for 24 h (B and D). Then, cells were harvested, and either their organization into capillary-like structures (A and B) or their chemotactic response to 20 ng/ml VEGF (C and D) was assayed. The prevention of EFNA3 down-modulation by EFNA3Δ expression inhibited capillary-like structure increase induced both by miR-210 transfection (A; n = 4) and by hypoxia (B;n = 8). Moreover, EFNA3Δ expression completely prevented the migration index increase induced both by miR-210 transfection (C;*, p < 0.007 versus all the other conditions; n = 3) and by hypoxia (D;*,p < 0.005 versus all the other conditions; n = 4).
DISCUSSION
In this study, we characterized miR-210 regulation and its functional relevance in EC response to hypoxia. It was found that miR-210 up-regulation was rapid and persistent, and it did not represent a general response to stress since no evidence was found that either intracellular pH decrease or increased reactive oxygen species played a role in miR-210 activation. We also found that neither growth factor deprivation nor osmotic stress elicited miR-210 increase,3 confirming the high specificity of the hypoxic signaling in miR-210 regulation. Moreover, miR-210 modulation by hypoxia is not restricted to EC. Thus, it is tempting to hypothesize a general role of miR-210 induction in the hypoxic response. In keeping with this interpretation, recent studies found that the identity of the miRNAs modulated by hypoxia differs widely according to the cell type used as much as the degree and the time of hypoxia (19–23). However, the activation of miR-210 does not seem linked to tight experimental conditions or to a specific cell type (19–21,23,24).
Like many other miRNAs, miR-210 biological information is limited almost exclusively to expression analysis. Thus, we focused on the investigation of the functional relevance of miR-210 regulation in EC response to hypoxia.
Evidence was provided that miR-210 up-regulation in normoxic conditions increases EC tubulogenesis and migration, whereas miR-210 blockade in the presence of hypoxia decreases capillary-like formation, EC migration, and EC survival and induces apoptosis. In keeping with this latter finding, it has been shown that miR-210 inhibition activates caspases (19,25). One may speculate that the inhibitory effects on migration and tubulogenesis are mediated by apoptosis. Although this is a very likely explanation, additional mechanisms may play a role as well. Indeed, both cell survival decrease and apoptosis induced by anti-miR-210 transfection were only partial, whereas the inhibition of tubulogenesis was almost complete. Moreover, anti-miR-210 transfected cells migrated spontaneously, thus exhibiting a behavior of living cells. The specific effect of miR-210 blockade was to decrease their ability to enhance their migration in response to VEGF.
One main hurdle that has limited the interpretation of many miRNA profiling studies is the difficulty of identifying miRNA targets (10,11). Indeed, the identity of only 102 targets has been experimentally proven in humans to date (as shown by the DIANA TarBase data base). To find genes directly regulated by miR-210, a very stringent strategy was adopted using four target prediction software programs and dismissing targets that were not identified by at least three algorithms. Four lines of evidence indicate that EFNA3 is a miR-210 target. 1) miR-210 overexpression induced the down-modulation of EFNA3 protein but not of EFNA3 mRNA. 2) Hypoxia induced EFNA3 protein demise, whereas EFNA3 mRNA was paradoxically increased. 3) miR-210 blocking by complementary Locked Nucleic Acid strategy prevented EFNA3 down-modulation induced by hypoxia. 4) EFNA3 3′-UTR containing a miR-210 binding sequence decreased the expression of a reporter luciferase gene upon hypoxia or miR-210 ectopic expression. Although all these evidences establish that miR-210 regulates EFNA3 expression directly, it is still possible that indirect mechanisms may increase the potency of miR-210 action.
Ephrin ligands and their Eph receptors have been shown to play a crucial role in the development of the cardiovascular system and in vascular remodeling (18). Specifically, numerous studies have shown the importance of EFNA1/EphA2 interaction in the regulation of angiogenesis and VEGF signaling (18,26). Although the specific role of EFNA3 in the regulation of angiogenesis is still unknown, EphA2 has been shown to bind EFNA3 as well as EFNA1 (27). We provided evidence that EFNA3 down-modulation is a necessary event of miR-210-mediated stimulation of capillary-like formation and EC chemotaxis in response to VEGF. Thus, it is tempting to speculate that miR-210-dependent down-modulation of EFNA3 expression may contribute to modulate the angiogenic response to ischemia. We can conclude that although further studies are needed, the modulation of miR-210 expression and/or activity may be a viable strategy to control angiogenesis, either positively or negatively.
Supplementary Material
Supplemental Data
Acknowledgments
The help of Gian-Luca Sarra Ferraris in experiments using the hypoxic incubator and of Paolo Sarti, “La Sapienza” University of Rome, in performing O2 concentration measurements is greatly appreciated.
*
This work was supported by the Ministero della Salute (Grants RC06/07-1.13, RF05-Conv.79.1, RF05-ISS 64D/F4, RF06-Conv.74.1; RF07Onc-26/1; and RO.06-M.-conv.29/07-1). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available athttp://www.jbc.org) contains supplemental text, three supplemental tables, and eight supplemental figures.
Footnotes
2
The abbreviations used are: EC, endothelial cell; HUVEC, human umbilical vein endothelial cells; miRNA, microRNA; qPCR, real-time PCR; pSUPER, p-SUPER.retro.puro vector; EFNA3, ephrin-A3; anti-miR-210, miR-210 complementary locked nucleic acid; VEGF, vascular endothelial growth factor; UTR, untranslated region.
3
P. Fasanaro, Y. D'Alessandra, M. C. Capogrossi, and F. Martelli, unpublished.
References
- 1.Giaccia, A. J., Simon, M. C., and Johnson, R. (2004) Genes Dev. 18 2183-2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pouyssegur, J., Dayan, F., and Mazure, N. M. (2006) Nature 441 437-443 [DOI] [PubMed] [Google Scholar]
- 3.Pugh, C. W., and Ratcliffe, P. J. (2003) Nat. Med. 9 677-684 [DOI] [PubMed] [Google Scholar]
- 4.Kulshreshtha, R., Davuluri, R. V., Calin, G. A., and Ivan, M. (2008) Cell Death Differ. 15 667-671 [DOI] [PubMed] [Google Scholar]
- 5.Bartel, D. P. (2004) Cell 116 281-297 [DOI] [PubMed] [Google Scholar]
- 6.Zamore, P. D., and Haley, B. (2005) Science 309 1519-1524 [DOI] [PubMed] [Google Scholar]
- 7.Vasudevan, S., Tong, Y., and Steitz, J. A. (2007) Science 318 1931-1934 [DOI] [PubMed] [Google Scholar]
- 8.Livak, K. J., and Schmittgen, T. D. (2001) Methods (Amst.) 25 402-408 [DOI] [PubMed] [Google Scholar]
- 9.Krek, A., Grun, D., Poy, M. N., Wolf, R., Rosenberg, L., Epstein, E. J., MacMenamin, P., da Piedade, I., Gunsalus, K. C., Stoffel, M., and Rajewsky, N. (2005) Nat. Genet. 37 495-500 [DOI] [PubMed] [Google Scholar]
- 10.Sethupathy, P., Megraw, M., and Hatzigeorgiou, A. G. (2006) Nat. Methods 3 881-886 [DOI] [PubMed] [Google Scholar]
- 11.Rajewsky, N. (2006) Nat. Genet. 38 (suppl.) S8-S13 [DOI] [PubMed] [Google Scholar]
- 12.Zaccagnini, G., Martelli, F., Fasanaro, P., Magenta, A., Gaetano, C., Di Carlo, A., Biglioli, P., Giorgio, M., Martin-Padura, I., Pelicci, P. G., and Capogrossi, M. C. (2004) Circulation 109 2917-2923 [DOI] [PubMed] [Google Scholar]
- 13.Mekhail, K., Khacho, M., Gunaratnam, L., and Lee, S. (2004) Cell Cycle 3 1027-1029 [PubMed] [Google Scholar]
- 14.Guzy, R. D., and Schumacker, P. T. (2006) Exp. Physiol. 91 807-819 [DOI] [PubMed] [Google Scholar]
- 15.Luo, F., Liu, X., Yan, N., Li, S., Cao, G., Cheng, Q., Xia, Q., and Wang, H. (2006) BMC Cancer 6 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubota, Y., Kleinman, H. K., Martin, G. R., and Lawley, T. J. (1988) J. Cell Biol. 107 1589-1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamakawa, M., Liu, L. X., Date, T., Belanger, A. J., Vincent, K. A., Akita, G. Y., Kuriyama, T., Cheng, S. H., Gregory, R. J., and Jiang, C. (2003) Circ. Res. 93 664-673 [DOI] [PubMed] [Google Scholar]
- 18.Kuijper, S., Turner, C. J., and Adams, R. H. (2007) Trends Cardiovasc Med. 17 145-151 [DOI] [PubMed] [Google Scholar]
- 19.Kulshreshtha, R., Ferracin, M., Wojcik, S. E., Garzon, R., Alder, H., Agosto-Perez, F. J., Davuluri, R., Liu, C. G., Croce, C. M., Negrini, M., Calin, G. A., and Ivan, M. (2006) Mol. Cell. Biol. 28 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebert, C., Norris, K., Scheper, M. A., Nikitakis, N., and Sauk, J. J. (2007) Mol Cancer 6 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua, Z., Lv, Q., Ye, W., Wong, C. K., Cai, G., Gu, D., Ji, Y., Zhao, C., Wang, J., Yang, B. B., and Zhang, Y. (2006) PLoS ONE 1 e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donker, R. B., Mouillet, J. F., Nelson, D. M., and Sadovsky, Y. (2007) Mol. Hum. Reprod. 13 273-279 [DOI] [PubMed] [Google Scholar]
- 23.Camps, C., Buffa, F. M., Colella, S., Moore, J., Sotiriou, C., Sheldon, H., Harris, A. L., Gleadle, J. M., and Ragoussis, J. (2008) Clin. Cancer Res. 14 1340-1348 [DOI] [PubMed] [Google Scholar]
- 24.Giannakakis, A., Sandaltzopoulos, R., Greshock, J., Liang, S., Huang, J., Hasegawa, K., Li, C., O'Brien-Jenkins, A., Katsaros, D., Weber, B. L., Simon, C., Coukos, G., and Zhang, L. (2007) Cancer Biol. Ther. 7 255-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng, A. M., Byrom, M. W., Shelton, J., and Ford, L. P. (2005) Nucleic Acids Res. 33 1290-1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandey, A., Shao, H., Marks, R. M., Polverini, P. J., and Dixit, V. M. (1995) Science 268 567-569 [DOI] [PubMed] [Google Scholar]
- 27.Davis, S., Gale, N. W., Aldrich, T. H., Maisonpierre, P. C., Lhotak, V., Pawson, T., Goldfarb, M., and Yancopoulos, G. D. (1994) Science 266 816-819 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data