Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells (original) (raw)
Abstract
The claudin-low subtype is a recently identified rare molecular subtype of human breast cancer that expresses low levels of tight and adherens junction genes and shows high expression of epithelial-to-mesenchymal transition (EMT) genes. These tumors are enriched in gene expression signatures derived from human tumor-initiating cells (TICs) and human mammary stem cells. Through cross-species analysis, we discovered mouse mammary tumors that have similar gene expression characteristics as human claudin-low tumors and were also enriched for the human TIC signature. Such claudin-low tumors were similarly rare but came from a number of distinct mouse models, including the p53 null transplant model. Here we present a molecular characterization of 50 p53 null mammary tumors compared with other mouse models and human breast tumor subtypes. Similar to human tumors, the murine p53 null tumors fell into multiple molecular subtypes, including two basal-like, a luminal, a claudin-low, and a subtype unique to this model. The claudin-low tumors also showed high gene expression of EMT inducers, low expression of the miR-200 family, and low to absent expression of both claudin 3 and E-cadherin. These murine subtypes also contained distinct genomic DNA copy number changes, some of which are similarly altered in their cognate human subtype counterpart. Finally, limiting dilution transplantation revealed that p53 null claudin-low tumors are highly enriched for TICs compared with the more common adenocarcinomas arising in the same model, thus providing a unique preclinical mouse model to investigate the therapeutic response of TICs.
Keywords: genetically engineered mouse model, gene profiling, array comparative genomic hybridization
Breast cancer (BC) is the second leading cause of cancer-related deaths among women in the United States (1). The large compendium of underlying genetic alterations and the resulting histological and molecular subtypes illustrate the heterogeneous nature of this disease. Both this intertumor heterogeneity and the cellular heterogeneity found within a breast tumor (intratumor heterogeneity) are major obstacles to effective treatments. One common feature of BC (and most cancers) is the loss of the tumor suppressor p53 function. p53 has been shown to be mutated in ≈40% of BCs, associated with poor clinical outcomes, and a higher frequency of mutations occurs in more-aggressive molecular subtypes, including the basal-like subtype of human BC (2).
Mice homozygous for p53 loss have been shown to develop lymphomas and sarcomas with a short latency (3, 4). When crossed into the BALB/c background, mammary tumors were observed in p53+/− mice (5). To circumvent the appearance of other tumor types that occurred with short latency, the model was further modified (6); namely, 6-wk-old p53−/− glands were removed and transplanted into 3-wk-old wild-type BALB/c recipients. These mice develop mammary tumors stochastically with an average latency of approximately 12 mo. Interestingly, the p53 null epithelium initially forms normal ductal growths, which exhibit few genetic changes compared with wild-type outgrowths (7). Unlike many transgenic mouse models, the p53 null tumor model exhibits histological heterogeneity reminiscent of human BCs, including a subset of the tumors expressing the estrogen receptor (ER). In addition antiestrogens are able to significantly delay tumor formation in this model (8). Last, these p53-deficient tumors exhibit genetic instability and/or aneuploidy, which likely play a critical role in progression (6).
Using gene expression profiling for classification we show that, like human tumors, p53 null mouse mammary tumors fall into multiple molecular groups, including basal-like, luminal, and claudin-low subtypes. The claudin-low tumors contain a majority of spindle-shaped cells, a histology originally described for carcinosarcomas, which are now called epithelial-to-mesenchymal transition (EMT) tumors (9). Like their human counterparts, the p53 null claudin-low tumors exhibit high expression of EMT inducers and a core EMT signature (10). Unlike many other mouse tumor models, p53 null tumors show extensive genomic instability. Accordingly, we determined that these different p53-deficient murine subtypes contain distinct genomic DNA copy number changes, as assessed by array comparative genomic hybridization (aCGH). We also show, by limiting dilution transplantation, that p53 null claudin-low tumors have a marked enrichment of functional tumor-initiating cells (TICs). These data show the utility of this heterogeneous tumor model and provide functional data further demonstrating the stem cell characteristics of the claudin-low subtype.
Results
p53 Null Tumors Show Variable Histology.
Previously, we hypothesized that the heterogeneity observed in human BC might arise not only because of activation of different oncogenes or loss of specific tumor suppressor genes, but might also be dependent on the cell of origin in which these genetic changes occur (11). Initially transplanted p53 null mammary epithelial cells gave rise to phenotypically normal ductal outgrowths, which then stochastically developed mammary tumors. We therefore hypothesized that the deletion of this single tumor suppressor gene might give rise to a spectrum of heterogeneous tumors, depending on the cell of origin in which additional stochastic changes occurred. To test this hypothesis, we collected 50 p53 null tumors that arose in wild-type BALB/c mice after transplantation of p53 null BALB/c mammary tissue into the cleared fat pads of 3-wk-old mice (6). Like some other genetically engineered mouse model (GEMM) mammary tumor models, the p53 null model gave rise to tumors with a diversity of histological phenotypes (Fig. S1 and Dataset S1) (9, 12). Approximately 10% of the tumors contained a majority of spindle-shaped cells, a histology originally described for carcinosarcomas, now called EMT tumors (9).
p53 Null Tumors Cluster into Distinct Tumor Subtypes.
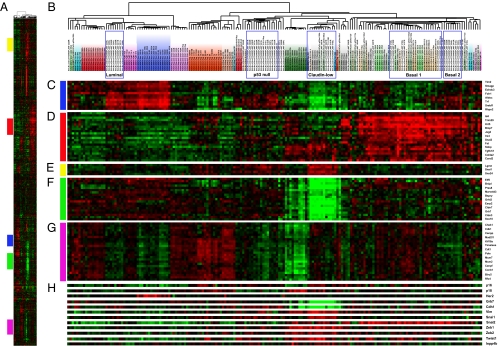
In a previous study we profiled 13 distinct mouse models, including the p53 null model (13). However, with only five p53 null tumor samples, we were not able to appreciate the full spectrum of molecular heterogeneity represented in this mouse model. Now, with a total of 50 tumors from the p53 null model, we see that these tumors cluster into five distinct tumor subtypes when performing hierarchical clustering analysis using our previously defined mouse intrinsic gene list (13) (Fig. 1); furthermore, we used SigClust (14) to assess the significance of this clustering and objectively determined that the p53 null model did populate multiple statistically significant groups/subtypes, as follows.
Fig. 1.
Intrinsic gene set clustering analysis of 50 p53 null tumors and 117 samples from 13 GEMM previously published in Herschkowitz et al. (13). (A) Overview of the complete cluster diagram. (B) Experimental sample-associated dendrogram. Boxes indicate the p53 null tumor subtypes based on SigClust analysis. (C) Luminal epithelial expression pattern that is highly expressed in luminal p53 null tumors, MMTV-Neu, and MMTV-PyMT tumors (D). Basal epithelial expression pattern including K5 and ID4, which are highly expressed in basal-like p53 null tumors. (E) Mesenchymal genes, including snail homolog 1. (F) Genes expressed at low levels in claudin-low tumors, including CLDN3, CLDN7, and ELF5. (G) Proliferation signature. (H) Individual genes discussed within the text.
Basal-like.
Two groups of basal-like mouse mammary tumors were observed (Fig. 1); in part, we define these groups as basal-like according to their high expression of known basal markers, including keratin 5 (K5), ID4, and TRIM29 (Fig. 1_D_) and selective high expression of the human basal-like tumor expression cluster (Fig. S2). Basal 1 tumors (5 of 50, 10%) clustered along with a group of other mouse basal-like tumors, including BRCA1-deficient and MMTV-Wnt1 tumors. Basal 2 tumors (8 of 50, 16%) clustered next to the Basal 1 tumors but showed a higher expression of the murine luminal cluster than did Basal 1 (Fig. 1_C_). Basal 1 p53 null tumors showed an increased proliferation signature separating them from Basal 2 and the other p53 null subtypes (Fig. 1_G_ and Fig. S2), and they also showed high p16 expression, which is a hallmark of impaired RB1 function (15). Basal tumors (eight of nine) tested stained positively for K5, as expected (Fig. S3 and Dataset S1); however, paradoxically, five of eight tested stained positively for the ER, of which four of five were of the Basal 2 subtype.
Luminal.
Eight of 50 p53 null tumors (16%) clustered close together and with the mouse luminal models MMTV-Neu and MMTV-PyMT. As we have seen for other luminal models, these tumors express luminal-specific genes like XBP1 but are missing ER and estrogen responsive genes; accordingly, only one of eight of the luminal tumors stained positively for ER. Interestingly, like human luminal tumors, p53 null luminal tumors showed lower levels of p18INK4C, and p18 null mice develop predominantly luminal-type mammary tumors (16).
Claudin-low.
Five of 50 p53 null tumors (10%) showed the murine claudin-low expression phenotype (Fig. 1_F_) and significantly clustered with the previously defined murine claudin-low tumors. These tumors had an EMT tumor histology (Fig. S1) and showed expression of the human claudin-low signature (Fig. S2). In agreement with the gene expression data and immunohistochemistry on human samples (17), we observed low to absent expression of CLDN3 and CDH1 by immunofluorescent staining (Fig. S3). Like human claudin-low tumors, these tumors highly express markers of EMT (17) and the previously determined EMT core signature (Fig. 2_A_) (10). All p53 null tumors tested stained positively for the luminal marker keratin 8 (K8), including the claudin-low tumors (thus suggesting an epithelial origin); however, they often exhibited comparatively less staining (Fig. S3).
Fig. 2.
p53 null claudin-low tumors have features of EMT. These tumors (A) express core EMT signature, EMT markers, and (B) showed marked down-regulation of miRNAs involved in negative regulation of stemness and EMT. Three technical replicates were averaged for each. 200a (P < 0.0001), 200b (P = 0.0002), 200c (P < 0.0001), 141 (P = 0.0005), 429 (P < 0.0001), 205 (P = 0.004), 182 (P = 0.004), 96 (P = 0.005), 183 (P < 0.0001), 203 (P = 0.04).
P53 null subtype.
Thirteen of 50 p53 null tumors (26%) clustered into a unique group made up exclusively of tumors from this BALB/c p53 null model. Tumors of this subtype seem to not show high expression of any of the other tumor subtype defining clusters. Last, 11 of 50 p53 null tumors clustered separately from these five groups and without a consistent group signature. Thus, at least five expression subtypes/phenotypes can be found from this single murine model, three of which mimic previously known human tumor subtypes (luminal, basal-like, and claudin-low).
miRNAs.
Because expression of a number of specific miRNAs has been associated previously with an EMT transition (18, 19), we took a candidate approach to identify miRNAs that were differentially expressed between p53 null claudin-low tumors and the other subtypes. First we evaluated the miR-200 family of miRNAs and miR-205, which are miRNAs that have been implicated in EMT and TICs (18, 20). Although a number of targets for these miRNAs have now been identified, important targets with respect to EMT are ZEB1 and ZEB2. ZEB1 and ZEB2 are expressed at high levels in claudin-low tumors (mouse and human), and as expected, these miRNAs were present at very low levels relative to the other p53 null tumors (Fig. 2_B_). Another cluster of miRNAs expressed at lower levels in both cancer and normal mammary stem cells contains miRNAs 182, 96, and 183 (20). Likewise, this cluster of miRNAs was expressed at low levels in murine p53 null claudin-low tumors. Additionally, miR-203, another stemness-inhibiting miRNA regulated by Zeb1 (21), was also expressed at low levels in claudin-low tumors. Marked decreases, however, were not seen for all miRNAs tested (e.g., miR-21).
It has been shown that human breast tumor subtype correlates with miRNA profiles (22, 23). We reanalyzed the Blenkiron et al. dataset (22) to determine which tumors contained the claudin-low gene expression pattern using the Prat claudin-low predictor (17). Using a supervised analysis, 17 miRNAs were identified that were significantly differentially expressed between claudin-low tumors vs. the other BCs (Dataset S2). This included seven of the miRNAs that we had observed, including miR-200a, 200b, 200c, 149, 182, 183, and 203. These results indicate that in addition to mRNA gene expression changes, mouse and human claudin-low tumors share common miRNA expression patterns.
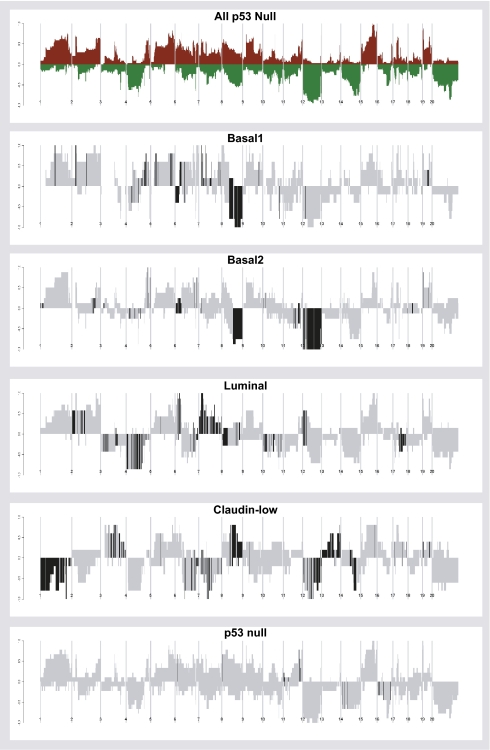
p53 Null Tumor Subtypes Display Distinct Copy Number Alterations.
Presumably stochastic genetic alterations selected during neoplastic progression collaborate with the loss of p53. It is also likely that different genetic events can cause tumors to show a given phenotype, or only sensitize one particular cell type to malignant transformation; thus, specific copy number and/or mutations may be highly enriched within a specific tumor subtype, as shown for human breast tumors (24). To investigate this on the genomic level, we performed aCGH on 44 p53 null tumors using Agilent 244,000 feature DNA microarrays to determine whether there were subtype-specific copy number alterations (CNAs) (Fig. 3). In comparison with many mouse models (25, 26) the p53 null mammary tumors contained a fair amount of genomic instability. Interestingly, all five tumor subtypes contain distinct CNAs, which are listed by subtype in Dataset S3. In the p53 null basal-like tumors (both Basal 1 and Basal 2 considered together), there was loss of the distal half of chromosome 8, including INPP4B, which has now been shown to be selectively lost in human basal-like/triple-negative tumors [4q31.22–q35.2(12)] (27, 28). p53 null luminal tumors showed loss of chromosome 4 and gain of chromosome 7. The p53 null unique subtype showed very few subtype-specific events; however, when converted to human genomic coordinates, these events identify amplification of human chromosome 17q12–q21.2(2), which is a common amplicon that is distal to the HER2 amplicon. Interestingly, one of these murine tumors (2304L) that clusters in the p53 null unique subtype, but that is not contained within the SigClust defined group (Fig. 1_B_), showed high Her2 mRNA and protein expression and was amplified for Her2 on mouse chromosome 11 (Fig. S4); thus, the p53 null model is even able to generate HER2-amplified tumors, albeit at a low frequency.
Fig. 3.
Tumor genomic DNA copy number landscape plots for mouse p53 null tumor classes. At top is the overall pattern for all 34 tumors considered together, and then below are the landscape plots for each of the five expression-defined subtypes. Gray shading indicates the overall frequency of aberrations seen in that subtype, and the black shading indicates the group-specific CNA (P value threshold 0.05).
The copy number landscape of human claudin-low tumors is not known, but the p53 null claudin-low tumors showed numerous subtype-enriched CNAs. These changes included the near-complete loss of mouse chromosome 1 and frequent but smaller losses on 7, 12, and 14. There were also specific gains on 3, 8, and 13. Many of these map to common regions of copy number changes in human BC; however, additional studies will be required to define the driving mutations/changes present in each region. Nonetheless, these claudin-low subtype-specific copy number changes do suggest the possible existence of driver mutations/changes.
The work of Bergamaschi et al. (24) identified numerous CNAs associated with some of the intrinsic subtypes. We, therefore, converted our mouse CNAs into human equivalent chromosome locations and determined that a number of significantly altered regions were in common between p53 null mouse tumors and human breast tumors (Dataset S4). Of note were the loss of two regions that occurred in both mouse and human basal-like tumors, human 4q31.22–q35.2(12) that contains INPP4B, and human 14q22.1–q23.1(4); the somatic loss of these two regions across species suggests that each contains a tumor suppressor(s) gene and that the loss of these genes may sensitize cells to become the basal-like subtype, similar to germline inactivation of BRCA1 (29).
p53 Null Claudin-Low Tumors Are Enriched for Tumor-Initiating Cells.
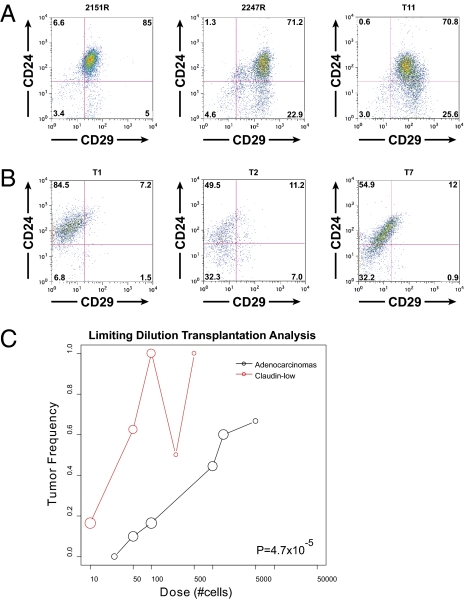
Similar to their utility in the isolation of mouse mammary stem cells (30), CD29 and CD24 have been used as markers that enrich for TICs in the p53 null tumor model, with the CD29+/CD24+ fraction showing the TIC capabilities (12). Furthermore, an EMT program has been shown to correlate with stem-like properties, and the loss of miR-200 expression as well as a “claudin-low” signature has been suggested to characterize both normal and cancer stem cells (20). By FACS analysis, in the p53 null claudin-low tumors tested, the percentage of double-positive cells was 70–85%, as compared with a maximum of 14% in the other p53 null tumors analyzed (Fig. 4 A and B) Interestingly, some luminal tumors exhibit very low levels of double-positive cells. This was suggestive, therefore, that there might be a high percentage of TICs in the claudin-low tumors.
Fig. 4.
p53 null claudin-low tumors express markers of stem cells and are enriched for tumor-initiating ability. (A) Claudin-low tumors have high percentages of double-positive (CD29+/CD24+) cells compared with (B) other p53 null tumors. (C) Limiting dilution transplantation of claudin-low vs. adenocarcinoma cells. Sample sizes are implied by the sizes of the circles (area is proportional to sample size).
To test this hypothesis, two different claudin-low p53 null tumors were FACS sorted for all four possible populations using CD29 and CD24, and limiting dilution transplantation was performed (Tables 1 and 2). The tumor-initiating frequency was similar between the CD29+/CD24+ and CD29+/CD24− fractions, and these two populations were highly enriched for TICs compared with the other two fractions. In addition, by transplanting FACS-sorted lineage-negative cells in limiting dilution, we determined that the tumor repopulating ability of these claudin-low phenotype tumors was >38 times greater than that of two other p53 null adenocarcinomas (T1 and T7) performed using the same methods (Fig. 4_C_) (31). Thus, these data indicate that an expanded population of TICs exists within these murine claudin-low tumors.
Table 1.
Limiting dilution transplantation of adenocarcinomas (T1 and T7)
| Cells injected | 5,000 | 1,500 | 1,000 | 100 | 50 | 25 | 10 |
|---|---|---|---|---|---|---|---|
| Lin−CD29HCD24H | 4/4 | 2/2 | 12/12 | 12/12 | 4/12 | 3/12 | |
| Lin−CD29HCD24L | 2/4 | 4/6 | 4/12 | 2/12 | 0/8 | 0/6 | |
| Lin−CD29LCD24H | 4/5 | 2/8 | 2/7 | 0/8 | 0/6 | 0/2 | 0/6 |
| Lin−CD29LCD24L | 2/6 | 2/8 | 0/7 | 0/8 | 0/6 | 0/2 | 0/6 |
| Lin− | 2/3 | 6/10 | 4/9 | 2/12 | 1/10 | 0/4 |
Table 2.
Limiting dilution transplantation of claudin-low tumors (T11 and 2247R)
| Cells injected | 500 | 250 | 100 | 50 | 10 |
|---|---|---|---|---|---|
| Lin−CD29HCD24H | 2/2 | 2/2 | 5/6 | 5/6 | 3/6 |
| Lin−CD29HCD24L | 1/2 | 2/2 | 5/6 | 5/6 | 3/4 |
| Lin−CD29LCD24H | 2/2 | 2/4 | 0/4 | 2/6 | 0/2 |
| Lin−CD29LCD24L | 0/2 | 3/4 | 2/4 | 2/6 | 0/2 |
| Lin− | 2/2 | 1/2 | 8/8 | 5/8 | 2/12 |
Discussion
GEMM have provided a rich resource for the study of different cancers; however, many individual models show significant molecular and histological heterogeneity (13). This heterogeneity complicates studies because multiple disease types may actually be present within a given model. One way to address this heterogeneity is to genomically characterize each tumor, then group tumors together according to important features and, most importantly, perform functional studies. The p53 null mammary transplant model is one such heterogeneous model, and we have taken advantage of this feature and identified transplantable lines that represent at least three human breast tumor subtypes. In addition, because all these tumors develop subsequent to the same initial loss of p53, the question is whether this heterogeneity is due to different collaborating oncogenes/tumor suppressors and/or different cells of origins. The cell type of origin in cancer is a highly debated topic (reviewed recently in ref. 32). Although specific genetic lesions clearly play a major role in determining the tumor phenotype, growing evidence indicates that cancers of different subtypes within an organ system may also reflect distinct cells of origin. However, it is not apparent whether a given oncogenic lesion actually dictates the cell of origin or, conversely, whether the cell of origin determines which oncogenic lesions can occur. Both of these possibilities most likely exist. There is evidence that tumors generated using the same oncogene targeted to different cell lineages can be phenotypically distinct (33). Recent studies have shown that BRCA1 mutant and basal-like human tumors are enriched in gene expression profiles and surface markers of luminal progenitors (34). Similarly, inactivation of BRCA1 (and p53) in the luminal or basal cell population of the mouse mammary gland showed that only the luminal cells gave rise to tumors histologically resembling those of BRCA1 mutation carriers (35). These results, however, fall short of actually proving that these tumor types originated in these cell types. Mouse models, like the heterogeneous one presented here, can provide an invaluable tool with which to decipher the cell of origin when genetics is combined with precise lineage tracing. At present we cannot definitively answer the cell-of-origin question because the necessary reagents are not yet available to perform the lineage tracing experiments, as has been done recently using mouse models of intestinal cancer (36). However, our experiments do provide several important insights: first, tumors of the basal-like, luminal, and claudin-low phenotypes clearly arise, although at different frequencies and with a predilection for basal-like; in particular, the Basal 1 group seems to most faithfully recapitulate human basal-like tumors, in that it shows high expression of basal gene expression features, of the proliferation signature, and of p16 (a hallmark of RB1 loss), all of which are features of human basal-like tumors (15). Second, the luminal tumors that do arise are largely ER negative (as are the vast majority of murine tumors from other GEMMs) and thus fundamentally more similar to a luminal B than the ER-positive luminal A human subtype.
Interestingly, claudin-low p53 null tumors were also seen, although at the lowest frequency (five total). As was shown for human claudin-low tumors and cell lines, these murine tumors lack tight junction proteins, including claudin 3 and E-cadherin, and show expression features of mesenchymal cells, normal mammary stem cells, and TIC. In addition to previously defined subtypes, we also identified a phenotype unique to this model and noted that nearly 20% of the tumors were scattered throughout the cluster, indicating even greater heterogeneity within this model. For example, tumor 2304L showed clear amplification and high expression of HER2, thus even somatically HER2 amplified tumors occur within this model.
The presence of specific CNAs in different subtypes of tumors arising in the p53 null model suggests that different gains and losses are important for tumor progression subsequent to p53 loss, and these are possibly occurring within different cell types. In the p53 null basal-like tumors, there was specific loss of the distal half of chromosome 8, which is in conserved synteny with human chromosome 4. Recently, loss of this region has been seen specifically with human basal-like/triple-negative BCs, and it is thought that the target of this loss is INPP4B. This gene is selectively low in human and murine basal-like tumors, thus suggesting that this approach of finding common regions of loss/gains across species can identify putative important tumor and/or subtype causative events. Interestingly, p53 null luminal tumors showed loss of chromosome 4. Chromosome 4 deletions and loss of heterozygosity have been reported in other luminal mouse models, including MMTV-Neu, MMTV-Myc, and MMTV-Ras (26, 37–39). Presumably there exist multiple luminal-specific tumor suppressor genes on chromosome 4. Although other subtypes showed gain of chromosome 1, p53 null claudin-low tumors showed large regions of loss on chromosome 1, which again hints at their uniqueness.
Several lines of evidence have suggested that claudin-low tumors are enriched in functional TICs, predominantly coming from expression analyses (Fig. 2_A_ and Fig. S2). However, because of their rarity and limitations in procurement of primary human claudin-low tumors, this hypothesis has not been tested functionally using human clinical samples. We have, however, herein identified a counterpart of human claudin-low tumors in the mouse. Accordingly, we have taken advantage of this mouse model to test by limiting dilution, the gold standard functional stem cell assay, whether these tumors are enriched in TICs compared with other tumors arising in the same model. With the p53 null model we also have the advantage of being able to transplant these tumors into syngeneic mice with an appropriate microenvironment complete with normal immune function. We showed that the claudin-low murine tumors were significantly more enriched for surface markers of TICs as well as functional TICs compared with other p53 null tumors. Recent studies have shown that minority subsets of tumors from MMTV-Myc and MMTV-MET tumors cluster with our claudin-low mouse tumors (40, 41). It has not been determined whether they too are enriched in functional TICs. However, the MMTV-Myc EMT-like/claudin-low tumors were reported to show an increase in metastasis.
The murine claudin-low tumors show large percentages of CD29+/CD24+ cells, MaSC-like mRNA, and miRNA expression profiles, as well as expression of other markers of MaSCs (e.g., high s-SHIP expression) (42). Therefore, it is conceivable that these tumors might have arisen from the MaSC population. Alternatively, they may have resulted from dedifferentiation of a progenitor or even a more differentiated cell. Lineage tracing experiments will be required to definitively resolve this issue.
To effectively target cancer stem cells or TICs, one pressing need is a genetically defined and renewable preclinical model to identify and test new stem cell targeted therapies. To address this need, we now have identified a mouse model that develops claudin-low tumors, in which the bulk of the tumors cells seem to be TICs. This is an example of a spontaneously occurring breast tumor with a high proportion of TICs. Thus, we now have appropriate and validated models for the investigation of important signaling pathways and therapeutics. Because of their transplantability into syngeneic hosts, this panel of tumors provides a valuable resource for preclinical testing of novel therapeutics. These tumors should serve as excellent models for both the general study of BC stem cells and preclinical models for testing stem cell targeted agents, enabling translation into the clinic. Finally, the finding that claudin-low tumors have an enrichment of functional TICs challenges the popular paradigm that TICs always need be a rare subpopulation (43).
Materials and Methods
Mice.
All animal protocols were reviewed and approved by the Animal Protocol Review Committee at Baylor College of Medicine and University of North Carolina, Chapel Hill.
Preparation of Single Cells.
Tumors were processed and digested into single cells as previously described (12). The cells were resuspended in HBSS (Invitrogen) containing 2% FBS and 10 mM Hepes buffer (Invitrogen) before labeling with antibodies.
Flow Cytometry.
Cells were labeled with antibodies (Dataset S5) at a concentration of 10 × 106 cells/mL under optimized conditions and were subjected to FACS analysis and sorting on an ARIA II sorter (BD Biosciences). Data analysis was performed using FlowJo (v9.1).
Transplantation.
Clearance of mammary epithelial cells and transplantation procedures were performed as previously described (44). After FACS, the designated number of cells were washed once with PBS and transplanted into the cleared fat pads of 21-d-old female BALB/c mice (Harlan).
Microarray Analysis.
Total RNA was collected from 45 murine tumors and purified using the Qiagen RNeasy Mini Kit using ≈25 mg tissue. Two micrograms of total RNA was reverse transcribed, amplified, and labeled with Cy5 using a Low RNA Input Amplification kit (Agilent). The common reference RNA (45) was reverse transcribed, amplified, and labeled with Cy3. They were then cohybridized overnight to Agilent Mouse Oligo 4x44K Microarrays. Finally, they were washed and scanned on an Agilent scanner (G2505B) and uploaded into the database, where a Lowess normalization is automatically performed. The genes for all analyses were filtered by requiring the Lowess normalized intensity values in both channels to be >10. The log2 ratio of Cy5/Cy3 was then reported for each gene. In the final dataset, only genes that reported values in 70% or more of the samples were included.
Supplementary Material
Supporting Information
Acknowledgments
We thank the Cytometry and Cell Sorting Core Facility at Baylor College of Medicine for flow cytometry assistance. This work was supported by National Cancer Institute (NCI) Grant Multi-PI RO1CA148761 (to J.M.R and C.M.P), NCI work contract N01-CN43308 (to C.M.P), the Breast Research Foundation (C.M.P), and Komen Postdoctoral Fellowship PDF0707744 (to J.I.H).
Footnotes
Conflict of interest statement: C.M.P. has an equity interest in University Genomics and BioClassifier LLC.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27101).
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 4.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 5.Kuperwasser C, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–2159. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jerry DJ, et al. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene. 2000;19:1052–1058. doi: 10.1038/sj.onc.1203270. [DOI] [PubMed] [Google Scholar]
- 7.Aldaz CM, et al. Serial analysis of gene expression in normal p53 null mammary epithelium. Oncogene. 2002;21:6366–6376. doi: 10.1038/sj.onc.1205816. [DOI] [PubMed] [Google Scholar]
- 8.Medina D, et al. Tamoxifen inhibition of estrogen receptor-alpha-negative mouse mammary tumorigenesis. Cancer Res. 2005;65:3493–3496. doi: 10.1158/0008.5472.CAN-04-3869. [DOI] [PubMed] [Google Scholar]
- 9.Cardiff RD. The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2010;15:225–233. doi: 10.1007/s10911-010-9184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15549–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76–R76.17. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Hayes DN, Nobel A, Marron JS. Statistical significance of clustering for high-dimension, low-sample size data. J Am Stat Assoc. 2008;103:1281–1293. [Google Scholar]
- 15.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei XH, et al. CDK inhibitor p18(INK4c) is a downstream target of GATA3 and restrains mammary luminal progenitor cell proliferation and tumorigenesis. Cancer Cell. 2009;15:389–401. doi: 10.1016/j.ccr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 19.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 20.Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 22.Blenkiron C, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214–R214.16. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greene SB, Herschkowitz JI, Rosen JM. Small players with big roles: MicroRNAs as targets to inhibit breast cancer progression. Curr Drug Targets. 2010;11:1059–1073. doi: 10.2174/138945010792006762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergamaschi A, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 25.Donehower LA, et al. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 26.Hodgson JG, et al. Copy number aberrations in mouse breast tumors reveal loci and genes important in tumorigenic receptor tyrosine kinase signaling. Cancer Res. 2005;65:9695–9704. doi: 10.1158/0008-5472.CAN-05-0755. [DOI] [PubMed] [Google Scholar]
- 27.Fedele CG, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci USA. 2010;107:22231–22236. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gewinner C, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proc Natl Acad Sci USA. 2010;107:3522–3527. doi: 10.1073/pnas.0910179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 33.Du Z, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci USA. 2006;103:17396–17401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim E, et al. kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 35.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 37.Montagna C, Andrechek ER, Padilla-Nash H, Muller WJ, Ried T. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene. 2002;21:890–898. doi: 10.1038/sj.onc.1205146. [DOI] [PubMed] [Google Scholar]
- 38.Radany EH, Hong K, Kesharvarzi S, Lander ES, Bishop JM. Mouse mammary tumor virus/v-Ha-ras transgene-induced mammary tumors exhibit strain-specific allelic loss on mouse chromosome 4. Proc Natl Acad Sci USA. 1997;94:8664–8669. doi: 10.1073/pnas.94.16.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver ZA, et al. A recurring pattern of chromosomal aberrations in mammary gland tumors of MMTV-cmyc transgenic mice. Genes Chromosomes Cancer. 1999;25:251–260. [PubMed] [Google Scholar]
- 40.Andrechek ER, et al. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci USA. 2009;106:16387–16392. doi: 10.1073/pnas.0901250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponzo MG, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci USA. 2009;106:12903–12908. doi: 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai L, Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24:1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 44.Medina D. The mammary gland: A unique organ for the study of development and tumorigenesis. J Mammary Gland Biol Neoplasia. 1996;1:5–19. doi: 10.1007/BF02096299. [DOI] [PubMed] [Google Scholar]
- 45.He XR, Zhang C, Patterson C. Universal mouse reference RNA derived from neonatal mice. Biotechniques. 2004;37:464–468. doi: 10.2144/04373RT02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information