The Acid Phosphatase AcpA Is Secreted In Vitro and in Macrophages by Francisella spp (original) (raw)
Abstract
Francisella tularensis is a remarkably infectious facultative intracellular pathogen that causes the zoonotic disease tularemia. Essential to the pathogenesis of F. tularensis is its ability to escape the destructive phagosomal environment and inhibit the host cell respiratory burst. F. tularensis subspecies encode a series of acid phosphatases, which have been reported to play important roles in Francisella phagosomal escape, inhibition of the respiratory burst, and intracellular survival. However, rigorous demonstration of acid phosphatase secretion by intracellular Francisella has not been shown. Here, we demonstrate that AcpA, which contributes most of the F. tularensis acid phosphatase activity, is secreted into the culture supernatant in vitro by F. novicida and F. tularensis subsp. holarctica LVS. In addition, both F. novicida and the highly virulent F. tularensis subsp. tularensis Schu S4 strain are able to secrete and also translocate AcpA into the host macrophage cytosol. This is the first evidence of acid phosphatase translocation during macrophage infection, and this knowledge will greatly enhance our understanding of the functions of these enzymes in Francisella pathogenesis.
INTRODUCTION
Francisella tularensis is a highly infectious bacterium that can cause tularemia in both humans and animals. As few as 10 F. tularensis subsp. tularensis (type A) bacteria can cause disease in humans via the pulmonary route, and if untreated, the infection results in high mortality (42). As intracellular pathogens, Francisella spp. are able to infect many cell types, including mononuclear phagocytes, epithelial cells, and hepatocytes (20). Among these cell types, macrophages are the major target for infection in vivo (26). Francisella spp. induce the formation of spacious pseudopod loops on macrophage surfaces which result in phagocytosis by these cells (17). Inside macrophages, _Francisella_-containing phagosomes go through a transient maturation process by sequential acquisition of early and late endosomal markers (16, 52, 53). However, Francisella spp. can remove themselves from the destructive endosomal system by rapid phagosomal escape, followed by replication within the host cell cytosol (16, 52, 53). Phagosomal escape is a critical step for Francisella sp. pathogenesis. Francisella sp. mutants that are defective or delayed in phagosomal escape have impaired intracellular replication and are also attenuated in vivo in mouse models (7, 15, 30, 54, 56, 57). Many of these attenuating mutations have been localized to genes (e.g., iglC, iglD, and pdpA) in a 30-kb chromosomal locus known as the Francisella pathogenicity island (FPI) (36). The expression of FPI genes is regulated by many transcriptional regulators (18), including MglA (29), SspA (12), FevR (8, 11), MigR (9), PmrA (6, 39), and Hfq (37, 45). The FPI is thought to encode a type VI secretion system that is required for phagosomal disruption and intracellular replication (5), but FPI proteins can also be secreted by FPI-independent mechanisms, including type I secretion (2, 24), a type IV pilus secretion system (25), and a Sec-dependent secretion system (33). In addition, Francisella spp. also produce outer membrane vesicles, which contain many of the virulence proteins, including IglA, IglB, IglC, PdpA, PdpB, and PdpD (44). However, the exact mechanisms involved in outer membrane vesicle production are not known.
Acid phosphatases are ubiquitous in nature and hydrolyze the phosphoryl groups of phosphomonoesters at an acid pH (58). These enzymes are essential in the production, acquisition, and mobilization of inorganic phosphate and in phosphorelay systems involved in signal transduction pathways in both prokaryotes and eukaryotes. Acid phosphatases from other pathogens have also been associated with inhibition of the respiratory burst, suggesting their involvement in pathogenesis (1, 3, 10, 27, 48, 50, 51). Francisella spp. have at least four acid phosphatases, AcpA, AcpB, AcpC, and Hap, among which AcpA is the major contributor of the acid phosphatase enzyme activity of Francisella spp. (14, 38, 41). AcpA has been extensively characterized biochemically and structurally (21, 22, 47). Initial work with AcpA demonstrated that it had the ability to inhibit the oxidative burst in porcine neutrophil extracts (47). The collective loss of all four acid phosphatases in F. novicida dramatically affected phagosomal escape, intramacrophage survival, and virulence while abrogating the innate ability of Francisella spp. (F. novicida, F. tularensis LVS, and F. tularensis Schu S4) to suppress the oxidative burst (38, 40, 41). AcpA also has direct phosphatase activity in vitro with purified, phosphorylated NADPH oxidase components p40phox and p47phox (40). Transcriptional analysis demonstrated that AcpA and Hap expression was induced rapidly after phagocytosis in macrophages (41). Thus, the collective data provide evidence that the Francisella sp. acid phosphatases, of which AcpA contributes the most activity, are important in the intracellular lifestyle of this organism by aiding intraphagosomal escape and/or survival against oxidative stresses.
Francisella sp. AcpA has been described to be an outer membrane protein (38). This makes the enzyme activity accessible to NADPH oxidase components in the host cytosol when the bacterium is released from phagosomes. However, inhibition of the host respiratory burst occurs rapidly following phagocytosis, while phagosomal escape does not take place until at least 30 min postinfection. This led us to hypothesize that AcpA is secreted and translocated to the host cytosol before the occurrence of phagosomal escape. Here, we show that Francisella sp. AcpA is secreted into the culture supernatant in vitro and is also secreted and translocated within macrophages across the phagosomal membrane into the host cell cytosol at an early stage by both F. novicida and the highly virulent type A strain Schu S4.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains, plasmids, and primers used in the present study are listed in Table 1. Escherichia coli DH5α was routinely cultured in Luria-Bertani (LB) medium (Difco Laboratories, Detroit, MI) supplemented with kanamycin (45 μg/ml) or tetracycline (15 μg/ml) when required. LVS was cultured on Chocolate II agar plates (Becton Dickinson, Sparks, MD). F. novicida U112 was routinely cultured on Chocolate II plates and in modified tryptic soy broth (TSB) (Difco Laboratories) containing 135 μg/ml ferric pyrophosphate and 0.1% cysteine hydrochloride. The virulent F. tularensis subsp. tularensis (type A) strain Schu S4 was routinely cultured on Chocolate II plate or modified Mueller-Hinton (MMH) plates with 0.1% glucose, 0.025% ferric pyrophosphate, and 2% IsoVitaleX (Becton Dickinson). When needed, the TSB or MMH medium was supplemented with tetracycline (10 μg/ml) or kanamycin (25 μg/ml). Plasmids were introduced into F. novicida U112 and F. tularensis subsp. tularensis Schu S4 by cryotransformation (38) or electroporation (31), respectively. All work with the type A Schu S4 strain was carried out in The Ohio State University BSL3 Select Agent Facility in accordance with national and local approved biosafety level 3 (BSL3) facility and safety plans. Plasmids conferring kanamycin resistance were used with Schu S4, as we have biosafety approval for the use of this antibiotic in BSL3-requiring strains.
Table 1.
Bacterial strains, plasmids, and primers_a_
| Strain, plasmid, or primer | Description or sequence (5′–3′) | Source or reference |
|---|---|---|
| Francisella strains | ||
| JSG1819 | Francisella novicida U112 | ATCC |
| JSG2660 | JSG1819 with Δ_acpA_::Kan | 38 |
| JSG3236 | JSG1819 with pFnAcpA-FLAG | This study |
| JSG3237 | JSG2660 complemented with pFnAcpA-FLAG | This study |
| FT4 | Francisella tularensis subsp. tularensis Schu S4 | R. Lyons (CDC) |
| FT11 | FT4 with Δ_acpA_::Kan | This study |
| FT43 | FT4 with pFtAcpA | This study |
| FT44 | FT11 complemented with pFtAcpA | This study |
| E. coli DH5α | F− 80d_lacZ_ΔM15 Δ(lacZYA-argF) U169 endA1 recA1 hsdR17 deoR thi-1 supE441 gyrA96 relA1 | BRL |
| Plasmids | ||
| pKEK894 | Francisella expression plasmid (Tetr) | 60 |
| pKEK1157 | pKEK894 expressing FLAG-VgrG | 5 |
| pKEK1191 | pKEK894 expressing ΔNBlaB-FLAG | 5 |
| pFnAcpA-FLAG | pKEK894 expressing _Fn_AcpA-FLAG | This study |
| pFnΔNAcpA-FLAG | pKEK894 expressing _Fn_ΔNAcpA-FLAG | This study |
| pLVSAcpA-FLAG | pKEK894 expressing LVS AcpA-FLAG | This study |
| pFNLTP6 | Francisella expression plasmid (Kanr) | 32 |
| pFtAcpA | pFNLTP6 expressing _Ft_AcpA | This study |
| Primers | ||
| JG1795 | ACGCGTCGACGGAGTTAGTGATTTAGTTGCAATAGGTGTTGC | This study |
| JG1796 | CGCGGATCCGCTTCATATGATACCTTTAGTTGTTAGATTCAAAGGAAATATTAATAAC | This study |
| JG1797 | CGCGGATCCAAATATTTACTCGGTAAGTTGCTTTAATCTAGTATTTTCGC | This study |
| JG1798 | TCCCCCCGGGGCTAAAGATAAGGGCATAAAGACTATCAAAGAGAG | This study |
| JG2007 | CGGAATTCCACTGCAGAGGAGGGTTTTTAATGAAGCTCAATAAAATTACTTTAGGAATTTTAAGTC | This study |
| JG2008 | CCGCTCGAGGTTTAATTTATCCACTACTAATCCTGTCTTAGG | This study |
| JG2191 | GGCCATGGATATGAAGCTCAATAAAATTACTTTAGG | This study |
| JG2192 | GCGGCATATGGTTTAATTTATCCATCACTAATCC | This study |
| JG2193 | GCGGCATATGGTTTAATTTATCCACTACTAATCC | This study |
| JG2422 | GGCCATGGATATGAATAATAGCAAACCAAATGA | This study |
The Schu S4 acpA mutant was constructed as described earlier for the LVS acpA mutant (35). Briefly, a 1-kb fragment upstream and a 1-kb fragment downstream of acpA were amplified with primers JG1795/JG1796 and JG1797/JG1798, respectively, and cloned into the SalI and BamHI, and BamHI and SmaI sites of pJC84, respectively. The resulting plasmid was transformed into Schu S4, and the acpA mutant was identified by sucrose selection and confirmed by PCR and Southern blot analysis.
Plasmids used in this study were derived from Francisella plasmid pKK214. pKEK1191 (Δ_NblaB_ [blaB gene whose product has its 17-amino-acid N-terminal secretion sequence removed]-FLAG) and pKEK1157 (FLAG-vgrG) were kind gifts from Karl Klose (University of Texas San Antonio, San Antonio, TX) and their constructions were described previously (5). The acpA gene from F. novicida U112 was PCR amplified using primers JG2191 and JG2192, and used to replace the Δ_NblaB_ insert by using NcoI and NdeI sites in pKEK1191 to make plasmid pFnAcpA-FLAG. pFnΔNAcpA-FLAG and pLVSAcpA-FLAG were constructed the same way by using primers JG2422/JG2192 and JG2422/JG2193, respectively. The plasmid expressing Schu S4 AcpA, pFtAcpA, was constructed by cloning the PCR product amplified from Schu S4 genomic DNA by using primers JG2007 and JG2008 and subsequent ligation into the EcoRI and XhoI sites of pFNLTP6.
Antibodies.
Mouse monoclonal anti-F. novicida lipopolysaccharide (LPS) primary antibody Fn8.2-producing cell lines were purchased from Immuno-Precise Antibodies Ltd. (Victoria, British Columbia, Canada). Antibody against F. tularensis LPS (FB11) was purchased from Abcam (Cambridge, MA). Anti-FLAG antibody and Cy3-conjugated anti-FLAG antibody were obtained from Sigma-Aldrich (St. Louis, MO). Anti-FopA antibody was a kind gift from Jason Huntley (University of Toledo Medical School). Rabbit anti-LAMP2 was purchased from Abcam (Cambridge, MA). A mouse anti-EEA1 monoclonal antibody from BD Biosciences (San Diego, CA) and a rabbit polyclonal antibody raised against the N-terminal domain of LAMP1 from Santa Cruz Biotechnology (Santa Cruz, CA) were used as controls in differential permeabilization assays.
Protein secretion.
For the in vitro protein secretion assay, wild-type F. novicida or LVS as well as acpA mutant or wild-type F. novicida carrying pKEK1157 (FLAG-VgrG), pKEK1191 (ΔNBlaB-FLAG), pFnAcpA-FLAG, pFnΔNAcpA-FLAG, or pLVSAcpA-FLAG was grown in modified TSB medium at 37°C at 200 rpm until the optical density at 600 nm (OD600) reached ∼1.0. Bacteria were pelleted by centrifugation at 10,000 × g for 20 min at 4°C, and the supernatants were passed through 0.2-μm filters. Bacterial pellets were lysed with 2× Laemmli sample buffer. As AcpA has been shown to localize primarily to the outer membrane (38), supernatants were subjected to ultracentrifugation at 150,000 × g for 2 h at 4°C to remove any outer membrane fragments and bacterial debris (43). Secreted proteins were then collected by precipitation with 10% trichloroacetic acid at 4°C and ultracentrifugation at 70,000 × g for 1 h. Protein pellets were washed with acetone and resuspended in 2× SDS-PAGE sample buffer, followed by SDS-PAGE and then Western immunoblot analysis with a mouse monoclonal anti-FLAG antibody (Sigma-Aldrich, St. Louis, MO).
Macrophage culture and infection.
The murine macrophage J774A.1 (ATCC, Manassas, VA) was maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and was seeded in 24-well plates with glass coverslips 24 h before Francisella infection.
Human monocyte-derived macrophages (hMDMs) were isolated from human blood via venipuncture from healthy donors following a protocol approved by the Ohio State University Institutional Review Board. Briefly, peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood as previously described (40, 55). PBMCs were then cultured in sterile screw-cap Teflon wells in RPMI 1640 plus l-glutamine with 20% autologous human serum at 37°C and 5% CO2 for 5 days. PBMCs were then recovered from Teflon wells by chilling Teflon wells on ice, and MDMs were allowed to attach to acid cleaned glass coverslips in 24-well tissue culture plates for 2 h at 37°C in 5% CO2, leaving MDM monolayers at a density of approximately 2.0 × 105 cells/well.
Francisella infection of macrophages was conducted as previously described (4). Briefly, Francisella spp. were cultured on chocolate agar, TSB, or MMH plates with appropriate antibiotics for 24 to 48 h and resuspended in phosphate-buffered saline (PBS). Viability of the bacteria prepared was >95%, as determined by counting in a Petroff-Hausser chamber and plating for CFU. The multiplicity of infection (MOI) was approximated by measuring the optical density of the bacterial suspension at 600 nm followed by plating on solid media for accurate enumeration. To synchronize infection, bacteria were centrifuged onto macrophage monolayers at 250 × g for 10 min at 4°C, and returned to 37°C for 30 min to allow for uptake. The monolayers were then washed and incubated for the desired time in culture medium and subsequently processed for immunofluorescence microscopy.
Immunofluorescence microscopy.
To examine the expression of AcpA (see Fig. 2), J774A.1 cells or hMDMs were infected with F. novicida or F. tularensis Schu S4 at specified MOIs until the desired time points. Monolayers were washed three times with PBS and fixed with 2% paraformaldehyde. Cells were permeabilized with 0.1% Triton X-100 in PBS at room temperature for 15 min, blocked with 5% goat serum and 0.5% bovine serum albumin (BSA) in PBS, incubated with the primary antibody for 1 h at room temperature, washed, and then followed with incubation with secondary antibody for 1 h at room temperature. For subsequent examination of bacterial protein secretion and translocation out of phagosomes, infected J774A.1 cells were sequentially permeabilized with digitonin and then saponin as described previously (13). In brief, the plasma membrane of infected cells was selectively permeabilized with 25 μg/ml digitonin at room temperature for 5 min and incubated with antibodies for 30 min to detect cytoplasmic proteins. Cells were fixed and then incubated with 0.1% saponin to permeabilize all host cell membranes, allowing detection of all intracellular bacteria and proteins regardless of their localization. The antibodies used for microscopy were described above. Host nucleic acid was stained with 0.05 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) at room temperature for 5 min. The coverslips were mounted on glass slides and viewed on an Olympus FluoView 1000 confocal microscope. For quantification, at least 300 bacteria per group were counted, and the results are presented as means ± standard deviations (SDs) of at least three independent experiments.
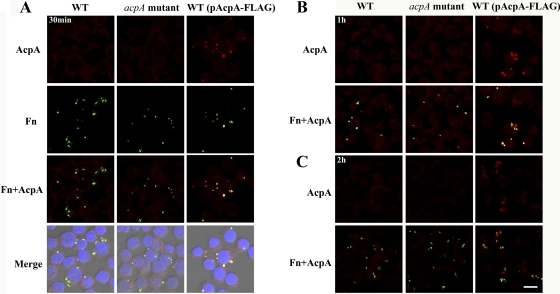
Fig 2.
AcpA colocalizes with Francisella in infected macrophages. J774A.1 cells were infected with the F. novicida (Fn) wild type (WT) or an acpA mutant with or without a plasmid expressing F. novicida AcpA for 30 min (A), 1 h (B), or 2 h (C). AcpA was visualized by fluorescence microscopy after Triton X-100 permeabilization with a rabbit polyclonal antibody against F. novicida AcpA and an AF 546-conjugated secondary antibody (red). F. novicida was detected with anti-F. novicida LPS antibody Fn8.2 and an AF 488-conjugated secondary antibody (green). Scale bar, 10 μm. Results shown are representative of 3 independent experiments.
RESULTS
AcpA is secreted into the culture supernatant in vitro.
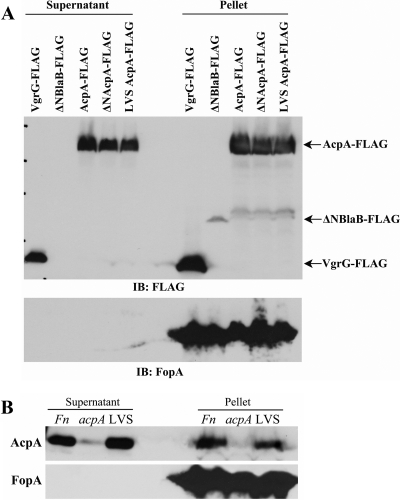
To test our hypothesis that AcpA is secreted by Francisella, a plasmid expressing C-terminally FLAG-tagged AcpA was introduced into wild-type F. novicida. We then examined the culture filtrates for the presence of secreted proteins. Because AcpA is an outer membrane protein, the culture filtrates were further subjected to an ultracentrifugation step to ensure the absence of outer membrane debris (43). VgrG, an FPI-encoded protein, has been shown to be secreted by F. novicida (5). As expected, it was detected at a relatively high level in culture supernatants of F. novicida expressing N-terminally FLAG-tagged VgrG. Consistent with our hypothesis, C-terminally FLAG-tagged AcpA was also detected in the culture supernatants. FLAG-tagged VgrG and FLAG-tagged AcpA were also detected in the bacterial pellet. In contrast, an N-terminally truncated β-lactamase (ΔNBlaB-FLAG), which lacks the 17-amino-acid N-terminal secretion sequence, was not detected in the culture supernatant, but it was present in the bacterial pellet (Fig. 1A). The above FLAG-tagged proteins were also examined in an acpA mutant background, which showed similar results (data not shown). The results demonstrate that AcpA is secreted by F. novicida into the culture medium under the in vitro growth conditions utilized in these experiments.
Fig 1.
Secretion of AcpA into the culture supernatant. (A) Wild-type F. novicida U112 carrying plasmids expressing FLAG-VgrG, ΔNBlaB-FLAG, F. novicida AcpA-FLAG (_Fn_AcpA-FLAG), _Fn_ΔNAcpA-FLAG, and LVS AcpA-FLAG were cultured in TSB medium for 24 h. Supernatants were collected after centrifugation, and bacterial pellets were lysed with 2× Laemmli sample buffer. Supernatants were subjected to further ultracentrifugation at 150,000 × g for 2 h to remove possible outer membrane contamination. Secreted proteins were then collected by trichloroacetic acid (TCA) precipitation and analyzed by SDS-PAGE and Western blotting together with bacterial lysates with a mouse monoclonal anti-FLAG antibody and a rat anti-FopA antibody. IB, immunoblot. (B) Culture supernatants of wild-type F. novicida, acpA mutant, and LVS strains were collected, and proteins were precipitated as described for panel A. Protein precipitates and bacterial lysates were subjected to SDS-PAGE and Western blot analysis with anti-AcpA and anti-FopA antibodies. Results shown are representative of at least 3 independent experiments.
By sequence analysis using SignalP (v3.0; http://www.cbs.dtu.dk/services/SignalP-3.0/), the first 21 amino acids of F. novicida and Schu S4 AcpA are predicted to be the secretion signal sequence, whereas LVS AcpA does not have this sequence (it contains a natural deletion of amino acids 2 to 24). To examine whether this predicted secretion signal is responsible for the AcpA secretion observed, we first checked the secretion of endogenous AcpA from F. novicida and LVS. The F. novicida acpA mutant was also included as a negative control. Consistent with the results obtained using bacteria with AcpA expressed from a plasmid, we detected the secretion of endogenous AcpA from wild-type F. novicida but not the acpA mutant. However, to our surprise, AcpA secretion was also detected for LVS, which lacks the predicted secretion signal sequence, suggesting that this part of the protein is not required for in vitro LVS AcpA secretion (Fig. 1B). To rule out any outer membrane contamination even after ultracentrifugation, we also examined FopA, a known outer membrane protein (that has not been shown to be secreted). FopA was detected in the pellet but not in the culture supernatant, indicating that the observed AcpA secretion from both F. novicida and LVS was not from outer membrane debris (Fig. 1A and B).
To further examine whether F. novicida AcpA secretion can occur in the absence of the predicted N-terminal secretion signal, we also constructed plasmids expressing C-terminal FLAG-tagged F. novicida AcpA with N-terminal deletion of the first 22 amino acids (ΔNAcpA-FLAG) or LVS AcpA-FLAG and examined their secretion. Consistent with endogenous LVS AcpA secretion, both ΔNAcpA-FLAG and LVS AcpA-FLAG were secreted, whereas FopA and ΔNBlaB-FLAG were detected only in the bacterial pellet and not in culture supernatants (Fig. 1A). These results demonstrated that both F. novicida and LVS were able to secret AcpA in vitro, and this secretion is not dependent on the N-terminal predicted secretion signal.
AcpA is secreted by intracellular F. novicida in macrophages.
We next examined if AcpA is secreted by F. novicida within macrophages. To do this, we first determined the expression and localization of AcpA after phagocytosis by J774A.1 cells. A previous study showed that AcpA transcripts increase dramatically 2 h after phagocytosis and gradually decrease over the next 22 h of infection (41). To detect intracellular AcpA, we first infected J774A.1 cells with F. novicida wild type or the acpA mutant with or without the plasmid expressing AcpA-FLAG, followed by gentamicin treatment to kill extracellular bacteria. Cells were incubated 0.5 to 2 h postinfection. AcpA was detected with a polyclonal antibody raised against F. novicida AcpA (38). As shown in Fig. 2, we were not able to detect AcpA expression in wild-type _F. novicida_-infected macrophages compared with the acpA mutant strain or with the use of isotype control sera (data not shown), likely because wild-type bacteria expressed AcpA at a level that cannot be detected by the techniques used here. However, we did detect a relatively high level of AcpA colocalized with intracellular bacteria expressing plasmid-borne AcpA-FLAG at 30 min and 1 h postinfection (Fig. 2A and B), which significantly decreased at 2 h postinfection (Fig. 2C). This was likely due to phagosomal membrane disruption at this time allowing secreted and phagosomally confined AcpA to enter the cytosol diffusing the fluorescence below the level of detection. Thus, in all subsequent studies, infections were carried out with wild-type or acpA mutant bacteria carrying pAcpA-FLAG.
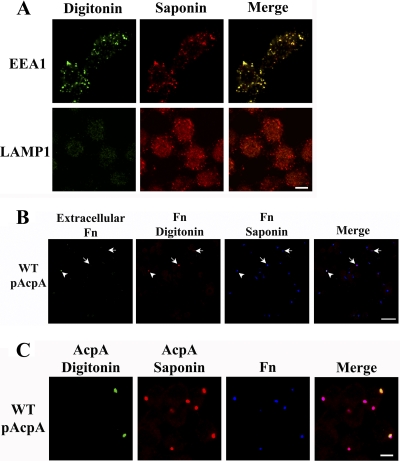
To maximize the chance that all intracellular bacteria were contained within intact phagosomes, we carried out the assay at 30 min postinfection. To detect proteins that were secreted and translocated out of phagosomes, we followed a differential permeabilization protocol (13), in which the host cell plasma membrane is first preferentially permeabilized with digitonin, allowing the detection of proteins in the cytosol, while intracellular vacuoles remain intact, followed by permeabilization of all membrane structures with saponin, allowing for the detection of all proteins regardless of their cellular location. To validate our assay, we first examined the detection of two endosomal membrane markers, EEA1 and LAMP1, by using a mouse monoclonal EEA1 antibody specific to a cytoplasmic epitope and a polyclonal antibody against the N terminus of LAMP1, which faces the endosomal lumen. Cytoplasmic proteins were detected with Alexa Fluor (AF) 488-conjugated secondary antibody after digitonin treatment, and total proteins (cytoplasmic and vacuole luminal) were detected after saponin treatment with AF 546-conjugated secondary antibody. As shown in Fig. 3A, consistent with the known localization of the epitopes, EEA1 was detected after digitonin treatment (green), whereas LAMP1-positive vacuoles were detected only after saponin treatment (red).
Fig 3.
Differential permeabilization to detect cytosolic F. novicida (Fn) and AcpA. (A) J774A.1 cells were sequentially treated with digitonin and saponin and labeled with antibodies against a cytosolic epitope of EEA1 or a luminal epitope of LAMP1. Cytosolic proteins were visualized with AF 488-conjugated secondary antibodies after digitonin treatment (green) and total proteins were visualized with AF 546-conjugated secondary antibodies after saponin treatment (red). Scale bar, 5 μm. Images are representative of 2 experiments. Identical microscope settings were used in the acquisition of all images. (B) J774A.1 cells were infected with wild-type (WT) F. novicida expressing AcpA-FLAG for 30 min. Extracellular bacteria were detected before permeabilization with anti-F. novicida LPS antibody and AF 488-conjugated secondary antibody (green; arrowhead). Cytosolic bacteria or those within compromised phagosomes were detected by digitonin treatment, with the same primary antibody and AF 546-conjugated secondary antibody (red; arrows). Cells were then permeabilized with saponin, and total bacteria were detected with the same primary antibody and AF 647-conjugated secondary antibody (blue). Scale bar, 20 μm. Results shown are representative of 3 independent experiments. (C) J774A.1 cells were infected as described for panel B. Cytosolic AcpA was detected with a polyclonal anti-AcpA antibody and an AF 488-conjugated secondary antibody (green) after digitonin treatment. Total AcpA was detected with the same anti-AcpA antibody and an AF 546-conjugated secondary antibody (red) after saponin treatment. F. novicida was detected with anti-F. novicida LPS monoclonal antibody Fn8.2 and an AF 647-conjugated anti-mouse antibody (blue) after saponin treatment. Scale bar, 5 μm. Images are representative of at least 3 independent experiments.
We next further validated the location of F. novicida after infection of J774.1 cells by using this method; only a small number of extracellular bacteria were detected without permeabilization (Fig. 3B, arrowhead). At 30 min postinfection, the majority (92.48% ± 2.35%) of the bacteria were localized within intact phagosomes. Only a small population (7.52% ± 2.35%) of bacteria were detected after digitonin but before saponin treatment, suggesting that these bacteria were localized either in the cytosol or within compromised phagosomes (Fig. 3B, arrows). In parallel experiments, we also further validated the location of AcpA after differential permeabilization of J774A.1 cells infected with wild-type F. novicida expressing AcpA-FLAG. As expected, AcpA expression was associated with all F. novicida bacteria after saponin treatment (red), whereas cytosolic AcpA was detected only for some of the bacteria after digitonin treatment (green) (Fig. 3C), which could represent AcpA secreted and translocated to the cytosol, AcpA associated with cytosol-localized bacteria (escaped from the phagosome by 30 min), or AcpA within a compromised phagosome.
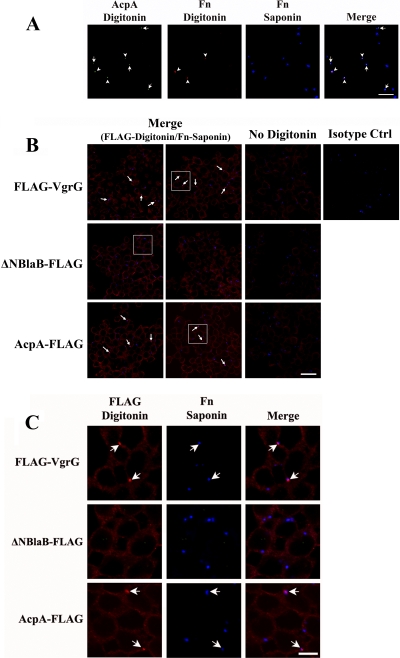
To explore these possibilities, we also visualized the bacteria after partial permeabilization with digitonin (green) together with cytosolic AcpA (red), and all bacteria were then detected after saponin permeabilization (blue). As shown in Fig. 4A, cytosolic AcpA was detected for bacteria localized within intact phagosomes (arrows indicate bacteria detected only after saponin permeabilization), as well as those in the cytosol or within compromised phagosomes (arrowheads indicate those detected after digitonin before saponin treatment). Together, these results indicate that AcpA can be secreted and translocated by F. novicida residing within intact phagosomes.
Fig 4.
AcpA is secreted by a small population of bacteria inside infected macrophages. (A) J774A.1 cells were infected with wild-type F. novicida (Fn) expressing AcpA-FLAG for 30 min. After digitonin treatment, cytosolic AcpA was visualized with an anti-AcpA antibody and AF 488-conjugated secondary antibody (green), and cytosolic bacteria were detected with anti-F. novicida LPS antibody and AF 546-conjugated secondary antibody (red). Total bacteria were detected after saponin treatment with anti-F. novicida LPS antibody and AF 647-conjugated secondary antibody (blue). Arrows indicate cytosolic AcpA produced by F. novicida residing within intact phagosomes, and arrowheads indicate cytosolic AcpA produced by cytosolic F. novicida or by those in compromised phagosomes. Scale bar, 20 μm. Images are representative of 2 independent experiments. (B) J774A.1 cells were infected with F. novicida strains carrying plasmids expressing FLAG-VgrG, ΔNBlaB-FLAG, or F. novicida AcpA-FLAG at an MOI of 100:1 for 30 min. Infected cells were sequentially permeabilized with digitonin (except for the “No Digitonin” lane) and saponin. Secreted FLAG-tagged proteins were detected with Cy3-conjugated monoclonal anti-FLAG antibody (red) after digitonin treatment. Bacteria were stained with an anti-F. novicida LPS monoclonal antibody Fn8.2 and an AF 647-conjugated anti-mouse antibody (blue) after saponin permeabilization. Bacteria with secreted VgrG-FLAG or AcpA-FLAG are marked with arrows. Scale bar, 20 μm. Ctrl, control. Enlarged images of the boxed areas are shown in panel C (scale bar, 5 μm). Results shown are representative of 4 experiments.
We then infected J774A.1 cells with wild-type F. novicida expressing FLAG-VgrG, ΔNBlaB-FLAG, or AcpA-FLAG. Secreted FLAG-tagged proteins were detected with Cy3-conjugated anti-FLAG antibody (red). F. novicida was detected (as in Fig. 3B and C and 4A) with a mouse monoclonal antibody directed against LPS and AF 647-conjugated secondary antibody (blue). As shown in Fig. 4B and C, although the majority of the bacteria localized within phagosomes (Fig. 3B), some of the F. novicida bacteria expressing FLAG-VgrG (26.67% ± 5.15%) and AcpA-FLAG (17.57% ± 2.56%) were associated with cytosolic VgrG/AcpA (arrows indicate those detected with Cy3-conjugated anti-FLAG antibody after digitonin treatment only). In contrast, no cytosolic ΔNBlaB-FLAG was detected. However, as both AcpA and VgrG can be secreted by bacteria grown in vitro, it was possible that bacteria with a positive FLAG signal were simply outside macrophages. To exclude this possibility, we carried out an experiment with parallel samples that were not permeabilized with digitonin, which labels extracellular bacteria but not intracellular bacteria. Without digitonin treatment, no FLAG-tagged proteins were detected (Fig. 4B), indicating that all of the bacteria with secreted VgrG or AcpA were intracellular rather than extracellular.
Intracellular F. novicida secrete AcpA out of LAMP2-positive phagosomes into the cytosol of macrophages.
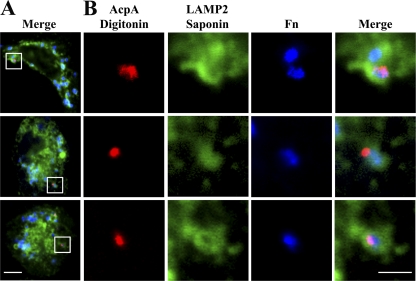
To further examine AcpA translocation and to explore it more in a relevant human cell type, we infected human monocyte-derived macrophages (hMDMs) with the wild type or acpA mutant F. novicida expressing pAcpA-FLAG. We detected cytosolic AcpA with Cy3-conjugated anti-FLAG antibody (red) and labeled the phagosome by an antibody against human LAMP2, a marker for the late endosome and early lysosome, and an AF 488-conjugated secondary antibody (green). Almost all (86.65% ± 6.37%) of the AcpA detected was associated with bacteria contained within LAMP2-positive phagosomes (13.35% ± 6.37% associated with phagosomes with incomplete LAMP2 staining). Obvious cytosolic AcpA was observed emanating from some bacteria, while each bacterium was clearly surrounded by LAMP2-positive staining (Fig. 5A and B). In most cases, AcpA is secreted in a localized pattern from the bacteria (Fig. 5B). This result demonstrated that intracellular F. novicida is able to secrete and translocate AcpA out of the phagosome at 30 min postinfection in human macrophages.
Fig 5.
AcpA is secreted and translocated out of the F. novicida (Fn) phagosome. (A) hMDMs were infected with wild-type F. novicida carrying a plasmid expressing F. novicida AcpA-FLAG at an MOI of 50:1. Infection was synchronized by centrifugation at 250 × g for 5 min at 4°C and then incubation at 37°C for 30 min. Secreted and translocated AcpA was detected using a Cy3-conjugated anti-FLAG antibody following digitonin treatment (red). Samples were then permeabilized with 0.1% saponin. Phagosomes were subsequently stained with an anti-LAMP2 monoclonal antibody and an AF 488-conjugated secondary antibody (green), and bacteria were stained with anti-F. novicida LPS monoclonal antibody Fn8.2 and an AF 647-conjugated anti-mouse antibody (blue). Scale bar, 5 μm. (B) Enlarged images of the boxed area in panel A. Scale bar, 2 μm. Results shown are representative of 3 experiments.
Intracellular human virulent F. tularensis subsp. tularensis Schu S4 secretes AcpA into the cytosol of macrophages.
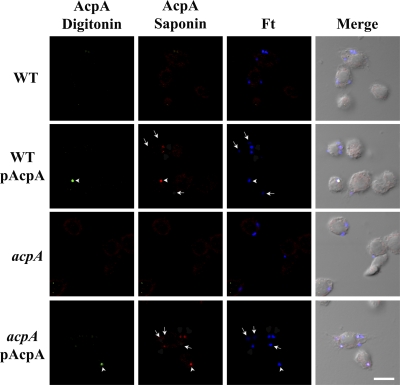
We have demonstrated that F. novicida and F. tularensis LVS are able to secrete AcpA in vitro and for F. novicida, also in vivo. We next examined whether AcpA is also secreted by the virulent type A strain Schu S4. The acpA locus is conserved between the F. novicida U112 and Schu S4 genomes, and the AcpA proteins share 98.1% identity (14). In vitro experiments previously demonstrated the secretion of AcpA into the Schu S4 culture supernatant (28). Wild-type Schu S4 or an acpA mutant carrying a complementing plasmid (expressing the Schu S4 acpA) was examined for intracellular secretion of AcpA. As with F. novicida, we were not able to detect AcpA with wild-type Schu S4 infection compared with the negative control, the acpA mutant (Fig. 6). However, we did detect AcpA when we infected J774A.1 cells with the strain expressing plasmid-borne acpA. The level of AcpA expression was much lower than that of F. novicida, with only 30.61% ± 3.47% of the wild type or 38.79% ± 9.53% of acpA mutant bacteria carrying the AcpA plasmid demonstrating detectable levels of AcpA (Fig. 6, arrows; 100% for F. novicida). Consistent with what we observed for F. novicida, only some of the intracellular bacteria, 11.46% ± 1.84% for the wild type and 8.95% ± 3.24% for the acpA mutant, secreted plasmid-borne AcpA into the cytosol (i.e., AcpA visualized following digitonin treatment; Fig. 6, arrowheads). This confirms that, as observed with F. novicida, the virulent Schu S4 strain can also secrete AcpA intracellularly and also likely translocate AcpA across the phagosomal membrane into the host cytosol.
Fig 6.
AcpA is secreted by intracellular F. tularensis subsp. tularensis Schu S4. J774A.1 cells were infected with a Schu S4 acpA mutant carrying a plasmid expressing F. tularensis (Ft) AcpA at an MOI of 200:1. Infection was synchronized by centrifugation at 250 × g for 5 min at 4°C and then incubation at 37°C for 60 min, followed by 30 min treatment with 50 μg/ml gentamicin. Infected cells were first treated with digitonin, and secreted AcpA was detected by rabbit anti-AcpA antibody and an AF 488-conjugated anti-rabbit secondary antibody (green; arrowheads). Samples were then permeabilized with 0.1% saponin. Total AcpA was subsequently stained with an anti-AcpA antibody and an AF 546-conjugated secondary antibody (red), and bacteria were stained with anti-F. tularensis LPS monoclonal antibody FB11 and an AF 647-conjugated anti-mouse antibody (blue). Arrows depict SchuS4 bacteria not expressing AcpA at detectable levels. Scale bar, 10 μm. Results shown are representative of 4 experiments. WT, wild type.
DISCUSSION
Acid phosphatases are a family of enzymes involved in the virulence of pathogens, including Leishmania donovani (48), Legionella micdadei (50), Bordetella bronchiseptica (27), and F. tularensis (47), primarily by inhibiting reactive oxygen species (ROS) production in host cells. Our previous work has shown that F. novicida acid phosphatases play a role in phagosomal escape, intramacrophage survival, and virulence in the tularemia mouse model (38, 41). In addition, our recent study showed that acid phosphatases can result in dephosphorylation of NADPH oxidase components in vitro and in vivo, with a quadruple mutant (AcpABC-Hap) incapable of respiratory burst suppression compared with wild-type F. novicida (40). ROS inhibition occurs rapidly during phagocytosis by human neutrophils or macrophages, but Francisella escape from phagosomes is generally thought not to occur until at least 30 min postinfection. These results led us to hypothesize that acid phosphatases may be secreted into host cell cytosol by Francisella spp. in the nascent phagosome. Thus, we investigated the secretion of AcpA, which is induced rapidly upon macrophage infection and contributes most of the acid phosphatase activity in both F. novicida and F. tularensis subsp. tularensis Schu S4.
We were able to detect endogenous AcpA in the culture supernatant of wild-type F. novicida or F. tularensis LVS, as well as plasmid-expressed AcpA from wild-type bacteria or from an acpA mutant. In addition, we show by immunofluorescence microscopy that early after macrophage phagocytosis, AcpA is not only secreted by some intracellular F. novicida and the virulent type A Schu S4 strain, it is also translocated across the phagosomal membrane into the host cytosol. These results support our hypothesis and further suggest the important role of AcpA for intracellular survival of Francisella spp.
Inside macrophages, AcpA colocalized with intracellular Francisella at an early stage of infection (30 min). However, this colocalization was significantly reduced later in infection (2 h). This correlated with the ability of Francisella spp. to escape the phagosome. It also suggested that at an early stage of infection, AcpA, if secreted, is largely confined by the phagosomal membrane. Consistent with this, only a small subset of intraphagosomal F. novicida bacteria could be observed to translocate AcpA across the phagosomal membrane and into the host cytosol as detected by differential membrane permeabilization and immunofluorescence microscopy. The mechanism underlying this is unclear, but interestingly, VgrG, an FPI-encoded protein secreted by an unknown mechanism (5), also had the same secretion and translocation pattern as AcpA. As AcpA has been shown to be released from the bacterium via outer membrane vesicles, it is possible that translocation happens by membrane fusion. However, another intriguing possibility is that through its phospholipase activity, secreted AcpA is able to modify the phagosomal membrane making it permeable to intraphagosomal proteins and contributing to Francisella phagosomal escape. Consistent with this, the loss of AcpA alone was shown to delay phagosomal escape and affect intramacrophage survival and virulence of F. novicida (38). Interestingly, when observed together with the phagosome marker LAMP2, cytosolic AcpA was found in a localized spot outside the phagosome, suggesting a spatially constrained translocation of AcpA. Further study is needed to explore this phenomenon.
During the course of our studies, we attempted to detect AcpA secretion by fusing AcpA to the Bordetella pertussis adenylate cyclase toxin (AcpA-CyaA). As the adenylate cyclase activity of CyaA requires the presence of calmodulin, a host cytosolic protein, an elevated cyclic AMP (cAMP) level should occur when AcpA-CyaA is secreted and translocated into the macrophage cytosol. However, we were not able to detect a significant increase in the cAMP level in macrophages upon infection with F. novicida expressing AcpA-CyaA (data not shown). This is likely because, as observed by immunofluorescence microscopy, only a small portion of intracellular bacteria translocate AcpA into the macrophage cytosol.
Secretion of acid phosphatases and their roles in pathogenesis have been described for other organisms, including Legionella pneumophila (1), Mycobacterium tuberculosis (51), and Staphylococcus aureus (19). In Legionella pneumophila, the secretion of acid phosphatases was further characterized as type II secretion or type IV pilus system dependent (1, 49). A recent proteomics study to characterize F. tularensis subsp. tularensis Schu S4 and F. tularensis LVS culture filtrate proteins found abundant AcpA in the culture supernatant of Schu S4, which corroborates our findings of AcpA secretion in vitro in the virulent strain (28). Another proteomics study on Francisella outer membrane vesicles found AcpA in these vesicles produced by F. novicida (44). However, in our study, we used ultracentrifugation at a g force that removed outer membrane vesicles. Many secretion systems have been described for Francisella spp., including type I, type II, and type IV pili and a putative type VI secretion system. Although AcpA has a predicted canonical signal peptide for bacterial protein secretion (59) at the N terminus, our data suggested that this signal peptide is not responsible for the observed AcpA secretion. It is not uncommon to find bacterial effectors secreted without this signal peptide. In fact, the secretion signal varies with different secretion systems (23). Peptide conformation, rather than sequence, and mRNA, rather than peptide sequence, have both been suggested or demonstrated as secretion signals (23). Specifically, putative effectors of type VI secretion systems, EvpP in Edwardsidlla tarda, and TssM in Burkholderia mallei can both be secreted despite the fact that they do not possess this canonical N-terminal signal peptide (46). Future experiments are required to identify the mechanism by which AcpA is secreted in Francisella spp.
Despite the numerous publications regarding the contribution of Francisella sp. acid phosphatases, particularly AcpA, in virulence-related phenotypes, their importance has been put into question by recent publications. Child et al. demonstrated the lack of a role for AcpA, -B, and -C in F. tularensis Schu S4 intramacrophage survival or mouse virulence (14). Additionally, a separate research group has reported that the loss of AcpA alone is dispensable for respiratory burst inhibition (34). While these disparate results may be attributable to differences in experimental procedures or host cells/donors or to strain-specific genetic differences known to occur in the francisellae, they are generally consistent with our previously published results that demonstrate a combined effect of all four Francisella acid phosphatases on virulence properties; single acid phosphatase deletions have only limited effects (41). This is, however, a bit at odds with the perceived importance of AcpA among the Francisella acid phosphatases, as AcpA transcription is induced in vivo (41), AcpA is the major contributor toward total acid phosphatase activity of the bacterium (14, 41), AcpA is secreted and translocated from the phagosome into the host cell cytosol (this study; 28, 44), and it plays a unique role in dephosphorylation of NADPH phox subunits in vitro (40). There are likely to be multiple intracellular targets for AcpA and potentially other factors (including other phosphatases) with redundant virulence/dephosphorylation-capable functions. Thus, the combined evidence suggests that AcpA has an evolutionary importance regarding _Francisella_-host interactions.
In summary, this study demonstrates that AcpA is secreted in vitro and in vivo by F. novicida and the highly virulent F. tularensis subsp. tularensis. Although much remains to be determined, the results of this study advance our understanding of AcpA function and bring us closer to a complete understanding of Francisella virulence factor secretion.
ACKNOWLEDGMENTS
We thank Karl Klose and Jean Celli for the generous contributions of plasmids and Jason Huntley for antibodies.
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research Program. We acknowledge membership within and support from the Region V Great Lakes Regional Center of Excellence (National Institutes of Health award 2-U54-AI-057153).
Footnotes
Published ahead of print 19 December 2011
REFERENCES
- 1.Aragon V, Kurtz S, Cianciotto NP. 2001. Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect. Immun. 69:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkins HS, et al. 2006. The identification and evaluation of ATP binding cassette systems in the intracellular bacterium Francisella tularensis. Res. Microbiol. 157:593–604 [DOI] [PubMed] [Google Scholar]
- 3.Baca OG, et al. 1993. Acid phosphatase activity in Coxiella burnetii: a possible virulence factor. Infect. Immun. 61:4232–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balagopal A, et al. 2006. Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect. Immun. 74:5114–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker JR, et al. 2009. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74:1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell BL, Mohapatra NP, Gunn JS. 2010. Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect. Immun. 78:2189–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bönquist L, Lindgren H, Golovliov I, Guina T, Sjostedt A. 2008. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76:3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brotcke A, Monack DM. 2008. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect. Immun. 76:3473–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchan BW, McCaffrey RL, Lindemann SR, Allen LA, Jones BD. 2009. Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect. Immun. 77:2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burtnick M, Bolton A, Brett P, Watanabe D, Woods D. 2001. Identification of the acid phosphatase (acpA) gene homologues in pathogenic and non-pathogenic Burkholderia spp. facilitates TnphoA mutagenesis. Microbiology 147:111–120 [DOI] [PubMed] [Google Scholar]
- 11.Charity JC, Blalock LT, Costante-Hamm MM, Kasper DL, Dove SL. 2009. Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog. 5:e1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charity JC, et al. 2007. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 3:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U. S. A. 103:14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Child R, Wehrly TD, Rockx-Brouwer D, Dorward DW, Celli J. 2010. Acid phosphatases do not contribute to the pathogenesis of type A Francisella tularensis. Infect. Immun. 78:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong A, et al. 2008. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76:5488–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemens DL, Horwitz MA. 2007. Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y. Acad. Sci. 1105:160–186 [DOI] [PubMed] [Google Scholar]
- 17.Clemens DL, Lee BY, Horwitz MA. 2005. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 73:5892–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai S, Morhapatra NP, Schlesinger LS, Gunn JS. 2011. Regulation of Francisella tularensis virulence. Front. Cell. Infect. Microbiol. 1:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.du Plessis EM, Theron J, Joubert L, Lotter T, Watson TG. 2002. Characterization of a phosphatase secreted by Staphylococcus aureus strain 154, a new member of the bacterial class C family of nonspecific acid phosphatases. Syst. Appl. Microbiol. 25:21–30 [DOI] [PubMed] [Google Scholar]
- 20.Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin. Microbiol. Rev. 15:631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felts RL, Reilly TJ, Tanner JJ. 2005. Crystallization of AcpA, a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. Biochim. Biophys. Acta 1752:107–110 [DOI] [PubMed] [Google Scholar]
- 22.Felts RL, Reilly TJ, Tanner JJ. 2006. Structure of Francisella tularensis AcpA: prototype of a unique superfamily of acid phosphatases and phospholipases C. J. Biol. Chem. 281:30289–30298 [DOI] [PubMed] [Google Scholar]
- 23.Filloux A. 2010. Secretion signal and protein targeting in bacteria: a biological puzzle. J. Bacteriol. 192:3847–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gil H, et al. 2006. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. U. S. A. 103:12897–12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hager AJ, et al. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227–237 [DOI] [PubMed] [Google Scholar]
- 26.Hall JD, et al. 2008. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 76:5843–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jungnitz H, West NP, Walker MJ, Chhatwal GS, Guzman CA. 1998. A second two-component regulatory system of Bordetella bronchiseptica required for bacterial resistance to oxidative stress, production of acid phosphatase, and in vivo persistence. Infect. Immun. 66:4640–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konecna K, et al. 2010. Comparative proteomic profiling of culture filtrate proteins of less and highly virulent Francisella tularensis strains. Proteomics 10:4501–4511 [DOI] [PubMed] [Google Scholar]
- 29.Lauriano CM, et al. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U. S. A. 101:4246–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindgren H, et al. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53:953–958 [DOI] [PubMed] [Google Scholar]
- 31.LoVullo ED, Sherrill LA, Perez LL, Pavelka MS., Jr 2006. Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology 152:3425–3435 [DOI] [PubMed] [Google Scholar]
- 32.Maier TM, et al. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 70:7511–7519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margolis JJ, et al. 2010. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl. Environ. Microbiol. 76:596–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCaffrey RL, et al. 2010. Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J. Leukoc. Biol. 88:791–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McRae S, et al. 2010. Inhibition of AcpA phosphatase activity with ascorbate attenuates Francisella tularensis intramacrophage survival. J. Biol. Chem. 285:5171–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meibom KL, Charbit A. 2010. The unraveling panoply of Francisella tularensis virulence attributes. Curr. Opin. Microbiol. 13:11–17 [DOI] [PubMed] [Google Scholar]
- 37.Meibom KL, et al. 2009. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun. 77:1866–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohapatra NP, Balagopal A, Soni S, Schlesinger LS, Gunn JS. 2007. AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect. Immun. 75:390–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohapatra NP, et al. 2007. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect. Immun. 75:3305–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohapatra NP, et al. 2010. Francisella acid phosphatases inactivate the NADPH oxidase in human phagocytes. J. Immunol. 184:5141–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohapatra NP, et al. 2008. Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect. Immun. 76:3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyston PC, Sjostedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2:967–978 [DOI] [PubMed] [Google Scholar]
- 43.Pegues DA, Hantman MJ, Behlau I, Miller SI. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169–181 [DOI] [PubMed] [Google Scholar]
- 44.Pierson T, et al. 2011. Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J. Proteome Res. 10:954–967 [DOI] [PubMed] [Google Scholar]
- 45.Postic G, et al. 2010. Identification of small RNAs in Francisella tularensis. BMC Genomics 11:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 47.Reilly TJ, Baron GS, Nano FE, Kuhlenschmidt MS. 1996. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J. Biol. Chem. 271:10973–10983 [DOI] [PubMed] [Google Scholar]
- 48.Remaley AT, et al. 1985. Leishmania donovani: surface membrane acid phosphatase blocks neutrophil oxidative metabolite production. Exp. Parasitol. 60:331–341 [DOI] [PubMed] [Google Scholar]
- 49.Rossier O, Cianciotto NP. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saha AK, et al. 1985. Properties of an acid phosphatase from Legionella micdadei which blocks superoxide anion production by human neutrophils. Arch. Biochem. Biophys. 243:150–160 [DOI] [PubMed] [Google Scholar]
- 51.Saleh MT, Belisle JT. 2000. Secretion of an acid phosphatase (SapM) by Mycobacterium tuberculosis that is similar to eukaryotic acid phosphatases. J. Bacteriol. 182:6850–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santic M, Al-Khodor S, Abu Kwaik Y. 2010. Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12:129–139 [DOI] [PubMed] [Google Scholar]
- 53.Santic M, Molmeret M, Klose KE, Abu Kwaik Y. 2006. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 14:37–44 [DOI] [PubMed] [Google Scholar]
- 54.Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7:969–979 [DOI] [PubMed] [Google Scholar]
- 55.Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA. 1990. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J. Immunol. 144:2771–2780 [PubMed] [Google Scholar]
- 56.Schmerk CL, Duplantis BN, Howard PL, Nano FE. 2009. A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology 155:1498–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmerk CL, et al. 2009. Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology 155:1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent JB, Crowder MW, Averill BA. 1992. Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem. Sci. 17:105–110 [DOI] [PubMed] [Google Scholar]
- 59.von Heijne G. 1990. The signal peptide. J. Membr. Biol. 115:195–201 [DOI] [PubMed] [Google Scholar]
- 60.Zogaj X, Chakraborty S, Liu J, Thanassi DG, Klose KE. 2008. Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology 154:2139–2150 [DOI] [PubMed] [Google Scholar]