Transcriptional mechanisms that regulate T helper 1 cell differentiation (original) (raw)
. Author manuscript; available in PMC: 2013 Apr 1.
Published in final edited form as: Curr Opin Immunol. 2012 Jan 10;24(2):191–195. doi: 10.1016/j.coi.2011.12.004
Abstract
Recent research has made great strides in uncovering the mechanisms by which the T helper 1 (Th1) cell gene expression program is established. In particular, studies examining the transcription factors T-bet, STAT1, and STAT4 have elucidated their roles in regulating Th1 signature genes, including Ifng, and have started to address their contributions to the epigenetic states in Th1 cells. Additionally, new findings have provided information about how the co-expression of T helper cell lineage-defining transcription factors impacts the phenotype of the cell. In this review, we will briefly highlight the research from the last few years examining the epigenetic states in T helper cells and the mechanisms by which they are established. We will then discuss how this new information contributes to our understanding of the flexibility of T helper cell genetic programs.
Introduction
The adaptive immune system has evolved to combat a diverse set of pathogens. At the time of their activation, CD4+ T helper cells are instructed by the cytokine environment to differentiate into a number of distinct subtypes in order to coordinate the immune response to clear the pathogenic insult. At present, several different T helper cell subtypes have been identified including T helper 1 (Th1), Th2, Th17, T follicular helper (Tfh), and induced T regulatory (iTreg) cells [1,2]. Each of these T helper cell subtypes is characterized by a specialized gene expression program, which includes signature cytokines, cell surface receptors, and regulatory factors. A few required lineage-defining transcription factors coordinate the induction of the individual T helper cell specific gene expression programs and the mechanisms by which they accomplish this task are a topic of much interest [2-4]. In this review, we will discuss our current views on how the T helper cell specific transcriptional programs, with a focus on Th1 cells, are established and how the epigenetic states in these cells contribute to our views on the flexibility versus stability of their phenotypes.
Transcription factors required for Th1 cell differentiation
There are several transcription factors that are required for Th1 cell differentiation, without which, the Th1 signature gene program cannot be properly expressed. In particular, STAT1, STAT4, and T-bet are the most recognizable and well-studied [5]. STAT1 is activated in response to IFNγ signaling and reinforces the Th1 phenotype in a positive feedback loop [6,7]. IL-12 signaling induces STAT4, which positively regulates many aspects of the Th1 genetic program [8,9]. STAT1 and STAT4 also contribute to the regulation of Tbx21 (the gene that encodes T-bet) expression [6,7,10,11]. T-bet is a T-box transcription factor that is required for the induction of many Th1 signature genes and is also needed for the repression of genes specific to alternative T helper cell fates [12-16]. A number of additional, more ubiquitously expressed regulatory proteins are also needed to impart a Th1 gene expression program, but STAT1, STAT4, and T-bet are required for Ifng expression and the Th1 phenotype [17,18]. To date, a great deal of research has focused on elucidating the identity of the required factors in Th1 development and now current studies are moving forward towards understanding the mechanisms that each of these factors utilize to regulate Th1 gene expression patterns. Here, we will discuss this topic in more detail, with a focus on the role for T-bet in Th1 cells.
Epigenetics and Th1 cell differentiation
One emerging question over the last several years has been the nature of the stability versus flexibility of the T helper cell phenotypes [4,19-22]. Historically, T helper cell differentiation was viewed through the lens of the Th1/Th2 paradigm, with each T helper cell type representing a developmentally stable lineage capable of expressing only one kind of signature cytokine [23,24]. However, recent years have dramatically altered our view of this concept. It is now widely accepted that there are several functionally distinct T helper cell subtypes represented by different signature cytokine and cell surface receptor expression profiles. Recognized T helper cell subtypes include the traditional Th1 and Th2 cells, which secrete IFNγ and IL-4, respectively, and have now extended to include Th17 cells that express IL-17, as well as Tfh cells that express IL-21 [1,4,5]. In addition, T regulatory (Treg) cells are responsible for keeping the immune response in check and preventing autoimmunity [25]. The predominant question in the field has now become whether these T helper cell subtypes actually represent distinct developmental lineages, or rather they are more appropriately categorized as subsets with the potential for flexibility to express at least portions of the gene programs characteristic of the opposing subtypes.
The epigenetic profiling of T helper cell subtypes, as well as new research examining the mechanisms by which these epigenetic patterns are established, have started to provide insight into this question [26-30]. Histone 3 lysine 4 trimethylation (H3K4me3) and H3K27me3 are the most commonly studied epigenetic modifications in T helper cell subtypes [29,31]. The H3K4me3 modification is associated with a permissive chromatin state and is generally centered around the transcription start site of active genes. This distinctive patterning is in part due to the association of H3K4-methyltransferase complexes with the basal transcription machinery [32-34]. This means that if a gene is being actively transcribed in a cell, there will be H3K4me3 at the promoter. In contrast, the H3K27me3 modification is catalyzed by the polycomb complex and is associated with a repressive epigenetic environment. Whereas the H3K4me3 modification is found at almost all active genes, the H3K27me3 modification is not ubiquitously associated with all inactive genes. Rather, the polycomb complex is selectively targeted to subsets of developmentally regulated genes to compact the epigenetic environment specifically surrounding these genes [35]. This means that not all of the repressed genes in T helper cells will be marked with the H3K27me3 modification, but that the genes containing this mark may be in a somewhat more permanently repressed state [29].
Our understanding of T helper cell, as well as Th1, differentiation has been aided in recent years by a combination of genome-wide studies examining epigenetic modifications and individual gene studies defining the mechanisms by which these epigenetic states are established [21,31,36]. The genome-wide experiments have in part served as confirmation of prior, targeted research studies examining T helper cell signature cytokine genes, including Ifng in Th1 cells and Il4 in Th2 cells [37-39]. Signature cytokine loci are found in a permissive state in the T helper cell type in which they are expressed [28,29,38-40]. In contrast, Ifng and Il4 are packaged into the repressive H3K27me3 state in alternative T helper cell fates when they are repressed [27,38,41]. These findings led to the viewpoint that T helper cell subtypes represented defined lineages, with the genes for alternative fates permanently extinguished as the cell differentiated. However, one of the most intriguing new insights to come out of the genome-wide analyses of helper T cell subtypes is that the vast majority of Th1 genes do not follow a strict epigenetic paradigm of a permanent polycomb-mediated repression (i.e. H3K27me3 modification) in alternative T helper cell fates [29]. One explanation for this may be that very few genes in the T helper cell subtypes are strictly expressed in a subtype-specific fashion. This may explain why they would not be regulated by a stable mechanism of epigenetic repression. Another surprise in the genome-wide analyses was that the lineage-defining transcription factors, such as T-bet for Th1 cells, are found in a bivalent (i.e. contains both H3K4me3 and H3K27me3) epigenetic state in alternative T helper cell fates [26,29]. In a developmental setting, it is thought that bivalent chromatin keeps genes poised to allow for their future expression [42]. The finding that the T helper cell lineage-defining transcription factors are not marked with epigenetic modifications indicative of permanent repression is now moving the field to the new viewpoint that T helper cells may be more accurately characterized as somewhat flexible subsets rather than true developmental lineages as we will now discuss.
So what does it mean that the signature cytokine genes for T helper cells are packaged into the stable H3K27me3 repressive epigenetic state in alternative T helper cell fates, but that the lineage-defining transcription factors are packaged into a poised chromatin state? To answer this question, we need to understand the mechanisms by which the T helper cell lineage-defining transcription factors regulate the signature cytokine genes. This information will then tell us what will happen if the lineage-defining transcription factors are re-expressed in an alternative T helper cell fate. Here, the research into the mechanisms by which T-bet regulates the Th1 signature cytokine Ifng have been enlightening. T-bet activates Th1 signature genes in part by recruiting chromatin remodeling complexes including Jmjd3, a Brg1-containing SWI/SNF-complex, and an H3K4me2-methyltransferase complex to its target genes (Figure 1) [27,43,44]. All of these complexes serve to establish a more permissive chromatin state conducive with transcriptional activation.
Figure 1.
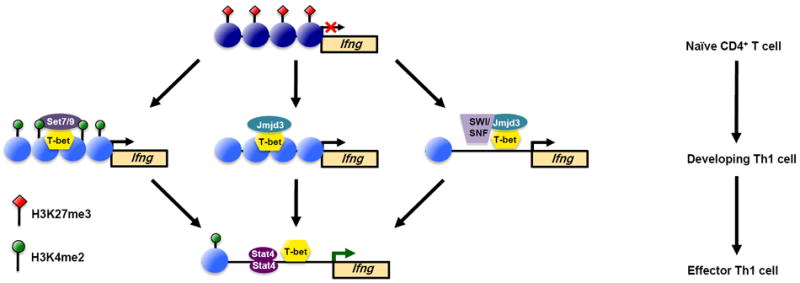
T-bet activates Th1 signature genes, including Ifng, during Th1 cell development by functionally regulating multiple aspects of chromatin remodeling. (a) In naïve CD4+ T helper cells, the nucleosomes encompassing the Ifng locus are marked by repressive H3K27me3 epigenetic modifications to keep the locus in a compacted chromatin state. (b-d) As naïve CD4+ T cells begin to express T-bet and differentiate towards the Th1 phenotype, T-bet physically recruits (b) an H3K4me2-methyltranserase complex containing Set7/9, (c) the H3K27-demethylase Jmjd3, and (d) a SWI/SNF-general chromatin remodeling complex through its association with Jmjd3. (e) Collectively, the T-bet-dependent remodeling events create a more permissive or accessible chromatin environment, which allows for the binding of additional transcriptional regulatory proteins, including STAT4, to activate gene transcription.
Important to the topic of the flexibility versus developmental stability of Th1 cells is the interaction between T-bet and Jmjd3, an H3K27-demethylase [27]. This interaction allows T-bet to effectively target the removal of the repressive H3K27me3 modification. Therefore, until T-bet is permanently extinguished, the cell will retain the capacity to re-express T-bet target genes that are configured in polycomb-mediated repressive chromatin, which includes the Th1 signature cytokine gene Ifng in alternative T helper cell subtypes. Coupling this information with what we now know about the poised epigenetic status of the Tbx21 locus (the gene that encodes T-bet) in the alternative T helper cell fates, it suggests that the flexibility to express a Th1-gene program is maintained longer than was previously appreciated. Thus, the new studies determining the genome-wide epigenetic state of T helper cells, along with those uncovering the mechanisms by which the key, required transcription factors act within the cell, have changed our inherent view of the stability of T helper cell phenotypes. At present, the data are more indicative of flexible subsets rather than truly stable developmental lineages.
Cooperation and antagonism of the T helper cell transcription factors
The poised epigenetic state of the loci encoding the T helper cell lineage-defining transcription factors maintains their potential to be expressed in more circumstances than had been anticipated. Previously, we viewed the expression of the key transcription factors such as T-bet, GATA3, RORγt, Foxp3, and Bcl-6 as being mutually exclusive, with each one restricted to the cell type where it defines the signature gene expression program [45]. Simplistically then, T-bet is solely expressed in Th1 cells, while GATA3, RORγt, Foxp3, and Bcl-6 are expressed in Th2, Th17, Treg, and Tfh cells, respectively. However, it has become clear that these transcription factors are not expressed in a strictly restricted pattern and in fact, in some cases, they are required for the proper functioning of what was traditionally thought of as an opposing T helper cell fate [15,28,29,46-48]. Once again, this concept is demonstrated by research examining the role for T-bet in establishing T helper cell gene expression patterns. Intriguingly, T-bet interacts with several other T helper cell lineage-defining transcription factors to mediate the repression of alternative cell fates [12-15]. In particular, in fully developed Th1 cells, T-bet interacts with Bcl-6 to repress a subset of genes that are important in opposing T helper cell fates [15]. This means that T-bet and Bcl-6 work together to establish the Th1 profile and their co-expression is not detrimental to the cell, but rather is important for it.
A similar need for the co-expression of two seemingly opposing transcription factors is found with the co-expression of T-bet and Foxp3 in Treg cells [47]. In this case, T-bet helps to impart a hybrid gene expression program to allow for the homing of the Treg cells to the same location as Th1 cells to dampen their response. This form of regulation is not exclusive to the expression of T-bet in Treg cells, but has also been observed for other key regulatory factors such as GATA3 and STAT3 to create Treg programs that specifically control each category of T helper cell response [49,50]. Taken together, it is clear that the T helper cell lineage-defining transcription factors are not as exclusively expressed as had been previously thought, and in fact, their co-expression in some settings is actually important for establishing the phenotype of the cell.
Current views and future directions
Ongoing research examining epigenetic events in T helper cells has brought with it a new appreciation for the flexibility of T helper cell phenotypes. Indeed, studies now suggest that changing environmental conditions impact the cytokine expression profile of the T helper cell [20,26]. This concept has broad implications in applications of adoptive T cell immunotherapy. For instance, if T helper cells are introduced into an environment that promotes an opposing regulatory program, the flexibility of the T helper cell to respond to this new environment may result in their conversion into the opposing phenotype. This change in phenotype would then prevent the therapeutically introduced cells from helping alleviate the pathogenic state, and instead they may actually exacerbate it. Thus, without true stability in the T helper cell phenotype, great caution will need to be taken with the introduction of T helper cells for the purpose of treating chronic pathogenic conditions. Future research will now be important to determine whether there are specific circumstances that promote the stable, terminal differentiation of the T helper cells or rather T helper cells always retain some degree of flexibility in their phenotype.
Highlights.
- T-bet, STAT1, and STAT4 are required for Th1 cell differentiation.
- Epigenetic studies have contributed to our understanding of stability versus flexibility in T helper cells.
- T-bet interacts with the H3K27-demethylase Jmjd3 to functionally remove repressive epigenetic modifications at its target genes.
- T-bet interacts with several chromatin remodeling complexes to create a permissive epigenetic state.
- The co-expression of T helper cell lineage-defining transcription factors contributes to establishing the phenotype of T helper cells.
Acknowledgments
We thank Weinmann lab members for enthusiastic discussions on this topic. Grants from the NIAID (AI061061 and AI07272) and the American Cancer Society (RSG-09-045-01-DDC) to A.S.W support the research in the authors’ lab.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crotty S. Follicular Helper CD4 T Cells (T(FH)) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller SA, Weinmann AS. Common themes emerge in the transcriptional control of T helper and developmental cell fate decisions regulated by the T-box, GATA and ROR families. Immunology. 2009;126:306–315. doi: 10.1111/j.1365-2567.2008.03040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 7.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 9.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 10.Schulz EG, Mariani L, Radbruch A, Hofer T. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity. 2009;30:673–683. doi: 10.1016/j.immuni.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood. 2007;110:2494–2500. doi: 10.1182/blood-2006-11-058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. Together with *13, **14, and **15 demonstrate that T-bet physically interacts with other transcription factors important for alternative T helper cell developmental decisions to functionally repress the opposing subtype specific gene expression programs and promote Th1 development. [DOI] [PubMed] [Google Scholar]
- 13*.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 14**.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 17.Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology. 2009;126:299–305. doi: 10.1111/j.1365-2567.2008.03026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol. 2010;11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oestreich KJ, Weinmann AS. Encoding Stability Versus Flexibility: Lessons Learned From Examining Epigenetics in T Helper Cell Differentiation. Curr Top Microbiol Immunol. 2011 doi: 10.1007/82_2011_141. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amsen D, Spilianakis CG, Flavell RA. How are T(H)1 and T(H)2 effector cells made? Curr Opin Immunol. 2009;21:153–160. doi: 10.1016/j.coi.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 25.Rudensky AY. Regulatory T cells and Foxp3. Immunol Rev. 2011;241:260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, Anderson SM, Wei L, Sun H, O’Shea JJ, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–632. doi: 10.1016/j.immuni.2011.07.015. Together with **28 and **29, these studies have examined the epigenetic states for multiple T helper cell subtypes and have provided new insight into the concept of flexibility in T helper cell gene expression programs. In particular, they have found that T helper cell lineage-defining transcription factor loci are contained within a bivalent chromatin environment in a number of T helper cell subtypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. Together with **44, demonstrate that T-bet has the ability to alter the chromatin structure by recruiting H3K27-demethylase, H3K4-methyltransferase, and Brg1-dependent chromatin remodeling complexes to its target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28**.Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 32.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 33.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 35.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirahara K, Vahedi G, Ghoreschi K, Yang XP, Nakayamada S, Kanno Y, O’Shea JJ, Laurence A. Helper T-cell differentiation and plasticity: insights from epigenetics. Immunology. 2011;134:235–245. doi: 10.1111/j.1365-2567.2011.03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 38.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W, Chang S, Aune TM. Long-range histone acetylation of the Ifng gene is an essential feature of T cell differentiation. Proc Natl Acad Sci U S A. 2004;101:2440–2445. doi: 10.1073/pnas.0306002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 41.Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Lewis MD, Miller SA, Miazgowicz MM, Beima KM, Weinmann AS. T-bet’s ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol Cell Biol. 2007;27:8510–8521. doi: 10.1128/MCB.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Lohning M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 47.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, O’Brien S, Blank R, Lamb E, Natarajan S, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]