Integrative genomic analysis of HIV-specific CD8+ T cells reveals that PD-1 inhibits T cell function by upregulating BATF (original) (raw)
. Author manuscript; available in PMC: 2012 Apr 16.
Published in final edited form as: Nat Med. 2010 Oct 3;16(10):1147–1151. doi: 10.1038/nm.2232
Introductory Paragraph
CD8+ T cells in chronic viral infections like HIV develop functional defects such as loss of IL-2 secretion and decreased proliferative potential that are collectively termed exhaustion1. Exhausted T cells express increased levels of multiple inhibitory receptors, such as Programmed Death 1 (PD-1)2,3 that contribute to impaired virus-specific T cell function. Reversing PD-1 inhibition is therefore an attractive therapeutic target, but the cellular mechanisms by which PD-1 ligation results in T cell inhibition are not fully understood. PD-1 is thought to limit T cell activation by attenuating T cell receptor (TCR) signaling4,5. It is not known whether PD-1 also acts by upregulating genes in exhausted T cells that impair their function. Here, we analyzed gene-expression profiles from HIV-specific CD8+ T cells in patients with HIV and show that PD-1 coordinately upregulates a program of genes in exhausted CD8+ T cells from humans and mice. This program includes upregulation of basic leucine transcription factor, ATF-like (BATF), a transcription factor in the AP-1 family. Enforced expression of BATF was sufficient to impair T cell proliferation and cytokine secretion, while BATF knockdown reduced PD-1 inhibition. Silencing BATF in T cells from chronic viremic patients rescued HIV-specific T cell function. Thus inhibitory receptors can cause T cell exhaustion by upregulating genes – such as BATF – that inhibit T cell function. Such genes may provide new therapeutic opportunities to improve T cell immunity to HIV.
We hypothesized that receptors like PD-1 function to inhibit T cells not only by reducing TCR signaling, but also by inducing the expression of genes that impair T cell function. To test this hypothesis, we queried gene expression profiles from HIV-specific CD8+ T cells for upregulation of PD-1 induced genes.
The majority of individuals infected with HIV show chronic elevation of viral load in the absence of anti-retroviral therapy (progressors), associated with defects in HIV-specific T cell cytokine secretion, proliferation and survival6,7. In contrast, spontaneous control of viral replication has been documented for a small minority of individuals (controllers)8. Analysis of CD8+ T cell responses to HIV in progressors and controllers therefore allows a comparison of populations of human antigen-specific T cells at the extremes of functional competence.
We sorted CD8+ T cells specific for epitopes from the Gag protein (hereafter termed HIV-specific CD8+ T cells) from 18 progressors and 24 controllers (Fig. 1a, Supplementary Fig. 1, and Supplementary Table 1). The gene expression profiles of HIV-specific CD8+ T cells from progressors showed marked differences to those from controllers (n = 518 genes, moderated t-statistic < −2.0, Fig. 1b and Supplementary Table 2). Genes upregulated in HIV-specific CD8+ T cells from progressors were enriched for those involved with the interferon response and MHC expression (Supplementary Table 3), consistent with a higher viral load in progressors. HIV-specific CD8+ T cells from controllers were enriched for genes involved in mRNA transcription and protein translation, consistent with prior observations of defects seen in the mouse model of chronic LCMV infection9 (Supplementary Table 4). We therefore compared the expression profiles of HIV-specific CD8+ T cells to exhausted LCMV-specific CD8+ T cells from the mouse model9. Using an analytic technique called gene set enrichment analysis (GSEA)10–12 (Supplementary Methods) we analyzed the expression profiles of murine virus-specific CD8+ T cells during infection with each of two strains of LCMV: clone 13 (Cl13), which gives rise to chronic infection with T cell exhaustion; and Armstrong (Arm), an acute infection that does not cause T cell exhaustion9,13. We found that the HIV progressor signature was significantly enriched in the profiles of exhausted LCMV-specific CD8+ T cells from Cl13 infection (P = 4.8 × 10−5, Supplementary Fig. 2), suggesting global similarity between the transcriptional profiles of exhausted CD8+ T cells in humans and in the mouse model.
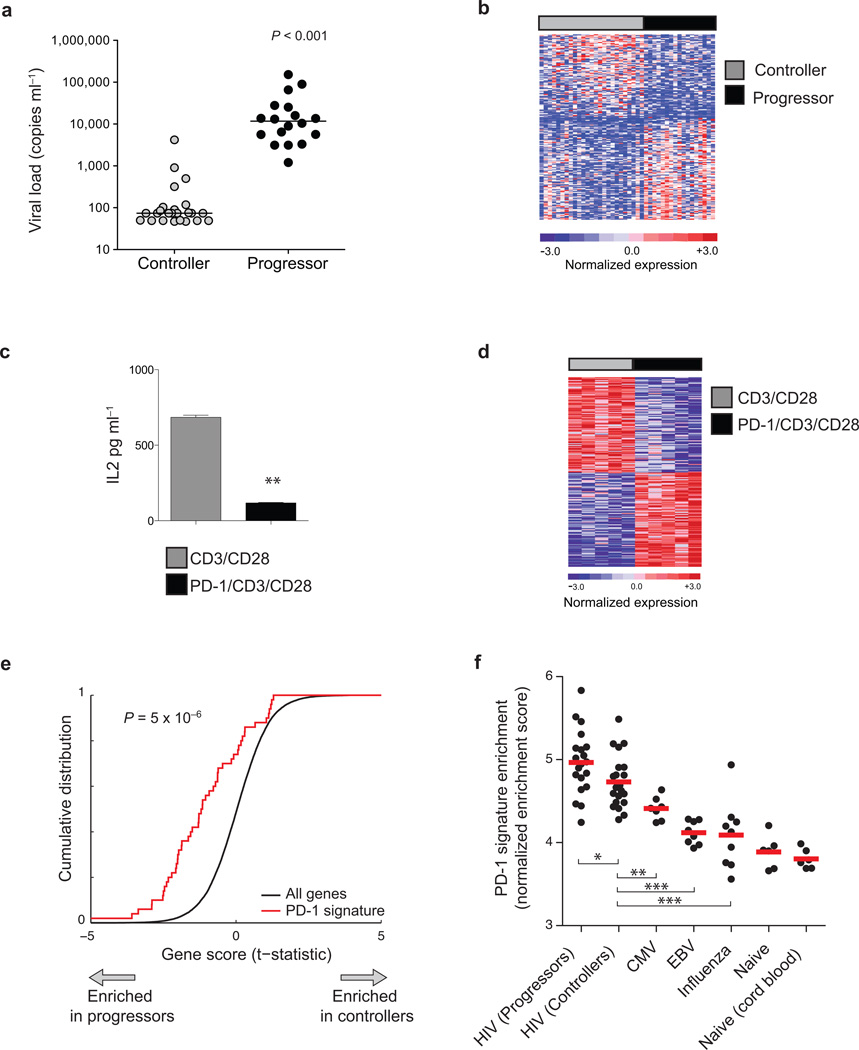
Figure 1. Transcriptional profiles of HIV-specific CD8+ T cells show coordinate upregulation of genes induced by PD-1 signaling.
(a) HIV viral load in controllers (grey circles) and progressors (black circles). Horizontal lines indicate median viral load in each cohort. (b) Genes differentially expressed in Gag-specific CD8+ T cells from controllers (grey bars) or progressors (black bars) ranked by moderated t–statistic. Each column represents an individual sample and each row an individual gene, colored to indicate normalized expression. The top 200 genes in either direction are shown. (c) IL-2 secretion from PD-1 expressing Jurkat cells cultured with inhibitory PD-1/CD3/CD28 beads (black bar) or control CD3/CD28 beads (grey bar) measured by ELISA (**P = 0.007). (d) Differentially expressed genes in PD-1 Jurkat cells cultured as in c. The top 100 differentially expressed genes from either condition are shown. (e) Enrichment analysis of PD-1 signature in HIV-specific CD8+ T cell profiles. The top 200 genes in the PD-1-specific signature were tested for enrichment in the rank-ordered list of genes differentially expressed in progressor vs. controller HIV-specific CD8+ T cells. X-axis indicates the t–statistic measured for each of the ~20,000 genes assayed in HIV-specific T cells, ranked in order of their differential expression in progressor vs. controller classes. Y-axis indicates the cumulative distribution of all genes (dotted lines) or of a set of 200 PD-1 signature genes (black line). Gene sets that are related to the class distinction on the X-axis would be expected to deviate from the dotted line (i.e. shifted towards the left if enriched in profiles of CD8+ T cells from progressors). (f) Enrichment of PD-1 signature in tetramer-sorted CD8+ T cells specific for different human viral pathogens. PD-1 signature genes were tested for enrichment by single-sample enrichment analysis in gene expression profiles from sorted tetramer+ CD8+ T cells specific for pathogens shown, or in naive CD8+ T cells. Each point represents the relative enrichment of PD-1 signature genes in an individual sample. Y-axis indicates normalized enrichment score (*indicates P < 0.05; **P < 0.01; ***P < 0.001 by Wilcoxon ranked-sum test).
We next asked if this exhausted CD8+ signature was influenced by PD-1 signaling. To do this, we first identified the genes upregulated following PD-1 ligation. We incubated PD-1 expressing Jurkat cells with beads coated with a cross-linking antibody to PD-1 together with antibodies to CD3 and CD28 (PD-1/CD3/CD28 beads); or with beads coated with equivalent amounts of control antibody together with CD3 and CD28 (CD3/CD28 beads). Incubation with PD-1/CD3/CD28 beads significantly decreased production of IL-2 compared to cells incubated with CD3/CD28 beads (P = 0.007, Fig. 1c) as previously observed14,15. Microarray analysis identified over one thousand genes that were significantly upregulated in cells functionally inhibited by PD-1 (n = 1179, t > 2.0, Fig. 1d and Supplementary Table 5). A similar number of genes was reduced in expression following PD-1 ligation (n = 1361, t < −2.0, Fig. 1d and Supplementary Table 5). We validated 13 representative genes that were upregulation in PD-1-ligated Jurkat cells. Incubation of human CD4+ T cells with PDL1-Ig/CD3/CD28 beads led to the coordinate upregulation of these representative PD-1 signature genes in a PDL1-Ig dose-dependent manner (Supplementary Fig. 3). Thus ligation of PD-1 in CD3/CD28 stimulated cells induces a specific transcriptional program in both Jurkat cells and primary human T cells.
We sought to determine whether the transcriptional program induced by PD-1 signaling defined in vitro could be detected in gene expression profiles from exhausted CD8+ T cells ex vivo. We therefore tested whether PD-1 induced genes were coordinately upregulated in HIV-specific CD8+ T cells from HIV Progressors. Using enrichment analysis, we found that a set of PD-1 signature genes was significantly upregulated in the HIV progressors compared with controllers (P = 5 × 10−6, Fig. 1e). Similarly, we found that PD-1 signature genes were significantly upregulated in exhausted LCMV-specific CD8+ T cells from Cl13 infection compared with Armstrong infection (P = 2 × 10−4, Supplementary Fig. 4). Thus PD-1 ligation results in upregulation of a consistent pattern of genes in exhausted CD8+ T cells in humans and mice.
The upregulation of PD-1 signature genes in exhausted CD8+ T cells contrasted with that seen in profiles of human virus-specific CD8+ T cells associated with functional T cell responses. Using single-sample GSEA (Supplementary Methods), we found that the PD-1 signature was significantly more enriched in HIV-specific CD8+ T cells than in antigen-specific CD8+ T cells specific for CMV (P < 0.01), EBV (P < 0.001), or influenza virus (P < 0.001) from healthy HIV-uninfected donors (Fig. 1f). Notably, the PD-1 signature was significantly more enriched in HIV-specific T cells than in EBV-specific T cells, despite the fact EBV-specific T cells express PD-116,17. This suggests that the upregulation of PD-1 signature genes may not occur in all cells that express PD-1, but may reflect increased strength and duration of PD-1 signaling experienced by CD8+ T cells in the setting of chronic infection.
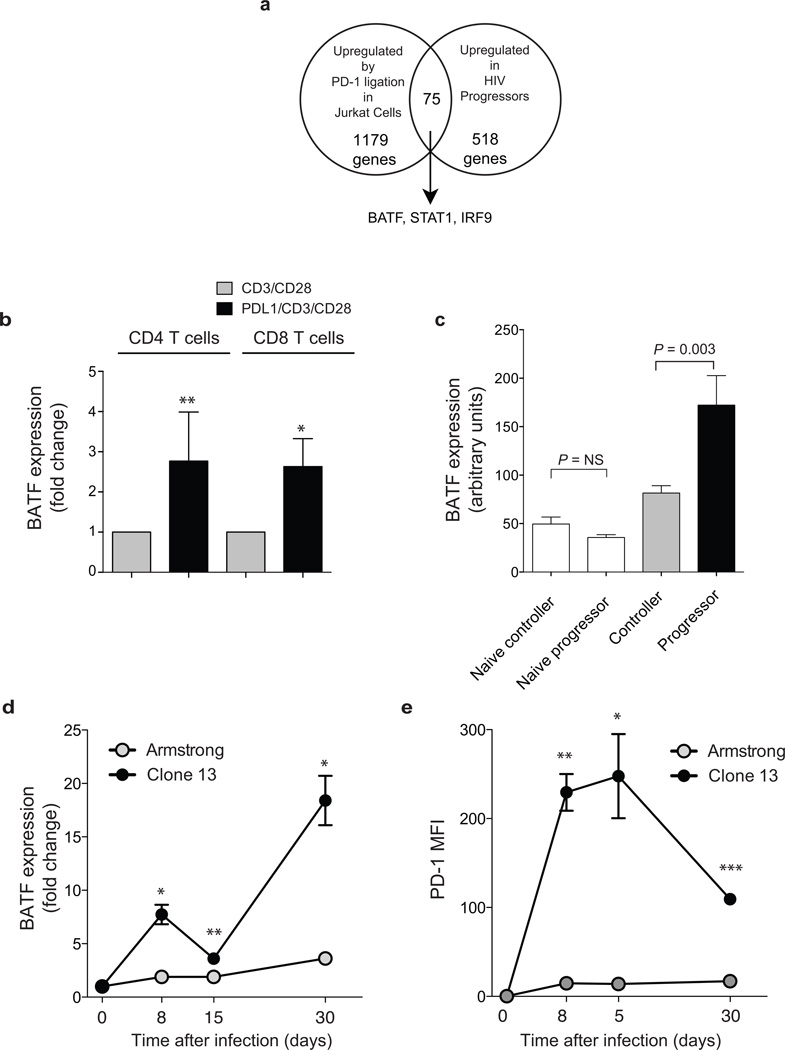
We reasoned that genes upregulated by PD-1 in exhausted CD8+ T cells might include those involved in the inhibition of T cell function. To refine the list of candidate genes, we identified genes that were both upregulated by PD-1 ligation in Jurkat cells and increased in HIV progressors (Fig. 2a and Supplementary Table 6). We focused on transcription factors because of their broad effect on the cell state. Of the 75 genes common to both gene sets, only three were transcription factors: BATF, STAT1 and IRF9 (Fig. 2a and Supplementary Table 6). We selected BATF for further analysis because it has been shown to function as a negative regulator of AP-1 activity18,19. Moreover, we have previously observed it to be upregulated during CD8+ memory differentiation in humans and mice20, suggesting that it may play a conserved role in regulating T cell function. BATF expression showed a 2 – 3 fold increase in both primary human CD4+ and CD8+ T cells after incubation with PDL1/CD3/CD28 beads compared with CD3/CD28 beads, indicating that BATF expression is increased by PD-1 ligation in vitro (P = 0.001 and P = 0.02, respectively, Fig. 2b).
Figure 2. Expression of BATF is upregulated by PD-1 and increased in exhausted T cells.
(a) Venn diagram representation of three transcription factors upregulated in Gag-specific T cells from HIV progressors and Jurkat cells after PD-1 ligation (t > 2.0). (b) BATF expression measured by real-time quantitative PCR in primary human CD4+ (left bars) and CD8+ (right bars) T cells cultured with CD3/CD28 beads (grey bars) or PDL1/CD3/CD28 beads (black bars) for 4 days. Data represent independent experiments with 4 – 10 normal donors and displayed as expression relative to CD3/28 condition (**P = 0.001; *P = 0.02; paired t test). (c) Relative BATF expression in arbitrary expression units from Affymetrix analysis of sorted naïve (CD62L+CD45RA+, white bars) or HIV gag-specific CD8+ T cell populations from controllers (grey bar) and progressors (black bar). (d) Batf expression measured by real-time quantitative PCR in LCMV-specific CD8+ T cells from mice infected with LCMV Armstrong (grey symbols), or LCMV clone 13 (black symbols) relative to naive (*P < 0.05; **P < 0.01). (e) PD-1 expression on LCMV-specific CD8+ T cells measured by flow cytometry following infection with the viruses indicated.
High BATF levels were seen in antigen-specific T cells with the greatest degree of dysfunction. BATF expression measured by microarray was significantly higher in exhausted HIV-specific CD8+ T cells from progressors than in HIV-specific T cells from controllers (P = 0.003, Fig. 2c). However BATF expression was not significantly different in naïve CD8+ T cells from controllers or progressors (P = NS, Fig. 2c). A significant correlation existed between microarray and RT-PCR measurements of BATF expression (Rs 0.53, P = 0.02, n = 18, Supplementary Fig. 5).
In order to define the kinetics of BATF expression following infection with a persistent virus, we compared BATF expression in murine virus-specific CD8+ T cell during acute and chronic infection (Fig. 2d). As early as day 8 post-infection, DbGP33-specific CD8+ T cells in Cl13 infection expressed significantly higher levels of BATF than in Arm infection (P = 0.02). BATF expression was maintained at higher levels in virus-specific cells in Cl13 infection at day 15 and by day 30 was ~7 fold higher than in DbGP33-specific CD8+ T cells generated during LCMV Arm infection (P = 0.02). The increased expression of BATF during acute and chronic infection was coincident with the upregulation of PD-1 as GP33-specific T cells showed increased levels of both PD-1 and BATF by day 8 (Fig. 2d). Increased BATF expression is therefore an early and persistent feature of exhausted CD8+ T cells in the setting of chronic viral infection in vivo and is temporally correlated with upregulation of PD-1.
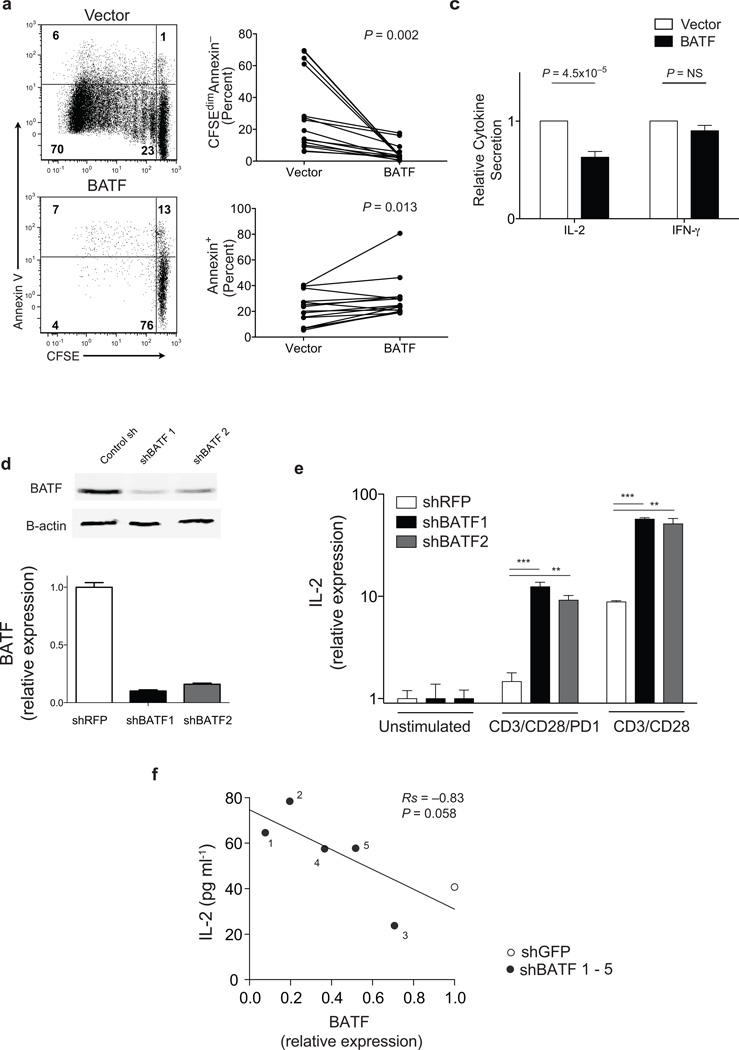
We next tested whether BATF could inhibit T cell function. Overexpression of BATF in primary human T cells (Supplementary Fig. 6a) markedly reduced proliferation in response to CD3/CD28 (P = 0.002, Fig. 3a,b). Apoptosis was also slightly increased in BATF overexpressing cells following stimulation (29% vs. 20%, P = 0.013, Fig. 3a,b), consistent with the previous defined role of PD-1 signaling in reducing cell survival21. However, the majority of BATF overexpressing cells were viable, suggesting that reduced proliferation was not simply from cell death. Overexpression of BATF also significantly reduced IL-2 secretion following CD3/CD28 stimulation, (P = 4.5 × 10−5, Fig. 3c), but was not overtly toxic because IFN-γ secretion was not significantly reduced compared with vector controls. Thus increased expression of BATF reduces proliferation and IL-2 secretion in primary human T cells.
Figure 3. BATF inhibits T cell function.
(a) CFSE-labeled primary human CD4+ or CD8+ T cells from healthy volunteers transduced with a lentivirus expressing BATF (lower plot) or with control vector (upper plot) and cultured for 4 days with CD3/CD28 beads. (b) Summary data of proliferation (percent CFSEdimAnnexin−, upper plot), and cell death (percent Annexin V+, lower plot) in primary human CD4+ or CD8+ T cells (n = 14) transduced as in (a) and cultured for 4 days with CD3/CD28 beads. (c) IL-2 (left bars, P = 4.5 × 10−5) and IFN-γ (right bars, _P_=NS) secretion by primary human CD4+ T cells (n = 10) transduced as in (a) and cultured with CD3/CD28 beads. Data is shown normalized to the empty vector condition. (d) BATF expression in PD-1 expressing Jurkat cells lentivirally transduced with shGFP (control) or two separate shBATF sequences measured by western blot (upper panel) or quantitative PCR (lower panel). (e) IL-2 expression by PD-1 Jurkat cells transduced with shRFP (control, white bar) or shBATF (black, grey bars) cultured with no beads or either PD-1/CD3/CD28 or CD3/CD28 beads as indicated for 18 hours. Data shows IL-2 expression normalized (+/− SEM) measured by quantitative PCR (***P < 0.001; **P = 0.01). Data are normalized to β-ACTIN and presented as fold change with respect to unstimulated conditions. (f) Correlation between BATF silencing and IL-2 secretion in PD-1 expressing Jurkat cells transfected with five sequence independent shBATF constructs (black symbols) or a control hairpin (grey symbol) and cultured with PD-1/CD3/CD28 beads. BATF expression was measured by quantitative PCR and presented as fold-change relative to control hairpin.
Enforced expression of BATF in vitro did not increase the expression of PD-1 itself, or of two other inhibitory receptors (CD244 or CD160) in CD8+ or CD4+ T cells (Supplementary Fig. 6b–e). This suggests that BATF does not mediate inhibition by modulating the expression of these inhibitory receptors. Future studies will be required to determine if BATF regulates other components of the PD-1 induced expression signature.
We next asked whether depletion of BATF enhanced T cell function, using shRNA-mediated gene-silencing (Fig. 3d,e). Compared with control hairpins, depletion of BATF in Jurkat cells with two different shRNA sequences (Fig. 3d) significantly increased IL-2 expression in cells cultured with PD-1/CD3/CD28 (P < 0.01, Fig. 3e), reversing inhibition to levels seen in CD3/CD28 stimulated cells. Testing additional hairpin sequences showed that there was a strong correlation between the extent of knockdown and degree of increase in IL-2 secretion, confirming the on-target specificity of BATF silencing (Rs −0.82, P = 0.056; Fig. 3f).
BATF silencing also increased IL-2 expression in cells stimulated with CD3/CD28 without exogenous PD-1 cross-linking (P < 0.01, Fig. 3e), suggesting that pathways in addition to PD-1 could inhibit cell activation via BATF. Silencing BATF may therefore have the effect of increasing IL-2 expression not only by relieving PD-1-mediated inhibition, but also by impairing other negative feedback pathways. Consistent with this, the expression of BATF across 42 samples of HIV-specific CD8+ T cells was significantly correlated with expression levels of several receptors with known or putative inhibitory function (Supplementary Fig. 7)22,23.
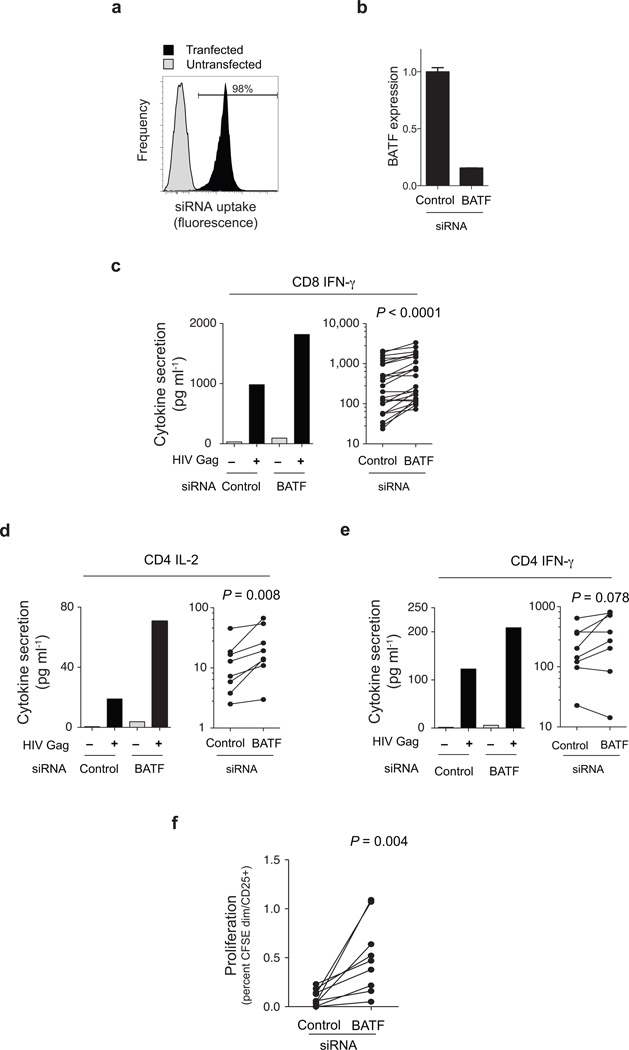
Finally, we tested whether silencing BATF would improve the function of HIV-specific T cells. HIV-specific T cell function after BATF knockdown (Fig. 4a, b) was assessed by measuring cytokine secretion or proliferation in response to Gag peptides. BATF knockdown caused a significant increase in CD8+ Gag-specific IFN-γ secretion (Fig. 4c) compared to a control siRNA pool, increasing IFN-γ secretion an average of 60% (P < 0.0001). Similar results were seen in HIV-specific CD4+ T cells where silencing BATF caused a two-fold increase in Gag-specific IL-2 secretion (P = 0.008, Fig. 4d) and a trend towards increase IFN-γ secretion (P = 0.078, Fig. 4e). HIV-specific CD8+ T cell proliferation was also increased by BATF knockdown, with a 5-fold increase in proliferating cells incubated with optimal Gag peptides (P = 0.004, Fig. 4f). Reducing BATF expression therefore increases the function of exhausted HIV-specific T cells.
Figure 4. BATF silencing improves HIV-specific T cell function.
(a) Efficacy of siRNA uptake in CD3+ T cells cultured with mixture of siRNA pool and fluorescent oligonucleotides (to monitor transduction) either with (black histogram) or without (grey histogram) electroporation. (b) Silencing of BATF by siRNA sequences targeting BATF in CD3+ T cells from a representative chronic progressor measured by quantitative PCR. Expression (mean, SEM) normalized to a housekeeping gene is presented as fold change relative to control siRNA (c–e) BATF silencing enhances HIV-specific cytokine secretion in CD8+ (c) and CD4+ (d,e) T cells from chronic progressors. PBMC depleted of CD4+ (a) or CD8+ (b,c) T cells were electroporated with siRNA pools targeting the genes indicated and cultured with or without HIV Gag peptides for four days, and IFN-γ (c,e) or IL-2 (d) measured using a highly-sensitive cytokine bead assay. In each figure the left panel shows a representative patient, and the right panel summary data (CD8+ responses, 26 HIV epitope responses in 4 subjects; CD4+ responses, HIV Gag peptide pool in 7 subjects). Cytokine levels shown are adjusted for background secretion, and statistical significance evaluated with the paired t test. (f) Proliferation of CFSE CD8+ T cells was measured by fraction of CFSEdim, CD25+ cells six days after transfection and peptide stimulation of PBMCs. Data represent nine HIV epitope-specific responses in 4 subjects.
We show that exhausted T cells specific for HIV in humans and LCMV in mice share a common expression signature that reflects the transcriptional consequences of PD-1 receptor ligation. Our data therefore suggest a model in which inhibitory receptors such as PD-1 may mediate T cell exhaustion not only by limiting TCR signaling, but also by inducing the expression of genes such as BATF that inhibit T cell function.
BATF is a highly conserved member of the AP-1/ATF family, a group of transcription factors that regulate many aspects of cellular function in the immune system24. Recent studies show that BATF is required for Th17 and follicular T helper cell differentiation25,26. BATF may therefore be one of a number of transcription factors, such as Blimp-127,28, that have distinct, context-dependent roles both in regulating the function of T cells responding to chronic viral infection and in CD4+ lineage decisions. Our studies do not identify the mechanism by which PD-1 ligation induces BATF upregulation, or whether this happens by direct or indirect pathways. However, our findings give impetus to further studies of how BATF regulates T cell state.
Blockade of PD-1:PDL-1 interactions partially reverses T cell dysfunction16,17 and improves control of viral replication2,29, indicating that the function of exhausted T cells can be rescued even in settings of viral persistence. BATF and the pathways that control its activity may provide new opportunities to reverse CD8+ T cell exhaustion. Integrated genomic analysis of the T cell response in humans may therefore provide a general approach to identifying novel regulators of T cell function that are potential therapeutic targets for improving T cell immunity in chronic infection.
Methods
Subjects
Subjects were recruited from outpatient clinics at local Boston hospitals, or referred from providers throughout the US, following institutional review board approval (Partners IRB) and written informed consent. HIV controllers included elite controllers (n = 20) with HIV RNA below the level of detection for the respective available ultrasensitive assay (< 75 copies ml−1 by DNA or < 50 copies ml−1 by ultrasensitive PCR); and viremic controllers (n = 4) with HIV RNA levels < 2000 copies ml−1. Chronic progressors (n = 18) were defined as having HIV RNA levels above 2,000 copies (Supplementary Table 1). All patients were off therapy and had detectable HIV-specific CD8+ T cells in the peripheral blood, allowing a median of 21,500 HIV Gag tetramer+ T cells (range 3,000 – 85,000 cells) to be isolated for microarray analysis from each patient.
Flow cytometry and sorting
PBMC were isolated via density centrifugation and were stained with a cocktail of antibodies to exclude irrelevant lineages and dead cells, anti-CD8 and MHC Class I HIV-Gag-specific tetramers to identify the antigen-specific populations, and antibodies against CD62L and CD45RA for memory phenotype characterization of the tetramer+ fraction. CD8+tetramer+ cells were sorted using a FACSAria Cell Sorter (BD Biosciences). All experiments examining proliferation via CFSE dilution and survival via Annexin V (BD Biosciences) staining were collected on a FC500 flow cytometer (Beckman Coulter). Analysis of flow cytometry data was carried out using FlowJo software (version 8.8.6, Tree Star)
Microarray data acquisition and analysis
Tetramer-sorted human CD8+ T cells or Jurkat cells following 18h of bead stimulation were pelleted and resuspended in TRIzol. RNA extraction was performed using the RNAdvance Tissue Isolation kit (Agencourt). Concentrations of total RNA were determined using a Nanodrop spectrophotometer or Ribogreen RNA quantitation kit (Molecular Probes/Invitrogen). RNA purity was determined by Bioanalyzer 2100 traces (Agilent Technologies). Total RNA was amplified using the WT-Ovation Pico RNA Amplification system (NuGEN) according to the manufacturer’s instructions. Following fragmentation and biotinylation, cDNA was hybridized to Affymetrix HT HG-U133A or HG-U133A2.0 microarrays. Microarray data for CMV, EBV, and influenza specific CD8+ T cells and for LCMV-specific T cells were obtained from previous studies9,20. Detailed description of microarray data analysis can be found in Supplementary Methods.
Quantitative PCR
Expression of BATF following in vitro stimulation of primary human T cells, shRNA and overexpression experiments was determined by real-time quantitative PCR using Taqman gene expression assays for BATF (assay #Hs00232390_m1) and β-ACTIN (Hs00357333_g1) which served as a loading and normalization control. For LCMV mouse experiments, RNA was converted to cDNA using a high capacity cDNA kit and RT-PCR primers were Taqman assays (Batf, Gapdh, Hprt-1; Applied Biosystems). Expression levels were compared using the relative quantification method, comparing Batf expression to either Gapdh or Hprt-1 housekeeping genes. Quantitative multiplex RT-PCR via ligation-mediated amplification was carried out as previously described30. PD-1 signature genes were selected for the multiplex validation panel based using criteria previously established30 and sequences for primer sets are available upon request.
Mouse model of LCMV infection
Groups of mice were infected with either LCMV Armstrong or LCMV clone-13. At 8, 15, or 30 days post-infection, spleens were harvested and splenocytes pooled prior to CD19 depletion and sorting. LCMV tetramer specific (DbGP33) CD8+ T cells were sorted using a BD FACSAria directly into siliconized 1.5 ml tubes containing Trizol LS, followed by RNA extraction. Separate aliquots of cells were stained for PD-1 expression as described previously9.
BATF siRNA knockdown in HIV samples
PBMCs from untreated, chronically HIV-infected individuals were isolated with density gradient centrifugation. Inhibition of BATF expression was achieved through siRNA transfection by electroporation on a Gene Pulser XCell (BioRad). Fifteen million cells were resuspended in 300 µl of Opti-MEM in a 2-mm cuvette and pulsed with 1 nmol of siRNA (ON-TARGET Non-Targeting pool or BATF ON-TARGETplus SMARTpool, Dharmacon). Pulse conditions were designed to maximize electroporation efficiency in T cells (a unique square wave with a pulse of 360 V and a duration of 5 ms), and transfection efficiency assayed with siGlo fluorescent oligonucleotides (Dharmacon). For assessment of CD4+ T cell cytokine responses, CD8+ T cell depleted PBMCs (RosetteSep CD8+ depletion reagents; StemCell) were stimulated with an HIV Gag peptide pool (1 µg ml−1 peptide−1), or left unstimulated. For HIV-specific CD8+ T cell cytokine responses, non-depleted PBMCs were stimulated with 0.2 µg ml−1 of HIV optimal epitopes. After a 96-hour incubation, IFN-γ and IL-2 levels were measured. Proliferation of CD8+ T cells was measured 6 days after transfection and stimulation using CFSE assay as published before18.
Additional methods
Detailed methodology is described in the Supplementary Methods.
Accession Codes
Gene Expression Omnibus
[data submitted, awaiting accession number]
Supplementary Material
1
Acknowledgments
The authors would like to thank the subjects for taking part in the study; E. Cutrell, B. Baker, K. Moss, A. Rathod and C. Brume for coordinating sample management; and A. Sharpe, T. Golub, G. Lauer, M. Altfeld, and H. Joffe for valuable discussions. This work was supported by National Institutes of Health grants AI082630, AI56299, HHSN26620050030C, HL092565, the International HIV Controllers Study (http://www.hivcontrollers.org), and the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative.
Footnotes
Author attributions
MQ designed and performed experiments, analyzed data and helped write the paper. FP designed the clinical components of the study. BN and JLJ designed and performed computational experiments. FP, DSK, JZ and DEK designed and performed siRNA experiments in samples from HIV patients. CF, QE, BJ, KB, SI, KR, IT, AP-T, DD, LF all performed experiments. GF designed experiments and developed PD-L1-Ig. JA, AC, HS, EJW designed and performed animal experiments, and analyzed data. WNH, BE and BDW conceived of the study and designed the experiments. WNH analyzed data and wrote the paper.
References
- 1.Zajac AJ, et al. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2005;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley JL. PD-1 signaling in primary T cells. Immunol. Rev. 2009;229:114–125. doi: 10.1111/j.1600-065X.2009.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- 6.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T-cells. Blood. 2006 doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 8.Pereyra F, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 9.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson B, Håkansson P, Johansson M, Nelander S, Fioretos T. Threshold-free high-power methods for the ontological analysis of genome-wide gene-expression studies. Genome Biol. 2007;8:R74. doi: 10.1186/gb-2007-8-5-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haining WN, Wherry EJ. Integrating genomic signatures for immunologic discovery. Immunity. 2010;32:152–161. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chemnitz JM, et al. RNA fingerprints provide direct evidence for the inhibitory role of TGFbeta and PD-1 on CD4+ T cells in Hodgkin lymphoma. Blood. 2007;110:3226–3233. doi: 10.1182/blood-2006-12-064360. [DOI] [PubMed] [Google Scholar]
- 15.Parry RV, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Biol. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 17.Trautmann L, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 18.Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–1763. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- 19.Williams KL, et al. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur. J. Immunol. 2001;31:1620–1627. doi: 10.1002/1521-4141(200105)31:5<1620::aid-immu1620>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Haining WN, et al. Identification of an evolutionarily conserved transcriptional signature of CD8 memory differentiation that is shared by T and B cells. J. Immunol. 2008;181:1859–1868. doi: 10.4049/jimmunol.181.3.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrovas C, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark GJ, Ju X, Tate C, Hart DN. The CD300 family of molecules are evolutionarily significant regulators of leukocyte functions. Trends Immunol. 2009;30:209–217. doi: 10.1016/j.it.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Foletta VC, Segal DH, Cohen DR. Transcriptional regulation in the immune system: all roads lead to AP-1. J. Leukoc. Biol. 1998;63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- 25.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009 doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 2010 doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velu V, et al. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature. 2009;458:206–210. doi: 10.1038/nature07662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haining WN, et al. High-throughput gene expression profiling of memory differentiation in primary human T cells. BMC Immunol. 2008;9:44. doi: 10.1186/1471-2172-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1