DDB2 promotes chromatin decondensation at UV-induced DNA damage (original) (raw)
In addition to its role in DNA lesion recognition, the damaged DNA-binding protein DDB2 elicits unfolding of large-scale chromatin structure independently of the CRL4 ubiquitin ligase complex.
Abstract
Nucleotide excision repair (NER) is the principal pathway that removes helix-distorting deoxyribonucleic acid (DNA) damage from the mammalian genome. Recognition of DNA lesions by xeroderma pigmentosum group C (XPC) protein in chromatin is stimulated by the damaged DNA-binding protein 2 (DDB2), which is part of a CUL4A–RING ubiquitin ligase (CRL4) complex. In this paper, we report a new function of DDB2 in modulating chromatin structure at DNA lesions. We show that DDB2 elicits unfolding of large-scale chromatin structure independently of the CRL4 ubiquitin ligase complex. Our data reveal a marked adenosine triphosphate (ATP)–dependent reduction in the density of core histones in chromatin containing UV-induced DNA lesions, which strictly required functional DDB2 and involved the activity of poly(adenosine diphosphate [ADP]–ribose) polymerase 1. Finally, we show that lesion recognition by XPC, but not DDB2, was strongly reduced in ATP-depleted cells and was regulated by the steady-state levels of poly(ADP-ribose) chains.
Introduction
Nucleotide excision repair (NER) is a versatile DNA repair pathway that removes a variety of structurally unrelated lesions from the genome, such as the UV light–induced pyrimidine-pyrimidone[6–4] photoproducts (6-4PPs) and cyclobutane pyrimidine dimers (CPDs). Inherited defects in NER give rise to the human disorder xeroderma pigmentosum (XP), which is characterized by extreme photosensitivity and high susceptibility to skin cancer (de Boer and Hoeijmakers, 2000). In mammalian cells, removal of photolesions by global genomic NER is initiated by the binding of the XP group C (XPC) protein to helix-distorting DNA lesions (Sugasawa et al., 1998; Volker et al., 2001). Although XPC has a high affinity for 6-4PPs, its binding to CPDs is rather weak, and efficient recognition of this type of lesion requires the presence of the damaged DNA-binding protein 2 (DDB2; Tang et al., 2000). Cells derived from XP-E patients, which lack functional DDB2, are deficient in CPD repair and show reduced 6-4PP repair (Hwang et al., 1999; Nichols et al., 2000; Tang et al., 2000; Rapić-Otrin et al., 2003; Moser et al., 2005). Genetic deletion of DDB2 in mice significantly impairs the repair of photolesions and causes hypersensitivity to UV-induced skin cancers, suggesting an important role for DDB2 in NER (Alekseev et al., 2005).
DDB2 is incorporated into a CUL4A–RING E3 ubiquitin ligase (CRL4) complex, consisting of CUL4A, RBX1, and DDB1, through its interaction with DDB1 (Groisman et al., 2003; He et al., 2006). CUL4A, DDB1, and DDB2 are rapidly recruited to UV-induced lesions, with similar association kinetics consistent with the binding of a preassembled CRL4–DDB2 complex (Luijsterburg et al., 2007; Alekseev et al., 2008). The ubiquitin ligase activity of the CRL4–DDB2 complex is transiently activated by UV irradiation and is specifically directed to chromatin at damaged sites (Groisman et al., 2003). Several proteins are ubiquitylated by the CRL4–DDB2 complex upon UV exposure, including the core histones H2A (Kapetanaki et al., 2006), H3 and H4 (Wang et al., 2006), XPC (Sugasawa et al., 2005), and DDB2 itself (Groisman et al., 2003; Sugasawa et al., 2005; Kapetanaki et al., 2006; Wang et al., 2006). Ubiquitylation of the core histones H3 and H4 by the CRL4–DDB2 complex weakens the interaction between the histones and DNA, which has been proposed to facilitate access of repair proteins to photolesions (Wang et al., 2006). Lesion recognition may be further enhanced by the CRL4–DDB2-mediated ubiquitylation of XPC, as this increases XPC’s affinity for DNA in vitro (Rapić-Otrin et al., 2002; Sugasawa et al., 2005). Finally, DDB2 itself is targeted for proteasomal degradation upon ubiquitylation by the CRL4–DDB2 complex, which may also enhance the binding of XPC to photolesions. Together, these studies suggest that the CRL4–DDB2 complex, through its ubiquitin ligase activity, initiates at least three simultaneous mechanisms that contribute to efficient recognition of photolesions by XPC.
In the present study, we identified a new role for DDB2, which involves the ATP- and poly(ADP-ribose) (PAR) polymerase (PARP)–dependent unfolding of higher-order chromatin structure at sites of DNA damage. Interestingly, this function of DDB2 is independent of its association with the CRL4 complex. Consistent with a role for DDB2-mediated chromatin unfolding in NER, we found that the recruitment of XPC, but not DDB2, to photolesions is ATP dependent and is regulated by the activity of PARP1. We propose that the DDB2-mediated chromatin decondensation establishes a local chromatin environment that promotes the recruitment of XPC to photolesions.
Results
Functional tethering of DDB2 to chromatin
To directly assess whether DDB2 can mediate changes in higher-order chromatin structure, we used a lactose repressor (LacR)–based system for tethering proteins to a defined chromosome region in vivo (Robinett et al., 1996). To this end, we fused full-length murine DDB2 to the LacR tagged with the RFP mCherry (mCherry-LacR; Fig. 1 A), which allows visualization and tethering of the fusion protein in mammalian cells carrying amplified lactose operator (LacO) sequences. Expression of mCherry-LacR-DDB2 in murine NIH2/4 cells, which contain an array of 256 copies of the LacO integrated in chromosome 3 (Soutoglou et al., 2007), resulted in localization of the fusion protein to the array (Fig. 1 B). Tethering of LacR-DDB2 resulted in enrichment of GFP-tagged DDB1 and CUL4A at the array, suggesting that the tethered DDB2 is part of the CRL4–DDB2 complex (Figs. 1 B and S1 A).
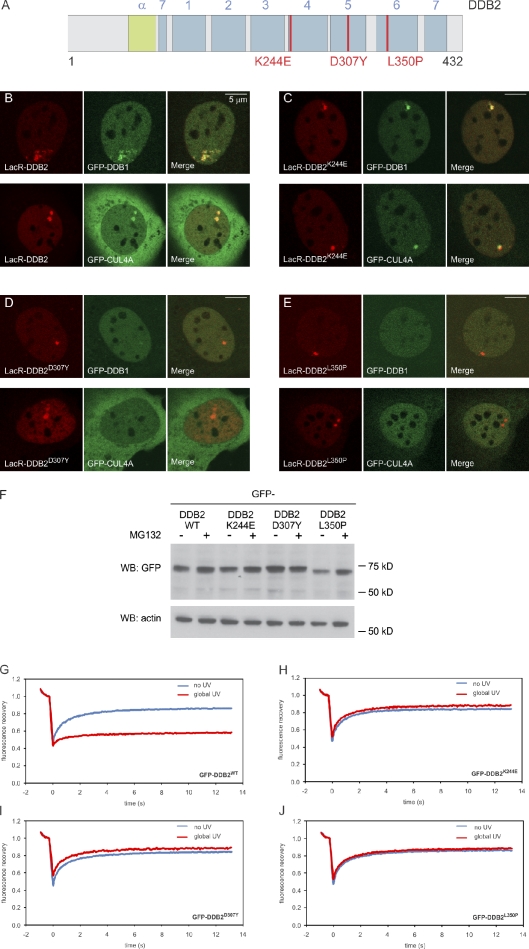
Figure 1.
Functional tethering of DDB2. (A) A schematic representation of murine DDB2 including mutations found in XP-E patients (in red). The seven WD40 β-propellers (1–7) and α-helical domain (α) are indicated. (B–E) Recruitment of GFP-DDB1 and GFP-CUL4A to tethered mCherry-LacR-DDB2 (B), mCherry-LacR-DDB2K244E (C), mCherry-LacR-DDB2D307Y (D), or mCherry-LacR-DDB2L350P (E). (F) Expression of GFP-DDB2 proteins in the absence (−) or presence (+) of proteasome inhibitor MG132 (10 µM). WB, Western blot; WT, wild type. (G–J) FRAP analysis of GFP-DDB2 (G), GFP-DDB2K244E (H), GFP-DDB2D307Y (I), or GFP-DDB2L350P (J) in unchallenged cells (blue lines) or globally (16 J/m2) UV-irradiated cells.
Next, we generated GFP fusion proteins of the naturally occurring DDB2 mutant proteins DDB2K244E, DDB2D307Y, and DDB2L350P (Fig. 1 A), which are found in the XP-E patients XP82TO, XP25PV, and GMO1389, respectively (Rapić-Otrin et al., 2003). Whereas the steady-state levels of GFP-tagged DDB2K244E and DDB2D307Y were comparable with the levels of wild-type DDB2, GFP-DDB2L350P was expressed at lower levels and was considerably stabilized by the proteasome inhibitor MG132 (Fig. 1 F), consistent with the reported destabilization of this mutant (Rapić-Otrin et al., 2003). Analysis of the mobility of the GFP-tagged DDB2 variants in living cells by FRAP showed that, in contrast to wild-type DDB2 (Fig. 1 G), the mutants DDB2K244E (Fig. 1 H), DDB2D307Y (Fig. 1 I), and DDB2L350P (Fig. 1 J) were not immobilized upon global UV exposure, suggesting that they are unable to bind photolesions, which may underlie the XP-E phenotype of cells expressing these mutant proteins.
Upon tethering mCherry-LacR fusion proteins of these mutants to chromatin, we observed that DDB2K244E (Figs. 1 C and S1 A) still targeted the CRL4 complex to the array, whereas DDB2D307Y (Figs. 1 D and S1 A) and DDB2L350P (Figs. 1 E and S1 A) failed to recruit DDB1 and CUL4A, consistent with previous biochemical experiments (Rapić-Otrin et al., 2003). Importantly, this panel of mCherry-LacR-DDB2 fusions that differ in their ability to recruit CUL4A and DDB1 provides us with a tool to dissect the contribution of ubiquitylation-dependent and -independent activities of DDB2 in living cells.
DDB2-mediated chromatin unfolding is independent of ubiquitylation and histone acetylation
To directly study the effect of DDB2 on chromatin structure, we tethered mCherry-LacR-DDB2 to a 90-Mbp heterochromatic locus in hamster AO3 cells consisting of ∼60 copies of a region encompassing ∼1,000 kbp of coamplified genomic DNA and ∼400 kbp of a tandemly arrayed 14-kbp plasmid, each of which contains 256 LacO repeats and a dihydrofolate reductase gene (Robinett et al., 1996; Tumbar et al., 1999; Ye et al., 2001; Nye et al., 2002). As reported previously, tethering of mCherry-LacR did not affect the heterochromatic nature of the array (Fig. 2 A; Tumbar et al., 1999). Conversely, tethering of mCherry-LacR-DDB2 caused extensive unfolding of the heterochromatic array into irregular, often fiberlike structures (Fig. 2 B), suggesting that prolonged binding or close proximity of DDB2 to chromatin triggers substantial decondensation. The array, which occupied ∼2% of the nuclear area after tethering mCherry-LacR, unfolded significantly to, on average, 14% of the nuclear area in cells expressing mCherry-LacR-DDB2 (Fig. 2 G, red bars). Tethering of DDB2 in murine NIH2/4 cells (Soutoglou and Misteli, 2008) harboring a small 256x LacO array (Figs. 2 H and S1 B) or in human U2OS 2–6-3 cells (Janicki et al., 2004) harboring a LacO-containing cassette (Figs. 2 I and S1 C) also resulted in extensive chromatin unfolding into fiberlike structures, suggesting that DDB2-mediated decondensation is a general phenomenon.
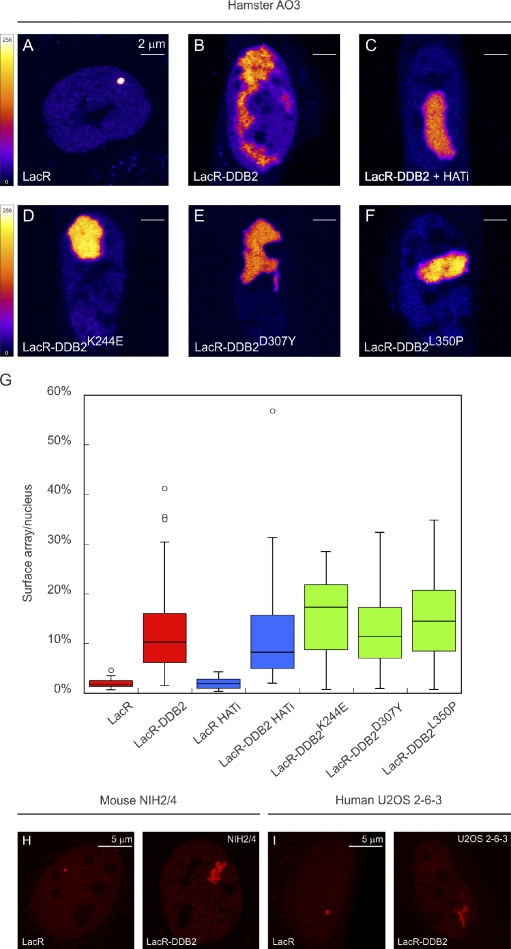
Figure 2.
DDB2 immobilization elicits extensive chromatin unfolding. (A–F) AO3 cells containing a 90-Mbp heterochromatic array were transfected with mCherry-LacR (A), mCherry-LacR-DDB2 in the absence (B) or presence (C) of HAT inhibitor (HATi), mCherry-LacR-DDB2K244E (D), mCherry-LacR-DDB2D307Y (E), or mCherry-LacR-DDB2L350P (F). Images show a confocal slice (1 Airy unit) of the nuclear distribution of the mCherry-LacR fusion proteins. The lookup table for the color coding is shown left of A and D. (G) Quantification of the relative array size (surface of the array/surface of the nucleus) is shown in a box plot (50–100 cells for each condition). Open circles are data points that are considered outliers, as their value is greater than the upper quartile, +1.5 the interquartile distance, or less than the lower quartile, −1.5 the interquartile distance. (H) Mouse NIH2/4 cells containing a 256x LacO array were transfected with mCherry-LacR (left) or mCherry-LacR-DDB2 (right). (I) Human U2OS 2–6-3 cells containing 200 copies of a LacO-containing cassette (∼4 Mbp) were transfected with mCherry-LacR (left) or mCherry-LacR-DDB2 (right).
It has been reported that DDB2 interacts with the p300 histone acetyltransferase (HAT; Datta et al., 2001), which prompted us to test whether an increase in histone acetylation could be responsible for the observed chromatin unfolding. However, tethering DDB2 in NIH2/4 mouse cells did not result in elevated levels of acetylated H3 (H3K4 and H3K9) or H4K16 at the decondensed array (Fig. S2). Moreover, chromatin unfolding in AO3 cells still occurred when LacR-DDB2 was tethered in the presence of the HAT inhibitor anacardic acid (Fig. 2, C and G [blue bars]).
The mutant DDB2K244E fails to bind both damaged and nondamaged DNA (Wittschieben et al., 2005), which allows us to assess whether chromatin unfolding involves DNA binding by DDB2. Tethering of DDB2K244E (Fig. 2 D) resulted in chromatin decondensation to the same extent as wild-type DDB2 (Fig. 2 G), suggesting that DDB2-mediated chromatin unfolding is independent of DNA binding by DDB2 and likely involves the recruitment of additional factors. To address whether the ubiquitin ligase activity of the CRL4–DDB2 complex is required to unfold chromatin, we used our panel of DDB2 mutants that differ in their ability to recruit CUL4A and DDB1. The DDB2D307Y (Fig. 2 E) and DDB2L350P mutants (Fig. 2 F), which fail to bind CUL4A and DDB1, triggered extensive unfolding of large-scale chromatin structure similar to wild-type DDB2 (compare red bars with green bars in Fig. 2 G). These results show that DDB2 elicits extensive higher-order chromatin unfolding independently of CRL4–DDB2-mediated ubiquitylation, DNA binding, or histone acetylation.
The first five blades of the DDB2 β-propeller are sufficient to mediate chromatin unfolding
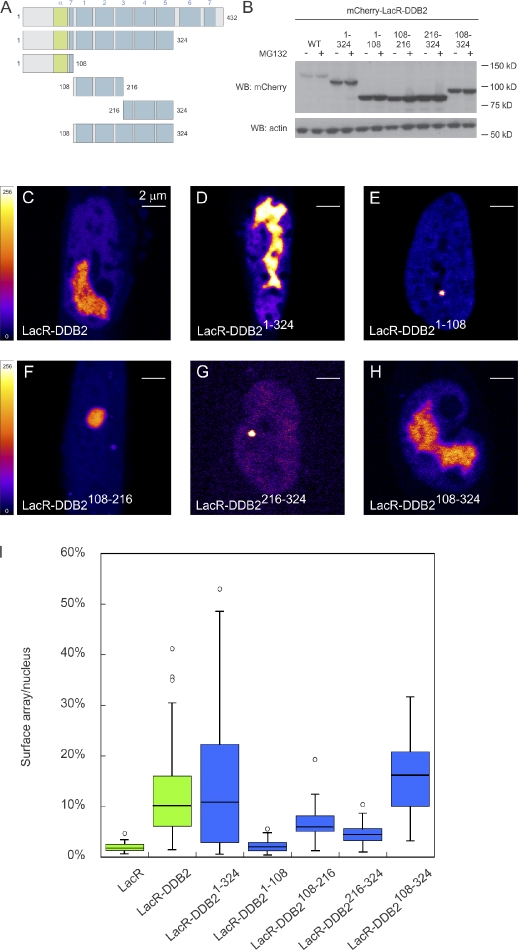
The structure of DDB2 consists of an N-terminal helix-loop-helix domain, which interacts with DDB1, followed by a seven-bladed WD40 β-propeller that is important for binding to UV-induced photolesions (Scrima et al., 2008). To determine whether a specific region of DDB2 is required for initiating chromatin unfolding, we created a series of deletion mutants fused to mCherry-LacR (Fig. 3 A). Importantly, the steady-state levels of these deletion mutants were not increased by proteasome inhibitors, suggesting that these mutants are relatively stable and not terminally misfolded (Fig. 3 B). Tethering of the deletion mutant DDB2(1–324) lacking the two C-terminal β-propeller blades resulted in unfolding of the LacO array, similar to wild-type DDB2 (Fig. 3, C, D, and I). To test which region of DDB2(1–324) is required to mediate chromatin decondensation, we generated three fragments spanning this region. The N-terminal DDB2(1–108) fragment, which is sufficient to recruit DDB1 (Fig. S3), failed to unfold chromatin (Fig. 3, E and I), whereas tethering the central fragment DDB2(108–216) (Fig. 3, F and I) as well as the C-terminal fragment of this region, DDB2(216–324) (Fig. 3, G and I), both resulted in partial unfolding. We then tested whether a region overlapping these deletion mutants can mediate efficient chromatin decondensation. Indeed, targeting of DDB2(108–324), which lacks the N-terminal helix-loop-helix domain but contains β-propeller blades 1–5, resulted in similar unfolding as full-length DDB2 (Fig. 3, H and I). Although capable of unfolding chromatin structure, DDB2(108–324) failed to interact with DDB1 (Fig. S3). Together, these results identify a region of the WD40 repeats spanning the first five β-propeller blades to be sufficient for chromatin unfolding. Moreover, our observation that the N-terminal domain of DDB2, which is sufficient for DDB1 binding, is not required to elicit chromatin unfolding provides another line of evidence for the CRL4-independent nature of this DDB2 activity.
Figure 3.
Minimal DDB2 domain that is sufficient for chromatin unfolding. (A) A schematic representation of DDB2 deletion mutants analyzed. (B) Expression of mCherry-LacR–tagged DDB2 proteins in the absence (−) or presence (+) of proteasome inhibitor MG132 (10 µM). WB, Western blot; WT, wild type. (C–H) Images show a confocal slice (1 Airy unit) of the distribution of the indicated mCherry-LacR fusion proteins in AO3 cells. The lookup table is shown. (I) Quantification of array decondensation in AO3 cells (in ratio array/nucleus) after tethering the indicated LacR-DDB2 fusion proteins shown in a box plot. Values represent the mean array decondensation of 50–100 cells collected in two independent experiments. Open circles are data points that are considered outliers, as their value is greater than the upper quartile, +1.5 the interquartile distance, or less than the lower quartile, −1.5 the interquartile distance.
UV-induced DNA lesions trigger ATP-dependent chromatin remodeling
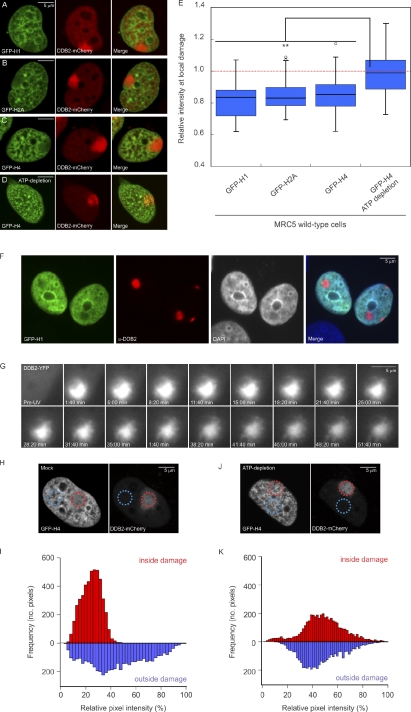
Having established that tethering of DDB2 to chromatin mediates extensive unfolding of chromatin structure, we subsequently addressed whether this phenomenon also occurs in the physiological context of the DNA damage response. For this purpose, we introduced photolesions by locally irradiating cells with UV-C light (Moné et al., 2001). At sites of local UV irradiation, marked by the accumulation of DDB2-mCherry, we observed a reduction in the density of GFP-tagged linker histone H1.0 (Fig. 4 A). As linker histones rapidly exchange within tens of minutes (Lever et al., 2000; Misteli et al., 2000), we wondered whether the distribution of the more immobile core histones also changes upon UV irradiation. Indeed, we observed a decrease in the density of the GFP-tagged core histones H2A (Fig. 4 B) and H4 (Fig. 4 C). Similar results were obtained when histone H4 was fused to super CFP (SCFP; Kremers et al., 2006) instead of GFP (Fig. S4). To exclude that the expression of DDB2-mCherry was responsible for the observed decrease in histone density, we locally UV irradiated cells expressing GFP-H1 and subsequently detected endogenous DDB2 using specific antibodies (Fig. 4 F). DNA damage sites marked by the accumulation of endogenous DDB2 showed a significant decrease in the density of GFP-H1 (Fig. 4 F), which was accompanied by a decrease in DAPI staining, indicating that the DNA density is also reduced (Figs. 4 F and S5). Likewise, local UV irradiation also triggered a local reduction in GFP-H4 density at sites marked by endogenous DDB2 accumulation (e.g., see Fig. 6 C). To quantify the loss of histones at sites of local UV-induced DNA damage, we measured the mean intensity in the irradiated area proportional to the mean nuclear intensity, which revealed an ∼15% reduction in histone density for each of the histones (H2A, H4, and H1.0; Fig. 4 E). Strikingly, depletion of ATP by the metabolic inhibitors 2-deoxyglucose and sodium azide (Kruhlak et al., 2006) prevented the reduction of GFP-H4 intensity upon local UV irradiation (Fig. 4, D and E), suggesting that chromatin changes in response to UV irradiation require ATP hydrolysis. Real-time imaging of living cells expressing DDB2-YFP after local UV irradiation (100 J/m2; Luijsterburg et al., 2007) revealed the appearance of a fiberlike structure emanating from within the locally damaged area (Fig. 4 G and Video 1). These DNA damage–induced fibers are reminiscent of the fibers caused by tethering DDB2 in the targeting system (see Fig. 2 B), suggesting that the appearance of fibers and the reduction in histone density are both physical manifestations of the same DDB2-mediated chromatin remodeling event. To analyze the histone reduction at damaged sites in more detail, we measured the pixel distribution of GFP-H4 signals at sites of local damage and compared it with the pixel distribution in a similarly sized region elsewhere in the nucleus. Our analysis shows that local UV exposure resulted in a reduction in the heterogeneity of chromatin structure at regions exposed to UV light (Fig. 4, H and I), suggesting that the chromatin structure at damaged sites is more uniform than elsewhere in the nucleus. This local loss of heterogeneity in chromatin density did not occur when cells were ATP depleted before local UV irradiation (Fig. 4, J and K). We conclude that UV exposure causes an ATP-dependent reorganization of chromatin in subnuclear regions containing DNA lesions.
Figure 4.
UV-induced DNA lesions trigger ATP-dependent chromatin remodeling. (A–C) Distribution of GFP-tagged histones H1.0 (A), H2A (B), or H4 (C) at sites of local damage (100 J/m2) in MRC5 cells marked by the accumulation of DDB2-mCherry. (D) GFP-H4 and DDB2-mCherry distribution at local UV damage after ATP depletion. (E) Quantification of the intensity of GFP-tagged histones at sites of UV-induced DNA damage shown in a box plot (∼50 cells for each condition). An intensity of 1 represents the mean intensity in the nucleus (excluding nucleoli), and intensities <1 represent a reduced histone density in the locally damaged area. The dotted line represents a ratio of 1, which means that the intensity in the local damage is the same as the intensity elsewhere in the nucleus. Asterisks indicate significant differences based on a t test (P < 0.05). Open circles are data points that are considered outliers, as their value is greater than the upper quartile, +1.5 the interquartile distance, or less than the lower quartile, −1.5 the interquartile distance. (F) Distribution of GFP-H1 and DNA stained by DAPI in U2OS cells after local damage (100 J/m2) through 5-µm pores. Cells were stained for endogenous DDB2. (G) Real-time imaging of DDB2-YFP in living cells after local UV irradiation (100 J/m2 through 5-µm pores at 37°C) revealed the appearance of a fiberlike structure emanating from within the locally damaged area. (H and I) A micrograph (H) and quantitative (I) analysis of the distribution of GFP-H4 pixel intensities (normalized between 0 and 100%) inside the locally UV-damaged area (red circle in H and red bars in the histogram shown in I) or inside a similarly sized nondamaged area in the nucleus (blue circle in H and blue bars in the histogram shown in I). (J and K) Similar analysis for an ATP-depleted cell.
Figure 6.
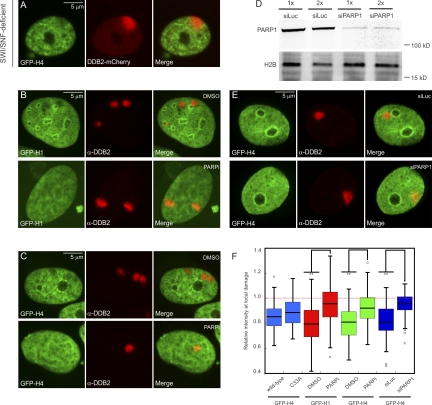
UV-induced chromatin changes require PARP1 activity. (A) Distribution of GFP-H4 at sites of local damage (100 J/m2 through 5-µm pores) in SWI/SNF-deficient C33A cells marked by the accumulation of DDB2-mCherry. (B and C) Distribution of GFP-H1 (B) or GFP-H4 (C) at sites of local damage (100 J/m2 through 5-µm pores) in U2OS that were either treated with DMSO (top rows) or 10 µM PARP inhibitor (PARPi) KU-0058948 for 3 h (bottom rows). (D) Western blot analysis of PARP1 expression levels after a single transfection or two consecutive transfections with siRNAs targeting luciferase (siLuc) or PARP1 (siPARP1) in U2OS cells. (E) Distribution of GFP-H4 at sites of local damage (100 J/m2 through 5-µm pores) in U2OS that were transfected twice with siLuc (top row) or siPARP1 (bottom row). (F) Quantification of the intensity of GFP-tagged histones at sites of UV-induced DNA damage shown in a box plot (between 50 and 100 cells for each condition in two independent experiments). The dotted line represents a ratio of 1, which means that the intensity in the local damage is the same as the intensity elsewhere in the nucleus. Asterisks indicate significant differences based on a t test (P < 0.05). Open circles are data points that are considered outliers, as their value is greater than the upper quartile, +1.5 the interquartile distance, or less than the lower quartile, −1.5 the interquartile distance.
UV-induced chromatin remodeling depends on DDB2
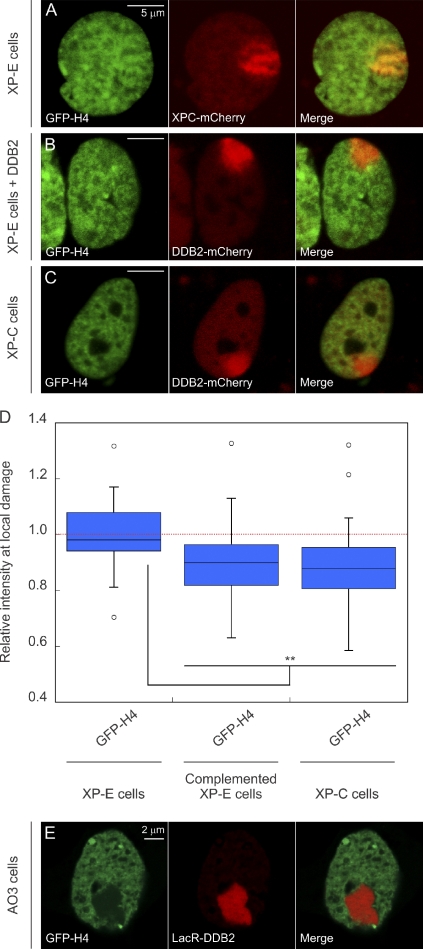
We subsequently used the XP23PV cell line, which is derived from an XP-E patient and hence lacks functional DDB2 (Rapić-Otrin et al., 2003), to address the role of DDB2 in ATP-dependent chromatin decondensation in response to UV exposure. In agreement with a role for DDB2 in UV-induced chromatin decondensation, we did not observe a reduction in GFP-H4 density in XP-E cells (Fig. 5, A and D). Strikingly, complementation of XP-E cells by ectopic expression of DDB2-mCherry restored the decrease in histone density at damaged sites (Fig. 5, B and D), confirming that the DNA damage–induced histone reduction strictly depends on the presence of functional DDB2 protein. The DDB2-dependent nature of chromatin decondensation in UV-damaged cells could also, however, be attributed to events that occur later in NER, such as preincision complex assembly, which is significantly stimulated by DDB2 (Moser et al., 2005; Nishi et al., 2009). To investigate this possibility, we analyzed the effect of UV exposure on the histone density in a cell line derived from an XP-C patient. Whereas XP-C cells still display recruitment of DDB2 to photolesions, the recruitment of XPC itself and the assembly of subsequent global genome repair complexes are severely impaired (Volker et al., 2001; Luijsterburg et al., 2007; Alekseev et al., 2008). The local reduction in GFP-H4 was not affected in XP-C cells (Fig. 5, C and D), showing that XPC and the subsequent binding of other global genome repair proteins are not required for UV-induced chromatin changes. These results demonstrate an essential role for DDB2 in mediating UV-induced chromatin remodeling independently of XPC and other NER factors. Importantly, we also observed that large-scale chromatin decondensation induced by the tethering of mCherry-LacR-DDB2 in AO3 cells was accompanied by a striking reduction in the levels of GFP-H4 at the array (Fig. 5 E). Together, our results provide direct evidence for DDB2-mediated large-scale chromatin remodeling at sites of UV-induced DNA lesions.
Figure 5.
UV-induced chromatin changes require DDB2. (A–C) Distribution of GFP-H4 at sites of local damage (100 J/m2 through 5-µm pores) in a DDB2-deficient XP-E cell line XP23PV (A), an XP-E cell line complemented by transient expression of DDB2-mCherry to restore the DDB2 deficiency (B), and an XP-C cell line (C). (D) Quantification of the intensity of GFP-tagged histones at sites of UV-induced DNA damage shown in a box plot (∼50 cells for each condition). The dotted line represents a ratio of 1, which means that the intensity in the local damage is the same as the intensity elsewhere in the nucleus. Asterisks indicate significant differences based on a t test (P < 0.05). Open circles are data points that are considered outliers, as their value is greater than the upper quartile, +1.5 the interquartile distance, or less than the lower quartile, −1.5 the interquartile distance. (E) Distribution of GFP-H4 at the LacO array in AO3 cells upon tethering LacR-DDB2.
UV-induced chromatin changes require the activity of PARP
We subsequently tested the involvement of SWI/SNF chromatin remodeling complexes in the DDB2-dependent reorganization of chromatin, given that these types of complexes have previously been linked to NER (Gong et al., 2008; Zhang et al., 2009; Zhao et al., 2009) and operate in an ATP-dependent manner. For this purpose, we used a C33A cervical cancer cell line that lacks both catalytic subunits of SWI/SNF complexes: BRM and BRG1 (Wong et al., 2000). Local UV irradiation of C33A cells expressing GFP-H4 revealed DDB2 recruitment and a local reduction in histone intensity similar to wild-type cells (Fig. 6, A and F), indicating that chromatin remodeling by SWI/SNF complexes is not involved in DDB2-mediated chromatin changes. We then tested a possible involvement of poly(ADP-ribosyl)ation in UV-induced chromatin changes, given that several chromatin remodeling events during the repair of DNA double-strand breaks are regulated by this modification (Ahel et al., 2009; Gottschalk et al., 2009; Chou et al., 2010; Polo et al., 2010). Strikingly, we found that chemical inhibition of poly(ADP-ribosyl)ation, using PARP inhibitor KU-0058948, significantly suppressed the reduction of linker histone GFP-H1 at sites of local UV irradiation, whereas a reduction in GFP-H1 was detected in untreated wild-type cells (Fig. 6, B and F). Similarly, the reduction of GFP-tagged core histone H4 at sites of local damage was also significantly suppressed by PARP inhibition, whereas control cells displayed clearly reduced H4 levels at sites of DNA damage (Fig. 6, C and F). To corroborate these findings, we reduced the levels of PARP1 by RNAi and measured how this affected UV-induced changes in chromatin structure. Western blot analysis confirmed that siRNAs directed against PARP1 efficiently lowered the PARP1 protein levels in U2OS cells (Fig. 6 D). Similar to chemical inhibition of PARP activity, we found that siRNA-mediated depletion of PARP1 significantly suppressed the reduction of GFP-H4 at sites of local UV damage compared with cells transfected with control siRNAs (Fig. 6, E and F). These findings show that poly(ADP-ribosyl)ation contributes to chromatin rearrangements after exposure to UV light.
DDB2-mediated chromatin unfolding is suppressed by ATP depletion and PARP inhibition
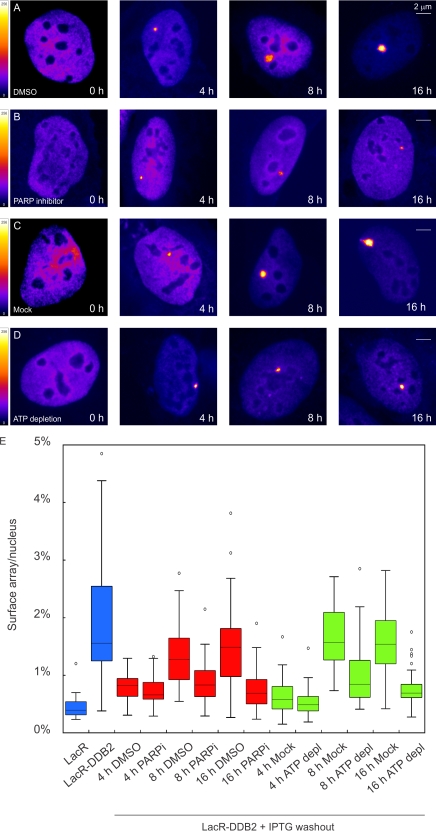
To test whether the activity of PARP enzymes and ATP hydrolysis is required for DDB2-mediated chromatin unfolding in the tethering system, we transfected U2OS 2–6-3 cells harboring a LacO array with LacR-tagged DDB2 in the presence of IPTG to prevent the newly synthesized LacR-DDB2 protein from binding to the array. We subsequently removed IPTG and released the cells in fresh medium in the absence (Fig. 7 A) or presence of PARP inhibitor (Fig. 7 B), after which cells were fixed, and the size of the arrays was quantified. Alternatively, cells were released in regular medium to allow unfolding of the array, after which cells were incubated for 30 min in control (Fig. 7 C) or ATP depletion medium (Fig. 7 D). Binding of LacR-DDB2 to the array was not detected until 4 h after IPTG washout, and clear unfolding of the array occurred between 8 and 16 h in control cells (Fig. 7, A, C, and E). However, either treatments with PARP inhibitor (Fig. 7 B) or ATP depletion (Fig. 7 D) significantly suppressed unfolding of the array at both 8 and 16 h after IPTG removal (Fig. 7 E). These experiments show that tethering of DDB2 to chromatin triggers an ATP-dependent and PAR-dependent rearrangement of higher-order chromatin structure, which is in line with our finding that the DDB2-mediated rearrangement of chromatin structure at sites of UV-induced DNA damage requires ATP hydrolysis (Fig. 4) and the activity of PARP1 (Fig. 6).
Figure 7.
Chromatin unfolding by tethered DDB2 requires PARP activity and ATP hydrolysis. (A–D) Human U2OS 2–6-3 cells containing 200 copies of a LacO-containing cassette (∼4 Mbp) were transfected with mCherry-LacR-DDB2 in the presence of IPTG. Cells were subsequently released for the indicated time points in medium containing DMSO (A) or 1 µM PARP inhibitor (B) or in regular medium followed by a 30-min incubation in mock medium (C) or ATP depletion medium (D). The lookup table for the color coding is shown. (E) Quantitative measurements on the relative size of the array (surface of the array/surface of the nucleus) in cells subjected to the indicated conditions (30–50 cells for each condition collected in two independent experiments) are shown in a box plot. Open circles are data points that are considered outliers, as their value is greater than the upper quartile, +1.5 the interquartile distance, or less than the lower quartile, −1.5 the interquartile distance. depl., depleted; PARPi, PARP inhibitor.
ATP depletion impairs recognition of UV lesions by XPC
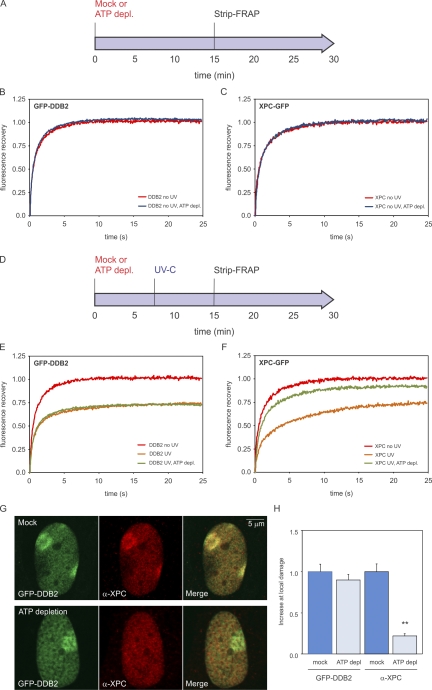
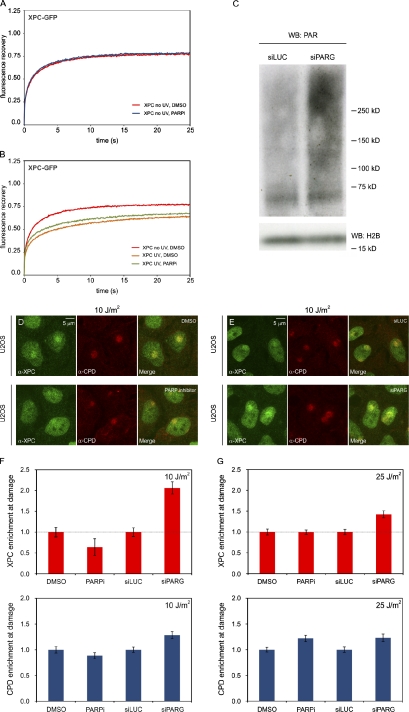
To gain insight in the role of ATP-dependent chromatin decondensation at sites of UV damage, we assessed whether the binding of XPC to photolesions requires ATP hydrolysis. For comparison, we also analyzed the ATP dependency of DDB2 recruitment to UV-induced DNA lesions. We first tested whether the mobility of XPC and DDB2 in unchallenged cells is ATP dependent by treating cell lines stably expressing a GFP-tagged version of these proteins with the metabolic inhibitors 2-deoxyglucose and sodium azide to deplete cells of ATP (Fig. 8 A; Kruhlak et al., 2006). FRAP analysis showed that the mobility of XPC-GFP or GFP-DDB2 in unchallenged cells was not affected by ATP depletion (Fig. 8, B and C). Global irradiation of the cells with UV-C light (8 J/m2; Fig. 8 D), which is known to significantly reduce the mobility of NER proteins as a result of their transient incorporation into chromatin-bound complexes engaged in repair (Houtsmuller et al., 1999; Hoogstraten et al., 2002, 2008; Rademakers et al., 2003; Zotter et al., 2006; Luijsterburg et al., 2007, 2010), led to a significant immobilization of GFP-DDB2 in both control and ATP-depleted cells (Fig. 8 E), suggesting that the binding of DDB2 to photolesions is ATP independent. Conversely, we found that ATP depletion severely compromised the UV-induced immobilization of XPC, consistent with a role of ATP-dependent chromatin remodeling in lesion recognition by XPC (Fig. 8 F). To validate these findings, we locally irradiated cells expressing GFP-DDB2 with UV-C light (50 J/m2) through 5-µm pores followed by the detection of endogenous XPC using specific antibodies to simultaneously monitor DNA damage–induced accumulation of GFP-DDB2 and XPC in the same cells (Fig. 8 G). Whereas mock-treated cells showed clear colocalization of GFP-DDB2 and endogenous XPC at sites of local UV-induced DNA lesions, ATP depletion led to significantly impaired XPC recruitment, whereas the GFP-DDB2 accumulation was not affected (Fig. 8 H). Our findings demonstrate that, in contrast to DDB2, efficient binding of XPC to photolesions is ATP dependent, suggesting an important role for DDB2-induced, ATP-dependent chromatin remodeling in XPC recruitment.
Figure 8.
Lesion recognition by XPC is ATP dependent. (A) Cells stably expressing GFP-DDB2 (VH10) or XPC-GFP (XP4PA) were mock treated or ATP depleted (depl.) and subjected to Strip-FRAP analysis, as indicated. (B and C) Quantitative analysis of the mobility of GFP-DDB2 (B) or XPC-GFP (C) in unchallenged cells that were either mock treated (red lines) or ATP depleted (blue lines). (D) Cells stably expressing GFP-DDB2 or XPC-GFP were mock treated or ATP depleted, globally UV-C irradiated (8 J/m2), and subjected to Strip-FRAP analysis, as indicated. (E and F) Quantitative analysis of the mobility of GFP-DDB2 (E) or XPC-GFP (F) in unchallenged cells (red lines) or globally UV-irradiated, mock-treated (orange lines) or ATP-depleted (green lines) cells. Values represent the mean of ∼20 cells. (G) Examples of GFP-DDB2–expressing cells that were mock treated (top row) or ATP depleted (bottom row), locally UV irradiated (50 J/m2), and subsequently stained for endogenous XPC. (H) A quantification of the recruitment of GFP-DDB2 and endogenous XPC in the same cells is shown (∼30 cells for each condition). Error bars represent the SD. Asterisks indicate significant differences based on a t test (P < 0.01).
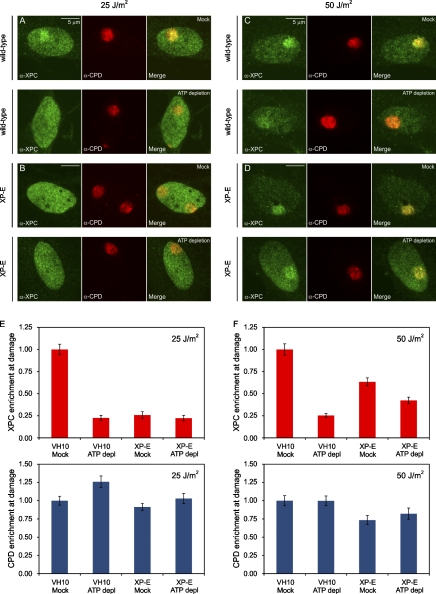
Previous studies revealed that the accumulation of XPC at UV-induced DNA lesions is significantly enhanced by DDB2 (Moser et al., 2005; Nishi et al., 2009). Our findings that ATP depletion impairs both XPC recruitment and UV-induced chromatin rearrangement, which is also dependent on DDB2, prompted us to assess whether the ATP-dependent association of XPC requires DDB2. To this aim, we locally irradiated (25 and 50 J/m2) wild-type VH10 cells or XP-E cells (GMO1389), which were either mock treated or ATP depleted and subsequently detected endogenous XPC and UV-induced DNA lesions (CPDs) using specific antibodies. Importantly, the accumulation of XPC was significantly suppressed by ATP depletion in VH10 wild-type cells at both 25 J/m2 (Fig. 9, A and E) and 50 J/m2 (Fig. 9, C and F), underscoring that XPC recruitment is indeed partially ATP dependent. Similarly, XPC accumulation was also significantly compromised in XP-E cells compared with wild-type cells (Fig. 9, E and F), showing that DDB2 contributes to efficient XPC recruitment, as previously reported (Moser et al., 2005; Nishi et al., 2009). Finally, the DDB2-independent accumulation of XPC (in XP-E cells) was not suppressed by ATP depletion at 25 J/m2 (Fig. 9, B and E) and only marginally suppressed at 50 J/m2 (Fig. 9, D and F). These data are consistent with the model that the ATP-dependent recruitment of XPC is, to a large extent, regulated through DDB2-mediated chromatin remodeling at UV-induced DNA lesions.
Figure 9.
The impact of DDB2 deficiency and ATP depletion on XPC recruitment to DNA damage. (A and B) Wild-type VH10 cells (A) or XP-E (GMO1389-TERT) cells (B) were either mock treated (top rows) or ATP depleted (bottom rows), locally UV irradiated (25 J/m2 through 8-µm pores), and subsequently stained for endogenous XPC (green) and CPDs (red). (C and D) Wild-type VH10 cells (C) or XP-E (GMO1389-TERT) cells (D) were either mock treated (top rows) or ATP depleted (bottom rows), locally UV irradiated (50 J/m2 through 8-µm pores), and subsequently stained for endogenous XPC (green) and CPDs (red). (E and F) The quantification of the relative accumulation of XPC (red bars) or the enrichment of CPDs (blue bars) is shown in E (25 J/m2) and F (50 J/m2). The signal represents the relative increase at the locally damaged site relative to the signal in the nondamaged nuclear region (∼50 cells for each condition collected in two independent experiments). Error bars represent the SD. depl, depleted.
PAR synthesis promotes recognition of UV lesions by XPC
Given that, in addition to ATP hydrolysis, the activity of PARP enzymes is also required for UV-induced chromatin decondensation (Fig. 6, B–F), we subsequently assessed whether the binding of XPC to photolesions involves poly(ADP-ribosyl)ation. FRAP analysis showed that the mobility of XPC-GFP in unchallenged cells was not affected by PARP inhibitors compared with mock-treated cells (Fig. 10 A). Whereas global irradiation with UV-C light (8 J/m2) led to a significant immobilization of XPC-GFP in mock-treated cells, we found that treatment with PARP inhibitors suppressed the UV-induced immobilization of XPC, suggesting that PAR synthesis promotes lesion recognition by XPC (Fig. 10 B). To validate these findings, we locally irradiated U2OS cells with UV-C light (10 and 25 J/m2) through 8-µm pores followed by the detection of endogenous XPC and UV-induced DNA lesions. Treatment with PARP inhibitor KU-0058948 significantly suppressed the recruitment of XPC at 10 J/m2 (Fig. 10, D and F), whereas XPC recruitment at 25 J/m2 was not affected (Fig. 10 G). We subsequently transfected cells with siRNAs targeting the PAR glycohydrolase PARG, which led to increased steady-state levels of PAR chains, as shown by Western blot analysis (Fig. 10 C). Importantly, the steady-state levels of bound XPC at sites of DNA damage were significantly higher after PARG knockdown, which was most pronounced at 10 J/m2 (Fig. 10, E–G), suggesting that PAR synthesis promotes the association of XPC with UV-induced DNA lesions. These findings reveal that efficient binding of XPC to photolesions is regulated through PAR synthesis.
Figure 10.
Lesion recognition by XPC is PARP dependent. (A and B) Cells stably expressing XPC-GFP (XP4PA) were DMSO treated or PARP inhibitor (PARPi) treated and subjected to Strip-FRAP analysis, as indicated. Quantitative analysis of the mobility of XPC-GFP in unchallenged cells that were treated with DMSO or PARP inhibitor (A) or XPC-GFP in UV-C–irradiated (8 J/m2) cells that were treated with DMSO or PARP inhibitor (B) is shown. Values represent the mean of ∼20 cells. (C) Western blot (WB) of high–molecular mass PAR chains after transfection with siRNAs targeting luciferase (siLuc) or PARG (siPARG) in U2OS cells. Histone H2B serves as a loading control. (D) U2OS cells were treated with DMSO or PARP inhibitor, locally UV irradiated (10 J/m2 through 8-µm pores), and subsequently stained for endogenous XPC (green) and CPDs (red). (E) U2OS cells were transfected with siLUC or siPARG, locally UV irradiated (10 J/m2 through 8-µm pores), and subsequently stained for endogenous XPC (green) and CPDs (red). (F and G) The quantification of the relative accumulation of XPC (red bars) or the enrichment of CPDs (blue bars) is shown in F (10 J/m2) and G (25 J/m2). The signal represents the relative increase at the locally damaged site relative to the signal in the nondamaged nuclear region (∼50 cells for each condition collected in two independent experiments). Accumulation of XPC is normalized to 1 for comparison. So, DMSO is set to 1 (and PARPi expressed relative to this value), and siLUC is set to 1 (and siPARG expressed relative to this value). Error bars represent the SD.
Discussion
The NER proteins XPC, TFIIH, XPA, XPG, RPA, ERCC1, and XPF are sufficient to reconstitute in vitro lesion recognition and dual incision on naked DNA (Aboussekhra et al., 1995). Although this represents the minimal number of proteins to perform NER on naked DNA, the efficiency of repair in a chromatin context is decreased to about only 10% (Wang et al., 1991; Hara et al., 2000; Ura et al., 2001). This shows that the packaging of genomic DNA into chromatin is a major obstacle for efficient NER and suggests that additional factors are required to mediate efficient DNA repair in vivo. Kinetic analysis of the NER system revealed that lesion recognition in chromatin is the rate-limiting step in this pathway, whereas subsequent repair complexes are rapidly assembled (Dinant et al., 2009; Luijsterburg et al., 2010). Electron microscopic studies suggest that lesion recognition by NER may be accompanied by changes in chromatin structure (Solimando et al., 2009). Although several chromatin remodeling activities have been linked to NER (Jiang et al., 2010; Lans et al., 2010), the underlying mechanisms are poorly understood. The observation that the lesion recognition protein DDB2 is nonessential for NER in vitro but is required for CPD repair and significantly promotes 6-4PP repair in vivo (Hwang et al., 1999; Tang et al., 2000; Moser et al., 2005) has led to the idea that this early NER factor may link chromatin remodeling to DNA repair (Jones et al., 2010; Palomera-Sanchez and Zurita, 2011).
To directly investigate the impact of DDB2 on higher-order chromatin structure, we used a system that allows visualization of chromatin unfolding triggered by a single repair factor in living cells (Tumbar et al., 1999; Ye et al., 2001). We show that tethering DDB2 to a chromosomal locus elicits extensive chromatin decondensation in hamster, mouse, and human cells, providing direct evidence for DDB2-mediated chromatin decondensation in living cells. Additionally, we show that chromatin unfolding triggered by tethered DDB2 is ATP dependent and requires the activity of PARP enzymes. As a complementary approach, we examined the effect of DNA damage on the density of core histones H2A and H4 and linker histone H1 at UV-irradiated regions in living human cells. We find that chromatin regions containing UV-induced DNA lesions undergo ATP-dependent chromatin decondensation that is strictly dependent on the presence of functional DDB2. Additionally, the DNA damage–induced reduction in histone density also required the activity of PARP1, which has recently been linked to chromatin remodeling in response to double-strand DNA breaks (Ahel et al., 2009; Gottschalk et al., 2009; Chou et al., 2010; Polo et al., 2010). Moreover, we found that efficient recruitment of XPC to DNA lesions is markedly ATP dependent, whereas DDB2 recruitment to UV-induced lesions does not require ATP hydrolysis. Additionally, efficient accumulation of XPC at UV-induced DNA lesions also required the synthesis of PAR chains, in agreement with an important role for PARP-dependent chromatin decondensation in promoting XPC recruitment to UV-induced DNA lesions in chromatin.
Our findings directly link lesion recognition by DDB2 to changes in higher-order chromatin structure and suggest that these chromatin changes may enable efficient lesion recognition by XPC and assembly of the NER complex. However, our data do not exclude the possibility that ATP-dependent and PARP-dependent events other than chromatin remodeling are involved in lesion recognition. How PARP is mechanistically linked to chromatin remodeling is also currently unclear. It is possible that DDB2, in concert with poly(ADP-ribosyl)ation, facilitates the recruitment of chromatin-modifying activities to promote structural rearrangement in UV-damaged chromatin. It is interesting to note that the ATPase activity of some chromatin remodeling enzymes is stimulated by PAR synthesis (Ahel et al., 2009; Gottschalk et al., 2009), which may explain the requirement for both ATP hydrolysis and PAR synthesis in DDB2-mediated chromatin decondensation and lesion recognition by XPC. However, establishing whether such a mechanism takes place in UV-damaged chromatin merits further study. We also noticed that the effect of ATP depletion on XPC recruitment is more pronounced than the impact of inhibiting PARP enzymes. This could indicate that these processes are not necessarily coupled. Alternatively, we cannot exclude the possibility that our ATP depletion protocol, in addition to suppressing UV-induced chromatin rearrangements, also suppresses the ubiquitin ligase activity of the DDB2–CRL4 complex, which could further impact lesion recognition by XPC (Sugasawa et al., 2005; Wang et al., 2006). As we found that SWI/SNF complexes are not required for local chromatin structural changes, we are currently undertaking an unbiased proteomics approach to identify novel DDB2-associated factors to gain insight into the mechanisms underlying DDB2-mediated chromatin remodeling (unpublished data).
When analyzing the ability of naturally occurring DDB2 mutants to mediate chromatin decondensation, we found that tethering DDB2L350P or DDB2D307Y to chromatin promoted extensive chromatin decondensation to the same extent as wild-type DDB2. As both of these mutants fail to form a complex with DDB1 and CUL4A (Rapić-Otrin et al., 2003; this study), these findings show that chromatin decondensation is a previously unrecognized activity of DDB2, which does not require the function of the CRL4–DDB2 ubiquitin ligase complex. In agreement with these findings, we show that a region encompassing the first five β blades of DDB2, but lacking the domain required for incorporation in the ubiquitin ligase complex, comprises the minimal domain that is sufficient to mediate chromatin unfolding. Even though these DDB2 mutants support chromatin decondensation in the targeting system, we also show that they fail to bind to UV-induced DNA lesions in living cells. Thus, although capable of modifying chromatin, this activity is absent at photolesions in cells expressing these mutant DDB2 proteins.
Previous studies have shown that DDB2 is essential to target DDB1 and CUL4A to chromatin, whereas DDB1 and CUL4A, in turn, are dispensable for DDB2 recruitment to photolesions (El-Mahdy et al., 2006; Li et al., 2006; Alekseev et al., 2008). However, reducing the levels of DDB1 or CUL4A by RNAi confers a CPD repair defect, despite the fact that DDB2 is still recruited to damaged sites under those conditions (El-Mahdy et al., 2006; Li et al., 2006), indicating that DDB2-mediated chromatin remodeling is not sufficient to promote repair and that CRL4–DDB2 complex–mediated activities are also required. Indeed, early studies have revealed that UV-induced ubiquitylation of XPC by the CRL4–DDB2 complex increases its affinity for DNA independently of the presence of helix-distorting lesions (Sugasawa et al., 2005), which may contribute to enhanced repair by NER. Additionally, the UV-induced ubiquitylation of the core histones H3 and H4 by CRL4–DDB2 may also enhance NER as a result of increased nucleosome destabilization (Wang et al., 2006). Another explanation for the requirement of the ubiquitin ligase function of the CRL4–DDB2 complex is that the levels of DDB2 need to be proteolytically lowered, which does not occur in DDB1/CUL4A knockdown cells (El-Mahdy et al., 2006; Li et al., 2006) because DDB2 may otherwise occupy potential binding sites for XPC. Indeed, the in vitro NER reaction using purified components is inhibited by the addition of excessive DDB2, which can be partially alleviated by the addition factors that support ubiquitylation (Sugasawa et al., 2005). Moreover, interference with DDB2 proteolysis in cells was found to compromise the repair of CPDs (Wang et al., 2005). Our present study suggests that, in addition to these catalytic activities of the CRL4–DDB2 complex, DDB2 also elicits local chromatin unfolding in a ubiquitylation-independent manner. It is tempting to speculate that the chromatin remodeling and ubiquitylation activities of the CRL4–DDB2 complex act synergistically to facilitate efficient recognition of photolesions by XPC. Consistent with a role for DDB2-dependent chromatin remodeling in NER, we show that lesion recognition by DDB2 in chromatin is independent of ATP, whereas lesion recognition by XPC is ATP dependent. An explanation for these findings is that DDB2 has access to lesions in chromatin structure, whereas the accessibility of XPC to lesions in chromatin is more limited and, thus, is significantly stimulated by the ATP-dependent and PARP-dependent opening of chromatin structure. This remodeling event, possibly together with DDB2-mediated histone acetylation and H3/H4 ubiquitylation, may create a local chromatin environment that is permissive to lesion recognition by XPC and thus amendable to repair.
Materials and methods
Cell lines
To establish VH10-TERT cells stably expressing human GFP-DDB2, DDB2 cDNA was cloned into pENTR4-GFP-C1. The resulting pENTR4-GFP-DDB2 plasmid was subsequently recombined with pLenti6.3 V5 M EST (Invitrogen) using Gateway recombination. Lentiviral particles were produced by cotransfection of 293T cells with pLenti6.3 GFP-DDB2 plasmid and the packaging/envelope plasmids pMDLg/pRRE, pRSV-REV, and pCMV-VSV-G using polyethylenimine (Polysciences, Inc.). Lentivirus-containing supernatant was collected 48 h after transfection and used to infect VH10-hTERT cells. Clones stably expressing GFP-DDB2 were selected with 5 µg/ml blasticidin S (Invitrogen). AO3 hamster cells, containing a 90-Mbp amplification of LacO sequences and flanking DNA (Tumbar et al., 1999), were cultured in a 1:1 mixture of DME/Ham’s F12 medium supplemented with antibiotics and 20% FCS. Mouse NIH2/4 cells (Soutoglou et al., 2007), human U2OS cells, U2OS 2–6-3 cells (Janicki et al., 2004), human C33A cervical cancer cells (Wong et al., 2000), human VH10-TERT GFP-DDB2 cells, VH10-TERT, GMO1389-TERT (XP-E), and SV40-immortalized human MRC5 cells XP4PA (XP-C) complemented with XPC-GFP (Hoogstraten et al., 2008; Solimando et al., 2009), XP4PA (XP-C), and XP23PV (XP-E) were cultured in DME supplemented with 10% FCS.
Transfection and drug treatments
Cells were transiently transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were typically imaged 24 h after transfection. 100 µM anacardic acid (Sigma-Aldrich) or 10 µM PARP inhibitor KU-0058948 was added to the culture medium for 3 h. For ATP depletion, cells were incubated in ATP depletion medium (137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 60 mM deoxyglucose, 30 mM NaAz, 20 mM Hepes, and 10% FCS), and mock-treated cells were incubated in microscopy medium (137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 20 mM d-glucose, 20 mM Hepes, and 10% FCS). The ATP depletion protocol was as follows (see also Fig. 8): cells were rinsed with PBS and incubated in microscopy medium or ATP depletion medium for 7.5 min at 37°. Cells were subsequently rinsed with PBS and either mock treated or either globally (8 J/m2) or locally (25 or 50 J/m2) UV-C irradiated and transferred to the microscope chamber or an incubator in microscopy medium or ATP depletion medium for 7.5 min at 37° to allow repair proteins to accumulate at DNA lesions. The binding of GFP-tagged NER factors to DNA lesions was subsequently analyzed by Strip-FRAP for 15 min, or cells were fixed and stained with the indicated antibodies. 5 mM IPTG was added to U2OS 2–6-3 cells before and during the transfection with the mCherry-LacR-DDB2 plasmid. IPTG was washed out 24 h after transfection, and cells were released in regular medium for 3.5, 7.5, or 15.5 h followed by a 30-min incubation in ATP depletion medium (137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 60 mM deoxyglucose, 30 mM NaAz, 20 mM Hepes, and 10% FCS) or microscopy medium (137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 20 mM d-glucose, 20 mM Hepes, and 10% FCS) before fixation. Alternatively, cells were released in medium containing 1 µM PARP inhibitor KU-0058948 or DMSO after IPTG washout and fixed after 4, 8, or 16 h.
RNAi
siRNA oligonucleotides (Thermo Fisher Scientific) were synthesized to target human PARP1 (5′-GAAAGUGUUCAACUAAU-3′), luciferase (5′-CGUACGCGGAAUACUUCGA-3′), or an siRNA smart pool targeting human PARG (Thermo Fisher Scientific). Cells were transfected with 40 nM siRNA using Lipofectamine 2000 at 0 and 36 h and were typically analyzed 60 h after the first transfection.
Western blotting
Cell extracts of U2OS cells after transfection with control siRNAs, PARP1 or PARG siRNAs, or cell extracts of NIH2/4 cells after transfection of GFP-tagged or mCherry-LacR–tagged DDB2 constructs were generated by cell lysis, boiled in Laemmli buffer, separated by SDS-PAGE, and transferred to polyvinylidene fluoride membranes (Millipore). Expression of PARP1 or PAR chains was analyzed by immunoblotting with mouse monoclonal anti-PARP1 antibodies at 1:1,000 (Abnova) or mouse α-PAR antibodies at 1:1,000 (Abcam) and rabbit monoclonal anti-H2B antibodies at 1:5,000 (Santa Cruz Biotechnology, Inc.) followed by secondary antibodies donkey anti–rabbit 700CW at 1:10,000 and donkey anti–mouse 800CW at 1:5,000 and detection using the Odyssey infrared imaging scanning system (LI-COR Biosciences). Expression of GFP-tagged or mCherry-LacR–tagged DDB2 proteins was analyzed by immunoblotting with rabbit anti-GFP at 1:5,000 (Abcam), rabbit anti-mCherry at 1:5,000 (a gift from J. Neefjes, Netherlands Cancer Institute, Amsterdam, Netherlands), and mouse anti–β-actin at 1:5,000 (Sigma-Aldrich) followed by secondary antibodies (goat anti–rabbit 1:5,000 and goat anti–mouse 1:5,000) and ECL detection.
Plasmids
To generate a LacR-tagged DDB2 fusion protein, murine DDB2 (Moser et al., 2005) was amplified using the following primers: 5′-GGTACCATGGCTCCCAAGAAATGCC-3′ and 5′-AGATCTTTATAGTCTTTCATGATCTTTCTGTGAC-3′. The product was inserted as a KpnI–BglII fragment into the KpnI–BamH1 site of the mCherry-LacR-NLS-KpnI plasmid (Kaiser et al., 2008; Soutoglou and Misteli, 2008). This generated mCherry-LacR-NLS-DDB2, which was subsequently used to generate mCherry-LacR fused to the naturally occurring mutants DDB2K244E, DDB2L350P, and DDB2D307Y using site-directed mutagenesis (Agilent Technologies). The following mutagenesis primers were used: 5′-CCGAATGCACAAGAAGGAAGTAGCCCACGTGGC-3′ and 5′-GCCACGTGGGCTACTTCCTTCTTGTGCATTCGG-3′ (K244E), 5′-ATTCACGGCACAACCCCATTGTTGTGGGCCG-3′ and 5′-CGGCCCACAACAATGGGGTTGTGCCGTGAAT-3′ (L350P), and 5′-GAGCTCGGCTCCTGACTACTTACCAGAACAATGAG-3′ and 5′-CTCATTGTTCTGGTAAGTAGTCAGGAGCCGAGCTC-3′ (D307Y). Truncation mutants of DDB2 consisting of amino acids 1–324, 1–108, 108–216, 216–324, and 108–324 were generated by PCR and inserted as KpnI–BglII fragments into the KpnI–BamH1 site of the mCherry-LacR-NLS-KpnI plasmid. In addition, wild-type and mutant DDB2 products were inserted into EGFP-C1, generating EGFP-DDB2, EGFP-DDB2K244E, EGFP-DDB2L350P, and EGFP-DDB2D307Y. All constructs were verified by sequencing, and expression was verified by Western blot analysis. The mCherry-DDB1 plasmid (Alekseev et al., 2008) was used to generate EGFP-DDB1 by replacing mCherry with EGFP (using AgeI and BsrGI). EGFP-CUL4A was generated by inserting EGFP (from GFP-C2) as an EcoRI–NheI fragment into pcDNA3.1-CUL4A, as previously described (Luijsterburg et al., 2007). The H1.0 cDNA was amplified by PCR using primers 5′-AGATCTATGACCGAGAATTCCACGTC-3′ and 5′-GGATCCTCACTTCTTCTTGCCGGCCC-3′ inserted into pEGFP-C1, yielding EGFP-H1. The cDNAs for H2A and H4 were inserted in pEGFP-C1 or SCFP-C1, yielding EGFP-H2A, EGFP-H4, and SCFP-H4.
Immunofluorescent labeling
Cells were fixed with 4% formaldehyde in PBS for 15 min at 4°C, permeabilized in 0.5% Triton X-100 (SERVA) in PBS for 5 min, and incubated with 100 mM glycine in PBS for 10 min to block unreacted aldehyde groups. For CPD staining, DNA was denatured with 0.1 M HCl for 10 min at 37°C, and cells were blocked in 10% BSA in phosphate buffer for 15 min. Cells were subsequently rinsed with PBS and equilibrated in wash buffer (PBS containing 0.5% BSA and 0.05% Tween 20; Sigma-Aldrich). Antibody steps and washes were performed in wash buffer. The primary antibodies were incubated overnight at 4°C. The primary antibodies used were mouse anti-H3ac at 1:100 (Abcam), mouse anti-H4K16ac at 1:100 (Abcam), rabbit anti-XPC at 1:500 (a gift from W. Vermeulen, Erasmus Medical Center, Rotterdam, Netherlands), mouse anti-DDB2 at 1:500 (MyBioSource), and mouse anti-CPD at 1:1,000 (a gift from O. Nikaido, Kanazawa University, Kanazawa, Japan). Detection was performed using goat anti–mouse or goat anti–rabbit Ig coupled to Alexa Fluor 488, 546, or 647 (1:1,000; Invitrogen). Samples were mounted in Mowiol, and images were acquired on a confocal microscope (LSM 510; Carl Zeiss) or a widefield microscope (Axioplan 2; Carl Zeiss; see Microscopy for details).
UV-C irradiation
UV lamp–induced damage was inflicted using a customized UV cross-linker (CL-1000; UVP, LLC) or with a UV source (TUV PL-S 9W; Philips) above the microscope stage. For induction of global UV damage, cells were rinsed with microscopy medium and irradiated with 8 or 16 J/m2. For induction of local UV damage, cells were UV irradiated through a polycarbonate mask (Millipore) with pores of 5 or 8 µm and subsequently irradiated with 10, 25, 50, or 100 J/m2.
FRAP
FRAP analysis was used to measure the mobility of GFP-DDB2 and naturally occurring GFP-tagged DDB2 mutants. FRAP analysis was performed by bleaching (five iterations) a narrow strip (512 × 40 pixels at zoom 8) spanning the nucleus with a maximal 488-nm laser intensity (acousto-optic tunable filter at 100%). The reequilibration of bleached and nonbleached molecules was monitored in a region of 512 × 50 pixels (zoom 8) with low laser intensity (acousto-optic tunable filter at 0.5%) for at least 700 images, with a 38-ms time interval between images. The data were normalized to pre- or postbleach intensity.
Microscopy
Live-cell imaging and analysis of fixed samples were performed on a confocal microscope (LSM 510 META; Carl Zeiss) equipped with a 37°C climate chamber, a 63× Plan-Apochromat (1.4 NA) oil immersion lens (Carl Zeiss), a 60-mW argon laser (488 and 514 nm), a 5-mW helium-neon 1 (543 nm) laser, a 15-mW helium-neon 2 (633) laser, two photomultiplier tubes, and a META detector. Images were recorded using LSM imaging software in multitrack mode (Carl Zeiss). Living cells were examined in culture medium at 37°C in an atmosphere of 5% CO2. An argon-ion laser (60 mW) was used for excitation at 488 nm and passed onto the sample by a 490-nm dichroic mirror, and emission light was filtered by a 505–550-nm emission filter. A helium-neon laser (5 mW) was used for excitation at 543 nm and passed onto the sample by a 543-nm dichroic mirror, and emission light was filtered by a 560–615-nm emission filter. A helium-neon laser (15 mW) was used for excitation at 633 nm and passed onto the sample by a 633-nm dichroic mirror, and emission light was filtered by a 650-nm long-pass emission filter. Real-time assembly of DDB2 was monitored on a widefield fluorescence microscope (Axiovert 200M; Carl Zeiss) equipped with a 100× Plan-Apochromat (1.4 NA) oil immersion lens and a xenon arc lamp with a monochromator (Cairn Research Limited). Images were recorded with a cooled charge-coupled device camera (CoolSNAP HQ; Roper Scientific) using MetaMorph imaging software (Molecular Devices). Images of fixed samples were captured using a widefield microscope (Axioplan 2) equipped with a Plan-NEOFLUAR 63× (1.25 NA) oil immersion lens (Carl Zeiss) and an HBO mercury lamp. Images were recorded with a cooled 12-bit camera (AxioCam MRm; Carl Zeiss) using AxioVision software (Carl Zeiss). All images were quantified using ImageJ software (National Institutes of Health).
Online supplemental material
Fig. S1 shows the quantification of the recruitment of DDB1 and CUL4A to LacR-tethered wild-type and mutant DDB2and shows that the LacR-DDB2 induces a comparable chromatin decondensation in NIH2/4 and U2OS 2–6-3 cells. Fig. S2 shows that LacR-DDB2 does not change acetylation of histone H3 and H4. Fig. S3 demonstrates that LacR tethering of an N-terminal fragment of DDB2, but not its central part, results in recruitment of DDB1. Fig. S4 shows that local UV irradiation causes a very similar reduction in density of GFP-tagged and SCFP-tagged histone H4. Fig. S5 shows that local UV irradiation causes a reduction in DNA content. Video 1 shows that local UV damage results in the appearance of fiberlike DDB2-YFP–positive structure emanating from within the locally damaged area. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106074/DC1.
Acknowledgments
The authors acknowledge Drs. W. Vermeulen, A.S. Belmont, T. Misteli, T. Chiba, K. Tanaka, J. Goedhart, A.K. Östlund Farrants, S. Janicki, J. Neefjes, and S. Dieckmann for sharing reagents and Dr. R. van Driel for reading the manuscript.
This work was supported by the Swedish Cancer Society (to N.P. Dantuma), the European Community Network of Excellence RUBICON (project no. LSHC-CT-2005-018683; to N.P. Dantuma), the Swedish Research Council (to N.P. Dantuma), the Netherlands Organization for Scientific Research (NWO-Rubicon and NWO-Veni to M.S. Luijsterburg, NWO-Vidi to H. van Attikum, and ZonMw 40-00812-98-08031 to L.H. Mullenders), the European Molecular Biology Organization (EMBO long-term fellowship to M.S. Luijsterburg), the Federation of European Biochemical Societies (FEBS long-term fellowship to M.S. Luijsterburg), and the Human Frontier Science Program (HFSP Career Development Award to H. van Attikum).
Footnotes
Abbreviations used in this paper:
CPD
cyclobutane pyrimidine dimer
HAT
histone acetyltransferase
NER
nucleotide excision repair
PAR
poly(ADP-ribose)
PARG
PAR glycohydrolase
PARP
PAR polymerase
SCFP
super CFP
XP
xeroderma pigmentosum
References
- Aboussekhra A., Biggerstaff M., Shivji M.K., Vilpo J.A., Moncollin V., Podust V.N., Protić M., Hübscher U., Egly J.M., Wood R.D. 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 80:859–868 10.1016/0092-8674(95)90289-9 [DOI] [PubMed] [Google Scholar]
- Ahel D., Horejsí Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P., et al. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 325:1240–1243 10.1126/science.1177321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseev S., Kool H., Rebel H., Fousteri M., Moser J., Backendorf C., de Gruijl F.R., Vrieling H., Mullenders L.H. 2005. Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res. 65:10298–10306 10.1158/0008-5472.CAN-05-2295 [DOI] [PubMed] [Google Scholar]
- Alekseev S., Luijsterburg M.S., Pines A., Geverts B., Mari P.O., Giglia-Mari G., Lans H., Houtsmuller A.B., Mullenders L.H., Hoeijmakers J.H., Vermeulen W. 2008. Cellular concentrations of DDB2 regulate dynamic binding of DDB1 at UV-induced DNA damage. Mol. Cell. Biol. 28:7402–7413 10.1128/MCB.01108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiácovo M.P., Elledge S.J. 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 107:18475–18480 10.1073/pnas.1012946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Bagchi S., Nag A., Shiyanov P., Adami G.R., Yoon T., Raychaudhuri P. 2001. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res. 486:89–97 [DOI] [PubMed] [Google Scholar]
- de Boer J., Hoeijmakers J.H. 2000. Nucleotide excision repair and human syndromes. Carcinogenesis. 21:453–460 10.1093/carcin/21.3.453 [DOI] [PubMed] [Google Scholar]
- Dinant C., Luijsterburg M.S., Höfer T., von Bornstaedt G., Vermeulen W., Houtsmuller A.B., van Driel R. 2009. Assembly of multiprotein complexes that control genome function. J. Cell Biol. 185:21–26 10.1083/jcb.200811080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mahdy M.A., Zhu Q., Wang Q.E., Wani G., Praetorius-Ibba M., Wani A.A. 2006. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 281:13404–13411 10.1074/jbc.M511834200 [DOI] [PubMed] [Google Scholar]
- Gong F., Fahy D., Liu H., Wang W., Smerdon M.J. 2008. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 7:1067–1074 10.4161/cc.7.8.5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A.J., Timinszky G., Kong S.E., Jin J., Cai Y., Swanson S.K., Washburn M.P., Florens L., Ladurner A.G., Conaway J.W., Conaway R.C. 2009. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. USA. 106:13770–13774 10.1073/pnas.0906920106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A.F., Tanaka K., Nakatani Y. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 113:357–367 10.1016/S0092-8674(03)00316-7 [DOI] [PubMed] [Google Scholar]
- Hara R., Mo J., Sancar A. 2000. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20:9173–9181 10.1128/MCB.20.24.9173-9181.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.J., McCall C.M., Hu J., Zeng Y., Xiong Y. 2006. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20:2949–2954 10.1101/gad.1483206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraten D., Nigg A.L., Heath H., Mullenders L.H., van Driel R., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 10:1163–1174 10.1016/S1097-2765(02)00709-8 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Bergink S., Ng J.M., Verbiest V.H., Luijsterburg M.S., Geverts B., Raams A., Dinant C., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. 2008. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J. Cell Sci. 121:2850–2859 (published erratum appears in J. Cell Sci. 2008. 121:3991) 10.1242/jcs.031708 [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers S., Nigg A.L., Hoogstraten D., Hoeijmakers J.H., Vermeulen W. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 284:958–961 10.1126/science.284.5416.958 [DOI] [PubMed] [Google Scholar]
- Hwang B.J., Ford J.M., Hanawalt P.C., Chu G. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA. 96:424–428 10.1073/pnas.96.2.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., Spector D.L. 2004. From silencing to gene expression: Real-time analysis in single cells. Cell. 116:683–698 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wang X., Bao S., Guo R., Johnson D.G., Shen X., Li L. 2010. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc. Natl. Acad. Sci. USA. 107:17274–17279 10.1073/pnas.1008388107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.L., Zhang L., Seldeen K.L., Gong F. 2010. Detection of bulky DNA lesions: DDB2 at the interface of chromatin and DNA repair in eukaryotes. IUBMB Life. 62:803–811 10.1002/iub.391 [DOI] [PubMed] [Google Scholar]
- Kaiser T.E., Intine R.V., Dundr M. 2008. De novo formation of a subnuclear body. Science. 322:1713–1717 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- Kapetanaki M.G., Guerrero-Santoro J., Bisi D.C., Hsieh C.L., Rapić-Otrin V., Levine A.S. 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA. 103:2588–2593 10.1073/pnas.0511160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers G.J., Goedhart J., van Munster E.B., Gadella T.W., Jr 2006. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry. 45:6570–6580 10.1021/bi0516273 [DOI] [PubMed] [Google Scholar]
- Kruhlak M.J., Celeste A., Dellaire G., Fernandez-Capetillo O., Müller W.G., McNally J.G., Bazett-Jones D.P., Nussenzweig A. 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172:823–834 10.1083/jcb.200510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H., Marteijn J.A., Schumacher B., Hoeijmakers J.H., Jansen G., Vermeulen W. 2010. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 6:e1000941 10.1371/journal.pgen.1000941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M.A., Th’ng J.P., Sun X., Hendzel M.J. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 408:873–876 10.1038/35048603 [DOI] [PubMed] [Google Scholar]
- Li J., Wang Q.E., Zhu Q., El-Mahdy M.A., Wani G., Praetorius-Ibba M., Wani A.A. 2006. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 66:8590–8597 10.1158/0008-5472.CAN-06-1115 [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., Goedhart J., Moser J., Kool H., Geverts B., Houtsmuller A.B., Mullenders L.H., Vermeulen W., van Driel R. 2007. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J. Cell Sci. 120:2706–2716 10.1242/jcs.008367 [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., von Bornstaedt G., Gourdin A.M., Politi A.Z., Moné M.J., Warmerdam D.O., Goedhart J., Vermeulen W., van Driel R., Höfer T. 2010. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 189:445–463 10.1083/jcb.200909175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881 10.1038/35048610 [DOI] [PubMed] [Google Scholar]
- Moné M.J., Volker M., Nikaido O., Mullenders L.H., van Zeeland A.A., Verschure P.J., Manders E.M., van Driel R. 2001. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2:1013–1017 10.1093/embo-reports/kve224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J., Volker M., Kool H., Alekseev S., Vrieling H., Yasui A., van Zeeland A.A., Mullenders L.H. 2005. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst.). 4:571–582 10.1016/j.dnarep.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Nichols A.F., Itoh T., Graham J.A., Liu W., Yamaizumi M., Linn S. 2000. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem. 275:21422–21428 10.1074/jbc.M000960200 [DOI] [PubMed] [Google Scholar]
- Nishi R., Alekseev S., Dinant C., Hoogstraten D., Houtsmuller A.B., Hoeijmakers J.H., Vermeulen W., Hanaoka F., Sugasawa K. 2009. UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair (Amst.). 8:767–776 10.1016/j.dnarep.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Nye A.C., Rajendran R.R., Stenoien D.L., Mancini M.A., Katzenellenbogen B.S., Belmont A.S. 2002. Alteration of large-scale chromatin structure by estrogen receptor. Mol. Cell. Biol. 22:3437–3449 10.1128/MCB.22.10.3437-3449.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomera-Sanchez Z., Zurita M. 2011. Open, repair and close again: Chromatin dynamics and the response to UV-induced DNA damage. DNA Repair (Amst.). 10:119–125 10.1016/j.dnarep.2010.10.010 [DOI] [PubMed] [Google Scholar]
- Polo S.E., Kaidi A., Baskcomb L., Galanty Y., Jackson S.P. 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29:3130–3139 10.1038/emboj.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers S., Volker M., Hoogstraten D., Nigg A.L., Moné M.J., Van Zeeland A.A., Hoeijmakers J.H., Houtsmuller A.B., Vermeulen W. 2003. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 23:5755–5767 10.1128/MCB.23.16.5755-5767.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapić-Otrin V., McLenigan M.P., Bisi D.C., Gonzalez M., Levine A.S. 2002. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 30:2588–2598 10.1093/nar/30.11.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapić-Otrin V., Navazza V., Nardo T., Botta E., McLenigan M., Bisi D.C., Levine A.S., Stefanini M. 2003. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum. Mol. Genet. 12:1507–1522 10.1093/hmg/ddg174 [DOI] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A., Konícková R., Czyzewski B.K., Kawasaki Y., Jeffrey P.D., Groisman R., Nakatani Y., Iwai S., Pavletich N.P., Thomä N.H. 2008. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 135:1213–1223 10.1016/j.cell.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solimando L., Luijsterburg M.S., Vecchio L., Vermeulen W., van Driel R., Fakan S. 2009. Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J. Cell Sci. 122:83–91 10.1242/jcs.031062 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Misteli T. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 320:1507–1510 10.1126/science.1159051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E., Dorn J.F., Sengupta K., Jasin M., Nussenzweig A., Ried T., Danuser G., Misteli T. 2007. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9:675–682 10.1038/ncb1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng J.M., Masutani C., Iwai S., van der Spek P.J., Eker A.P., Hanaoka F., Bootsma D., Hoeijmakers J.H. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 2:223–232 10.1016/S1097-2765(00)80132-X [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., Hanaoka F. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 121:387–400 10.1016/j.cell.2005.02.035 [DOI] [PubMed] [Google Scholar]
- Tang J.Y., Hwang B.J., Ford J.M., Hanawalt P.C., Chu G. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell. 5:737–744 10.1016/S1097-2765(00)80252-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Sudlow G., Belmont A.S. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354 10.1083/jcb.145.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura K., Araki M., Saeki H., Masutani C., Ito T., Iwai S., Mizukoshi T., Kaneda Y., Hanaoka F. 2001. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J. 20:2004–2014 10.1093/emboj/20.8.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M., Moné M.J., Karmakar P., van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J.H., van Driel R., van Zeeland A.A., Mullenders L.H. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 8:213–224 10.1016/S1097-2765(01)00281-7 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhai L., Xu J., Joo H.Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 22:383–394 10.1016/j.molcel.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Wang Q.E., Wani M.A., Chen J., Zhu Q., Wani G., El-Mahdy M.A., Wani A.A. 2005. Cellular ubiquitination and proteasomal functions positively modulate mammalian nucleotide excision repair. Mol. Carcinog. 42:53–64 10.1002/mc.20065 [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Wu X.H., Friedberg E.C. 1991. Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem. 266:22472–22478 [PubMed] [Google Scholar]
- Wittschieben B.O., Iwai S., Wood R.D. 2005. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J. Biol. Chem. 280:39982–39989 10.1074/jbc.M507854200 [DOI] [PubMed] [Google Scholar]
- Wong A.K., Shanahan F., Chen Y., Lian L., Ha P., Hendricks K., Ghaffari S., Iliev D., Penn B., Woodland A.M., et al. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171–6177 [PubMed] [Google Scholar]
- Ye Q., Hu Y.F., Zhong H., Nye A.C., Belmont A.S., Li R. 2001. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 155:911–921 10.1083/jcb.200108049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang Q., Jones K., Patel M., Gong F. 2009. The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle. 8:3953–3959 10.4161/cc.8.23.10115 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Wang Q.E., Ray A., Wani G., Han C., Milum K., Wani A.A. 2009. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J. Biol. Chem. 284:30424–30432 10.1074/jbc.M109.044982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotter A., Luijsterburg M.S., Warmerdam D.O., Ibrahim S., Nigg A., van Cappellen W.A., Hoeijmakers J.H., van Driel R., Vermeulen W., Houtsmuller A.B. 2006. Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced dna damage depends on functional TFIIH. Mol. Cell. Biol. 26:8868–8879 10.1128/MCB.00695-06 [DOI] [PMC free article] [PubMed] [Google Scholar]