High Expression Hampers Horizontal Gene Transfer (original) (raw)
Abstract
Horizontal gene transfer (HGT), the movement of genetic material from one species to another, is a common phenomenon in prokaryotic evolution. Although the rate of HGT is known to vary among genes, our understanding of the cause of this variation, currently summarized by two rules, is far from complete. The first rule states that informational genes, which are involved in DNA replication, transcription, and translation, have lower transferabilities than operational genes. The second rule asserts that protein interactivity negatively impacts gene transferability. Here, we hypothesize that high expression hampers HGT, because the fitness cost of an HGT to the recipient, arising from the 1) energy expenditure in transcription and translation, 2) cytotoxic protein misfolding, 3) reduction in cellular translational efficiency, 4) detrimental protein misinteraction, and 5) disturbance of the optimal protein concentration or cell physiology, increases with the expression level of the transferred gene. To test this hypothesis, we examined laboratory and natural HGTs to Escherichia coli. We observed lower transferabilities of more highly expressed genes, even after controlling the confounding factors from the two established rules and the genic GC content. Furthermore, expression level predicts gene transferability better than all other factors examined. We also confirmed the significant negative impact of gene expression on the rate of HGTs to 127 of 133 genomes of eubacteria and archaebacteria. Together, these findings establish the gene expression level as a major determinant of horizontal gene transferability. They also suggest that most successful HGTs are initially slightly deleterious, fixed because of their negligibly low costs rather than high benefits to the recipient.
Keywords: horizontal gene transfer, expression level, evolution, prokaryotes
Introduction
Horizontal gene transfer (HGT) refers to the process by which an organism acquires genetic material from another organism without being the offspring of that organism. HGT occurs through three cellular mechanisms: transformation, conjugation, and transduction (Thomas and Nielsen 2005). In transformation, a cell absorbs naked DNA directly from its environment. In conjugation, DNA is transferred from one cell to another by direct cell–cell contact or through a bridge-like connection. In transduction, virus mediates the transfer of DNA between cells. HGT allows acquisitions of foreign genes, a major mechanism for prokaryotic adaptation to their environments (Lawrence 1999; Ochman et al. 2000; Koonin et al. 2001; Boucher et al. 2003; Gogarten and Townsend 2005; Pal et al. 2005; Fournier and Gogarten 2008; Zhaxybayeva and Doolittle 2011). Although the exact extent of HGT in prokaryotic evolution is debatable (Doolittle 1999; Daubin et al. 2003; Kurland et al. 2003), there is no doubt that it is widespread, frequent, and important (Koonin et al. 2001; Nakamura et al. 2004; Lerat et al. 2005; Ciccarelli et al. 2006; Choi and Kim 2007; Sorek et al. 2007; Dagan et al. 2008; Popa et al. 2011; Zhaxybayeva and Doolittle 2011). However, what determines the probability with which a gene can be horizontally transferred, compared with other genes in the same genome, is not well understood. Extensive studies in the last 15 years resulted in two rules (Rivera et al. 1998; Jain et al. 1999) that are widely although not universally (Wellner and Gophna 2008; Omer et al. 2010; Cohen et al. 2011; Gophna and Ofran 2011) accepted. The first rule, derived from empirical observations, states that genes involved in information processing such as DNA replication, transcription, and translation are less transferable than genes involved in cellular operations such as metabolism (Rivera et al. 1998). Because this rule mainly concerns the distinction between two classes of protein functions, we will call it the protein function rule. The underlying mechanism of the first rule is described by the second rule, hereby referred to as the protein complexity rule (Jain et al. 1999). This rule asserts that proteins with more protein interaction partners tend not to have proper functions after HGT and therefore exhibit lower rates of successful HGTs. Because the protein products of informational genes form large protein complexes (e.g., the ribosome) more often than those of operational genes (Jain et al. 1999), the protein complexity rule provides a mechanistic basis for the protein function rule.
Although the above two rules offer some explanations of the variation in HGT rates among genes, it is unlikely that they are the only rate determinants. More importantly, it is unclear whether they are the primary rate determinants. Based on five considerations, we propose that gene expression level also impacts HGT rates and that highly expressed genes are less transferable than lowly expressed ones. First, expressing an unnecessary gene wastes energy and reduces fitness (Dekel and Alon 2005; Wagner 2005; Stoebel et al. 2008). Acquisition of a highly expressed gene imposes a greater fitness cost to the recipient cell than that of a lowly expressed gene. Second, because different species prefer different synonymous codons (Hershberg and Petrov 2009), a transferred gene may use codons that are unpreferred by the recipient. Because using unpreferred codons may increase translational errors (Akashi 2003; Stoletzki and Eyre-Walker 2007; Drummond and Wilke 2008), which can cause cytotoxic protein misfolding (Drummond and Wilke 2008; Geiler-Samerotte et al. 2011), acquisition of a strongly expressed gene leads to more misfolded protein molecules and a lower fitness than that of a weakly expressed gene. In addition, even correctly translated proteins may occasionally misfold and cause harm, and the total harm from such events increases with the expression level of the gene (Yang et al. 2010). Third, the expression of a foreign gene with a codon usage that is suboptimal in the recipient cell intensifies ribosomal sequestering that reduces the overall translational efficiency (Qian et al. 2012) and the fitness of the recipient (Kudla et al. 2009), and this fitness reduction is amplified when the foreign gene is strongly expressed. Consistent with the second and third mechanisms, a recent experiment showed that expressing a gratuitous gene in Escherichia coli at a high level decreases cellular growth and that the reduction in growth is positively correlated with the fraction of unpreferred codons in the gene (Kudla et al. 2009). Consistent with the third mechanism, mismatches between the codon usage of a foreign gene and the corresponding tRNA concentrations of the recipient cell decreases the transferability of the gene (Tuller et al. 2011). Fourth, a protein may interact with other proteins that it should not normally interact with and such misinteractions can be deleterious (Vavouri et al. 2009; Yang et al. 2012). Compared with a lowly expressed transferred gene, a highly expressed transferred gene induces more misinteractions (Yang et al. 2012) and is thus more deleterious to the recipient cell and less likely to be fixed. Fifth, acquisition of a foreign gene may also impact the recipient because of the specific function of the acquired gene. For instance, when the foreign gene is functionally similar to an endogenous gene, the HGT effectively raises the dose of the endogenous protein, which could be deleterious. The damage caused is expected to rise with the expression level of the foreign gene relative to the endogenous gene. Alternatively, when a foreign gene bestows a new function to the recipient, the new function could be deleterious to the recipient by disturbing the normal physiology. In such situations, the deleterious effect is expected to increase with the expression level of the transferred gene. Given these five considerations, we set out to test whether high gene expression indeed hinders HGT. Below, we first examine laboratory and natural HGTs to E. coli and then expand the analysis to HGTs to other prokaryotes. We show that gene expression level predicts HGT rates better than the two established rules. These findings also shed light on the population genetic forces dictating the fixation of HGTs.
Materials and Methods
Genome Sequences
We retrieved all publicly available prokaryotic genome sequences and associated annotations from the Integrated Microbial Genomes (IMG) system (http://genome.jgi-psf.org/programs/bacteria-archaea/index.jsf) (Markowitz et al. 2009).
Horizontally Transferred Genes
We used three large data sets of HGTs. The first data set (Sorek et al. 2007) included genes that can and cannot be transformed into E. coli in laboratory. The second data set (Lercher and Pal 2008) described genes that were naturally transferred into E. coli at different evolutionary times, inferred from the presence/absence of genes across species. The inference was based on the DELTRAN algorithm, with relative penalties of 2:1 for HGTs and gene losses (Lercher and Pal 2008), as in a recent study (Gophna and Ofran 2011). We identified the likely donor species of each horizontally transferred gene in this data set by Blasting the gene with an E value cutoff of 10−6 in all 1,127 finished Bacteria and Archaea genomes in IMG that are outside the family Enterobacteriaceae, to which E. coli belongs (fig. 2_A_). The genome harboring the best basic local alignment search tool (Blast) hit is considered the donor of the transferred gene. Reciprocal Blast searches are unnecessary, because the best Blast hit of the identified donor gene in E. coli will be 1) either the original gene under investigation or 2) a paralog of the original gene under investigation. But, because the gene under investigation was identified by phylogenetic analysis to be horizontally transferred to E. coli rather than a recent paralog of another gene in E. coli, (2) is not possible. Thus, the only possibility is (1), which makes it unnecessary to Blast the E. coli genome using the identified donor gene as the query. Furthermore, errors in donor identification are expected to be random, which would weaken the true signal but not bias our result. The third data set included relatively recent HGTs identified from 171 recipient genomes by nucleotide composition-based Bayesian inference (Nakamura et al. 2004). We discarded 38 of these genomes because of the lack of any annotation of ribosomal protein genes that are required for determining the preferred codons for codon adaptation index (CAI) estimation.
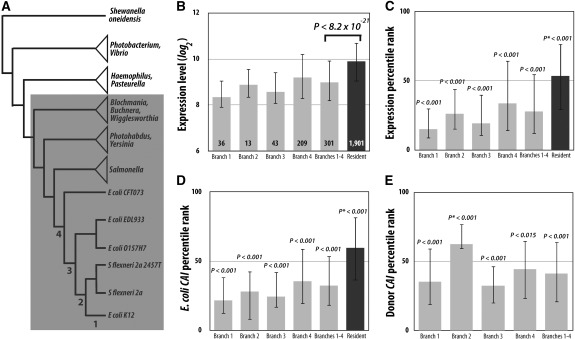
FIG. 2.—
Recently transferred genes to Escherichia coli have lower expressions than the resident genes in E. coli. (A) A phylogeny of E. coli K12 and related strains and species that was used to identify the HGTs (Lercher and Pal 2008) analyzed here. Only those HGTs that occurred in branches 1–4 are considered recent HGTs to E. coli K12. All other genes in E. coli K12 are considered resident genes. The shaded clade is the family Enterobacteriaceae mentioned in Materials and Methods. (B) Microarray expression levels of horizontally acquired genes are lower than those of resident genes in E. coli. The numbers of genes analyzed are indicated inside bars. (C) Percentile ranks of microarray expression levels of horizontally acquired genes and resident genes in E. coli. Percentile ranks range from 0 for the gene with the lowest expression to 100 for the gene with the highest expression. (D) Percentile ranks of CAIs of horizontally acquired genes and resident genes in E. coli, calculated using the codon usage pattern of E. coli. Percentile ranks range from 0 for the gene with the lowest CAI to 100 for the gene with the highest CAI. (E) Percentile ranks of CAIs of transferred genes, calculated using the codon usage patterns of their respective likely donors. The percentile rank of a transferred gene ranges from 0 for the lowest CAI to 100 for the highest CAI in its donor genome. In (B_–_E), median values are presented, with the error bars indicating 25% and 75% quartiles. In (B), the P value is from the Mann–Whitney U test. In (C–E), P values show the probabilities that the median percentile ranks are lower than 50 (or higher than 50 for P*), determined by bootstrapping the genes 10,000 times.
Genome-Wide Gene Expression Data
We used published E. coli gene expression data from the log growth phase obtained from a high-density oligonucleotide tiling array experiment (Cho et al. 2009). To download all publicly available microarray expression data from other prokaryotes, we used the Stanford Microarray Database (Hubble et al. 2009) that houses hundreds of expression data sets based on cDNA microarrays. Expression data from six species (Bacillus subtilis, ID: 66211; Campylobacter jejuni, ID: 28770; Helicobacter pylori, ID: 16576; Mycobacterium tuberculosis, ID: 14047; Salmonella typhimurium, ID: 23956; and Vibrio cholerae: ID 66211) were used in our analysis. We also used the NCBI Gene Expression Omnibus and downloaded the microarray data of Dehalococcoides ethenogenes (GSE 10185), Geobacter sulfurreducens (GSE 22511), Listeria monocytogenes (GSE 16336), and Streptococcus agalactiae (GSE 21564).
Synonymous Codon Usage Bias
To calculate the relative synonymous codon usage (RSCU) in a species (Sharp and Li 1986), we used ribosomal protein genes, which are generally among the most highly expressed genes in a genome (Sharp et al. 1986). Based on the RSCU values, the CAI was calculated for each gene in a genome (Sharp and Li 1987). Briefly, CAI of a gene is the geometric mean of RSCU of all codons divided by the highest possible geometric mean of RSCU given the same amino acid sequence.
Classification of Informational Genes and Operational Genes
Following an earlier study (Jain et al. 1999), we regarded genes annotated with “transcription,” “translation,” “DNA replication,” or any of their subterms in Gene Ontology (Ashburner et al. 2000) as informational genes. All other genes were considered operational genes.
Protein–Protein Interactions
The E. coli protein–protein interaction data were retrieved from a recent publication (Hu et al. 2009), in which 5,993 nonredundant pairwise physical interactions among 1,757 proteins were identified by an affinity-based method and genomic context-based inferences.
Statistical Analysis
We estimated the relative contributions of all predictors to the total variance in gene transferability by calculating the relative contribution of variability explained (RCVE) for each predictor using RCVE=1−Rreduced2/Rfull2, where Rfull2 and Rreduced2 are the _R_2 (square of the correlation coefficient) for the full linear model and the model without the predictor of interest, respectively (Park and Makova 2009). To diagnose multicollinearity of each predictor, variance inflation factors (VIFs) (Kutner et al. 2005) were calculated. All predictors in the model used had VIFs below 2, suggesting that multicollinearity did not adversely affect our model. Linear multiple regression analysis was performed in the R statistical package.
Results
Laboratory HGTs to E. coli
We test the impact of gene expression level on the rate of HGT by first using a data set of laboratory HGTs to E. coli that was compiled based on microbial genome sequencing (Sorek et al. 2007). Briefly, when sequencing a microbial genome, researchers typically randomly shear its genomic DNA, clone the DNA fragments into a plasmid, and transform the plasmid to E. coli for DNA amplification and shotgun sequencing. Genes that cannot be transferred to E. coli leave gaps in the assembled genome that are later filled by a clone-independent procedure. Thus, these gaps in shotgun assemblies can be used to infer genes nontransferable to E. coli via plasmid mediated transformation. Among the 79 donor genomes (246,045 genes in total) analyzed (Sorek et al. 2007), 14 genomes are amenable to statistical analysis because they each contain at least 30 so-called nontransferable genes. Of these 14 species, four have publicly available microarray-based genome-wide gene expression data (see Materials and Methods). In all four cases, expression levels are significantly higher for nontransferable genes than for transferable genes (fig. 1_A_). The median expression level of nontransferable genes is 1.6–5.3 times that of transferable genes (fig. 1_A_).
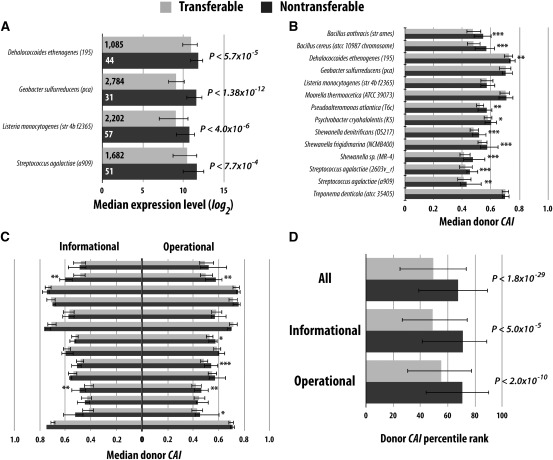
FIG. 1.—
Nontransferable genes have higher expressions than transferable genes in laboratory HGTs to Escherichia coli. (A) Median microarray expression levels of transferable and nontransferable genes in donor species. The numbers of genes used are indicated inside the bars. (B) Median CAIs of transferable and nontransferable genes in donor species. (C) Median CAIs of transferable and nontransferable genes in donor species when informational genes are separated from operational genes. (D) Median CAI percentile ranks of transferable and nontransferable genes from all 14 donor species. The CAI percentile rank of a gene is based on the rank of its CAI relative to those of all genes in the same donor genome. In all panels, error bars show 25% and 75% quartiles in the sample. All P values are from the Mann–Whitney U test. In (B) and (C), *P < 0.05, **P < 0.01, and ***P < 0.001.
Within a genome, the codon adaptation index (CAI) (Sharp and Li 1987) of a gene is highly positively correlated with the expression level of the gene and can thus be used as a proxy for gene expression level. It has even been argued that CAIs reflect the relative expression levels in an organism's natural environment better than the actual expression levels measured in laboratory conditions (Fraser et al. 2004). We calculated the CAIs of all genes in each of the 14 donor species. The median CAI is higher for nontransferable genes than transferable genes in 12 of the 14 donors (fig. 1_B_), significantly more than the random expectation of 7 (P = 0.006, one-tail binomial test). Ten species show a significant difference in median CAI between transferable and nontransferable genes, all in the predicted direction (fig. 1_B_).
One caveat in the above analysis is that the CAI of a gene estimated based on the codon usage of its host species may not represent its true expression level if the gene was only recently acquired by the species via HGT because it takes time for the CAI of a gene to evolve and adapt to a new cellular environment. We thus repeated the above analysis after removing from the 14 species those genes that were identified in an earlier study (Nakamura et al. 2004) to be recently acquired by HGT. However, the results remain qualitatively unchanged (supplementary fig. S1, Supplementary Material online).
To exclude the possibility that the above observation is a byproduct of the protein function rule, we separately analyzed informational genes and operational genes. Because of the reduction in sample size, the statistical power of the analysis is decreased. Yet, the general pattern of higher expressions or higher CAIs of nontransferable genes than transferable genes remains valid for both informational genes and operational genes (fig. 1_C_). For example, when only informational genes are considered, 13 of the 14 species show higher CAIs for nontransferable genes than transferable genes, significantly more than random expectation (P < 0.001, one-tail binomial test). For operational genes, 11 species show this pattern (P < 0.03). Two and five species show significant differences in CAIs between transferable and nontransferable genes among informational genes and operational genes, respectively, and all of these significant differences are in the predicted direction (fig. 1_C_). These results indicate that the impact of gene expression level on HGT rates is not a byproduct of the protein function rule. Because of the lack of protein interactome data for the 14 species, we cannot evaluate the impact of the protein complexity rule here. Nonetheless, the similarity between the protein function rule and complexity rule (Jain et al. 1999) suggests that our results are unlikely caused by the confounding factor of the protein complexity rule.
Because of the relatively small number of nontransferable genes from each of the 14 species, we conducted a combined analysis of all 14 species. We first converted the CAIs of all genes in a genome to percentile ranks; the highest CAI has a percentile rank of 100 and the lowest has a percentile rank of 0. We then combined all the genes from the 14 species. We observed significantly higher CAI percentile ranks for nontransferable genes than transferable genes (fig. 1_D_), and this pattern is true for both informational genes and operational genes (fig. 1_D_).
Because the impact of expression level on the fixation of an HGT occurs after the gene is transferred to the recipient cell, one wonders whether the expression level measured in the donor species is relevant. We believe the answer is yes for both laboratory and natural HGTs. In the laboratory HGTs considered here, a gene is likely to be cloned into a plasmid together with its promoter and thus is likely controlled by its own promoter even in the recipient. For this reason, expression levels in the donor and recipient are expected to be positively correlated, although the transcriptional machinery (i.e., _trans_-factors) may differ between the donor and recipient. The same argument can be made for all three mechanisms of HGTs and thus applies to natural HGTs. The fact that some of the nontransferable genes become transferable when only the coding regions but not the promoters are transferred to E. coli (Sorek et al. 2007) supports our view.
Natural HGTs to E. coli
It is important to confirm in natural HGTs the patterns observed from laboratory HGTs because the laboratory HGTs were based on only one mechanism—plasmid mediated transformation, while natural HGTs occur by three mechanisms. In addition, laboratory conditions are different from the nature in many aspects, which may influence HGT rates. We thus analyze genes that have been naturally transferred to E. coli K12 since its divergence from Salmonella ∼100 Ma (Battistuzzi et al. 2004). These genes, previously identified by a phylogenetic analysis of gene gains and losses (Lercher and Pal 2008), are divided into four temporal groups according to the dates of the transfers (fig. 2_A_). We focused on these recently transferred genes because inferring recent HGTs is much more reliable than inferring ancient HGTs.
We first compared these recently acquired genes with the resident genes in the E. coli genome, which include genes that were acquired by E. coli before its divergence from Salmonella. If foreign genes have been continuously transferred into E. coli to replace its endogenous genes and different genes have different transferabilities, the recently transferred genes should be enriched with highly transferable genes. We thus assume that the recently transferred genes have higher HGT rates than the resident genes of E. coli as well as the rest of the genes in various potential donor species. Using E. coli microarray gene expression data (Cho et al. 2009), we found the expression levels of the recently acquired genes to be significantly lower than those of resident genes, with a difference of ∼2-fold in median expression (fig. 2_B_). The same can be seen in the comparison of expression percentile ranks, after the expression levels are converted to percentile ranks (fig. 2_C_). For example, the recently acquired genes, either separated into four age groups or combined, have median expression percentile ranks significantly below 50, whereas the resident genes have a median percentile rank significantly above 50 (fig. 2_C_). Analysis of percentile ranks of CAIs calculated based on E. coli codon usage gives a similar result (fig. 2_D_), suggesting that CAI percentile ranks are good proxies of expression percentile ranks.
The difference in expression level between the recently acquired genes and the resident genes (fig. 2_B_) can have only two nonmutually exclusive explanations. The first explanation is our hypothesis that highly expressed genes are less transferable than lowly expressed genes. As a result, foreign genes that were recently acquired by E. coli tend to be lowly expressed. Second, it is also possible that all genes have reduced expressions when transferred into new hosts, compared with the expressions in their original hosts, because of potential mismatches between the promoters of the transferred genes and the transcriptional machinery (including _trans_-regulatory factors) of the recipient (Lercher and Pal 2008) and/or host defense (Navarre et al. 2006; Marraffini and Sontheimer 2010). If the expression difference between transferred and resident genes in figure 2_B_ is entirely caused by the second reason, the transferred genes should not be biased toward low expressions in their original hosts. We identified the most likely donor species of each recent HGT to E. coli (see Materials and Methods) and then calculated the CAI percentile rank of the transferred gene among all genes in the donor by considering the codon usage in the donor. Clearly, the horizontally transferred genes have relatively low CAIs among all genes in their donors (fig. 2_E_). Thus, the expression difference observed in figure 2_B_ must be caused, at least in part, by the first reason that high expression hampers HGT. Note that, for the horizontally transferred genes, their expression percentile ranks in the recipient (fig. 2_C_) appear lower than their CAI percentile ranks in the donor (fig. 2_E_), suggesting that the aforementioned second reason is likely at work too. Interestingly, the CAI percentile ranks of the horizontally acquired genes in the recipient (fig. 2_D_) are slightly closer than the corresponding expression ranks (fig. 2_C_) to the CAI percentile ranks in the donor (fig. 2_E_), suggesting that CAI percentile ranks in the recipient (fig. 2_D_) is at least as good a proxy as expression percentile ranks in the recipient (fig. 2_C_) for CAI percentile ranks in the donor (fig. 2_E_). This finding allows the use of CAI percentile ranks in the recipient as a proxy for those in the donor, which becomes necessary when the donor is unknown, as in the case of natural HGTs to other prokaryotes presented in a later section.
To compare the relative importance of the protein function rule, protein complexity rule, and gene expression level in determining the rate of natural HGTs to E. coli, we conducted several regression analyses. Assigning a transferability score of 1 to the recently acquired genes and 0 to the resident genes in the E. coli genome, we found, consistent to the result in figure 2_B_, a significantly negative correlation between transferability and expression level (Spearman's rank correlation ρ = −0.283, P = 0.0001; rank biserial correlation rrb = −0.499, P = 0.0001; table 1). Assigning a function score of 1 to informational genes and 0 to operational genes, we found no correlation between function and transferability (ρ = −0.001, P = 0.9976; Phi correlation φ = −0.001, P = 0.9999; table 1). Using systematically annotated E. coli protein interactions (Hu et al. 2009), we found a significant negative correlation between the transferability of a gene and its number of protein interaction partners (ρ = −0.250, P = 0.0001; rrb = −0.432, P = 0.0001; table 1). It has been observed that genes acquired by HGT tend to have low frequencies of guanine (G) and cytosine (C) (Syvanen 1994; Lawrence and Ochman 1997; Navarre et al. 2007). We thus considered the GC% of a gene as an additional factor potentially impacting HGT. Indeed, we found a significantly negative correlation between gene transferability and GC% (ρ = −0.147, P = 0.0001; rrb = −0.259, P = 0.0001; table 1). Note, however, that this correlation may be in part a byproduct of the correlation between expression level and transferability because highly expressed proteins tend to use metabolically cheap amino acids (Akashi and Gojobori 2002), which are encoded by GC-rich codons (Akashi and Gojobori 2002).
Table 1.
Relative Contributions of Protein Function (Informational vs. Operational), Complexity (Number of Protein Interaction Partners), GC%, and Expression Level on Gene Transferability in Natural HGTs to Escherichia coli
| Factors Considered | Rank Correlations with Gene Transferabilitya | Multiple Linear Regressionb | ||||
|---|---|---|---|---|---|---|
| Correlation | P Value | Partial Correlationc | P Value | RCVEd | P Valuee | |
| Expression level | −0.283 (−0.499)f | 0.0001 (0.0001) | −0.195 | 0.0001 | 0.337 | 0.0001 |
| Number of protein interactions | −0.250 (−0.432)f | 0.0001 (0.0001) | −0.148 | 0.0001 | 0.191 | 0.0001 |
| Informational/operationalg | −0.001 (−0.001)h | 0.9976 (0.9999) | 0.014 | 0.5280 | 0.002 | 0.5281 |
| GC% | −0.147 (−0.259)f | 0.0001 (0.0001) | −0.055 | 0.0120 | 0.026 | 0.0121 |
| Totali | 0.105 |
After the controls of protein function, complexity, and GC%, the partial rank correlation between gene expression and transferability remains significant (ρ = −0.195, P = 0.0001; table 1), indicating that expression level affects gene transferability independent of the other three factors. Among the four factors examined here, expression level has the strongest correlation with transferability (table 1).
Because several factors studied above might be interrelated, we also conducted a multiple regression analysis to assess the relative contributions of the four factors in explaining the total variability in transferability among genes. This multiple regression model explains ∼10% of the total variance in gene transferability and all predictors except protein function remain significant after the Bonferroni correction for multiple tests (table 1). Gene expression level is the best predictor, explaining at least ∼34% of the variance explained by the model (table 1).
Natural HGTs to Other Prokaryotes
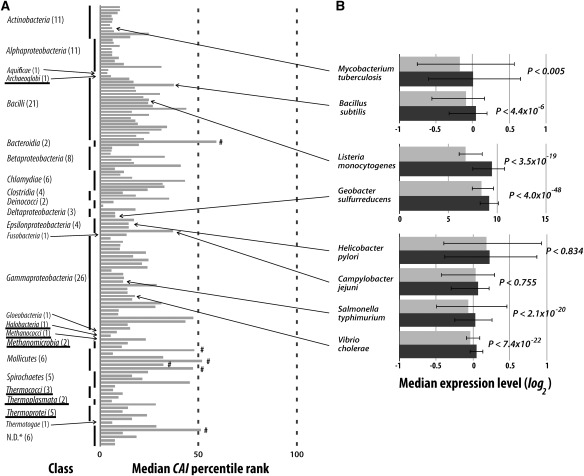
To confirm that the patterns observed in laboratory and natural HGTs to E. coli are not unique to E. coli, we examined HGTs to many other prokaryotes. We analyzed 133 prokaryotic genomes, where recently acquired genes were previously identified based on abnormal nucleotide compositions (Nakamura et al. 2004). We found the horizontally acquired genes to have a median CAI percentile rank significantly below 50 in 127 species and insignificantly below 50 in three species (fig. 3_A_). Although the median CAI percentile rank of horizontally transferred genes exceeds 50 in the remaining three species, none exceeds 50 significantly (fig. 3_A_). An important caveat is that the low CAIs of the transferred genes observed here may be an artifact due to HGT identification by abnormal nucleotide compositions. To exclude this possibility, we reanalyzed it using gene expression data. Eight of the 133 species have publicly available microarray expression data (see Materials and Methods). In all eight species, expression levels of horizontally acquired genes are lower than those of resident genes, and the difference is significant in six of the eight species (fig. 3_B_). Because the 133 species examined here include a diverse set of eubacteria and archaebacteria (fig. 3_A_) and because the CAI-based and microarray-based analyses are largely concordant (fig. 3), we conclude that the phenomenon of lower HGT rates for more highly expressed genes is general among prokaryotes. While the list of horizontally transferred genes used here (Nakamura et al. 2004) may contain some errors due to the non–phylogeny-based identification, we note that such errors are expected to be random and to only blur the distinction between resident genes and horizontally acquired genes, which makes our results more conservative.
FIG. 3.—
Recently horizontally acquired genes have lower expressions than resident genes in most recipient species. (A) Median CAI percentile ranks of horizontally acquired genes in 133 recipient species examined. The percentile rank of a horizontally acquired gene relative to all other genes in the recipient genome ranges from 0 for the lowest CAI to 100 for the highest CAI. All are significantly different from 50, except those indicated with “#.” Statistical significance was determined by bootstrapping the genes 10,000 times. Class names are indicated, with the numbers of genomes examined shown in parentheses. N.D.*: not defined by taxonomic classes. Underlined class names indicate archaebacteria, while the rest belong to eubacteria. Information about the individual genomes analyzed here is provided in supplementary table S1 (Supplementary Material online). (B) Median expression levels of horizontally acquired genes (light gray) and resident genes (dark gray) in recipient species with publicly available microarray gene expression data. Error bars show 25% and 75% quartiles. The microarray data of the eight species came from different sources and the gene expression levels of different species are not comparable. P values are from the Mann–Whitney U test. Arrows connect the same genomes in the two panels.
Discussion
Examining laboratory and natural HGTs to E. coli and natural HGTs to many other prokaryotes, we showed that high expression hinders HGT. Furthermore, we found gene expression level to be a more important determinant of gene transferability than three known factors: protein function (i.e., informational vs. operational), protein complexity (i.e., number of protein interaction partners), and GC%. We proposed that high expression hampers HGT because the fitness cost of an HGT to the recipient arising from 1) energy expenditure in transcription and translation, 2) cytotoxic protein misfolding, 3) reduction in cellular translational efficiency, 4) detrimental protein misinteraction, and 5) disturbance of the optimal protein concentration or cell physiology all increases with the expression level of the transferred gene. Which of the five mechanisms plays the most important role in reducing the HGT rates of highly expressed genes? This question is difficult to address at this time for three reasons. First, key parameters in several of the above mechanisms, such as the probability of protein misinteraction, are difficult to estimate accurately (Yang et al. 2012). Second, a suitable HGT data set with large numbers of both transferable and nontransferable genes to the same recipient species is required. Third, an ideal HGT data set should contain genes with quantitatively different transferabilities to a recipient so that the quantitative impact of a factor can be detected. For example, the laboratory HGT data contain only absolutely nontransferable genes, which are lethal or almost lethal to the recipient when transferred. Such data do not allow the test of factors that have quantitative rather than qualitative effects. This is also why Sorek and colleagues inferred from these data that the barrier to HGT is the toxicity of the transferred gene to the recipient (Sorek et al. 2007), which belongs to our fifth mechanism. While Sorek et al.'s finding that reducing the expression levels of a few toxic genes increases their transferability to E. coli supports our hypothesis about the impact of expression level on HGT, our hypothesis goes well beyond the mechanism of cytotoxicity and the small number of toxic genes. In theory, our hypothesis applies to all genes in a genome and all prokaryotes, as has been demonstrated here in natural HGTs to E. coli and more than 100 other prokaryotes.
In yeast, deleting a highly expressed gene affects the fitness more than deleting a lowly expressed gene (Zhang and He 2005). The same may be expected in gene acquisition. That is, acquisition of a highly expressed gene is expected to have a greater fitness effect than that of a lowly expressed gene. If most gene acquisitions by HGT are beneficial, strongly expressed genes would confer higher benefits and be more transferable. The observation that it is weakly expressed genes that are more transferable suggests that most HGTs are not beneficial. In other words, most HGTs are fixed not because their benefits to the recipients are high, but because their costs are negligibly low. In an analogy, HGT is like moving a family to a new neighborhood. Lowly expressed genes, like quiet families, disturb their new neighborhood less and are therefore more likely to be accepted.
Although an HGT may be neutral or slightly deleterious to the recipient and gets fixed by genetic drift, the transferred gene must be useful to the recipient for it to be stably retained in the recipient's genome during evolution. When the transferred gene is functionally equivalent to an endogenous gene in the recipient, the endogenous gene may by chance pseudogenize, permitting the stable retention of the transferred gene. Alternatively, when the transferred gene brings in a new function that is initially useless or even deleterious to the recipient, the new function may become beneficial when the environment or the genetic background is altered. These processes explain how a horizontally acquired gene, even with a nearly neutral origin via HGT, can later become indispensable to the recipient and/or facilitate its adaptation.
A gene can evolve in three broad aspects: its product function, its expression level and pattern, and its genomic environment. HGT is a common mechanism for gene evolution in the last-named aspect. Compared with lowly expressed genes, highly expressed genes are known to be slower in coding sequence evolution (Pal et al. 2001) and expression-profile evolution (Liao and Zhang 2006). The present study showed that highly expressed genes are also slower in HGT. Thus, high expression constrains gene evolution in all three broad aspects. It would be interesting to examine whether the mechanisms of the impact of expression level on these three aspects of gene evolution are similar or distinct (Drummond and Wilke 2008; Cherry 2010; Gout et al. 2010; Yang et al. 2010; Yang et al. 2012).
Supplementary Material
Supplementary figure S1 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Rotem Sorek, Martin Lercher, and Yoji Nakamura for sharing their HGT data, Uri Gophna and members of the Zhang laboratory for discussion, and Tal Pupko, Wenfeng Qian, and two anonymous reviewers for valuable comments. This work was supported by a research grant from the US National Institutes of Health (R01GM067030) to J.Z.
References
- Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164:1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y, et al. Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- Cherry JL. Expression level, evolutionary rate, and the cost of expression. Genome Biol Evol. 2010;2:757–769. doi: 10.1093/gbe/evq059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, et al. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IG, Kim SH. Global extent of horizontal gene transfer. Proc Natl Acad Sci U S A. 2007;104:4489–4494. doi: 10.1073/pnas.0611557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Cohen O, Gophna U, Pupko T. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol. 2011;28:1481–1489. doi: 10.1093/molbev/msq333. [DOI] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Moran NA, Ochman H. Phylogenetics and the cohesion of bacterial genomes. Science. 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier GP, Gogarten JP. Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J Bacteriol. 2008;190:1124–1127. doi: 10.1128/JB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Wall DP, Eisen MB. Coevolution of gene expression among interacting proteins. Proc Natl Acad Sci U S A. 2004;101:9033–9038. doi: 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, et al. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci U S A. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Gophna U, Ofran Y. Lateral acquisition of genes is affected by the friendliness of their products. Proc Natl Acad Sci U S A. 2011;108:343–348. doi: 10.1073/pnas.1009775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout JF, Kahn D, Duret L. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 2010;6:e1000944. doi: 10.1371/journal.pgen.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. General rules for optimal codon choice. PLoS Genet. 2009;5:e1000556. doi: 10.1371/journal.pgen.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubble J, et al. Implementation of GenePattern within the Stanford Microarray Database. Nucleic Acids Res. 2009;37:D898–D901. doi: 10.1093/nar/gkn786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Canback B, Berg OG. Horizontal gene transfer: a critical view. Proc Natl Acad Sci U S A. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner MH, Nachtsheim CJ, Neter J, Li W. Applied linear statistical models. New York: McGraw-Hill; 2005. [Google Scholar]
- Lawrence JG. Gene transfer, speciation, and the evolution of bacterial genomes. Curr Opin Microbiol. 1999;2:519–523. doi: 10.1016/s1369-5274(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Pal C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol. 2008;25:559–567. doi: 10.1093/molbev/msm283. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2009;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- Navarre WW, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Omer S, Kovacs A, Mazor Y, Gophna U. Integration of a foreign gene into a native complex does not impair fitness in an experimental model of lateral gene transfer. Mol Biol Evol. 2010;27:2441–2445. doi: 10.1093/molbev/msq145. [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- Park C, Makova KD. Coding region structural heterogeneity and turnover of transcription start sites contribute to divergence in expression between duplicate genes. Genome Biol. 2009;10:R10. doi: 10.1186/gb-2009-10-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci U S A. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics. 2008;178:1653–1660. doi: 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Syvanen M. Horizontal gene transfer: evidence and possible consequences. Annu Rev Genet. 1994;28:237–261. doi: 10.1146/annurev.ge.28.120194.001321. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- Tuller T, et al. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res. 2011;39:4743–4755. doi: 10.1093/nar/gkr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Wellner A, Gophna U. Neutrality of foreign complex subunits in an experimental model of lateral gene transfer. Mol Biol Evol. 2008;25:1835–1840. doi: 10.1093/molbev/msn131. [DOI] [PubMed] [Google Scholar]
- Yang J-R, Liao B-Y, Zhuang S-M, Zhang J. Protein-misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci U S A. 2012;109:E831–E840. doi: 10.1073/pnas.1117408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JR, Zhuang SM, Zhang J. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol. 2010;6:421. doi: 10.1038/msb.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, He X. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol Biol Evol. 2005;22:1147–1155. doi: 10.1093/molbev/msi101. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Doolittle WF. Lateral gene transfer. Curr Biol. 2011;21:R242–246. doi: 10.1016/j.cub.2011.01.045. [DOI] [PubMed] [Google Scholar]