Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation (original) (raw)
Abstract
Recent identification of intestinal epithelial stem cell (ISC) markers and development of ISC reporter mice permit visualization and isolation of regenerating ISCs after radiation to define their functional and molecular phenotypes. Previous studies in uninjured intestine of Sox9-EGFP reporter mice demonstrate that ISCs express low levels of Sox9-EGFP (Sox9-EGFP Low), whereas enteroendocrine cells (EEC) express high levels of Sox9-EGFP (Sox9-EGFP High). We hypothesized that Sox9-EGFP Low ISCs would expand after radiation, exhibit enhanced proliferative capacities, and adopt a distinct gene expression profile associated with rapid proliferation. Sox9-EGFP mice were given 14 Gy abdominal radiation and studied between days 3 and 9 postradiation. Radiation-induced changes in number, growth, and transcriptome of the different Sox9-EGFP cell populations were determined by histology, flow cytometry, in vitro culture assays, and microarray. Microarray confirmed that nonirradiated Sox9-EGFP Low cells are enriched for Lgr5 mRNA and mRNAs enriched in Lgr5-ISCs and identified additional putative ISC markers. Sox9-EGFP High cells were enriched for EEC markers, as well as Bmi1 and Hopx, which are putative markers of quiescent ISCs. Irradiation caused complete crypt loss, followed by expansion and hyperproliferation of Sox9-EGFP Low cells. From nonirradiated intestine, only Sox9-EGFP Low cells exhibited ISC characteristics of forming organoids in culture, whereas during regeneration both Sox9-EGFP Low and High cells formed organoids. Microarray demonstrated that regenerating Sox9-EGFP High cells exhibited transcriptomic changes linked to p53-signaling and ISC-like functions including DNA repair and reduced oxidative metabolism. These findings support a model in which Sox9-EGFP Low cells represent active ISCs, Sox9-EGFP High cells contain radiation-activatable cells with ISC characteristics, and both participate in crypt regeneration.
Keywords: enteroendocrine cells, quiescent intestinal stem cells
continuous renewal of the adult small intestinal epithelium is driven by intestinal epithelial stem cells (ISCs), which are located at or near the base of the crypt (2, 46, 54). Current evidence suggests that these cells self-renew and give rise to daughter cells, termed progenitors or transit-amplifying cells, which actively divide and then differentiate into four terminally differentiated intestinal epithelial cell (IEC) lineages. Until recently, it was difficult to directly evaluate or isolate ISCs because of lack of valid biomarkers. Recent studies have used lineage tracing and in vitro culture approaches to identify markers of self-renewing ISCs in normal adult small intestinal epithelium that exhibit multipotency for all differentiated lineages. These biomarkers include Lgr5 (3), Bmi1 (54), Hopx (61), and Sox9 (17, 19, 21).
ISC reporter mouse models, which express fluorescent reporter genes under the control of genetic regulatory sequences of ISC biomarker genes, permit both visualization and isolation of ISCs. A recently characterized Sox9-EGFP (enhanced green fluorescent protein) bacterial artificial chromosome (BAC) transgenic reporter mouse offers the opportunity to distinguish and isolate four different cell populations from small intestinal epithelium on the basis of distinct levels of EGFP expression (21). In adult mouse small intestinal epithelium, cells that express low levels of the Sox9-EGFP transgene (Sox9-EGFP Low) were shown to reside at the same location as Lgr5-expressing ISCs, also termed crypt base columnar cells (CBCs) (12, 17). Sox9-EGFP Low cells were also shown to be highly enriched for Lgr5 mRNA and to exhibit the stem cell properties of self-renewal and multipotency in culture (21). Cells expressing high levels of Sox9-EGFP (Sox9-EGFP High) were shown to exhibit morphological features and express biomarkers of terminally differentiated enteroendocrine cells (EECs) (17, 21). Sox9-EGFP High cells are distributed throughout the base of the crypt but a majority of these cells localize to the +4 to +5 cell position from the crypt base (17). A third level of Sox9-EGFP expression termed “Sublow” was shown to mark proliferating cells in the typical location of progenitors. Sox9-EGFP Negative IECs (cells that do not express Sox9-EGFP) were found to express markers of terminally differentiated enterocytes (17, 21). The present study used the Sox9-EGFP reporter mouse to more fully define the molecular phenotypes of these different Sox9-EGFP-expressing cell populations and examine their functional and molecular characteristics during crypt regeneration after crypt loss due to high-dose radiation.
Ablation of small intestinal crypts and ISCs by high-dose whole body irradiation (WBR) has been a “gold standard” model to study ISC-mediated crypt regeneration (44, 47). After this type of injury, a small number of surviving ISCs clonally expand to regenerate crypts and the entire intestinal epithelium (45, 46, 48). Most prior studies of crypt regeneration after high-dose WBR have typically focused on early time points (day 1 to day 4 after WBR) because of high animal mortality from complications of intestinal damage, i.e., gastrointestinal syndrome (5, 7, 13, 23, 31, 38). To circumvent this problem, we used a high-dose (14 Gy) abdominal irradiation model that resulted in 100% survival of animals for 9 or more days after irradiation and permitted the characterization of Sox9-EGFP-expressing cells over longer periods of crypt regeneration than after similar doses of WBR.
Current views suggest that the small intestine may contain two ISC populations. One is an actively cycling population that corresponds to Lgr5-expressing CBCs and Sox9-EGFP Low cells. The other is a slower cycling or quiescent cell population that is thought to reside above CBCs and Paneth cells and has been termed the +4 ISCs because of their predominant location at approximately four cells from the base of the crypt (33, 46). Recent evidence suggests that the +4 or quiescent ISC population may be marked by Bmi1 (54), Hopx (61), doublecortin and CaM kinase-like-1 (DCAMKL-1) (39), or EEC markers (57). Furthermore, a bidirectional lineage relationship between active CBCs and +4 ISCs has been recently demonstrated (61, 64). In this study, we hypothesized that, following crypt and ISC ablation after high-dose radiation, crypt regeneration would involve expansion, hyperproliferation, and altered molecular phenotype of Sox9-EGFP Low cells, which correspond to active ISCs. To test our hypothesis, we characterized changes in Sox9-EGFP cell populations after irradiation using histology and flow cytometry. We also assessed the ability of Sox9-EGFP-expressing cells isolated during irradiation-induced regeneration to form organoids in vitro (21, 55) and used microarray to define gene expression changes exhibited by each Sox9-EGFP cell population during crypt regeneration. Our findings support a major role of Sox9-EGFP Low ISCs in crypt regeneration but also provide evidence for an additional ISC population contained within Sox9-EGFP High cells that is activated to proliferate during irradiation-induced crypt regeneration.
MATERIALS AND METHODS
Animals
Mice expressing a BAC transgene with ∼226.5 kb of Sox9 genomic regulatory region driving EGFP expression were established and maintained at the University of North Carolina (Chapel Hill, NC) as previously described (17, 21). Sox9-EGFP mice are on the outbred CD-1 strain and were maintained as heterozygotes by breeding with wild-type CD-1 strain mice. Genotyping was performed as in Refs. 17 and 21. All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina.
Abdominal Irradiation
Mice were given a single dose of 14 Gy irradiation by using an XRad 320 (Precision X-Ray, East Haven, CT) (Filter: 2 mm Al; 47 cm; 320 kV/s, 10 mA; 2.8 Gy/min). All radiation experiments were performed under isoflurane anesthesia, and mice were placed in the radiator so that only the abdomen lay in the radiated zone. Body weight was recorded every day. Typically, mice lost up to 25% body weight until day 7 postirradiation when body weight started to increase toward normal. Pilot studies indicated that CD-1 strain mice survived and began to gain weight even after losing the maximum 15% body weight typically used to warrant euthanasia. Our approved IACUC protocol therefore permitted an exception to euthanize mice only if they lost more than 25% body weight. We observed 100% survival up to 9 days after abdominal irradiation, which was the latest time point studied in these experiments.
Tissue Harvest for Histology or Flow Cytometry
To visualize and quantify Sox9-EGFP cells during crypt regeneration following radiation, Sox9-EGFP mice (6–10 wk old) were euthanized with a lethal dose of Nembutal (150 μg/g body wt) for tissue collection at days 3, 4, 5, 7, and 9 after abdominal radiation. At each time point, nonirradiated control mice were also euthanized. To mark cells in S-phase, 5-ethynyl-2′deoxyuridine (EdU, Sigma, St. Louis, MO) was administered to all animals 90 min prior to euthanasia via intraperitoneal injection at a dose of 100 μg/25 g body wt. For each animal, entire jejunum (≈ the middle two-thirds of the small intestine) was dissected on ice and divided in three equal segments. The middle segment was used for histological analyses and the proximal and distal segments were pooled for IEC isolation and flow cytometry.
Histological Analyses
Jejunal segments were flushed with ice-cold 1× phosphate-buffered saline (PBS; 0.137 M NaCl, 3 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), opened longitudinally, and fixed in in fresh 4% paraformaldehyde in 1× PBS overnight at 4°C. Tissues were then rinsed in 1× PBS and incubated sequentially in 10% sucrose and 30% sucrose overnight at 4°C. Tissues were embedded in optimal cutting temperature medium (OCT), frozen on dry ice, and stored at −80°C prior to cryosectioning. Thin sections (≈7 μm) were cut on a cryostat and placed on positively charged microscope slides. Sections were stained with hematoxylin and eosin to visualize crypt and villus morphology. For immunofluorescence, frozen sections were brought to room temperature and rinsed with 1× PBS to remove the OCT. The Sox9-EGFP transgene expression was directly visualized on sections stained with bisbenzamide (1:40,000 dilution in 1× PBS and 10-min incubation at room temperature) to stain nuclei. When describing cells with distinct levels of Sox9-EGFP expression observed in histological analyses, we respectively use the terminology “Sox9-EGFP High,” “Sox9-EGFP Low,” or “Sox9-EGFP Sublow” for cells with intense, moderate, or very low EGFP expression, respectively. We recognize that the High, Low, and Sublow designation was defined by specific EGFP intensity observed by flow cytometry/fluorescence-activated cell sorting (FACS) and cannot be directly extrapolated to histology. However, we use this terminology to avoid multiple nomenclatures and we include a qualification that this is an extrapolation in the appropriate figure legends. For studies colocalizing Sox9-EGFP with the proliferation marker EdU, DCAMKL-1, or the EEC marker chromogranin-A (ChgA), an anti-GFP antibody was utilized. The fidelity of the anti-GFP antibody for visualizing and reproducing the distinct intensities of fluorescence, observed when Sox9-EGFP was directly visualized, was verified by direct comparisons of relative numbers of Sox9-EGFP High or Low cells per crypt section in 30 crypts/animal, three animals/condition, by two different observers. For triple staining, frozen sections were brought to room temperature and rinsed with 1× PBS to remove the OCT medium. Heat-induced epitope retrieval was performed by use of a decloaking chamber at 120°C for 30 s followed by 90°C for 10 s. Sections were then incubated for 30 min in blocking buffer (Dako, X0909, Glostrup, Denmark). EdU staining was then performed by using the Click-iT EdU Alexa Fluor 594 kit according to the manufacturer's instructions (Invitrogen; Carlsbad, CA). Sections were then washed twice in blocking buffer (Dako, X0909) and subsequently incubated with primary antibodies in Dako background reducing diluent buffer at 4°C overnight. Dilutions were as follows: anti-GFP (1:300; chicken; Aves Labs, Tigard, OR) and anti-ChgA (rabbit; 1:200; Abcam, Cambridge, MA). Sections were then rinsed extensively with 1× PBS prior to incubation with secondary antibodies (anti-chicken-Alexa Fluor 488; goat; 1:300; and anti-rabbit-Dylight 649; goat; 1:300; Jackson ImmunoResearch Laboratories, West Grove, PA) in Dako background reducing diluent buffer at room temperature for 2.5 h. After being rinsed with 1× PBS, slides were covered with mounting medium containing 4,6-diamidino-2-phenylindole (DAPI; Fluoro-Gel II with DAPI, Electron Microscopy Sciences, Hatfield, PA). DCAMKL-1 was colocalized with Sox9-EGFP following the same protocol using primary antibodies anti-DCAMKL-1 (rabbit; 1:500; Abcam) and anti-GFP (1:300; chicken; Aves Labs) and secondary antibodies anti-chicken (Alexa Fluor 488; goat; 1:300; Invitrogen) and anti-rabbit (Cy3; goat; 1:500; Jackson ImmunoResearch Laboratories). Images were captured on an inverted fluorescence microscope (Olympus IX81, Tokyo, Japan) fitted with a digital camera (ORCA-03G, Hamamatsu, Japan). The objective lenses used were ×20 and ×40 with numerical apertures of 0.45 and 0.6, respectively (LUC Plan FLN, Olympus, Tokyo, Japan). Confocal images were obtained using a DMI400B microscope (Leica, Wetzlar, Germany) equipped with a TCS SPE confocal microscope system (Leica). The objective lens used was ×40 with numerical aperture of 0.6 (HCX PL FLUOTAR, Leica).
Preparation of Dissociated Single Intestinal Epithelial Cells for Flow Cytometry
Intestinal segments were flushed with ice-cold 1× PBS, cut open longitudinally, and placed in 30 mM EDTA-1.5 mM DTT-PBS over ice for 15 min and then incubated in 30 mM EDTA-PBS at 37°C for 8 min. Jejunal tissue was shaken vigorously and intact tissue was discarded. Remaining cells were pelleted at 1,750 rpm for 5 min at 4°C and washed with 1 × PBS. Cells were pelleted and resuspended in Hanks' buffered saline solution and 0.3 U/ml dispase (Collaborative Biomedical Products, Bedford, MA) at 37°C. Samples were shaken vigorously every 2 min for 10 min, and fetal bovine serum (FBS, 10% vol/vol) (Gemini, West Sacramento, CA) and 100 μg/ml DNase I (Roche, Basel, Switzerland) were subsequently added. For flow cytometry, samples were sequentially passed through 100-μm, 70-μm, and 40-μm filters to remove any intact crypts or cell aggregates. Cells were pelleted and resuspended in 3 ml DMEM- 10% FBS-100 μg DNaseI-10 μM Y27632 (Sigma, St. Louis, MO). Verification that the cell suspension was dissociated into single cells was performed by placing 200 μl of the preparation into a well of a 96-well plate and observing the cells under a fluorescent microscope as well as under bright-field optics. Quantification of cells expressing distinct Sox9-EGFP levels (High, Low, and Sublow or Negative) was performed by using a Cyan flow cytometer, Summit v4.3 software, and parameters previously described (21). Propidium iodide staining (Sigma, St. Louis, MO) was used to gate out dead cells. Immune cells were excluded by forward-side scatter gating and doublets were discriminated by use of both forward scatter and side scatter height-width plots. For each flow cytometry run, the proportion of each cell population at each time point after radiation was compared with the proportion of the respective cell population in nonirradiated control mice studied in the same run.
Tissue Harvest for FACS of Different Sox9-EGFP-Expressing Cell Populations
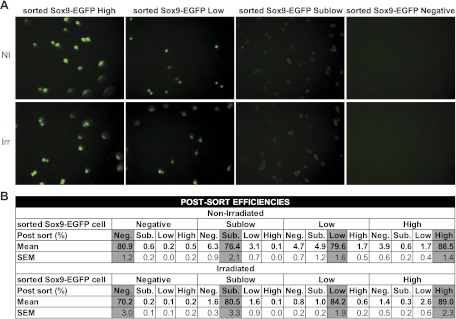
These studies focused on nonirradiated mice and mice at 5 days after irradiation when histology revealed maximum crypt regeneration/cell proliferation. FACS was used to isolate cells for testing in the ISC/organoid culture system and for gene microarray. For these analyses, 6- to 10-wk-old nonirradiated Sox9-EGFP mice or Sox9-EGFP mice at day 5 after abdominal radiation were euthanized with a lethal dose of Nembutal and the entire jejunum was placed on ice and processed for IEC isolation and dissociation into single cells as described above. Prior to FACS, the dissociated cells were sequentially passed through 100-μm, 70-μm, and 30-μm filters. The cells were pelleted and placed in 3 ml Advanced DMEM F12–10% FBS-100 μg DNaseI-10 μM Y27632 (Sigma). Sorting of Sox9-EGFP High, Low, Sublow, and Negative cells was performed by using a MoFlo XDP FACS machine (Dako/Cytomation, Carpinteria, CA) and Summit v4.3 software. Dead and immune cells were excluded by forward-side scatter gating and doublets were discriminated by using both forward scatter and side scatter height-width plots. For each sort of a preparation of dissociated cells from an irradiated mouse, gates delimiting the distinct Sox9-EGFP populations were defined by use of a nonirradiated control littermate sorted in the same run. Postsort analysis indicated that the different Sox9-EGFP cell populations were correctly isolated on the basis of EGFP status (Fig. 1_A_). In addition, postsort efficiencies were assessed by flow cytometry after each run for each sample to verify minimal contamination by other Sox9-EGFP cell types (Fig. 1_B_). Results demonstrated high postsort efficiencies and negligible cross-contamination of Sox9-EGFP High, Low, Sublow, and Negative cells with other cell populations (Fig. 1_B_).
Fig. 1.
Postsort analysis of the different fluorescence-activated cell sorting (FACS) isolated Sox9-EGFP cell populations. A: representative photographs of the 4 Sox9-EGFP cell fractions isolated by FACS demonstrate that they express appropriate intensities of enhanced green fluorescence protein (EGFP) fluorescence (magnification ×40). B: data from flow cytometry used to assess the postsort efficiencies and potential cross-contamination of cells that do not express Sox9-EGFP and those expressing sublow, low, and high levels of Sox9-EGFP (Sox9-EGFP Negative, Sublow, Low, and High cells, respectively) with other Sox9-EGFP cell populations. Data are expressed as means and SE of the postsort percentages of corresponding cells collected for each sort (n ≥ 9 in each group). Note that each sorted cell population contains more than 70% of the appropriate cell type and presents minimal contamination with other cells. Importantly, the postsort efficiencies and cross-contamination values do not significantly differ in cells from nonirradiated and irradiated mice.
Organoid Culture on FACS Isolated Cells
Organoid culture was carried out following methods originally described in Sato et al. (55) and adapted to IEC isolated from Sox9-EGFP mice by Gracz and colleagues (21). Briefly, sorted Sox9-EGFP cells (Sox9-EGFP Negative, Sox9-EGFP Sublow, Sox9-EGFP Low, and Sox9-EGFP High cells) from nonirradiated mice or mice at 5 days after irradiation were immediately resuspended at a density of 20,000 cells in 50 μl per well (24-well plate) in Matrigel (BD Biosciences, San Jose, CA) supplemented with 1 μM Jagged-1 peptide (AnaSpec, San Jose, CA), 50 ng/ml EGF (R&D, Minneapolis, MN), 100 ng/ml Noggin (PeproTech, Rocky Hill, NJ), and 1 μg/ml R-Spondin 1 (R&D). After total polymerization, each formed droplet was overlaid with 500 μl Advanced DMEM/F12 containing N2 supplement (Invitrogen), B27 supplement minus vitamin A (Invitrogen), 10 mM HEPES (Invitrogen), and 10 μM Y27632 (Sigma). Growth factors were added every other day at the same initial concentrations, except R-Spondin 1 was reduced to 500 ng/ml following the initial plating. Medium was replaced every 4 days. Y27632 was withdrawn from medium after 4 days of culture. Number of organoids was counted every other day for 12 days by an observer blinded to treatment groups and using previously reported methods (21, 55). Representative photographs of organoids were collected via an inverted microscope (Olympus IX81) fitted with a digital camera (ORCA-03G, Hamamatsu, Japan). The objective lens used was ×10 with numerical aperture of 0.3 (U Plan FLN, Olympus, Japan).
Statistical Analyses
Data were expressed as means ± SE. Unpaired _t_-test, Mann-Whitney test, one-way ANOVA or two-way ANOVA were performed to compare different groups as indicated in the results or figure legends. A P value of less than 0.05 was considered statistically significant.
Microarray on FACS Isolated Cells
Gene expression analysis was performed by using Agilent Mouse GE 4 × 44K v2 microarray (Agilent; Santa Clara, CA). Total RNA was extracted from FACS-isolated Sox9-EGFP Negative, Sublow, Low, and High cells obtained from jejunum of nonirradiated controls or mice at 5 days postirradiation. Total RNA was also extracted from the corresponding nonsorted total IEC preparation from nonirradiated mice. Four independent nonirradiated mice were initially studied to define specific gene signatures of the four Sox9-EGFP cell types. To study transcriptomic changes induced by irradiation in Sox9-EGFP cells, cells from three irradiated mice and from three independent nonirradiated mice processed in parallel and used as controls to set the gates during FACS isolation were analyzed. RNA extraction was performed with RNeasy Mini Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's recommendations. RNA integrity was analyzed by the Genomics and Bioinformatics core of the University of North Carolina (UNC) Lineberger Comprehensive Cancer Center (UNC, Chapel Hill, NC) using the Agilent RNA 6000 Nano microfluidic chips and the Agilent 2100 Bioanalyzer platform (Agilent, Santa Clara, CA). Subsequent RNA amplification and labeling were performed by using Agilent Low Input QuickAmp Labeling Kit (Two Color), which generates fluorescent complementary RNA. Total RNA (0.1–1 μg) was amplified and labeled (Cy5 for samples and Cy3 for the reference). Hybridization of the microarrays was performed by use of Agilent Microarray Hybridization equipment and protocol. Each individual sample was compared with a reference pool consisting of transcripts of nonsorted total IEC preparation from nonirradiated mice. Hybridized slides were scanned with Agilent Microarray Scanner and Agilent Scan Control software. Quality control of the arrays was performed by the UNC Lineberger Comprehensive Cancer Center (UNC, Chapel Hill, NC) following Agilent recommendations. Data were processed with Agilent Feature Extraction Software version 10.7.3.1. Sample-to-reference ratios (Cy5/Cy3) were calculated and Log2 transformed.
Gene Microarray Analyses
Our data analyses strategy had several goals as follows: 1) to identify genes specifically enriched in Sox9-EGFP Low, Sublow, High, and Negative cells from nonirradiated mice to obtain a molecular signature of each of these cell types and identify potential biomarkers of those cells; 2) to identify genes up- or downregulated specifically in Sox9-EGFP Low or Sublow cells vs. other cell types during peak crypt regeneration at day 5 after irradiation, a time we found to be associated with dramatic expansion of Sox9-EGFP Low cells; and 3) to identify genes up- or downregulated specifically in Sox9-EGFP High cells since histology and organoid culture data suggested changes in functional phenotype within this cell population after irradiation. Since our studies provided gene microarray data on seven independent samples of sorted cells from nonirradiated mice, each of these replicates was used in comparisons with the three replicates from irradiated mice to maximize statistical power.
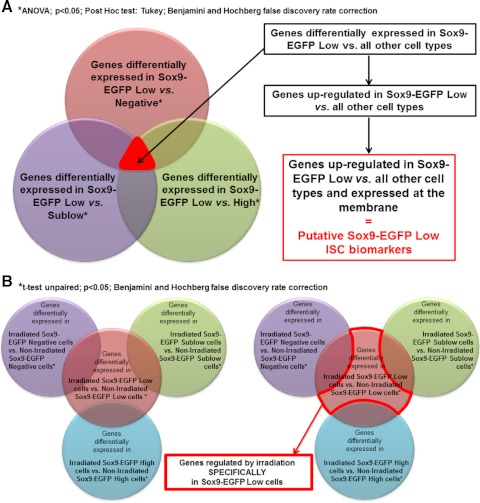
To identify differentially expressed genes, gene expression Log2 (ratios) were analyzed with Genespring GX v10.0 software (Agilent; Santa Clara, CA). Data were normalized 1) with Lowess normalization and 2) to median of control samples, i.e., for each probe, the median of the Log2 (ratio) of the individual nonsorted nonirradiated IEC control samples (hybridized against the reference pool, which is a mix of these control samples) is first computed and then used for the baseline transformation of all samples. The identification of genes specifically upregulated in Sox9-EGFP Low cells from nonirradiated mice compared with all other Sox9-EGFP cells in nonirradiated jejunum was performed in two steps. First, the genes differentially expressed between Sox9-EGFP Low cells vs. Sox9-EGFP Negative, Sublow, or High cells were identified by ANOVA followed by a Tukey post hoc test and Benjamini and Hochberg false discovery rate correction with a significance threshold of 0.05. Then genes that were significantly upregulated in Sox9-EGFP Low cells vs. all of the other Sox9-EGFP cell populations (i.e., Sox9-EGFP Negative, Sox9-EGFP Sublow, and Sox9-EGFP High cells) were identified (Fig. 2_A_). We then adapted this same analysis strategy to identify genes specifically upregulated in Sox9-EGFP Sublow, High, or Negative cells vs. all of the other cell populations. The identification of genes regulated specifically in Sox9-EGFP Low cells during crypt regeneration after irradiation was performed in two steps. First, the genes differentially expressed between Sox9-EGFP Low cells from irradiated mice vs. Sox9-EGFP Low cells from nonirradiated mice were identified by unpaired _t_-test and Benjamini and Hochberg false discovery rate correction with a significance threshold of 0.05. Identical analyses were performed to identify genes significantly regulated after radiation in Sox9-EGFP Negative, Sublow, and High cells. We then identified genes whose expression was regulated after irradiation only and specifically in Sox9-EGFP Low cells (Fig. 2_B_). We adapted this same analysis strategy to identify genes regulated after irradiation only and specifically in Sox9-EGFP Sublow or High cells vs. all other cell types.
Fig. 2.
Schematization of approaches used to analyze the microarray data. A: analysis approach used to identify new putative Sox9-EGFP Low intestinal epithelial stem cell (ISC) biomarkers. Briefly, statistical analysis identified genes significantly differentially regulated in Sox9-EGFP Low cells vs. Negative cells (red), Sublow cells (purple), or High cells (green) (ANOVA; P < 0.05; Tukey; Benjamini-Hochberg multiple testing comparison). By selecting genes that were common to each of these 3 analyses, we isolated genes that were differentially regulated in Sox9-EGFP Low cells vs. all other cell types. Genes that were upregulated in Sox9-EGFP Low ISCs vs. all other cell types and that encode membrane proteins were selected as putative Sox9-EGFP Low ISC biomarkers (see Tables 5 and 6). This same analysis strategy was used to identify new putative Sox9-EGFP Sublow progenitor biomarkers (see Tables 5 and 6). B: analysis approach used to identify genes whose expression was regulated by irradiation specifically in Sox9-EGFP Low ISCs. Briefly, statistical analysis identified genes significantly differentially regulated in Sox9-EGFP Low cells isolated from mice at day 5 postirradiation vs. Sox9-EGFP Low cells from nonirradiated mice (red). Similar analysis was performed for Sox9-EGFP Negative (purple), Sublow cells (green), and High cells (blue) (_t_-test unpaired; P < 0.05; Benjamini-Hochberg multiple testing comparison). We then selected genes that were regulated after irradiation exclusively in Sox9-EGFP Low ISCs. This same analysis strategy was used to identify genes regulated by irradiation specifically in Sox9-EGFP Sublow or High cells.
Ingenuity Pathway Analysis (IPA) was also used to identify signaling pathways and cellular functions related to the genes specifically and significantly up- or downregulated in nonirradiated Sox9-EGFP Low or Sublow cells or in Sox9-EGFP Low, Sublow, or High cells after irradiation.
Microarray data were uploaded to GEO database http://www.ncbi.nlm.nih.gov/geo/ and are available under the accession number GSE32227.
Real-Time Quantitative PCR on FACS Isolated Cells
Total RNA was extracted from FACS-isolated Sox9-EGFP Negative, Sublow, Low, and High cells obtained from jejunum of nonirradiated controls or mice at 5 days postirradiation. RNA was extracted by using RNeasy Mini Kit (Qiagen) according to the manufacturer's recommendations. 0.5 μg of total RNA was processed for reverse transcription by use of the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). PCR amplifications were performed using Platinum Quantitative PCR SuperMix-UDG (Invitrogen) and TaqMan probes for Sox9 (Mm00448840_m1) and Dclk1 (Mm00444950_m1) (Applied Biosystems). eGFP mRNA expression was quantified by using the primer pair 5′AGTCCGCCCTGAGCAAAGA3′ (eGFP-F) and 5′TCCAGCAGGACCATGTGATC3′ (eGFP-R) and the TaqMan probe, 5′6-FAM-CCCAACGAGAAGCG-MGB3′ (Applied Biosystems). Gene expression data were normalized with Rps6 (Mm02342456_g1) (Applied Biosystems).
RESULTS
Discrete Levels of Sox9-EGFP Transgene Expression Mark Four Different Cell Populations that Exhibit Distinct Gene Expression Signatures by Microarray
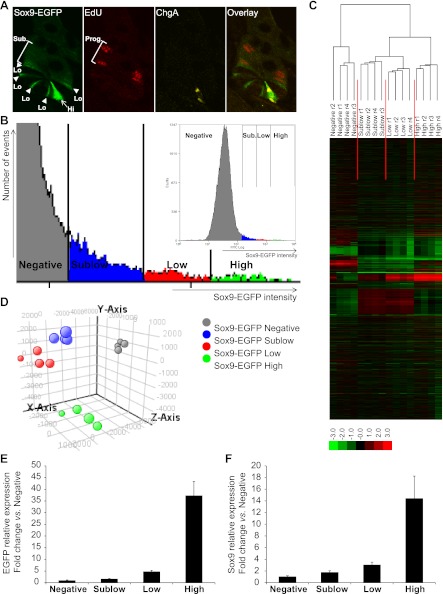
We first aimed to confirm and strengthen previously reported evidence that IEC populations expressing different levels of the Sox9-EGFP transgene are enriched for phenotypically distinct cells (17, 21). These prior studies demonstrated that the Sox9-EGFP transgene is expressed at low/moderate levels (Sox9-EGFP Low; Fig. 3_A_) in cells with ISC characteristics (21), intercalated between Sox9-EGFP Negative Paneth cells (17), and at higher/more intense levels (Sox9-EGFP High; Fig. 3_A_) in cells that colabel with the EEC marker ChgA (17, 21). As illustrated in Fig. 3_A_, this expression pattern is maintained in independent animals derived from breeding this model across multiple generations. In addition, Fig. 3_A_ demonstrates that a short 90-min pulse of the S-phase marker EdU predominantly labels cells located higher in the crypt than Sox9-EGFP Low and High cells where rapidly dividing progenitors reside, and these cells express very low or what have been termed Sublow levels of Sox9-EGFP (Sox9-EGFP Sublow) (21).
Fig. 3.
Sox9-EGFP transgene marks 4 different cell populations that exhibit distinct gene expression signatures. A: confocal microscopy on a jejunal crypt from a nonirradiated Sox9-EGFP mouse illustrates that the Sox9-EGFP transgene (green) is expressed at different levels (Hi, high/intense staining, arrow; Lo, low/moderate staining, arrowhead; Sub., sublow/very low staining). Note that in this and subsequent histological figures, we use the “High, Low, and Sublow” terminology to describe cells with intense, moderate, and very low intensities of Sox9-EGFP fluorescence, respectively. 5-Ethynyl-2′deoxyuridine (EdU)-positive cells (red) are predominantly located in the progenitor (Prog.) compartment and express sublow levels of Sox9-EGFP. Chromogranin-A (ChgA; yellow) is localized to Sox9-EGFP High cells (magnification ×40). B: representative graph illustrates gating used for flow cytometry and FACS-based isolation of Sox9-EGFP-expressing cells from single cell preparations of IECs from nonirradiated mouse jejunum. Four levels of expression of the transgene (Sox9-EGFP intensity) can be distinguished: Negative (gray), Sublow (blue), Low (red), and High (green). Hierarchical clustering (C) and principal component analysis (PCA; D) on gene expression data from the 4 Sox9-EGFP cell populations isolated from 4 independent nonirradiated Sox9-EGFP mice reveal 4 distinct and consistent gene expression signatures (hierarchical clustering on genes and conditions, Spearman rank correlation, single linkage, r, replicate; PCA on conditions, 4 principal components, mean centered). E: real-time quantitative PCR (qPCR) data showed that the expression profile of EGFP was consistent with the 4 different Sox9-EGFP sorted groups (i.e., Negative, Sublow, Low, and High). Data are expressed as fold change (mean ± SE) vs. Sox9-EGFP Negative cells (n = 6). F: real-time qPCR data demonstrated that the expression of the endogenous Sox9 gene matched the expression levels of the Sox9-EGFP transgene in the different sorted cell populations. Data are expressed as fold change (mean ± SE) vs. Sox9-EGFP Negative cells (n = 6).
Illustrative flow cytometry data demonstrate the gradient of expression of the Sox9-EGFP transgene in dissociated single cell preparations of IECs obtained from nonirradiated Sox9-EGFP mice (Fig. 3_B_). Based on distinct levels of Sox9-EGFP, four different cell populations, i.e., Sox9-EGFP Negative, Sublow, Low, and High cells, were distinguished and isolated by FACS by using gating procedures previously reported (21) (Fig. 3_B_; also see Fig. 1_A_). Gene microarray was performed on Sox9-EGFP Negative, Sublow, Low, and High cells isolated from jejunum of four independent animals. Unsupervised principal component analysis and hierarchical clustering of gene expression data clearly demonstrated that Sox9-EGFP Negative, Sublow, Low, and High cells each exhibit a unique gene expression signature, which is consistent across individual mice (Fig. 3, C and D). Real-time quantitative PCR analysis validated that the expression profile of EGFP in FACS isolated cells matched with the four different Sox9-EGFP groups (Fig. 3_E_). Also, endogenous Sox9 expression paralleled EGFP mRNA abundance (Fig. 3_F_), confirming that the Sox9-EGFP reporter gene faithfully recapitulates expression patterns of endogenous Sox9 (17, 21, 49).
Sox9-EGFP Low, Sublow, High, and Negative Cell Populations Exhibit Specific Gene Expression Signatures Consistent with Distinct Cellular Phenotypes
Gene microarray data obtained for the four Sox9-EGFP cell populations were analyzed according to the strategy described in Fig. 2_A_ to identify genes upregulated specifically in Sox9-EGFP Low, Sublow, High, or Negative cells. The genes specifically and significantly upregulated in each of these cell populations relative to all of the other cell populations contained relevant genes that provide strong evidence that Sox9-EGFP Low cells are enriched for ISCs, Sox9-EGFP Sublow cells for progenitors, Sox9-EGFP High cells for EECs, and Sox9-EGFP Negative cells for differentiated IEC lineages (Tables 1–4).
Table 1.
Genes upregulated specifically in Sox9-EGFP Low cells
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| LGR5 | leucine-rich repeat containing G protein-coupled receptor 5 | 11.72 | 14160 |
| SCN2B | sodium channel, voltage-gated, type II, beta | 10.44 | 72821 |
| TNFRSF19 | tumor necrosis factor receptor superfamily, member 19 | 10.08 | 29820 |
| NKD1 | naked cuticle homolog 1 (Drosophila) | 8.43 | 93960 |
| SLCO3A1 | solute carrier organic anion transporter family, member 3A1 | 7.01 | 108116 |
| ASCL2 | achaete-scute complex homolog 2 (Drosophila) | 6.86 | 17173 |
| PHTPRO | protein tyrosine phosphatase, receptor type, O | 6.73 | 19277 |
| RGMB | RGM domain family, member B | 6.55 | 68799 |
| AQP4 | aquaporin 4 | 5.63 | 11829 |
| CDC42EP1 | CDC42 effector protein (Rho GTPase binding) 1 | 5.27 | 104445 |
| CRLF1 | cytokine receptor-like factor 1 | 4.56 | 12931 |
| ZBTB12 | zinc finger and BTB domain containing 12 | 3.45 | 193736 |
| CITED4 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | 2.76 | 56222 |
| FGFRL1 | fibroblast growth factor receptor-like 1 | 2.62 | 116701 |
| RNF43 | ring finger protein 43 | 2.50 | 207742 |
| SLC23A3 | solute carrier family 23 (nucleobase transporters), member 3 | 2.42 | 22626 |
| LAMB3 | laminin, beta 3 | 2.26 | 16780 |
| ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | 2.03 | 23872 |
| TNS3 | tensin 3 | 1.86 | 319939 |
| SP5 | Sp5 transcription factor | 1.85 | 64406 |
| PDGFA | platelet-derived growth factor alpha polypeptide | 1.44 | 18590 |
Table 4.
Genes upregulated specifically in Sox9-EGFP Negative cells
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| FABP2 | fatty acid binding protein 2, intestinal | 27.38 | 14079 |
| FABP1 | fatty acid binding protein 1, liver | 26.30 | 14080 |
| TREH | trehalase (brush-border membrane glycoprotein) | 20.63 | 58866 |
| LCT | lactase | 18.54 | 226413 |
| ALPI | alkaline phosphatase, intestinal | 10.37 | 76768 |
| ANPEP | alanyl (membrane) aminopeptidase | 9.61 | 16790 |
| SLC2A5 | solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | 6.37 | 56485 |
| MGAM | maltase-glucoamylase (alpha-glucosidase) | 3.37 | 232714 |
| Muc3 | mucin 3, intestinal | 6.64 | 666339 |
| Muc2 (mouse) | mucin 2 | 6.63 | 17831 |
| TFF3 | trefoil factor 3 (intestinal) | 5.05 | 21786 |
| MUC20 | mucin 20, cell surface associated | 2.68 | 224116 |
| CSF2 | colony stimulating factor 2 (granulocyte-macrophage) | 24.58 | 12981 |
Sox9-EGFP Low cells and Lgr5-ISC phenotype.
Lgr5 and Ascl2, which are validated ISC markers (3, 65), were specifically enriched in Sox9-EGFP Low cells, as were several genes previously reported to be significantly enriched in ISCs expressing high levels of Lgr5-EGFP (Lgr5-EGFPhi ISCs) compared with progenitors expressing low levels of Lgr5-EGFP (Lgr5-EGFPlo progenitors) (65) (Table 1). These gene expression data strengthen previous evidence (17, 21) that Sox9-EGFP Low cells are highly enriched for ISCs/CBCs.
Sox9-EGFP Sublow cells and progenitor phenotype.
Prior studies reported genes enriched in Lgr5-EGFPhi ISCs vs. Lgr5-EGFPlo progenitors (65); however, to our knowledge there are no comprehensive datasets on genes specifically enriched in small intestinal progenitors vs. all other cell types. Thus genes specifically enriched in Sox9-EGFP Sublow cells, which comprise progenitor cells, provide new potential biomarkers. Sox9-EGFP Sublow cells showed significantly higher expression of a number of genes associated with progenitors in other organs (Table 2). Among these genes is MYCN, which is known to be critical for the maintenance of proliferating progenitors in other organs (8, 42, 43, 68) (Table 2). Also MET is modestly but significantly enriched in Sox9-EGFP Sublow progenitors, consistent with known roles of the HGF/c-met axis in regulating proliferation, survival, differentiation, and migration of progenitors in various organs (10, 15, 53, 62) (Table 2). HDAC2 and HDAC8, which encode class I histone deacetylases (HDAC), were upregulated in Sox9-EGFP Sublow cells, consistent with other studies suggesting that expression levels of class I HDAC regulate cell fate decision of progenitors in various organs (52, 67). Several genes encoding cell division cycle (CDC) and cyclin-dependent kinase (CDK) proteins were also upregulated, further supporting the progenitor phenotype of the Sox9-EGFP Sublow cells (21).
Table 2.
Genes upregulated specifically in Sox9-EGFP Sublow cells
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| MYCN | v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) | 3.14 | 18109 |
| HDAC2 | histone deacetylase 2 | 2.53 | 15182 |
| CDC45 | cell division cycle 45 homolog (S. cerevisiae) | 2.33 | 12544 |
| CDC40 | cell division cycle 40 homolog (S. cerevisiae) | 1.72 | 71713 |
| CDC73 | cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | 1.56 | 214498 |
| HDAC8 | histone deacetylase 8 | 1.55 | 70315 |
| CDK7 | cyclin-dependent kinase 7 | 1.45 | 12572 |
| MET | met proto-oncogene (hepatocyte growth factor receptor) | 1.28 | 17295 |
Sox9-EGFP High cells, EEC phenotype, and +4-ISC phenotype.
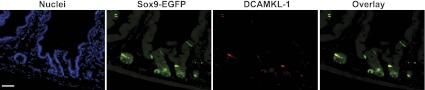
Sox9 mRNA was highly enriched in Sox9-EGFP High cells vs. all other Sox9-EGFP cell populations (+12.03-fold change vs. Sox9-EGFP Negative cells; P = 1.52E-16) (Table 3 and Fig. 3_F_). Sox9-EGFP High cells were also highly enriched (+3.46-+101.8-fold change vs. Sox9-EGFP Negative cells) in multiple genes encoding gastrointestinal hormones including neurotensin, substance P, ChgA, chromogranin-B, cholecystokinin, glucagon, secretin, gastric inhibitory peptide, and ghrelin (Table 3). These findings strongly support prior data (17, 21) that the Sox9-EGFP High cells are highly enriched for EECs (Table 3). Unexpectedly, microarray data also revealed that BMI1 and HOPX, which mark +4 slowly cycling or quiescent ISCs (54, 61), were significantly and exclusively enriched in Sox9-EGFP High cells vs. all other Sox9-EGFP cell types (+2.65- and +4.15-fold change vs. Sox9-EGFP Negative cells, respectively; P = 4.04E-11 and P = 9.25E-15, respectively) (Table 3). Several lines of evidence suggest that Dclk1, also called DCAMKL-1, may represent another putative marker of slowly cycling/quiescent ISCs (39). Immunofluorescence studies clearly demonstrated that DCAMKL-1 colocalized with cells expressing high levels of Sox9-EGFP (Fig. 4) and quantitative PCR revealed that DCAMKL-1 mRNA was dramatically enriched in Sox9-EGFP High cells (+147.0 ± 61.9-fold change vs. Sox9-EGFP Negative cells; P < 0.05).
Table 3.
Genes upregulated specifically in Sox9-EGFP High cells
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| NTS | neurotensin | 101.80 | 67405 |
| TAC1 | tachykinin, precursor 1 | 60.44 | 21333 |
| CHGB | chromogranin B (secretogranin 1) | 56.17 | 12653 |
| CCK | cholecystokinin | 24.96 | 12424 |
| GCG | glucagon | 23.36 | 14526 |
| CHGA | chromogranin A (parathyroid secretory protein 1) | 22.31 | 12652 |
| SCT | secretin | 17.95 | 20287 |
| GIP | gastric inhibitory polypeptide | 16.23 | 14607 |
| GHRL | ghrelin/obestatin prepropeptide | 3.46 | 58991 |
| SOX9 | SRY-box containing gene 9 | 12.03 | 20682 |
| BMI1 | Bmi1 polycomb ring finger oncogene | 2.65 | 12151 |
| HOPX | HOP homeobox | 4.15 | 74318 |
Fig. 4.
DCAMKL-1 colocalizes with Sox9-EGFP High cells. Immunostaining for Sox9-EGFP (green) and DCAMKL-1 (red) demonstrates that in intestinal epithelial crypts, DCAMKL-1 colocalizes with high levels of Sox9-EGFP [nuclei staining, 4,6-diamidino-2-phenylindole (DAPI), blue; scale bar 50 μm].
Sox9-EGFP Negative cells and phenotype of differentiated IEC lineages.
A number of genes that were specifically expressed at significantly higher levels in Sox9-EGFP Negative cells vs. all other Sox9-EGFP cell populations are known markers of absorptive enterocytes including trehalase, lactase, aminopeptidase N, alkaline phosphatase, and glucoamylase, which are all enterocyte brush border enzymes (Table 4). Furthermore, Sox9-EGFP Negative cells exhibited specific upregulation of FABP1, FABP2, and SLC2A5 genes, coding for liver and intestinal fatty acid-binding protein and GLUT5, which are all well-accepted markers of mature enterocytes (Table 4). Several genes known to be highly expressed in goblet cells were specifically upregulated in Sox9-EGFP Negative cells including genes encoding mucins 3, 2, and 20 and trefoil factor 3 (Table 4). In addition, the gene CSF2 coding for granulocyte-macrophage colony-stimulating factor, which is exclusively expressed in Paneth cells (18), was markedly upregulated specifically in Sox9-EGFP Negative cells (Table 4). Together these data strengthen the prior evidence (21) that Sox9-EGFP Negative cells are enriched for differentiated IEC lineages.
Microarray Data Reveal New Potential Biomarkers of Sox9-EGFP Low ISCs and Sox9-EGFP Sublow Progenitors
Since previously published work suggested that Sox9-EGFP Low and Sublow cells correspond to ISCs and progenitors respectively (17, 21), we initially focused in-depth analyses of microarray data from nonirradiated mice on these two cell types. First, we identified the total number of IPA-annotated genes differentially upregulated in Sox9-EGFP Low or Sublow cells vs. all other Sox9-EGFP cell types (Fig. 2_A_). From these data, we extracted genes coding for membrane proteins that might represent useful new biomarkers for future downstream applications such as cell sorting.
Genes uniquely expressed in Sox9-EGFP Low cells.
A total of 164 genes were significantly upregulated in Sox9-EGFP Low cells compared with all other cells, i.e., Sox9-EGFP Sublow, High, and Negative cells (Supplemental Table S1; supplemental material for this article is available online at the Journal website). Thirty genes encoding membrane proteins were identified as specifically enriched in Sox9-EGFP Low cells (Table 5). Of these, 8 have been reported as enriched in Lgr5-expressing ISCs but 22 have not, to our knowledge, been reported as enriched in ISCs (Table 5). Evidence from the literature linking these genes to stem cells or pathways functionally relevant to stem cells in other organs adds support to the concept that some of these 22 genes may represent ISC biomarkers. For example, CXCR7 encodes a receptor of stromal cell-derived factor 1 (SDF-1), which is a major regulator of retention, migration, and mobilization of hematopoietic stem cells and endothelial progenitor cells in normal conditions and upon injury (35, 60). GAS1 is a major regulator of cell cycle progression and Hedgehog signaling (36), which has recently been shown to be enriched in hair follicle stem cells (51).
Table 5.
Putative membrane biomarkers of Sox9-EGFP Low ISC
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| PCDH8 | protocadherin 8 | 22.12 | 18530 |
| LGR5* | leucine-rich repeat containing G protein-coupled receptor 5 | 11.72 | 14160 |
| SCN2B* | sodium channel. voltage-gated. type II. beta | 10.44 | 72821 |
| TNFRSF19* | tumor necrosis factor receptor superfamily. member 19 | 10.08 | 29820 |
| OR1S1 | olfactory receptor. family 1. subfamily S. member 1 | 8.58 | 258991 |
| GPM6B | glycoprotein M6B | 8.51 | 14758 |
| CXCR7 | chemokine (C-X-C motif) receptor 7 | 7.94 | 12778 |
| SLCO3A1* | solute carrier organic anion transporter family. member 3A1 | 7.01 | 108116 |
| Olfr361 | olfactory receptor 361 | 6.96 | 258365 |
| PTPRO* | protein tyrosine phosphatase. receptor type. O | 6.73 | 19277 |
| RGMB* | RGM domain family. member B | 6.55 | 68799 |
| FAIM2 | Fas apoptotic inhibitory molecule 2 | 6.20 | 72393 |
| AQP4* | aquaporin 4 | 5.63 | 11829 |
| KCNA3 | potassium voltage-gated channel. shaker-related subfamily. member 3 | 4.52 | 16491 |
| Vmn1r171 (includes others) | vomeronasal 1 receptor 171 | 4.50 | 100043103 |
| TBC1D9 | TBC1 domain family. member 9 (with GRAM domain) | 4.18 | 71310 |
| PODXL2 | podocalyxin-like 2 | 3.30 | 319655 |
| OR1B1 | olfactory receptor. family 1. subfamily B. member 1 | 3.18 | 259053 |
| CD320 | CD320 molecule | 3.01 | 54219 |
| ADAM8 | ADAM metallopeptidase domain 8 | 2.73 | 11501 |
| FGFRL1* | fibroblast growth factor receptor-like 1 | 2.62 | 116701 |
| Olfr190/Olfr192 | olfactory receptor 192 | 2.60 | 258392 |
| GAS1 | growth arrest-specific 1 | 2.51 | 14451 |
| PARD6G | par-6 partitioning defective 6 homolog gamma (C. elegans) | 2.29 | 93737 |
| TRIB2 | tribbles homolog 2 (Drosophila) | 2.29 | 217410 |
| TNFRSF25 | tumor necrosis factor receptor superfamily. member 25 | 2.23 | 85030 |
| SIGLEC7/SIGLEC9 | sialic acid binding Ig-like lectin 7 | 2.20 | 83382 |
| OFD1 | oral-facial-digital syndrome 1 | 1.80 | 237222 |
| Vmn1r188 (includes others) | vomeronasal 1 receptor 194 | 1.71 | 171257 |
| TMPRSS4 | transmembrane protease. serine 4 | 1.51 | 214523 |
Genes uniquely expressed in Sox9-EGFP Sublow progenitors.
By adapting the analysis approach described in Fig. 2_A_ to the Sox9-EGFP Sublow cells, we identified 290 genes that were significantly upregulated in Sox9-EGFP Sublow cells compared with all other Sox9-EGFP cell populations (Supplemental Table S2). Among these genes, 16 genes encode membrane proteins and therefore may represent novel putative membrane biomarkers of progenitors (Table 6). Interestingly, among them was FZD5, which has been shown to regulate neural potential of progenitors in developing nervous system (66), and L1CAM, which codes for cell adhesion molecule L1, a protein previously localized to proliferating progenitors of intestinal crypts (63) (Table 6).
Table 6.
Putative membrane biomarkers of Sox9-EGFP Sublow progenitors
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| CLCN1 | chloride channel 1 skeletal muscle | 3.45 | 12723 |
| L1CAM | L1 cell adhesion molecule | 3.16 | 16728 |
| IL13RA2 | interleukin 13 receptor alpha 2 | 2.84 | 16165 |
| Olfr307 | olfactory receptor 307 | 2.12 | 258610 |
| GP9 | glycoprotein IX (platelet) | 1.95 | 54368 |
| TMEM5 | transmembrane protein 5 | 1.85 | 216395 |
| MTNR1A | melatonin receptor 1A | 1.72 | 17773 |
| GJB3 | gap junction protein beta 3.31 kDa | 1.70 | 14620 |
| SLC1A1 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter. system Xag member 1) | 1.61 | 20510 |
| FZD5 | frizzled homolog 5 (Drosophila) | 1.53 | 14367 |
| SH3BGRL2 | SH3 domain binding glutamic acid-rich protein like 2 | 1.52 | 212531 |
| STT3A | STT3. subunit of the oligosaccharyltransferase complex. homolog A (S. cerevisiae) | 1.44 | 16430 |
| STX17 | syntaxin 17 | 1.35 | 67727 |
| MOBKL1B | MOB1. Mps One Binder kinase activator-like 1B (yeast) | 1.33 | 232157 |
| MET | met proto-oncogene (hepatocyte growth factor receptor) | 1.28 | 17295 |
| DLG1 | discs. large homolog 1 (Drosophila) | 1.15 | 13383 |
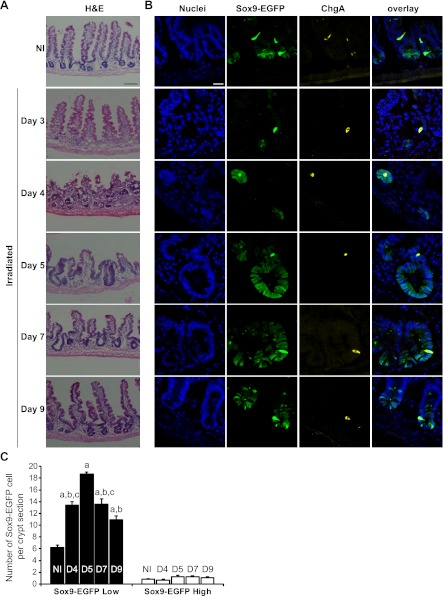
Histological Analyses of Jejunal Morphology and Sox9-EGFP-Expressing Cells Between 3–9 Days After 14 Gy Abdominal Irradiation
We next sought to study Sox9-EGFP-expressing cells during crypt regeneration after radiation-induced crypt loss and focused analyses on days 3-9 after high-dose abdominal radiation. Hematoxylin and eosin staining (Fig. 5_A_) revealed consistent and essentially complete crypt loss in jejunum by day 3 after radiation, followed by the appearance of microcolonies at day 4, which are believed to represent clonogens derived from surviving ISCs (45, 48). By days 5 and 7 we observed hyperplastic regenerating crypts followed by a return toward normal crypt and villus morphology by day 9 (Fig. 5_A_). The 3- to 9-day period after high-dose irradiation therefore seemed ideal to assess Sox9-EGFP-expressing cells over the entire course of regeneration, i.e., from crypt loss to normalization of the intestinal epithelium.
Fig. 5.
Sox9-EGFP Low ISCs expand during crypt regeneration following radiation. A: hematoxylin and eosin (H&E) staining of jejunum from nonirradiated (NI) and irradiated mice at days 3, 4, 5, 7, and 9 after 14 Gy abdominal irradiation demonstrates crypt loss at day 3, formation of regenerating microcolonies at day 4; crypt hyperplasia at days 5 (D5) and 7 (D7) and a trend toward baseline crypt and villus morphology by day 9. Scale bar 100 μm. B: immunostaining for Sox9-EGFP (green) and ChgA (yellow) demonstrates that regenerating microcolonies and hyperplastic crypts are highly enriched in Sox9-EGFP Low cells whereas Sox9-EGFP High cells colabeled with ChgA did not expand after radiation (nuclei staining, DAPI, blue; confocal images; scale bar 25 μm). C: histogram shows means ± SE for the number of Sox9-EGFP High or Low cells per crypt section. At least 30 crypt sections were analyzed per animal by 3 independent blinded observers (n ≥ 3 independent mice at each time point; a_P_ < 0.001 vs. nonirradiated; b_P_ < 0.001 vs. day 5; c_P_ < 0.05 vs. day 9).
Sox9-EGFP Low cells expand during crypt regeneration.
At day 3, despite total crypt loss, we observed rare, individual Sox9-EGFP Low cells and rare Sox9-EGFP High cells that colocalized with ChgA (Fig. 5_B_). The numbers of these cells at day 3 postradiation were too low to quantify; however, the dramatic loss of Sox9-EGFP Low cells at day 3 after radiation is consistent with prior reports that high-dose radiation results in massive apoptosis of CBCs/ISCs (3). By day 4, microcolonies were populated almost entirely with Sox9-EGFP Low cells, but also with rare Sox9-EGFP High cells (Fig. 5_B_). By days 5 and 7, hyperplastic crypts contained large numbers of Sox9-EGFP Low cells and by day 9, crypt morphology was returning toward basal state with Sox9-EGFP Low cells at the base of the crypt (Fig. 5_B_).
Quantification of the numbers of Sox9-EGFP Low cells per crypt section confirmed marked expansion of Sox9-EGFP Low cells during radiation-induced crypt regeneration, with peak expansion at day 5 (Fig. 5_C_). At each time point studied, there were relatively few Sox9-EGFP High cells per crypt section that were interspersed with Sox9-EGFP Low cells and colocalized with ChgA (Fig. 5_B_). Statistically, the number of Sox9-EGFP High cells per crypt section did not change significantly between days 4 and 9 after irradiation (Fig. 5_C_).
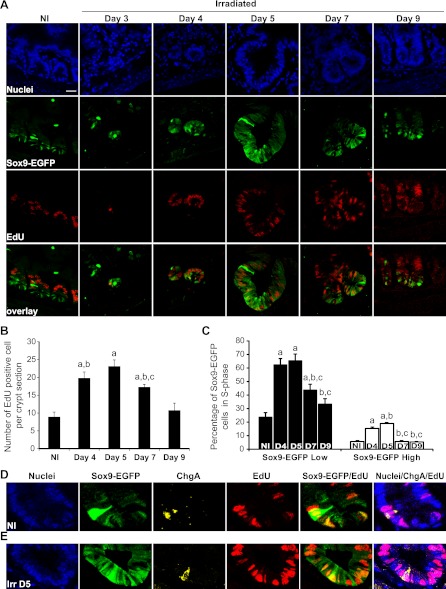
Proportions of EdU-labeled Sox9-EGFP Low and High cells are increased during crypt regeneration.
To determine whether Sox9-EGFP cells were actively proliferating, we measured the percentage of Sox9-EGFP Low or High cells colocalized with EdU, which was given as a single pulse 90 min before tissue harvest. In normal jejunum, EdU-positive cells were located mainly in the progenitor zone and only a few Sox9-EGFP Low cells at the crypt base were colabeled with EdU, suggesting that they are more slowly cycling than progenitor cells (Figs. 3_A_ and 6_A_). This is consistent with prior studies reporting slower proliferation of ISCs than progenitors (3, 41, 46). Quantification indicated that in nonirradiated jejunum there were 8.9 ± 1.3 EdU-positive cells per crypt section and that 23.6 ± 3.6% of Sox9-EGFP Low cells were EdU positive (Figs. 6, B and C). At day 3, rare single EdU-positive cells were detected but cell numbers were too few to be quantifiable (Fig. 6_A_). At day 4, almost all cells within regenerating microcolonies were EdU positive and 62.5 ± 4.7% of Sox9-EGFP Low cells were EdU positive. Hyperplastic crypts observed at days 5 and 7 were almost entirely composed of EdU-positive cells with maximum numbers (65.5 ± 4.7%) of Sox9-EGFP Low cells colabeled with EdU at day 5 followed by a decline at day 7 (Fig. 6, A–C). By day 9, the number of EdU-positive cells per crypt section as well as the percentage of Sox9-EGFP Low cells positive for EdU did not differ significantly from values in crypt sections from nonirradiated controls (Fig. 6, A–C).
Fig. 6.
Increase in proliferating Sox9-EGFP Low and High cells during crypt regeneration. A: EdU (red) and Sox9-EGFP (green) colocalization reveals that a large proportion of cells in regenerating microcolonies at day 4 and hyperplastic crypts observed at days 5 and 7 were positive for EdU and illustrates increased numbers of Sox9-EGFP Low cells that are EdU positive during crypt regeneration (nuclei staining, DAPI, blue; scale bar 25 μm). B: histogram shows mean ± SE of the number of EdU-positive cells per crypt section. At least 30 crypts were studied per animal per time point (n = 3 at each time point; a_P_ < 0.01 vs. nonirradiated; b_P_ < 0.05 vs. day 9; c_P_ < 0.05 vs. day 5). C: means ± SE of the percentage of Sox9-EGFP Low (black bars) and High (white bars) cells colabeled with EdU. Early phases of regeneration after radiation involve significant increases in the proportion of Sox9-EGFP Low and High cells in S-phase (n = 3 at each time point; a_P_ < 0.05 vs. nonirradiated; b_P_ < 0.05 vs. day 4; c_P_ < 0.05 vs. day 5). D and E: confocal images of a crypt from jejunum of a nonirradiated Sox9-EGFP mouse (D) and at 5 days postirradiation (Irr D5) (E) illustrating Sox9-EGFP High cells (green) expressing ChgA (yellow) and colabeled with EdU (red) (nuclei staining, DAPI, blue; magnification ×40).
In nonirradiated jejunal crypts there was typically one Sox9-EGFP High cell per crypt section and ∼1 in 20 (5.9 ± 0.6%) of these rare Sox9-EGFP High cells was colocalized with EdU (Fig. 6, C and D), whereas none on the villi colocalized with EdU (data not shown). This is consistent with prior reports (17, 21) that Sox9-EGFP High cells in the crypts are predominantly nonproliferating cells. However, the observation that a small proportion of Sox9-EGFP High cells were colabeled with EdU demonstrates that a subset of these cells are in S-phase under normal conditions. Between days 4 and 5 after irradiation, there was a significant increase in the percentage of Sox9-EGFP High cells colabeled with EdU (Fig. 6, C and E). The EdU-positive Sox9-EGFP High cells in nonirradiated and irradiated crypts coexpressed ChgA, indicating that they retain EEC phenotype (Fig. 6, D and E). Thus early phases of crypt regeneration after irradiation are associated with significant increases in the number of actively proliferating Sox9-EGFP High cells expressing EEC markers as well as increases in proliferating Sox9-EGFP Low cells.
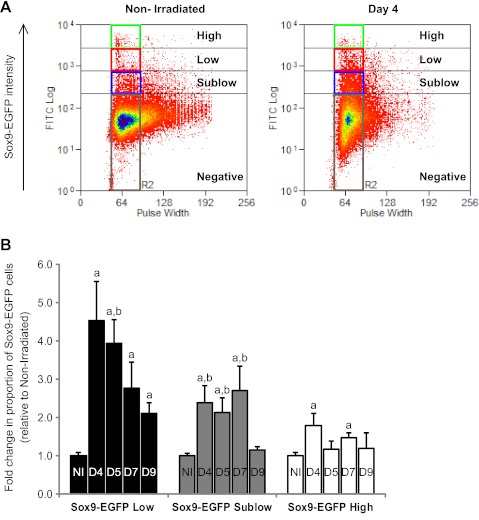
Flow Cytometry Quantitatively Demonstrates Changes in the Distinct Sox9-EGFP Cell Populations After Irradiation
Flow cytometry was performed as an independent method to quantify changes in proportions of Sox9-EGFP Low and High cells after radiation. This method also permits assessment of the changes in proportions of Sox9-EGFP Sublow cells, which are difficult to quantify by histology because of the very low level of fluorescence in this population. Additionally, flow cytometry measures the proportion of Sox9-EGFP Low, Sublow, and High cells relative to the total number of cells in the IEC preparation, whereas histology quantifies numbers of Sox9-EGFP Low or High cells per individual crypt section. Irradiation induced a massive increase in the proportion of Sox9-EGFP Low cells, which peaked at day 4 (4.5 ± 1.0-fold increase vs. nonirradiated) and day 5 and then progressively decreased, although it remained significantly elevated at day 9 (Fig. 7, A and B). The proportion of Sox9-EGFP Sublow cells was also increased after radiation reaching a peak at day 7 before returning to basal state by day 9 (Fig. 7_B_). There were significant increases in the proportion of Sox9-EGFP High cells at day 4 and day 7, although much less dramatic than for Sox9-EGFP Low cells (Fig. 7_B_).
Fig. 7.
Irradiation induces distinct changes in the proportion of Sox9-EGFP High, Low, and Sublow cells. A: representative flow cytometry data show the proportion of cells expressing High (green rectangle), Low (red rectangle), and Sublow (blue rectangle) levels of Sox9-EGFP (Sox9-EGFP intensity) isolated from a nonirradiated mouse or at day 4 postradiation. Note the marked and obvious increase in the proportion of Sox9-EGFP Low and Sublow cells at day 4 after radiation. B: histograms show the quantification of the flow cytometry data expressed as mean fold change ± SE in the proportion of Sox9-EGFP High, Low or Sublow cells at each time point postradiation vs. nonirradiated controls (n ≥ 3 at each time point; a_P_ < 0.05 vs. nonirradiated; b_P_ < 0.05 vs. day 9).
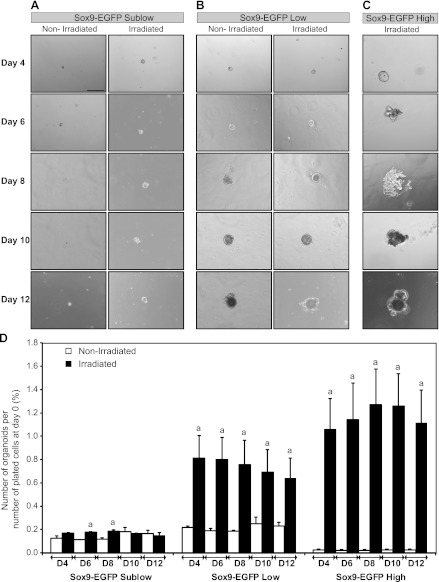
Sox9-EGFP Low and High Cells Isolated at 5 Days After Radiation Exhibit Enhanced Ability To Form Organoids In Vitro
A recently developed ISC/organoid 3D culture system permits assays of the growth capacity of isolated putative ISCs on the basis of their ability to survive and expand to form organoids (55). Prior studies on cells isolated from uninjured intestine demonstrated that only Sox9-EGFP Low cells survived and expanded to form organoids, indicative of ISC characteristics (21). In the present study, this culture system was used to study the growth capacities of Sox9-EGFP High, Low, Sublow, and Negative cells isolated from jejunum of mice at day 5 postirradiation compared with cells from nonirradiated mice. The 5-day postradiation time point was chosen to specifically assess the functional phenotype of Sox9-EGFP cells isolated during maximal regeneration in vivo. Sox9-EGFP Negative cells isolated from either nonirradiated or irradiated mice did not demonstrate the capacity to form organoids (data not shown). Consistent with previous studies (21), Sox9-EGFP Sublow cells from nonirradiated jejunum formed only small structures composed of one or a few cells that did not increase in size throughout 12 days in culture, and this was also the case for Sox9-EGFP Sublow cells isolated after irradiation (Fig. 8_A_). Sox9-EGFP Low cells from nonirradiated mice also formed few organoids but, consistent with prior findings (21), these continued to grow throughout the entire 12 days of the experiment when they began to develop cryptlike structures (Fig. 8_B_). Also consistent with prior studies (21), the organoids obtained from nonirradiated Sox9-EGFP Low cells contained cells expressing low/moderate and high/intense levels of Sox9-EGFP (Fig. 9). Strikingly, a greater percentage of Sox9-EGFP Low cells formed organoids when isolated at 5 days postirradiation compared with Sox9-EGFP Low cells from nonirradiated mice. This was evident as early as 4 days after plating and persisted to day 12 after plating (Fig. 8_D_). Although more organoids were formed by irradiated Sox9-EGFP Low cells, individual organoids showed the same rate of growth and morphology as organoids derived from nonirradiated Sox9-EGFP Low cells (Fig. 8_B_). Furthermore, organoids grown from irradiated Sox9-EGFP Low cells also comprised cells expressing low/moderate and high/intense levels of Sox9-EGFP (Fig. 9). These data demonstrate that Sox9-EGFP Low cells isolated during maximal regeneration after radiation exhibit enhanced in vitro abilities to survive and proliferate to form organoids.
Fig. 8.
Sox9-EGFP Low and High cells isolated at 5 days postirradiation possess enhanced ability to form organoids in vitro. Organoid cultures were performed on FACS-isolated cells from nonirradiated mice or at day 5 postradiation and were monitored at 2-day intervals between day 4 (D4) and day 12 (D12) after plating. Representative photographs show Sox9-EGFP Sublow cells from nonirradiated or irradiated mice (A), Sox9-EGFP Low cells from nonirradiated or irradiated mice (B), and Sox9-EGFP High cells isolated at day 5 postradiation (C) (bright field; scale bar 200 μm). D: histograms show quantitative data (mean ± SE) for the number of organoids formed by the 3 different Sox9-EGFP-sorted cell populations at each time point divided by the initial number of plated cells ×100 (n ≥ 3; a_P_ < 0.05 vs. nonirradiated). Sox9-EGFP Low and High cells isolated from irradiated mice exhibited significantly greater ability to form organoids compared with nonirradiated controls.
Fig. 9.
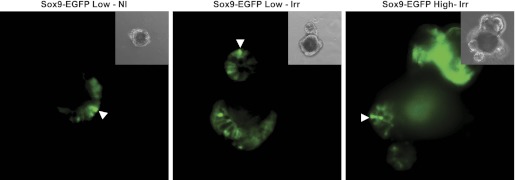
Organoids grown from nonirradiated and irradiated Sox9-EGFP Low cells and from Sox9-EGFP High cells isolated at 5 days postirradiation each express both low and high levels of Sox9-EGFP. Representative photographs shows EGFP fluorescence of organoids obtained from nonirradiated Sox9-EGFP Low cells (left; Sox9-EGFP Low-NI), Sox9-EGFP Low cells isolated at 5 days postradiation (middle; Sox9-EGFP Low-Irr), and Sox9-EGFP High cells isolated at 5 days postradiation (right, Sox9-EGFP High-Irr). Organoids contained discrete cells expressing Low/moderate or High/intense levels of Sox9-EGFP. Arrowheads show cells expressing High/intense levels of the Sox9-EGFP transgene. Note: High-intensity green in upper right corner of the photograph illustrating EGFP fluorescence of organoid obtained from Sox9-EGFP High cells isolated at 5 days postradiation is autofluorescence from an aggregate of dead cells.
Consistent with prior findings (21), Sox9-EGFP High cells from nonirradiated mice did not form detectable organoids (data not shown). Unexpectedly, Sox9-EGFP High cells isolated from jejunum 5 days after radiation formed many organoids (Fig. 8, C and D). Additionally, the percentage of Sox9-EGFP High cells isolated postradiation that formed organoids was comparable to that observed for irradiated Sox9-EGFP Low cells (Fig. 8_D_), and the organoids generated by irradiated Sox9-EGFP High cells rapidly formed cryptlike structures (Figs. 8_C_) and expressed low/moderate and high/intense levels of Sox9-EGFP (Fig. 9). These results suggest that Sox9-EGFP High cells isolated from mice during maximal crypt regeneration contain a population of cells with ISC properties.
Genes and Pathways Regulated in Sox9-EGFP Cells During Radiation-Induced Crypt Regeneration
To identify genes regulated specifically in Sox9-EGFP Low, Sublow, or High cells during radiation-induced crypt regeneration, we compared gene expression signatures between Sox9-EGFP Low, Sublow, High, and Negative cells isolated from irradiated vs. nonirradiated mice. We then excluded irradiation-regulated genes common to cell populations expressing different levels of Sox9-EGFP (Fig. 2_B_). IPA analyses of the genes specifically regulated in regenerating Sox9-EGFP Low, Sublow, or High cells identified major signaling pathways and cellular functions regulated significantly and exclusively in each of these Sox9-EGFP cell populations during crypt regeneration after radiation.
Genes linked to PKA signaling and Cdx2 are regulated specifically in regenerating Sox9-EGFP Low cells.
We found that 567 genes were differentially regulated specifically in Sox9-EGFP Low cells isolated during regeneration at day 5 postirradiation (Supplemental Table S3). Although most of these genes have not to our knowledge been linked to radiation, regeneration, or stem cells, IPA analysis identified protein kinase A (PKA) signaling as the most significant canonical pathway regulated in regenerating Sox9-EGFP Low cells (P = 0.001), with multiple PKA-linked genes showing small but significant changes in expression. Interestingly, CDX2, which is known to control IEC proliferation and differentiation (14), was downregulated specifically in Sox9-EGFP Low cells after radiation (Supplemental Table S3). Since loss of Cdx2 confers resistance to radiation-induced apoptosis in colonic epithelium (6), specific downregulation of Cdx2 in small intestinal Sox9-EGFP Low cells at times associated with peak expansion after radiation may contribute to their survival during crypt regeneration. Furthermore, CASP12, which encodes caspase 12, was also markedly downregulated specifically in regenerating Sox9-EGFP Low cells (Supplemental Table S3).
Genes linked to ephrin receptor signaling are specifically regulated in Sox9-EGFP Sublow cells during crypt regeneration.
Nine hundred thirty-three genes were differentially regulated specifically in Sox9-EGFP Sublow cells after radiation (Supplemental Table S4). IPA analysis identified ephrin receptor signaling as significantly modulated by radiation specifically in Sox9-EGFP Sublow cells (data not shown). Interestingly, ephrin receptors and especially EphB receptors are major coordinators of proliferation and migration of cells located within the ISC/progenitor compartment as well as positioning of IECs along the crypt-villus axis (4, 25). In addition, several genes coding for markers of IEC differentiation were also upregulated specifically in Sox9-EGFP Sublow cells such as alkaline phosphatase, fatty acid binding proteins 1 and 2, and zonula occludens 2 (Supplemental Table S4). This might reflect a more rapid induction of differentiation in Sox9-EGFP Sublow progenitors after radiation as part of a mechanism to restore functional epithelium.
Postirradiation Sox9-EGFP High cells undergo a shift in transcriptome consistent with proliferative or ISC-like phenotype.
Two thousand seventy-one genes were regulated specifically in Sox9-EGFP High cells isolated at 5 days after radiation (Supplemental Table S5). IPA analysis revealed that the top three molecular and cellular functions regulated specifically in Sox9-EGFP High cells during irradiation-induced regeneration were cell cycle (199/2,071), DNA replication, recombination and repair (166/2,071), and cellular assembly and organization (108/2,071) (Supplemental Table S6).
Several cyclins and cyclin-dependent kinases, such as CCND1 and CDK1, were upregulated in Sox9-EGFP High cells after irradiation as well as other cell cycle division-associated proteins (Supplemental Tables S5 and S6), indicating an activation of proliferation in Sox9-EGFP High cells during crypt regeneration. Among them, CCNE2, which encodes cyclin E2 and is required for quiescent cells to reenter the cell cycle (20), was markedly and exclusively upregulated in Sox9-EGFP High cells during crypt regeneration (+6.73-fold increase vs. Sox9-EGFP High cells isolated from nonirradiated mice; P = 0.03) (Supplemental Table S5). Strikingly, IPA analysis revealed that, after irradiation, Sox9-EGFP High cells showed specific upregulation of many genes associated with DNA repair (Supplemental Tables S5 and S6). Irradiation also induced specifically in Sox9-EGFP High cells an upregulation of many genes associated with assembly of mitotic spindles, organization of centrosomes, and segregation of chromosomes (Supplemental Tables S5 and S6) concomitantly with a downregulation of genes regulating cellular adhesion or motility including several cadherin and myosin genes (Supplemental Tables S5 and S6). Overall, these data indicate an activation of pathways in Sox9-EGFP High cells that would favor successful ISC and crypt regeneration.
Recent studies provided evidence for lower mitochondrial mass and oxidative phosphorylation in undifferentiated, pluripotent stem cells and instead a preferential use of nonoxidative glycolysis as a major source of energy (50). Furthermore, inhibition of mitochondrial oxidation appears to promote “stemness” in adult and embryonic stem cells (50). IPA analysis identified oxidative phosphorylation as the top canonical pathway regulated specifically in regenerating Sox9-EGFP High cells (Table 7). Strikingly, the majority of the identified genes involved in oxidative phosphorylation were downregulated, indicating a metabolic shift toward lower oxidative phosphorylation. IPA analysis also revealed that p53 signaling was significantly regulated specifically in regenerating Sox9-EGFP High cells. The pattern of regulation of expression of the identified p53 signaling-related genes suggests a gene expression profile that would favor enhanced survival and proliferation (Table 8). Among these genes, TOPBP1, a gene critical to genome integrity (58), which is normally repressed by p53 (28), and CHEK2, a gene recently linked to DNA damage response in human induced pluripotent stem cells and embryonic stem cells (40), were significantly upregulated. Together, these data provide evidence that during crypt regeneration after radiation, Sox9-EGFP High cells contain a population of cells enriched for genes associated with active division, metabolic profiles, and DNA damage response of stem cells.
Table 7.
Oxidative phosphorylation-related genes regulated by irradiation specifically in Sox9-EGFP High cells
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| COX7A1 | cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) | −2.95 | 12865 |
| ATP6V1B2 | ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B2 | −2.87 | 11966 |
| ATP6V0E2 | ATPase, H+ transporting V0 subunit e2 | −2.53 | 76252 |
| ATP6V0B | ATPase, H+ transporting, lysosomal 21 kDa, V0 subunit b | −2.51 | 114143 |
| ATP6V0A1 | ATPase, H+ transporting, lysosomal V0 subunit a1 | −2.32 | 11975 |
| ATP6V1G1 | ATPase, H+ transporting, lysosomal 13 kDa, V1 subunit G1 | −2.26 | 66290 |
| ATP6AP1 | ATPase, H+ transporting, lysosomal accessory protein 1 | −2.18 | 54411 |
| ATP6V1E1 | ATPase, H+ transporting, lysosomal 31 kDa, V1 subunit E1 | −2.00 | 11973 |
| ATP6V0D1 | ATPase, H+ transporting, lysosomal 38 kDa, V0 subunit d1 | −1.95 | 11972 |
| ATP6V1H | ATPase, H+ transporting, lysosomal 50/57 kDa, V1 subunit H | −1.84 | 108664 |
| ATP6V1G2 | ATPase, H+ transporting, lysosomal 13 kDa, V1 subunit G2 | −1.78 | 66237 |
| ATP6V1F | ATPase, H+ transporting, lysosomal 14 kDa, V1 subunit F | −1.56 | 66144 |
| ATP5G1 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) | −1.43 | 11951|100041835 |
| NDUFB7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18 kDa | −1.38 | 66916 |
| ATP5B | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | −1.34 | 11947 |
| TCIRG1 | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 | −1.29 | 27060 |
| NDUFB9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22 kDa | −1.28 | 66218 |
| NDUFS5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15 kDa (NADH-coenzyme Q reductase) | −1.28 | 595136 |
| NDUFB10 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22 kDa | −1.27 | 68342 |
| UQCRQ | ubiquinol-cytochrome c reductase, complex III subunit VII, 9.5 kDa | −1.25 | 22272 |
| UQCRB | ubiquinol-cytochrome c reductase binding protein | −1.24 | 67530 |
| NDUFB3 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12 kDa | −1.19 | 66495 |
| NDUFA5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13 kDa | −1.13 | 68202 |
| COX7B | cytochrome c oxidase subunit VIIb | 1.21 | 66142 |
| NDUFA8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19 kDa | 1.23 | 68375 |
| CYC1 | cytochrome c-1 | 1.36 | 66445 |
| NDUFC2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5 kDa | 1.41 | 68197 |
| COX6B2 | cytochrome c oxidase subunit VIb polypeptide 2 (testis) | 1.48 | 333182 |
| NDUFA10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42 kDa | 1.51 | 67273 |
| NDUFAB1 | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8 kDa | 1.64 | 100043472|70316 |
Table 8.
p53 Signaling-related genes regulated by irradiation specifically in Sox9-EGFP High cells
| Symbol | Gene Name | Fold Change | ID |
|---|---|---|---|
| GADD45A | growth arrest and DNA-damage-inducible, alpha | −3.32 | 13197 |
| MAPK14 | mitogen-activated protein kinase 14 | −2.63 | 26416 |
| CCND2 | cyclin D2 | −2.21 | 12444 |
| TP63 | tumor protein p63 | −1.96 | 22061 |
| CASP6 | caspase 6, apoptosis-related cysteine peptidase | −1.95 | 12368 |
| PLAGL1 | pleiomorphic adenoma gene-like 1 | −1.89 | 22634 |
| RRM2B | ribonucleotide reductase M2 B (TP53 inducible) | −1.84 | 382985 |
| HDAC1 | histone deacetylase 1 | −1.61 | 433759 |
| HIPK2 | homeodomain interacting protein kinase 2 | −1.35 | 15258 |
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | 1.23 | 11651 |
| CCNK | cyclin K | 1.29 | 12454 |
| STAG1 | stromal antigen 1 | 1.33 | 20842 |
| CDK4 | cyclin-dependent kinase 4 | 1.71 | 12567 |
| LRDD | leucine-rich repeats and death domain containing | 1.77 | 57913 |
| APAF1 | apoptotic peptidase activating factor 1 | 1.90 | 11783 |
| CCND1 | cyclin D1 | 2.43 | 12443 |
| TNFRSF10A | tumor necrosis factor receptor superfamily, member 10a | 2.76 | 21933 |
| TOPBP1 | topoisomerase (DNA) II binding protein 1 | 2.93 | 235559 |
| CHEK2 | CHK2 checkpoint homolog (S. pombe) | 3.04 | 50883 |
Intriguingly, several markers of cell fate specification were regulated at 5 days postradiation exclusively in Sox9-EGFP High cells (Table 9). Upregulated genes included ATOH1, which drives cell fate toward the secretory lineages (70), NEUROG3, which is essential for EEC differentiation (27), HES5, which promotes goblet cell fate (73), and SPDEF, which is necessary for Paneth and goblet cell maturation (22). In addition, KLF7, HES2, and NEUROG1, whose roles in cell fate determination have not been well-documented in the mammalian intestine but have already been reported in other organs (32, 34, 59), were specifically downregulated in Sox9-EGFP High cells during radiation-induced crypt regeneration. Furthermore, it is important to note that these gene transcripts were not enriched in Sox9-EGFP High cells in uninjured intestine (data not shown). These findings suggest that transcriptional networks driving cell fate toward different secretory cell lineages are specifically activated in Sox9-EGFP High cells (or a subset of these cells) during radiation-induced crypt regeneration.
Table 9.
Cell fate/early progenitor markers regulated by irradiation in Sox9-EGFP High cells
| Symbol | Description | Fold Change | P Value |
|---|---|---|---|
| KLF7 | Kruppel-like factor 7 (ubiquitous) | −2.20 | 0.028 |
| HES2 | hairy and enhancer of split 2 (Drosophila) (Hes2) | −1.55 | 0.014 |
| NEUROG1 | neurogenin 1 | −1.29 | 0.006 |
| HES5 | hairy and enhancer of split 5 (Drosophila) (Hes5) | 1.68 | 0.042 |
| ATOH1 | atonal homolog 1 (Drosophila) (Atoh1) | 2.73 | 0.023 |
| SPDEF | SAM pointed domain containing ets transcription factor (Spdef) | 3.68 | 0.018 |
| NEUROG3 | neurogenin 3 (Neurog3) | 3.77 | 0.020 |
DISCUSSION
Recent lineage tracing studies have defined two populations of ISCs in the normal adult small intestinal epithelium. One comprises rapidly cycling Lgr5-expressing ISCs/CBCs intercalated between Paneth cells at the crypt base (3, 12), and the other, more slowly cycling ISCs located above Paneth cells at the +4 position from the crypt base expressing Bmi1 and Hopx (54, 61). Although recent studies have demonstrated a bidirectional lineage relationship between these two ISC populations (61, 64), their respective roles during crypt regeneration and mucosal repair after injury have not been reported. Using a Sox9-EGFP reporter mouse, the present studies provide novel in vivo and in vitro evidence that after high-dose irradiation two distinct Sox9-EGFP-expressing cell populations are activated to proliferate during crypt regeneration and possess the ISC ability to form organoids in culture. Our data also define novel genes and pathways regulated specifically in each of these cell populations during crypt regeneration.
Low levels of Sox9-EGFP expression were previously shown to mark cells enriched for Lgr5 and with ISC characteristics of self-renewal and multipotency for all IEC lineages in vitro (17, 21). The present studies demonstrate that these cells isolated from normal jejunum exhibit upregulation of genes previously shown to be enriched in Lgr5-expressing ISCs (65) and are also enriched in c-Myb mRNA, which has recently been linked to Lgr5 promoter activation (11) (data not shown). In addition, our microarray data identified multiple previously unreported mRNAs encoding membrane proteins enriched in Sox9-EGFP Low cells. These genes provide new putative ISC biomarkers that may be useful to isolate ISCs and may be functionally relevant to ISC biology. Of particular interest among these enriched genes are PCDH8, a protocadherin linked to cell adhesion and embryogenesis (29), CXCR7, a receptor linked to cancer growth in other organs (30), FGFRL1, a newly discovered member of the FGF receptor family (74), GAS1, a newly described critical mediator of Hedgehog signaling (26), PARD6G, a regulator of cell polarity in early embryogenesis (1), and TRIB2, a regulator of FOXO transcription factors and cancer cell growth (72). Together these putative markers may represent new regulators of ISC proliferation and differentiation worthy of future study.
A major goal of our study was to assess whether Sox9-EGFP Low cells contribute to crypt regeneration after injury. Our data demonstrate massive expansion of actively cycling Sox9-EGFP Low cells during early phases of irradiation-induced crypt regeneration. In vitro studies recapitulated previous results that only Sox9-EGFP Low cells, and not other Sox9-EGFP-expressing cells from nonirradiated jejunum, are able to grow and form organoids in culture (21). Importantly, Sox9-EGFP Low cells isolated at times of maximal expansion in vivo during crypt regeneration postradiation showed enhanced in vitro ability to form organoids, suggesting that regenerating Sox9-EGFP Low ISCs are activated to exhibit enhanced survival and/or growth capacity. This is further supported by microarray data demonstrating proapoptotic genes specifically repressed in regenerating Sox9-EGFP Low ISCs including CDX2, which normally promotes differentiation (14) and increases sensitivity to radiation-induced apoptosis (6), and CASP12, whose downregulation enhances crypt mucosal repair in a colitis model (16). IPA analysis identified multiple PKA signaling-related genes as modestly but significantly and specifically regulated in Sox9-EGFP Low cells at times of maximal expansion of these cells, suggesting that PKA signaling may be relevant to ISC-mediated crypt regeneration after injury. Consistent with this concept, recent proteomics data indicate a role for PKA signaling-related proteins in mediating the response of fibroblasts to radiation (69), suggesting that radiation affects PKA signaling at both mRNA and protein levels. Many of the 567 genes specifically regulated in Sox9-EGFP Low cells during radiation-induced crypt regeneration have not been previously linked to stem cell biology or radiation response and set the stage for interesting future directions to define their functional roles in regulating ISC-mediated crypt regeneration after injury.
A major and unexpected finding of this study was that during crypt regeneration postradiation there was a significant increase in the percentage of Sox9-EGFP High cells in S-phase. Observations that Sox9-EGFP High cells isolated during crypt regeneration exhibit abilities to form organoids, whereas Sox9-EGFP High cells from nonirradiated mice fail to generate organoids, strongly suggest that crypt regeneration involves activation of a subset of normally nonproliferating cells within Sox9-EGFP High cells. This surprising gain of ISC-like functions induced by radiation in Sox9-EGFP High cells was further supported by our microarray data, which demonstrated a drastic shift in gene expression profile toward a proliferative/stem cell-like gene signature, as well as the regulation of genes linked to activation of quiescent cells in other systems. Several genes and pathways induced by radiation in Sox9-EGFP High cells are functionally linked to remodeling of the cytoskeleton concomitant with an activation of mitosis- and DNA repair-related pathways, consistent with a prosurvival advantage and enhanced growth capacities observed in vitro. IPA analysis also indicated that, after radiation, Sox9-EGFP High cells exhibit regulation of genes linked to a shift toward nonoxidative metabolism, a hallmark of stemness (50). Several genes, promoting cell fate specification toward different secretory epithelial cell lineages and normally expressed at relative low levels in Sox9-EGFP High cells from nonirradiated intestine, were also specifically upregulated in Sox9-EGFP High cells, providing further evidence that during radiation-induced crypt regeneration, cells within the Sox9-EGFP High cell population are activated to adopt a stem/progenitor cell-like phenotype. Our findings were surprising because under normal conditions, Sox9-EGFP High cells appear to be primarily nonproliferating EECs, usually considered a terminally differentiated cell type. However, the concomitant expression of ISC and EEC markers has been previously reported (24, 57). Furthermore, prior reports using lineage tracing have demonstrated that a subset of cells expressing the EEC precursor marker neurogenin 3 can give rise to entire crypt-villus units containing all differentiated cell lineages (56). In addition, it has been hypothesized that after injury, a subset of postmitotic differentiated cells can adopt or dedifferentiate to a “stem cell-like” state (75). The existence of such cells, termed facultative stem cells (FSCs) because of their ability to acquire multipotency abilities upon injury, is well accepted in urodele amphibians (9). In mammals, the FSC paradigm has been suggested in the liver and the pancreas (71), and current views suggest the existence of quiescent ISCs that may be activated during repair after injury (33, 46). Our findings provide evidence that FSCs or an activatable/quiescent ISC population is contained within Sox9-EGFP High cells.
Several models are worthy of consideration when interpreting our data on the Sox9-EGFP High cell population during radiation-induced crypt regeneration. The model we favor is that, in normal intestine, a subset of Sox9-EGFP High cells expressing EEC markers are quiescent or slowly cycling ISCs, and these cells are activated to proliferate during crypt regeneration postradiation. This model is supported by our data demonstrating that, in normal crypts, a very small fraction of ChgA-positive Sox9-EGFP High cells colabel with EdU, indicating proliferative potential of some EECs, and our findings that the percentage of EdU-labeled Sox9-EGFP High cells is increased during crypt regeneration in vivo. Gene microarray data indicate that in nonirradiated animals, Sox9-EGFP High cells are significantly enriched in mRNAs encoding +4 ISC markers Bmi1 and Hopx when compared with Sox9-EGFP Low cells or any other Sox9-EGFP cell type, further suggesting that this fraction is enriched in slowly cycling or quiescent +4 ISCs previously validated by lineage tracing (54, 61). Microarray data also indicate that during crypt regeneration, Sox9-EGFP High cells retain their EEC identity since they are still highly enriched for EEC marker mRNAs, including substance P, ChgA, and cholecystokinin genes (69- to 155-fold increase vs. irradiated Sox9-EGFP Negative cells). This same cell population also exhibits upregulation of many cyclins and cyclin-dependent kinases linked to proliferation during radiation-induced crypt regeneration, notably cyclin E2 which is essential to escape quiescence (20). Altogether our data strongly suggest that at least a subset of Sox9-EGFP High EECs represent slowly cycling/quiescent ISCs that are activated during crypt regeneration. A limitation of our study is that the Sox9-EGFP model does not permit lineage tracing to define the relationship between the putative quiescent ISCs within the Sox9-EGFP High cell population and the Sox9-EGFP Low ISCs. At present, we favor a model where the Sox9-EGFP High cell divides to self-renew and generate a Sox9-EGFP Low cell. This model is based on findings that during irradiation-induced crypt regeneration, there is an increase in the percentage of EdU-labeled Sox9-EGFP High cells but no increase in numbers of Sox9-EGFP High cells per crypt section, whereas there is a marked increase in the number of Sox9-EGFP Low cells per crypt section. This model is also consistent with a recent study that demonstrates that slowly cycling or quiescent Bmi1-ISCs can give rise to active Lgr5-ISCs (64). It is tempting to further speculate that downregulation of the level of Sox9 expression, which correlates with the level of Sox9-EGFP expression, might participate in driving the cell from a slowly cycling/quiescent to an active stemness state, and inversely. This is consistent with previous work showing that overexpression of Sox9 in IECs inhibits proliferation (17) and recent findings demonstrating that Sox9 directly binds and activates the promoter of Bmi1 (37), a quiescent ISC marker that our microarray data identified as enriched specifically in Sox9-EGFP High cells. Our microarray data on genes specifically enriched in Sox9-EGFP High cells could be worthy of consideration for future studies aiming at identifying genes that contain Sox9-DNA binding elements or are responsive to Sox9.
In summary, the present study provides strong evidence that two distinct ISC populations, which express either low or high levels of Sox9-EGFP, are activated to proliferate during radiation-induced crypt regeneration, possess functional characteristics of ISCs in vitro after irradiation, and exhibit distinct patterns of gene expression in situations of normal renewal vs. crypt regeneration. The radiation model developed in Sox9-EGFP mice and the gene expression data for these two ISC populations provide some new and useful tools to use in future studies aimed at defining the mechanisms controlling ISC-mediated crypt regeneration after injury and potentially identifying and testing strategies to promote ISC-mediated crypt regeneration.
GRANTS
This work was supported by a grant from the North Carolina Biotechnology Center, and National Institute of Diabetes and Digestive and Kidney Diseases Grants DK40247 (P. K. Lund) and K01-DK080181-01 (S. T. Magness).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
LVL designed the study, performed the experiments, analyzed the data, and wrote the paper; MAS took part in histology analysis; AEK participated in flow cytometry experiments; ATM took part in microarray experiments; JJD helped with radiation; ADG provided training and expertise in FACS-based experiments and in vitro culture assays; BPS generated preliminary data; KMN performed immunofluorescence experiments; STM took part in the project design and writing the paper; PKL designed the study, coordinated the project, and participated in all aspects of data analysis and manuscript preparation. All authors reviewed and approved the paper.
Supplementary Material
Supplemental Tables
ACKNOWLEDGMENTS
The work was assisted by services from the UNC Flow Cytometry core facility, the Genomics and Bioinformatics core of the UNC Lineberger Comprehensive Cancer Center, the Mouse Cardiovascular Models Core Lab (UNC McAllister Heart Institute), and the Histology Research Core Facilities of the Center for Gastrointestinal Biology and Disease (P30-DK034987) and the Department of Cell and Molecular Physiology (UNC). Technical assistance of Eliane Wauthier, Barry Udis, Jackie Kylander, Peter Starback, and Eric Blue as well as useful discussions with Susan J. Henning and Lola Reid are gratefully acknowledged.
REFERENCES
- 1.Alarcon VB. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol Reprod 83: 347–358, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev 22: 1856–1864, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111: 251–263, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Bhanja P, Saha S, Kabarriti R, Liu L, Roy-Chowdhury N, Roy-Chowdhury J, Sellers RS, Alfieri AA, Guha C. Protective role of R-spondin1, an intestinal stem cell growth factor, against radiation-induced gastrointestinal syndrome in mice. PLoS One 4: e8014, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonhomme C, Duluc I, Martin E, Chawengsaksophak K, Chenard MP, Kedinger M, Beck F, Freund JN, Domon-Dell C. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut 52: 1465–1471, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnaud S, Niaudet C, Legoux F, Corre I, Delpon G, Saulquin X, Fuks Z, Gaugler MH, Kolesnick R, Paris F. Sphingosine-1-phosphate activates the AKT pathway to protect small intestines from radiation-induced endothelial apoptosis. Cancer Res 70: 9905–9915, 2010 [DOI] [PubMed] [Google Scholar]
- 8.Braydich-Stolle L, Kostereva N, Dym M, Hofmann MC. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev Biol 304: 34–45, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science 310: 1919–1923, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Cacci E, Salani M, Anastasi S, Perroteau I, Poiana G, Biagioni S, Augusti-Tocco G. Hepatocyte growth factor stimulates cell motility in cultures of the striatal progenitor cells ST14A. J Neurosci Res 74: 760–768, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Cheasley D, Pereira L, Lightowler S, Vincan E, Malaterre J, Ramsay RG. Myb controls intestinal stem cell genes and self-renewal. Stem Cells 29: 2042–2050, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat 141: 537–561, 1974 [DOI] [PubMed] [Google Scholar]
- 13.Cohn SM, Schloemann S, Tessner T, Seibert K, Stenson WF. Crypt stem cell survival in the mouse intestinal epithelium is regulated by prostaglandins synthesized through cyclooxygenase-1. J Clin Invest 99: 1367–1379, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crissey MA, Guo RJ, Funakoshi S, Kong J, Liu J, Lynch JP. Cdx2 levels modulate intestinal epithelium maturity and Paneth cell development. Gastroenterology 140: 517–528, e518, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C. The role of SF/HGF and c-Met in the development of skeletal muscle. Development 126: 1621–1629, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, Vallance BA, Saleh M. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 32: 367–378, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuzawa H, Sawada M, Kayahara T, Morita-Fujisawa Y, Suzuki K, Seno H, Takaishi S, Chiba T. Identification of GM-CSF in Paneth cells using single-cell RT-PCR. Biochem Biophys Res Commun 312: 897–902, 2003 [DOI] [PubMed] [Google Scholar]
- 19.Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43: 34–41, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell 114: 431–443, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol 298: G590–G600, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregorieff A, Stange DE, Kujala P, Begthel H, van den Born M, Korving J, Peters PJ, Clevers H. The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology 137: 1333–1345, e1–3, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer 3: 117–129, 2003 [DOI] [PubMed] [Google Scholar]
- 24.Gulati AS, Ochsner SA, Henning SJ. Molecular properties of side population-sorted cells from mouse small intestine. Am J Physiol Gastrointest Liver Physiol 294: G286–G294, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Holmberg J, Genander M, Halford MM, Anneren C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisen J. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 125: 1151–1163, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Izzi L, Levesque M, Morin S, Laniel D, Wilkes BC, Mille F, Krauss RS, McMahon AP, Allen BL, Charron F. Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev Cell 20: 788–801, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, Jensen J, Kedinger M, Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J 21: 6338–6347, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KH, Yoo HY, Joo KM, Jung Y, Jin J, Kim Y, Yoon SJ, Choi SH, Seol HJ, Park WY, Nam do H. Time-course analysis of DNA damage response-related genes after in vitro radiation in H460 and H1229 lung cancer cell lines. Exp Mol Med 43: 419–426, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Nonclustered protocadherin. Cell Adh Migr 5: 97–105, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollmar O, Rupertus K, Scheuer C, Nickels RM, Haberl GC, Tilton B, Menger MD, Schilling MK. CXCR4 and CXCR7 regulate angiogenesis and CT26. WT tumor growth independent from SDF-1. Int J Cancer 126: 1302–1315, 2010 [DOI] [PubMed] [Google Scholar]
- 31.Komarova EA, Kondratov RV, Wang K, Christov K, Golovkina TV, Goldblum JR, Gudkov AV. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene 23: 3265–3271, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Laub F, Lei L, Sumiyoshi H, Kajimura D, Dragomir C, Smaldone S, Puche AC, Petros TJ, Mason C, Parada LF, Ramirez F. Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol Cell Biol 25: 5699–5711, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science 327: 542–545, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20: 469–482, 1998 [DOI] [PubMed] [Google Scholar]
- 35.Maksym RB, Tarnowski M, Grymula K, Tarnowska J, Wysoczynski M, Liu R, Czerny B, Ratajczak J, Kucia M, Ratajczak MZ. The role of stromal-derived factor-1—CXCR7 axis in development and cancer. Eur J Pharmacol 625: 31–40, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinelli DC, Fan CM. The role of Gas1 in embryonic development and its implications for human disease. Cell Cycle 6: 2650–2655, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KS, Skotheim RI, Lothe RA, Lopez de Munain A, Briscoe J, Serrano M, Lovell-Badge R. Oncogenicity of the developmental transcription factor Sox9. Cancer Res 72: 1301–1315, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 26: 630–637, 2008 [DOI] [PubMed] [Google Scholar]
- 39.May R, Sureban SM, Hoang N, Riehl TE, Lightfoot SA, Ramanujam R, Wyche JH, Anant S, Houchen CW. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells 27: 2571–2579, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Momcilovic O, Knobloch L, Fornsaglio J, Varum S, Easley C, Schatten G. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS One 5: e13410, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montgomery RK, Breault DT. Small intestinal stem cell markers. J Anat 213: 52–58, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okubo T, Knoepfler PS, Eisenman RN, Hogan BL. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development 132: 1363–1374, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, Wickramasinghe R, Scott MP, Wechsler-Reya RJ. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci USA 100: 7331–7336, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potten CS. Extreme sensitivity of some intestinal crypt cells to X and gamma irradiation. Nature 269: 518–521, 1977 [DOI] [PubMed] [Google Scholar]
- 45.Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: the mucosal governor. Int J Exp Pathol 78: 219–243, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potten CS, Gandara R, Mahida YR, Loeffler M, Wright NA. The stem cells of small intestinal crypts: where are they? Cell Prolif 42: 731–750, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potten CS, Hendry JH. Differential regeneration of intestinal proliferative cells and cryptogenic cells after irradiation. Int J Radiat Biol Relat Stud Phys Chem Med 27: 413–424, 1975 [DOI] [PubMed] [Google Scholar]
- 48.Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development 110: 1001–1020, 1990 [DOI] [PubMed] [Google Scholar]
- 49.Ramalingam S, Daughtridge GW, Johnston MJ, Gracz AD, Magness ST. Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. Am J Physiol Gastrointest Liver Physiol 302: G10–G20, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rehman J. Empowering self-renewal and differentiation: the role of mitochondria in stem cells. J Mol Med 88: 981–986, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rittie L, Stoll SW, Kang S, Voorhees JJ, Fisher GJ. Hedgehog signaling maintains hair follicle stem cell phenotype in young and aged human skin. Aging Cell 8: 738–751, 2009 [DOI] [PubMed] [Google Scholar]
- 52.Rossig L, Urbich C, Bruhl T, Dernbach E, Heeschen C, Chavakis E, Sasaki K, Aicher D, Diehl F, Seeger F, Potente M, Aicher A, Zanetta L, Dejana E, Zeiher AM, Dimmeler S. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med 201: 1825–1835, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci USA 107: 75–80, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009 [DOI] [PubMed] [Google Scholar]
- 56.Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol 270: 443–454, 2004 [DOI] [PubMed] [Google Scholar]
- 57.Sei Y, Lu X, Liou A, Zhao X, Wank SA. A stem cell marker-expressing subset of enteroendocrine cells resides at the crypt base in the small intestine. Am J Physiol Gastrointest Liver Physiol 300: G345–G356, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sokka M, Parkkinen S, Pospiech H, Syvaoja JE. Function of TopBP1 in genome stability. Subcell Biochem 50: 119–141, 2010 [DOI] [PubMed] [Google Scholar]
- 59.Solter M, Locker M, Boy S, Taelman V, Bellefroid EJ, Perron M, Pieler T. Characterization and function of the bHLH-O protein XHes2: insight into the mechanisms controlling retinal cell fate decision. Development 133: 4097–4108, 2006 [DOI] [PubMed] [Google Scholar]
- 60.Sun X, Cheng G, Hao M, Zheng J, Zhou X, Zhang J, Taichman RS, Pienta KJ, Wang J. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev 29: 709–722, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeda N, Jain R, Leboeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science 334: 1420–1424, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tesio M, Golan K, Corso S, Giordano S, Schajnovitz A, Vagima Y, Shivtiel S, Kalinkovich A, Caione L, Gammaitoni L, Laurenti E, Buss EC, Shezen E, Itkin T, Kollet O, Petit I, Trumpp A, Christensen J, Aglietta M, Piacibello W, Lapidot T. Enhanced c-Met activity promotes G-CSF-induced mobilization of hematopoietic progenitor cells via ROS signaling. Blood 117: 419–428, 2011 [DOI] [PubMed] [Google Scholar]
- 63.Thor G, Probstmeier R, Schachner M. Characterization of the cell adhesion molecules L1, N-CAM and J1 in the mouse intestine. EMBO J 6: 2581–2586, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478: 255–259, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912, 2009 [DOI] [PubMed] [Google Scholar]
- 66.Van Raay TJ, Moore KB, Iordanova I, Steele M, Jamrich M, Harris WA, Vetter ML. Frizzled 5 signaling governs the neural potential of progenitors in the developing Xenopus retina. Neuron 46: 23–36, 2005 [DOI] [PubMed] [Google Scholar]
- 67.Wada T, Kikuchi J, Nishimura N, Shimizu R, Kitamura T, Furukawa Y. Expression levels of histone deacetylases determine the cell fate of hematopoietic progenitors. J Biol Chem 284: 30673–30683, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum 9: 537–547, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang F, Waters KM, Miller JH, Gritsenko MA, Zhao R, Du X, Livesay EA, Purvine SO, Monroe ME, Wang Y, Camp DG, 2nd, Smith RD, Stenoien DL. Phosphoproteomics profiling of human skin fibroblast cells reveals pathways and proteins affected by low doses of ionizing radiation. PLoS One 5: e14152, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294: 2155–2158, 2001 [DOI] [PubMed] [Google Scholar]
- 71.Yanger K, Stanger BZ. Facultative stem cells in liver and pancreas: fact and fancy. Dev Dyn 240: 521–529, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zanella F, Renner O, Garcia B, Callejas S, Dopazo A, Peregrina S, Carnero A, Link W. Human TRIB2 is a repressor of FOXO that contributes to the malignant phenotype of melanoma cells. Oncogene 29: 2973–2982, 2010 [DOI] [PubMed] [Google Scholar]
- 73.Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of postmitotic intestinal epithelial cells. Genes Dev 19: 1686–1691, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhuang L, Villiger P, Trueb B. Interaction of the receptor FGFRL1 with the negative regulator Spred1. Cell Signal 23: 1496–1504, 2011 [DOI] [PubMed] [Google Scholar]
- 75.Zipori D. The nature of stem cells: state rather than entity. Nat Rev Genet 5: 873–878, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables