The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 26.
Abstract
Prions are self-templating protein conformers that are naturally transmitted between individuals and promote phenotypic change. In yeast, prion-encoded phenotypes can be beneficial, neutral or deleterious depending upon genetic background and environmental conditions. A distinctive and portable ‘prion domain’ enriched in asparagine, glutamine, tyrosine and glycine residues unifies the majority of yeast prion proteins. Deletion of this domain precludes prionogenesis and appending this domain to reporter proteins can confer prionogenicity. An algorithm designed to detect prion domains has successfully identified 19 domains that can confer prion behavior. Scouring the human genome with this algorithm enriches a select group of RNA-binding proteins harboring a canonical RNA recognition motif (RRM) and a putative prion domain. Indeed, of 210 human RRM-bearing proteins, 29 have a putative prion domain, and 12 of these are in the top 60 prion candidates in the entire genome. Startlingly, these RNA-binding prion candidates are inexorably emerging, one by one, in the pathology and genetics of devastating neurodegenerative disorders, including: amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U), Alzheimer’s disease and Huntington’s disease. For example, FUS and TDP-43, which rank 1st and 10th among RRM-bearing prion candidates, form cytoplasmic inclusions in the degenerating motor neurons of ALS patients and mutations in TDP-43 and FUS cause familial ALS. Recently, perturbed RNA-binding proteostasis of TAF15, which is the 2nd ranked RRM-bearing prion candidate, has been connected with ALS and FTLD-U. We strongly suspect that we have now merely reached the tip of the iceberg. We predict that additional RNA-binding prion candidates identified by our algorithm will soon surface as genetic modifiers or causes of diverse neurodegenerative conditions. Indeed, simple prion-like transfer mechanisms involving the prion-like domains of RNA-binding proteins could underlie the classical non-cell-autonomous emanation of neurodegenerative pathology from originating epicenters to neighboring portions of the nervous system.
Prions: unusual protein-based genetic elements
Even under physiological conditions, it is now clear that certain primary sequences enable proteins to adopt a range of alternative structures that are each capable of conformational self-replication via templating the conversion of other copies of the same protein (Alberti et al., 2009; Gendoo and Harrison, 2011; Goldschmidt et al., 2010; Halfmann et al., 2011; Sawaya et al., 2007; Toombs et al., 2010; Wiltzius et al., 2009). Typically, this conversion to a self-templating form radically alters protein function. Thus, a dramatic change in phenotype idiosyncratic to the function of the specific protein in question can rapidly ensue as self-templating forms deplete other conformers from the population. Sometimes these self-templating protein conformers can be naturally transmitted between individuals and promote phenotypic change. In these cases, the self-templating structures are termed prions (Colby and Prusiner, 2011; Cushman et al., 2010; Halfmann and Lindquist, 2010; Shorter, 2010; Weissmann et al., 2011).
Prions are perhaps most infamous as the etiological agents of infectious neurodegenerative diseases in mammals, including bovine spongiform encephalopathy, which can even traverse species barriers via the food chain and cause variant Creutzfeldt-Jakob disease in humans (Colby and Prusiner, 2011; Collinge and Clarke, 2007; Weissmann et al., 2011). Indeed, it is now possible to induce prion disease in wild-type mice by simply inoculating recombinant prion protein (PrP) that has been previously folded into a self-templating form in the presence of poly-anions and lipid in vitro (Wang et al., 2010; Wang et al., 2011a; Wang et al., 2011b). This simple transforming principle helps establish the unfamiliar view of self-templating protein structures as genetic material (Fink, 2005).
As self-replicating entities, prions are protein-based genetic elements, which are inescapably bound by the laws of natural selection. Thus, the concentration of specific self-templating forms will ebb and flow depending upon their intrinsic ability to self-replicate conformation in the prevailing environmental conditions (Duennwald and Shorter, 2010; Ghaemmaghami et al., 2009; Li et al., 2010a; Li et al., 2011; Roberts et al., 2009; Shorter, 2010; Wang et al., 2008a; Weissmann et al., 2011). In this sense, prion disorders can be viewed as a conflict between levels of selection. The initiation of selfish prion replication launches a microevolutionary process in which the prion replicator initially prospers and amplifies but ultimately destroys the host. The mammalian nervous system is particularly vulnerable to this conflict and can become severely and selectively devastated by prionogenesis (Shorter, 2010; Weissmann et al., 2011).
Increased awareness of prion-related phenomena in neurodegenerative disease
In recent years, awareness has increased that a similar microevolutionary process might be at work in other neurodegenerative diseases connected with protein misfolding, including Alzheimer’s disease, Parkinson’s disease and Huntington’s disease (Shorter, 2010). Indeed, it now appears probable that these devastating disorders are also underpinned by the spread of self-templating protein conformers. Here, self-templating forms spread from cell to cell within contiguous regions of the brains of afflicted individuals, thereby spreading the specific neurodegenerative phenotypes distinctive to the protein being converted to the self-templating form (Brundin et al., 2010; Cushman et al., 2010; Dunning et al., 2011; Goedert et al., 2010; Polymenidou and Cleveland, 2011; Prusiner, 1984; Walker et al., 2006). In these instances, transmission is usually restricted to within a tissue or within an individual. Transmission between individuals does not seem to occur naturally, but can be induced in experimental model systems (Clavaguera et al., 2009; Desplats et al., 2009; Eisele et al., 2010; Meyer-Luehmann et al., 2006). This type of phenomena has been termed prion-like and Adriano Aguzzi has even coined the term ‘prionoid’ to distinguish these self-templating conformers from bona fide prions (Aguzzi, 2009; Aguzzi and Rajendran, 2009).
Prion and prionoid semantics aside, there is a great deal of interest in defining whether these types of self-templating cascades are invariably associated with pathology or whether they have been captured by cells during evolution and exploited for adaptive purposes. Another burning question concerns the definition of primary sequence elements that confer the ability to populate self-templating prion or prionoid forms. In this review, we will focus on these questions as they relate to an unusual class of emergent RNA-binding proteins.
Yeast prions: good or evil or both?
As ever, answers to these critical questions have been rapidly gleaned from the best-characterized model organism on the planet, the baker’s yeast: Saccharomyces cerevisiae (Gitler, 2008). In yeast, multiple proteins can form prions that confer specific heritable phenotypes, which are passed from mother to daughter and typically segregate in a dominant non-Mendelian fashion (Chien et al., 2004; Shorter and Lindquist, 2005; Tuite and Serio, 2010; Wickner et al., 2007). These phenotypes can be advantageous, benign or deleterious depending on the genetic background and environmental conditions (Alberti et al., 2009; Eaglestone et al., 1999; McGlinchey et al., 2011; Nakayashiki et al., 2005; Namy et al., 2008; True and Lindquist, 2000; True et al., 2004). Thus, some authors have suggested that prions are adaptive bet-hedging devices or evolutionary capacitors that empower survival in intermittently stressful and fluctuating environments (Halfmann et al., 2010; Halfmann and Lindquist, 2010; Lancaster et al., 2010; Masel and Bergman, 2003; Masel and Griswold, 2009; Shorter and Lindquist, 2005; Shorter, 2010; Tuite and Serio, 2010). Conversely, others contend that yeast prions are molecular degenerative diseases more akin to mammalian neurodegenerative disorders (Wickner et al., 2007; Wickner et al., 2011). However, the fact that yeast prions can confer strong selective advantages under defined conditions separates them from simple degenerative disorders that are invariably deleterious.
Regardless of this still controversial debate, the specific heritable phenotypes can be established in yeast de novo, by transforming prion-free cells with pure self-templating conformers of the specific prion protein in question; for example, Sup35, Ure2, Rnq1 or Mot3 (Alberti et al., 2009; Brachmann et al., 2005; King and Diaz-Avalos, 2004; Patel and Liebman, 2007; Shorter and Lindquist, 2006; Tanaka et al., 2004). Typically, a loss-of-function phenotype idiosyncratic to the prion protein in question arises because the self-templating conformation limits functionality (Baxa et al., 2002). However, for some prion proteins, a gain of function occurs (Rogoza et al., 2010). Indeed, some evidence suggests that a gain of function (increased affinity for RNA) of a prion conformer formed by the RNA-binding protein, Cytoplasmic Polyadenylation Element Binding protein (CPEB), which also harbors two RNA recognition motifs (RRMs), might even play an adaptive role in long-term memory formation in metazoa (Fiumara et al., 2010; Heinrich and Lindquist, 2011; Keleman et al., 2007; Shorter and Lindquist, 2005; Si et al., 2003; Si et al., 2010). Recently, it has become clear that this unusual tie between RNA-binding modalities and prion formation could contribute to neurodegenerative disease (Cushman et al., 2010; Fuentealba et al., 2010; Gitler and Shorter, 2011; Udan and Baloh, 2011).
Distinctive, portable prion domains encode yeast prion behavior
A unifying feature of the majority of known yeast prion proteins is the presence of a distinctive prion domain that is enriched in uncharged polar amino acids (particularly asparagine, glutamine and tyrosine) and glycine (Alberti et al., 2009; Toombs et al., 2010). Typically, yeast prion domains are at least 60 amino acids in length and are primary sequences of low complexity that are predicted to be intrinsically unfolded (Alberti et al., 2009; Toombs et al., 2010). Variations on this theme are beginning to appear. For example, Swi1, which accesses a prion conformation that underpins the non-Mendelian state [_SWI+_] (Du et al., 2008), harbors a large predicted N-terminal prion domain (amino acids 1–385) (Alberti et al., 2009). It appears, however, that only the N-terminal 37 amino acids, which lack glutamine but are enriched for asparagine and threonine are required to drive Swi1 prionogenesis (Crow et al., 2011). Yeast prion domains can switch between an intrinsically unfolded conformation (non-prion form) and an infectious cRoss-β conformation (prion form) (Alberti et al., 2009; Brachmann et al., 2005; Patel and Liebman, 2007; Serio et al., 2000; Sondheimer and Lindquist, 2000; Tanaka et al., 2004; Taylor et al., 1999). Overexpression of this domain induces the prion state and deletion of this domain renders the protein unable to access the prion conformation (Masison and Wickner, 1995; Masison et al., 1997; Ter-Avanesyan et al., 1993; Ter-Avanesyan et al., 1994).
Importantly, yeast prion domains are portable (Wickner et al., 2000). For example, appending the prion domain of Sup35 to innocuous reporter proteins like beta-galactosidase or GFP enables them to access prion states (Li and Lindquist, 2000; Osherovich and Weissman, 2001; Tyedmers et al., 2010). This type of prion domain is not found in mammalian PrP (Colby and Prusiner, 2011) or in HET-s (Saupe, 2007), a prion protein from Podospora anserina, which suggests that other primary sequences can encode prion behavior (Taneja et al., 2007). Nonetheless, the presence of such a distinctive prion domain that confers prionogenicity in a portable manner stimulated the development of bioinformatic algorithms designed to detect these domains in genomes.
Algorithms designed to detect yeast prion domains
Characterization of the first prion proteins to be identified in yeast, Sup35 and Ure2, revealed the importance of their unusual N-terminal glutamine and asparagine-rich domain for prion behavior (Masison and Wickner, 1995; Masison et al., 1997; Ter-Avanesyan et al., 1993; Ter-Avanesyan et al., 1994). An initial algorithm that simply detected stretches that were enriched for glutamine or asparagine (at least 30 residues in an 80 amino acid stretch must be glutamine or asparagine) revealed that this type of domain might be relatively common in eukaryotic genomes (~100–400 per genome), but rare in prokaryotes (Michelitsch and Weissman, 2000). A later algorithm used binomial probabilities to identify regions biased for high glutamine and asparagine content, and also to filter results based on subsidiary biases towards glycine, serine, and tyrosine, and against charged or hydrophobic residues (Harrison and Gerstein, 2003). Initial surveys found numerous glutamine/asparagine-rich domains, which suggested that prion-like phenomena based on these determinants might be widespread in eukaryotic clades (Harrison and Gerstein, 2003; Michelitsch and Weissman, 2000). By contrast, the distinctive HET-s prion domain is an evolutionary innovation restricted to Sordariomycetes and is not found broadly in eukaryotes (Gendoo and Harrison, 2011).
The simple types of algorithm outlined above enabled the identification of the yeast prion protein, New1 (Santoso et al., 2000), and the potential CPEB prion in Aplysia (Si et al., 2003). Simple BLAST searches with the Sup35 and Ure2 prion domains helped to uncover the Rnq1 prion protein (Sondheimer and Lindquist, 2000). However, while these simple bioinformatic approaches successfully identified candidates with obvious similarities to Sup35 and Ure2, relatively few new prions were revealed in this way (Du et al., 2008; Patel et al., 2009; Rogoza et al., 2010).
BLAST searches in particular do not exploit the key observation that the amino acid composition of the yeast prion domain, rather than any precise linear stretch of primary sequence determinants per se, is largely responsible for prion formation and propagation (Ross et al., 2004; Ross et al., 2005). Subsequently, a refined algorithm was developed that used a hidden Markov model able to identify regions that have the unusual amino acid composition characteristic of known yeast prions (Alberti et al., 2009; Cushman et al., 2010). This approach provides a unified probabilistic framework for biases for or against any amino acid type, and it parses proteins into sharply defined prion-like and non-prion-like regions. Prion-like domains of length ≥60 residues were ranked with a prion-domain score, defined as the maximum log-likelihood for the prion-like state versus the non-prion-like state over any 60 consecutive amino acids within the regions. This algorithm returned ~200 proteins in the yeast genome with a candidate prion domain. An extensive experimental analysis of the top 100 candidates found that 19 domains were able to confer prion behavior in yeast, whereas ~69% of these candidates were aggregation-prone upon overexpression (Alberti et al., 2009). Thus, although the algorithm successfully identifies many aggregation-prone proteins, these candidates may not be capable of accessing a self-perpetuating prion form in yeast (Alberti et al., 2009). Regardless, the identification of 19 novel prion domains, some of which enable advantageous prion behavior, suggests that prions provide yeast with deep reservoirs of unplumbed heritable phenotypic variation that might increase the adaptability and evolvability of yeast populations in the face of diverse and fluctuating environments (Alberti et al., 2009; Halfmann et al., 2010; Halfmann and Lindquist, 2010; Shorter, 2010).
Two interesting questions naturally ensue from these observations. First: what distinguishes prion domain candidates that confer aggregation-prone behavior from those that do not? Second: what distinguishes prion domain candidates that encode prions from those that confer only aggregation-prone behavior? To answer the first question, aggregation-prone prion domains were found to be enriched for asparagine, whereas non-aggregating prion domains contained more glutamines, charged residues and prolines (Alberti et al., 2009). This bias for asparagine over glutamine was unexpected, because they had previously been considered equipotent in promoting prion formation (Harrison and Gerstein, 2003; Michelitsch and Weissman, 2000; Sondheimer and Lindquist, 2000). The second question is more difficult to answer. However, it appears that the spacing of charged residues and prolines with the prion domain plays a critical role (Alberti et al., 2009). Moreover, simultaneously replacing asparagines with glutamines, and, glutamines with asparagines reveals opposing roles for these two uncharged polar residues in prion domains. Thus, glutamines promote the formation of toxic oligomeric species and asparagines promote the formation of self-templating prions and reduce proteotoxicity (Halfmann et al., 2011). This finding could have important implications for predicting a priori functional prions from aggregation-prone proteins that cause disease.
In an effort to more accurately predict which prion domain candidates encode prion behavior, Ross and colleagues have developed a method that scores amino acid sequences using experimentally-derived prion propensities rather than their inherent similarity to known prions (Maclea and Ross, 2011; Toombs et al., 2010). Specifically, a portion of a scrambled version of the Sup35 prion domain was substituted with random sequences to generate a library of mutants. By comparing the frequencies of the substituted amino acids in the mutants that retained prionogenecity in yeast to those that did not, a prion propensity score was assigned to each specific amino acid (Toombs et al., 2010). Candidate domains that actually encoded prion behavior were distinguished by positive average prion propensity scores across extended disordered regions, as predicted by FoldIndex (Prilusky et al., 2005). Remarkably, by averaging scores for 41 overlapping windows (each of 41 amino acids) this method was able to separate with high accuracy the candidate domains that encode prion behavior from those that do not (Toombs et al., 2010). Interestingly, this strategy also revealed that hydrophobic residues, which are typically under-represented in prion domains, can greatly enhance prion propensity (Toombs et al., 2010).
An abundance of human RNA-binding proteins with prion-like domains
With these improved prion domain algorithms in hand it is of massive interest to scour the human genome for potential prion candidates. Thus, we have identified prion-like regions of 60 amino acids or longer using the hidden Markov model described above (Alberti et al., 2009; Couthouis et al., 2011; Cushman et al., 2010). Among the 21,873 human genes analyzed (Ensembl GrCh37.59), 246 had prion-like regions and were ranked by prion-domain score (Couthouis et al., 2011). Thus, ~1% of human protein-coding genes harbor a candidate prion domain. Of this 1%, there is a striking ~12-fold enrichment for proteins that harbor a canonical RNA recognition motif (RRM; PFAM ID PF00076.15) (Haider et al., 2009; Kenan et al., 1991). Indeed, ~1% of human protein-coding genes contain an RRM (210 genes). Yet, ~11.7% of human protein-coding genes that harbor a candidate prion domain also contain an RRM. Thus, 29 human RRM-bearing proteins also harbor a putative prion domain (Table 1, Figure 1), and 12 of these are in the top 60 prion candidates. Curiously, human CPEB isoforms were not among these 29, which might suggest that they are not prone to prion behavior in the same way as Aplysia CPEB (Couthouis et al., 2011). Indeed, perhaps other RRM-prion candidates play important roles in long-term memory formation in humans. Nonetheless, the striking over-representation of RRM-bearing proteins among prion candidates suggests that prion-like phenomena or aggregation-prone behavior might be rampant among this distinctive class of human RNA-binding proteins.
Table 1.
Human RNA-binding proteins with prion-like domains.
| Protein | Priondomainrank(wholegenome)(Alberti et al., 2009) | Priondomainrank(RRMproteins)(Alberti et al., 2009) | Priondomain(core)residues(Alberti et al., 2009) | Priondomaincentralresidues(Toombs et al., 2010) | Prionpropensityscore(FoldIndex)(Toombs et al., 2010) | Yeastoverexpressionphenotype(toxicity &localization)(Couthouis et al., 2011) |
|---|---|---|---|---|---|---|
| FUS | 12 | 1 | 1–237 (118–177) | 40–80 | 0.101 (−0.211) | Highly toxic, cytoplasmic aggregates |
| TAF15 | 22 | 2 | 1–152 (33–92) | 33–73 | 0.126 (−0.268) | Mildly toxic, cytoplasmic aggregates |
| EWSR1 | 25 | 3 | 1–280 (205–264) | 209–249 | 0.057 (−0.277) | Mildly toxic, cytoplasmic aggregates |
| HNRPDL | 27 | 4 | 316–420 (341–400) | 353–393 | 0.117 (−0.29) | Not toxic, cytoplasmic aggregates |
| HNRNPD | 29.5 | 5 | 262–355 (281–340) | 292–332 | 0.164 (−0.291) | Mildly toxic, diffuse nuclear |
| HNRNPA2B1 | 32 | 6 | 197–353 (276–335) | 274–314 | 0.043 (−0.208) | Highly toxic, cytoplasmic aggregates |
| HNRNPA1 | 38 | 7 | 186–372 (266–325) | 278–318 | 0.093 (−0.092) | Highly toxic, cytoplasmic aggregates |
| HNRNPAB | 39 | 8 | 235–327 (235–294) | 253–293 | 0.123 (−0.327) | ND |
| HNRNPA3 | 41 | 9 | 207–378 (287–346) | 302–342 | 0.057 (−0.194) | No expression |
| TDP-43 | 43 | 10 | 277–414 (301–360) | 361–401 | 0.043 (0.001) | Highly toxic, cytoplasmic aggregates |
| TIA1 | 53 | 11 | 292–386 (292–351) | 307–347 | 0.115 (−0.079) | Highly toxic, cytoplasmic aggregates |
| HNRNPA1L2 | 57 | 12 | 198–320 (243–302) | 227–267 | 0.052 (−0.091) | ND |
| HNRNPH1 | 63 | 13 | 382–472 (388–447) | 407–447 | 0.137 (0.039) | ND |
| SFPQ | 79 | 14 | 41–104 (41–100) | 638–678 | −0.077 (0.054) | ND |
| HNRNPA0 | 81 | 15 | 206–305 (206–265) | 228–268 | 0.079 (−0.03) | Highly toxic, cytoplasmic aggregates |
| HNRNPH2 | 101 | 16 | 382–449 (388–447) | 400–440 | 0.069 (−0.023) | ND |
| DAZ2 | 119 | 17 | 211–410 (235–294) | 390–430 | 0.067 (−0.014) | Highly toxic, cytoplasmic aggregates |
| RBM14 | 122 | 18 | 264–576 (362–421) | 328–368 | 0.006 (0.117) | Highly toxic, cytoplasmic aggregates |
| CSTF2 | 126 | 19 | 203–288 (203–262) | 491–531 | −0.024 (0.085) | ND |
| DAZ3 | 144.5 | 20.5 | 211–410 (235–294) | 390–430 | 0.067 (−0.014) | Mildly toxic, cytoplasmic aggregates |
| DAZ4 | 144.5 | 20.5 | 211–382 (283–342) | 148–188 | 0.002 (0.021) | No expression |
| DAZ1 | 148 | 22 | 541–716 (565–624) | 696–736 | 0.067 (−0.014) | Highly toxic, cytoplasmic aggregates |
| HNRNPH3 | 151 | 23 | 268–346 (276–335) | 306–346 | 0.079 (−0.037) | ND |
| CSTF2T | 153 | 24 | 476–568 (509–568) | 532–572 | −0.016 (0.085) | No expression |
| CELF4 | 156 | 25 | 241–305 (241–300) | 405–445 | −0.04 (0.066) | ND |
| TIAL1 | 162 | 26 | 301–392 (314–373) | 309–349 | 0.11 (−0.097) | ND |
| RBM33 | 178 | 27 | 591–707 (591–650) | 873–913 | −0.083 (−0.113) | No expression |
| DAZAP1 | 203 | 28 | 346–407 (346–405) | 214–254 | −0.028 (0.026) | Highly toxic, cytoplasmic aggregates |
| PSPC1 | 231 | 29 | 414–523 (415–474) | 479–519 | −0.121 (−0.103) | Not toxic, cytoplasmic aggregates |
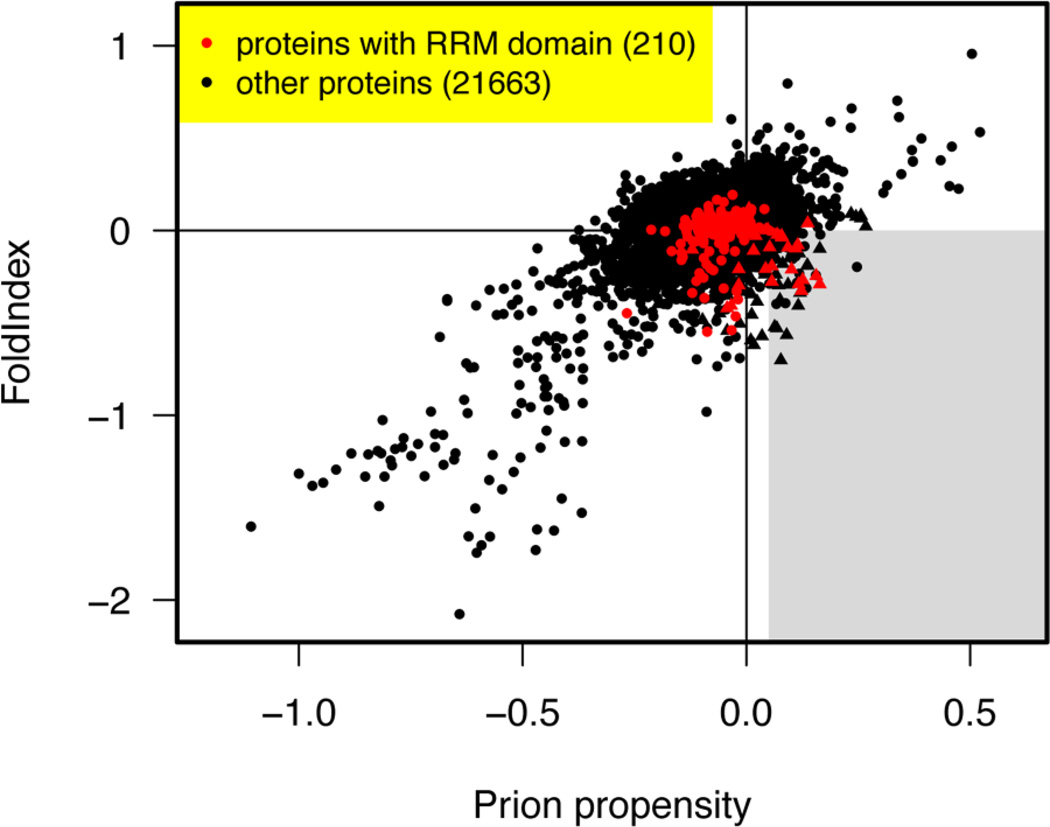
Figure 1. Human RNA-binding proteins with prion-like domains.
All human proteins from Ensembl release GRCh37.59 (78928 proteins including variant isoforms) were scanned for prion-like domains. The FoldIndex (Prilusky et al., 2005) and prion propensity scores (Toombs et al., 2010) are plotted for each human protein. Only the highest scoring protein isoform mapping to any single Ensembl gene ID is shown. RRM-containing proteins are indicated in red, and other proteins in black. Prion candidates contain regions that satisfy both conditions in a way that places them in the grey shaded sweet spot in the lower right. Both the FoldIndex and prion propensity scores represent averages of scores for 41 consecutive 41 amino acid (AA) windows (Toombs et al., 2010). The plotted scores for each protein are based on the consecutive windows that maximize the signed distance to the boundary of the grey region, which is positive for regions satisfying both conditions and negative otherwise. Proteins containing a region with prion-like amino acid composition are indicated by triangles (Alberti et al., 2009). These are defined as positive log-likelihood ratio when averaged over the 41 consecutive windows, based on the hidden Markov model of Alberti et al. (2009) but without imposing a hard minimum length requirement of 60 residues in the Viterbi parse. The prion-like amino acid frequencies were set to the average for 19 experimentally verified prion-like domains in S. cerevisiae (Alberti et al., 2009), and the background amino acid frequencies were set to the average of the proteome-wide amino acid frequencies in S. cerevisiae and H. sapiens. The RRM proteins that satisfy the Alberti et al. (2009) criteria are listed and ranked in Table 1.
Next, we asked how many of these 29 RRM-bearing prion candidates also pass the prion propensity and predicted disorder requirements of the Toombs et al. algorithm. Remarkably, 17 of 29 also passed this test, and a number of others were so exceptionally close to passing that a single point mutation could take them past the threshold (Table 1, Figure 1). Note also that these thresholds should not be regarded as absolute: they were chosen to discriminate the candidate yeast genes that passed four assays for prionogenicity in Alberti et al. (2009) from those that passed none, and most candidates narrowly missing the thresholds did pass some of the assays (Toombs et al., 2010). Moreover these assays were performed under controlled conditions in yeast, and it is likely that other factors influence the misfolding and aggregation of native proteins in human cells. The prion domain predictions for all 29 RNA-binding proteins can be found in the supplement (Supplemental material). Taken together, these data suggest that, at a minimum, this class of RNA-binding proteins is likely to be aggregation prone, and in addition a further subset could even access prion-like forms. Disturbingly, however, the misfolding and aberrant homeostasis of these RNA-binding proteins is beginning to emerge in connection with a series of devastating and presently incurable neurodegenerative disorders (Couthouis et al., 2011; Kwiatkowski et al., 2009; Neumann et al., 2006; Neumann et al., 2011; Sreedharan et al., 2008; Vance et al., 2009). We suggest that the RNA-binding prion candidates that have not yet emerged in neurodegenerative disease should be investigated as potential causative agents as soon as possible (Table 1).
TDP-43: the first of many?
TDP-43 was the first RNA-binding protein with a prion-like domain (amino acids 277–414, Figure 2) to emerge in connection with neurodegenerative disease (Arai et al., 2006; Neumann et al., 2006). The TDP-43 prion domain passes the Alberti algorithm, ranking 10th among RRM-bearing prion candidates, and narrowly misses the thresholds for the Toombs algorithm (Figure 2, Table 1) (Cushman et al., 2010). TDP-43 is a predominantly nuclear protein, which shuttles in and out of the nucleus, and functions in transcriptional regulation and RNA processing (Buratti and Baralle, 2008; Buratti and Baralle, 2010). Pathology and genetics now connect TDP-43 misfolding with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U) (Chen-Plotkin et al., 2010; Da Cruz and Cleveland, 2011; Neumann et al., 2006). In both these disorders, TDP-43 is found in cytoplasmic inclusions and depleted from the nucleus in afflicted neurons (Chen-Plotkin et al., 2010; Da Cruz and Cleveland, 2011). Prominent TDP-43 pathology is also evident in Perry syndrome and inclusion body myopathy and Paget disease of the bone (Chen-Plotkin et al., 2010). Remarkably, TDP-43 pathology is a secondary feature of several other neurodegenerative disorders including Alzheimer’s disease (over 50% of cases), Parkinson’s disease and Huntington’s disease (Chen-Plotkin et al., 2010). These findings suggest that TDP-43 misfolding likely contributes to neurodegeneration very broadly.
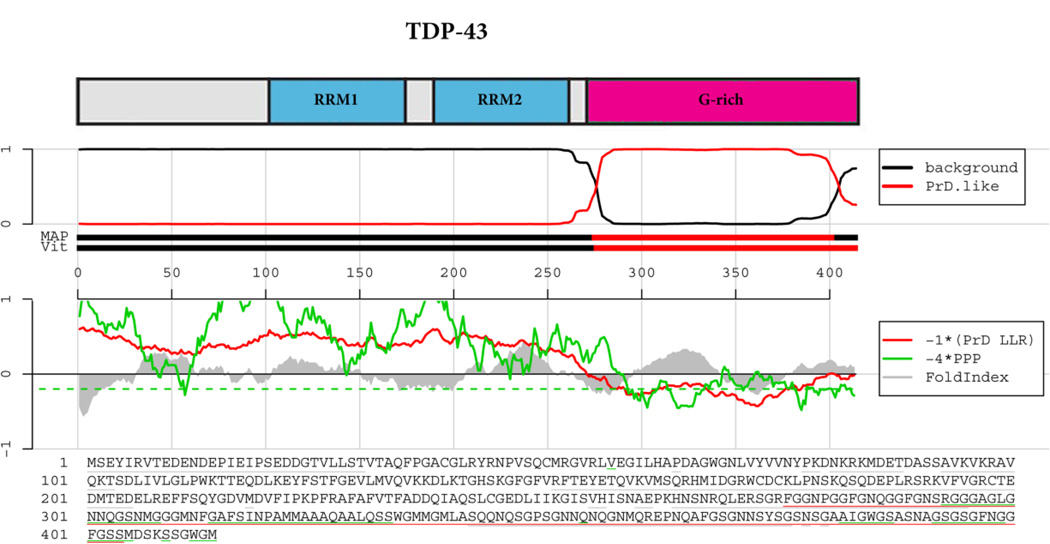
Figure 2. TDP-43 prion domain prediction.
The top panel shows the domain architecture of TDP-43. RRM=RNA-recognition motif; G-rich=Glycine-rich domain. Below the cartoon the probability of each residue belonging to the hidden Markov model state prion domain or ‘background’ is plotted; the tracks ‘MAP’ and ‘Vit’ illustrate the Maximum a Posteriori and the Viterbi parses of the protein into the prion domain or non-prion domain (Alberti et al., 2009). The plots in the middle panel show the log-likelihood ratio scores (PrD LLR) from the Alberti et al. algorithm in red (Alberti et al., 2009), the predicted prion propensity (PPP) log-odds ratio scores from the Toombs et al. algorithm in green (Toombs et al., 2010) and FoldIndex scores in grey (Prilusky et al., 2005), each averaged over sliding windows of 41 residues. Note that the curves are rescaled to give similar ranges, and so that negative scores are suggestive of both disorder and prion propensity; the rescaled cutoff corresponding to PPP > 0.05 is indicated by the dashed green line. The lower part of the panel shows the primary sequence of TDP-43. The Alberti prion domain is underlined in red (Alberti et al., 2009), the Toombs prion domain in underlined in green (Toombs et al., 2010), and the cyan residues represent the regions that satisfy these requirements of disorder and prion propensity of the Toombs algorithm (Toombs et al., 2010) as well as the amino acid composition requirement of the Alberti algorithm (Alberti et al., 2009). Note the lack of cyan residues for TDP-43.
Importantly, the prion-like domain of TDP-43 plays a critical role in TDP-43 misfolding. Aggregated C-terminal fragments of TDP-43 containing the prion-like region are biochemical signatures of ALS (Lee et al., 2011; Neumann et al., 2006). In isolation, TDP-43 is intrinsically aggregation-prone, and deleting the prion-like domain eliminates this behavior (Johnson et al., 2009). Indeed, deletion of just one short segment (amino acids 311–320) of the prion-like domain can prevent aggregation in vitro (Saini and Chauhan, 2011). Deletion of the entire prion-like domain prevents aberrant TDP-43 misfolding events and toxicity in several model systems (Ash et al., 2010; Johnson et al., 2008). Conversely, elevated expression of C-terminal fragments of TDP-43 that contain the prion-like domain elicits toxicity and cytoplasmic TDP-43 aggregation in diverse settings (Ash et al., 2010; Caccamo et al., 2012; Johnson et al., 2008; Pesiridis et al., 2011; Yang et al., 2010; Zhang et al., 2009). Remarkably, over forty ALS-linked mutations in TDP-43 have been reported and all but three of these are located in the C-terminal prion-like domain (Da Cruz and Cleveland, 2011). These ALS-linked TDP-43 variants can be divided into two classes. First, some mutations, including G294A, do not accelerate TDP-43 misfolding in vitro and do not promote toxicity in yeast (Johnson et al., 2009). These data suggest that some ALS-linked TDP-43 variants may not impact misfolding events directly. Second, some mutations, including Q331K and M337V, accelerate TDP-43 misfolding in vitro and enhance TDP-43 toxicity in yeast (Johnson et al., 2009). Importantly, Q331K is also much more toxic than wild-type TDP-43 in Drosophila (Elden et al., 2010). Indeed, several groups have observed similar effects of ALS-linked mutations on TDP-43 in diverse experimental systems ranging from cell culture, flies, chicken embryos, mouse, and rat (Barmada et al., 2010; Guo et al., 2011; Kabashi et al., 2010; Li et al., 2010b; Ritson et al., 2010; Sreedharan et al., 2008; Zhang et al., 2009). Collectively, these data suggest that some ALS-linked TDP-43 variants might cause disease via a gain-of-toxic function mechanism (Gitler and Shorter, 2011).
However, does TDP-43 access a prion or prionoid like form? A striking feature of ALS is the spread of pathology from initiating epicenters to neighboring regions of the brain, which involves multiple cell types and might be underpinned by a prion or prionoid (Cushman et al., 2010; Ravits and La Spada, 2009; Udan and Baloh, 2011). For yeast prions, the self-templating form is undoubtedly a cross-beta amyloid conformer, although not all amyloid conformations encode prions (Cushman et al., 2010; Salnikova et al., 2005). Short, synthetic TDP-43 peptides derived from the prion-like domain can access toxic amyloid forms (Chen et al., 2010; Guo et al., 2011). However, the physiological releVance of these short peptides that do not occur naturally is unclear, and practically all proteins harbor short peptides that can adopt the amyloid form in isolation (Goldschmidt et al., 2010). By contrast, full-length TDP-43 purified under native conditions does not appear to access a classic amyloid form in isolation (Johnson et al., 2009). This finding is consistent with ALS pathology, which is strikingly devoid of amyloid structures recognized by diagnostic dyes such as Congo Red or Thioflavin-T (Kwong et al., 2008). Importantly, in isolation, TDP-43 rapidly populates small pore-like oligomers and short fibrils, which cluster together to form large complex aggregates that bear remarkable ultrastructural resemblance to TDP-43 inclusions in the degenerating motor neurons of ALS patients (Couthouis et al., 2011; Johnson et al., 2009; Sun et al., 2011). The small pore-shaped oligomers formed by TDP-43 resemble toxic oligomers formed by Aβ42 and α -synuclein, which are highly neurotoxic (Kayed et al., 2003; Lashuel et al., 2002). Thus, TDP-43 might get trapped in this particularly toxic oligomeric form and cause neurodegeneration.
In contrast to yeast prions, it is less clear whether infectious forms of mammalian PrP must invariably be amyloid, even though mammalian prions can form amyloid and seed amyloid assembly (Colby and Prusiner, 2011; Shorter and Lindquist, 2005). Indeed, mammalian prion disease can present without abundant amyloid deposits (Colby and Prusiner, 2011). For example, PrP amyloid plaques are usually not present in Creutzfeldt-Jakob disease though PrP immunohistochemistry will nearly always be positive (Bell et al., 1997; Budka et al., 1995). Moreover, bona fide synthetic mammalian prions can adopt amyloid and non-amyloid forms (Colby et al., 2009; Colby et al., 2010; Legname et al., 2004; Piro et al., 2011). Thus, could ALS be akin to mammalian prion disorders that do not present with gross amyloid pathology? Intriguingly, TDP-43 and C-terminal TDP-43 fragments (193–414) purified under denaturing conditions can assemble into fibrillar forms that do not appear to be classic amyloid, in that they do not bind Thioflavin-T (Furukawa et al., 2011). Yet, the fibrillar species formed by TDP-43 (193–414) appear to be able to seed TDP-43 aggregation in vitro and in cell culture (Furukawa et al., 2011). Thus, TDP-43 might populate an unusual self-templating form that is not a classic amyloid (Furukawa et al., 2011; Johnson et al., 2009), but perhaps shares features with synthetic mammalian prions that also do not appear to be classic amyloid (Colby et al., 2010; Piro et al., 2011).
Finally, it is important to note that simple TDP-43 misfolding per se is insufficient to cause toxicity. Rather, TDP-43 must be competent to engage RNA and aggregate for toxicity (Elden et al., 2010; Johnson et al., 2008; Voigt et al., 2010). Thus, TDP-43 aggregates might sequester essential RNA molecules and promote neurodegeneration (Polymenidou et al., 2011; Tollervey et al., 2011). Indeed, an interesting possibility is that aggregation might cause TDP-43 to bind RNA more avidly as is the case with Aplysia CPEB (Si et al., 2003). Alternatively, or in addition, RNA might stabilize or divert TDP-43 to adopt specific misfolded forms that are highly toxic. Indeed, different RNAs could enable TDP-43 to take on different forms or ‘strains’. Further studies are needed to distinguish these possibilities and to understand TDP-43 misfolding trajectories in fine detail.
It is interesting to note that RNA can enable mammalian PrP to adopt an infectious fold (Deleault et al., 2003; Wang et al., 2010; Wang et al., 2011a; Wang et al., 2011b). Thus, perhaps RNA enables TDP-43 to access self-templating forms. Curiously, a massive expansion of a noncoding GGGGCC hexanucleotide repeat in the first intron of the C9ORF72 gene has recently been identified as the major cause of familial FTD (11.7%) and ALS (23.5%) (Al-Sarraj et al., 2011; DeJesus-Hernandez et al., 2011; Gijselinck et al., 2012; Murray et al., 2011; Renton et al., 2011), and might even be connected to AD (Majounie et al., 2012). The transcribed GGGGCC hexanucleotide repeat forms nuclear foci (DeJesus-Hernandez et al., 2011). This non-coding RNA might promote the misfolding of RNA-binding proteins with prion-like domains into self-templating forms. One interesting candidate is hnRNP A2/B1, which ranks 6th among human RRM-bearing prion candidates (Table 1), is predicted to engage GGGGCC RNA, and is sequestered in RNA foci in the fragile X tremor ataxia syndrome (FXTAS) (Iwahashi et al., 2006; Sofola et al., 2007). Future studies will reveal how the GGGGCC hexanucleotide repeat might perturb RNA-binding proteostasis. A suggested starting point would be to analyze all of the prion-domain containing RRM proteins in Table 1 for mislocalization in c9FTD/ALS.
FUS, another RRM-bearing prion candidate implicated in neurodegeneration
Soon after the discovery of TDP-43’s role in neurodegeneration, the number 1 ranked RRM-bearing prion candidate, FUS, was connected via genetics and pathology with diverse neurodegenerative diseases. The FUS prion-like domain (amino acids 1–238) passes both the Alberti and Toombs algorithms (Figure 3, Table 1) (Cushman et al., 2010). Curiously, FUS harbors an additional region (amino acids 391–407) that almost satisfies the Alberti algorithm (Figure 3) (Cushman et al., 2010; Gitler and Shorter, 2011; Sun et al., 2011). Like TDP-43, FUS is a predominantly nuclear protein, which shuttles in and out of the nucleus, and functions in transcriptional regulation and RNA homeostasis (Bertolotti et al., 1996; Kasyapa et al., 2005; Zinszner et al., 1997). Mutations in FUS cause familial ALS (Da Cruz and Cleveland, 2011; Kwiatkowski et al., 2009; Vance et al., 2009). Additional FUS mutations have now also been connected with sporadic ALS and with FTLD-U (Belzil et al., 2009; Blair et al., 2010; Broustal et al., 2010; Corrado et al., 2010; Da Cruz and Cleveland, 2011; DeJesus-Hernandez et al., 2010; Drepper et al., 2011; Hewitt et al., 2010; Mackenzie et al., 2010; Neumann et al., 2009; Rademakers et al., 2010; Urwin et al., 2010). In these cases, FUS is found aggregated in the cytoplasm of degenerating neurons, whereas TDP-43 localization is not affected (Mackenzie et al., 2010). FUS aggregation, involving the wild-type protein, is connected with several neurodegenerative disorders, including: juvenile ALS, basophilic inclusion body disease, some cases of FTLD-U (now called FTLD-FUS), Huntington’s disease, and the spinocerebellar ataxias (Doi et al., 2010; Huang et al., 2010; Munoz et al., 2009; Urwin et al., 2010; Woulfe et al., 2010). Thus, FUS misfolding contributes broadly to neurodegeneration.
Figure 3. FUS prion-like domain prediction.
The top panel shows the domain architecture of FUS. QGSY-rich=Glutamine, glycine, serine and tyrosine-rich domain; RRM=RNA-recognition motif; G-rich=Glycine-rich domain; RRM=RNA-recognition motif; RGG=RGG domain, a domain with repeated Gly-Gly dipeptides interspersed with Arg and aromatic residues. Zn=Zinc finger motif. Below the cartoon the probability of each residue belonging to the Hidden Markov Model state prion domain or ‘background’ is plotted; the tracks ‘MAP’ and ‘Vit’ illustrate the Maximum a Posteriori and the Viterbi parses of the protein into the prion domain or non-prion domain (Alberti et al., 2009). The plots in the middle panel show the log-likelihood ratio scores (PrD LLR) from the Alberti et al. algorithm in red (Alberti et al., 2009), the predicted prion propensity (PPP) log-odds ratio scores from the Toombs et al. algorithm in green (Toombs et al., 2010) and FoldIndex scores in grey (Prilusky et al., 2005), each averaged over sliding windows of residues. Note that the curves are rescaled to give similar ranges, and so that negative scores are suggestive of both disorder and prion propensity; the rescaled cutoff corresponding to PPP > 0.05 is indicated by the dashed green line. The lower part of the panel shows the primary sequence of TDP-43. The Alberti prion domain is underlined in red (Alberti et al., 2009), the centers of windows satisfying the disorder and prion propensity criteria of Toombs are underlined in grey and green (Toombs et al., 2010), and the cyan residues represent the centers of regions that satisfy both Toombs criteria as well as the amino acid composition requirement of the Alberti algorithm.
Importantly, the prion-like domain of FUS plays a critical role in FUS misfolding. Purified FUS is extremely aggregation-prone and aggregates more rapidly than TDP-43 (Couthouis et al., 2011; Sun et al., 2011). FUS rapidly forms pore-like oligomeric species similar to toxic oligomers formed by other proteins connected with neurodegenerative disease (Couthouis et al., 2011; Sun et al., 2011). The FUS prion-like domain is more enriched for glutamine (18.1%) than asparagine (3.4%), which might render it more prone to becoming trapped in toxic oligomeric forms (Halfmann et al., 2011). However, pure FUS quickly accesses filamentous structures that closely resemble the ultrastructure of FUS aggregates in degenerating motor neurons of ALS patients (Baumer et al., 2010; Couthouis et al., 2011; Huang et al., 2010; Sun et al., 2011). Thus, all the information needed to assemble these structures is encoded in the primary sequence of FUS. Deleting the prion-like domain of FUS eliminates this behavior (Sun et al., 2011). However, unlike TDP-43, FUS fragments that harbor the prion-like domain (amino acids 1–238) do not aggregate, unless they also contain a C-terminal RGG domain (amino acids 374–422) (Sun et al., 2011). Intriguingly, this RGG domain contains a short region (amino acids 391–407) that is detected by the Alberti algorithm, but does not quite reach significance (Figure 3). Thus, compared to TDP-43 and to yeast prion proteins, FUS misfolding and aggregation is a more complex multidomain process, which requires communication between N- and C-terminal portions of the protein (Gitler and Shorter, 2011; Sun et al., 2011). This complex set of domain requirements is also required for the cytoplasmic aggregation and toxicity of FUS in yeast (Fushimi et al., 2011; Ju et al., 2011; Kryndushkin et al., 2011; Sun et al., 2011).
Does FUS access self-templating prion or prionoid forms? More experiments are needed to address this question, but like TDP-43, FUS does not appear to access a classic amyloid form (Fushimi et al., 2011; Ju et al., 2011; Kryndushkin et al., 2011; Sun et al., 2011). However, the requirement for N- and C-terminal domains for FUS misfolding hints that an intermolecular domain swap might promote polymerization. Intermolecular domain swapping is a common mechanism that usually involves domains at the N- and C-terminal ends of proteins and can promote the polymerization of filamentous structures in various designed and natural proteins (Guo and Eisenberg, 2006; Lee and Eisenberg, 2003; Liu and Eisenberg, 2002; Nelson and Eisenberg, 2006; Ogihara et al., 2001). Such a process could in principle yield seeding behavior without necessitating an amyloid form. Further experiments are needed to test this proposed mechanism of FUS polymerization.
The majority of ALS-linked FUS mutations cluster at the extreme C-terminal region (Da Cruz and Cleveland, 2011; Kwiatkowski et al., 2009; Vance et al., 2009) and many of these are predicted to disrupt a conserved PY-nuclear localization signal (NLS), which is decoded by karyopherin beta2 (Lee et al., 2006; Suel et al., 2008). Indeed, nuclear localization of FUS is disrupted by some of these mutations, (e.g. P525L) and the severity of mislocalization correlates with the severity of the ALS phenotype (Dormann et al., 2010; Dormann and Haass, 2011). Importantly, the C-terminal ALS-linked FUS variants do not accelerate FUS misfolding in vitro and do not promote aggregation or toxicity in yeast, which fail to decode even the wild-type FUS PY-NLS (Ju et al., 2011; Sun et al., 2011). These data suggest that C-terminal mutations promote FUS accumulation in the cytoplasm rather than FUS misfolding per se. Thus, even though FUS and TDP-43 are similar RNA-binding proteins, the mechanisms by which ALS-linked mutations contribute to pathogenesis might be distinct for either protein. However, a large number of FUS mutations connected with ALS and FTLD-U have now been uncovered in the N-terminal and C-terminal prion-like portions of FUS (Da Cruz and Cleveland, 2011). It will be important to determine whether these mutations accelerate FUS misfolding just as some ALS-linked mutations in the prion-like domain of TDP-43 accelerate misfolding (Johnson et al., 2009).
Like TDP-43, FUS must aggregate and engage RNA to promote toxicity in yeast (Sun et al., 2011). Thus, RNA might enable FUS to access specific toxic or self-templating conformers. Alternatively, or in addition, FUS might sequester or deplete essential RNAs and promote toxicity. Interestingly, recent studies in mammalian cells suggest that FUS appears to bind RNA, including most cell-expressed mRNAs, at high frequency, and recognizes AU-rich stemloops (Hoell et al., 2011). The repertoire of RNAs engaged by FUS shifts dramatically in ALS-linked variants that are mislocalized to the cytoplasm (Hoell et al., 2011). This change in repertoire might contribute to FUS toxicity (Hoell et al., 2011). Curiously, and in contrast to TDP-43 (Polymenidou et al., 2011; Tollervey et al., 2011), no specific RNA elements recognized by FUS have emerged (Hoell et al., 2011).
It remains uncertain if TDP-43 and FUS misfolding elicit motor neuron degeneration via common or divergent pathways. Studies in Drosophila indicate that FUS and TDP-43 might function together in a common genetic pathway in neurons (Wang et al., 2011c). Surprisingly, however, genome-wide deletion and overexpresssion screens in yeast revealed remarkably little overlap in genetic modifiers of TDP-43 and FUS toxicity (Sun et al., 2011). These data suggest that TDP-43 and FUS might cause toxicity by different mechanisms.
TAF15 emerges in ALS and FTLD-U
Remarkably, RRM-bearing prion candidates continue to emerge in connection with neurodegeneration. In 2011, TAF15, the second ranked RRM-bearing prion candidate, has been connected to ALS and FTLD-U (Couthouis et al., 2011; Neumann et al., 2011; Ticozzi et al., 2011). FUS together with EWSR1 and TAF15 form a protein family (FET), which share a common domain architecture (Tan and Manley, 2009). TAF15 harbors a prominent N-terminal prion-like domain (amino acids 1–149), which passes both Alberti and Toombs algorithms (Figure 4, Table 1) (Alberti et al., 2009; Couthouis et al., 2011; Toombs et al., 2010). The TAF15 prion-like domain is more enriched for glutamine (22.3%) than asparagine (5.4%), which might render it more prone to becoming trapped in toxic oligomeric forms (Halfmann et al., 2011). All FET family proteins are nuclear proteins that associate with the transcription factor II D complex and RNA polymerase II (Tan and Manley, 2009). We recently uncovered TAF15 in a simple yeast screen as a RNA-binding protein with similar properties to TDP-43 and FUS (Couthouis et al., 2011). Thus, TAF15 aggregates in the cytoplasm and is toxic to yeast (Couthouis et al., 2011). TAF15 is intrinsically aggregation prone in vitro and rapidly assembles in to pore-shaped oligomers and filamentous structures (Couthouis et al., 2011). In isolation, TAF15 aggregates more rapidly than TDP-43, but less rapidly than FUS (Couthouis et al., 2011). Thus, the relative aggregation kinetics of FUS, TAF15 and TDP-43 were foreshadowed by the prion domain algorithm, which ranks FUS above TAF15 and TAF15 above TDP-43 (Alberti et al., 2009; Cushman et al., 2010).
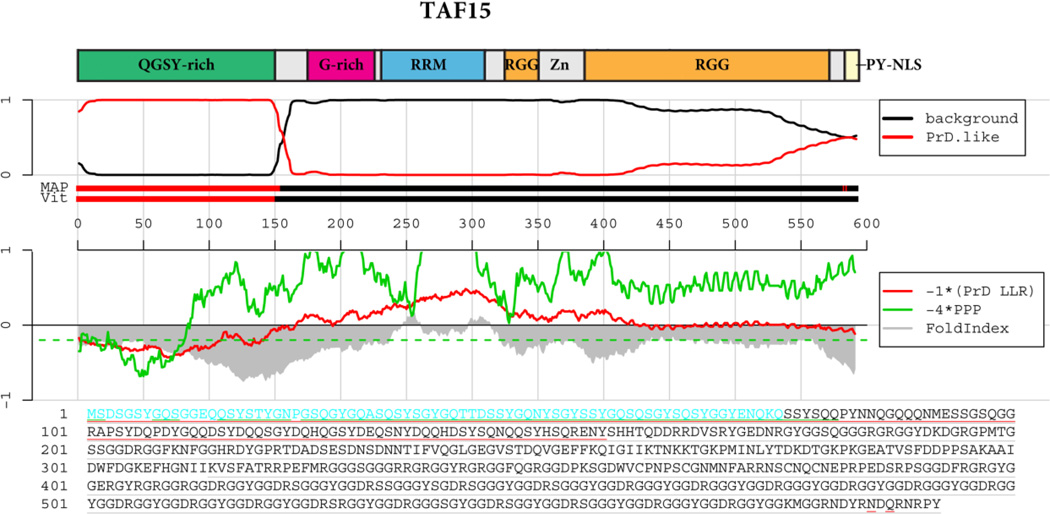
Figure 4. TAF15 prion-like domain prediction.
The top panel shows the domain architecture of TAF15. QGSY-rich=Glutamine, glycine, serine and tyrosine-rich domain; RRM=RNA-recognition motif; G-rich=Glycine-rich domain; RRM=RNA-recognition motif; RGG=RGG domain, a domain with repeated Gly-Gly dipeptides interspersed with Arg and aromatic residues. Zn=Zinc finger motif. Below the cartoon the probability of each residue belonging to the Hidden Markov Model state prion domain or ‘background’ is plotted; the tracks ‘MAP’ and ‘Vit’ illustrate the Maximum a Posteriori and the Viterbi parses of the protein into the prion domain or non-prion domain (Alberti et al., 2009). The plots in the middle panel show the log-likelihood ratio scores (PrD LLR) from the Alberti et al. algorithm in red (Alberti et al., 2009), the predicted prion propensity (PPP) log-odds ratio scores from the Toombs et al. algorithm in green (Toombs et al., 2010) and FoldIndex scores in grey (Prilusky et al., 2005), each averaged over sliding windows of 41 residues. Note that the curves are rescaled to give similar ranges, and so that negative scores are suggestive of both disorder and prion propensity; the rescaled cutoff corresponding to PPP > 0.05 is indicated by the dashed green line. The lower part of the panel shows the primary sequence of TDP-43. The Alberti prion domain is underlined in red (Alberti et al., 2009), the centers of windows satisfying the disorder and prion propensity criteria of Toombs are underlined in grey and green (Toombs et al., 2010), and the cyan residues represent the centers of regions that satisfy both Toombs criteria as well as the amino acid composition requirement of the Alberti algorithm.
Remarkably, sequencing TAF15 in sporadic ALS patients revealed several variants: M368T, G391E, R408C, G452E and G473E, that are not found in thousands of control samples. Further examination of G391E and R408C revealed that they aggregated more rapidly than wild-type TAF15 in vitro (Couthouis et al., 2011). Furthermore, elevated expression of TAF15 caused neurodegeneration in Drosophila and G391E or R408C elicited a more severe phenotype (Couthouis et al., 2011). Moreover, TAF15 localized to the nucleus when expressed in rat motor neurons in culture, whereas M368T, G391E, R408C and G473E formed numerous cytoplasmic inclusions (Couthouis et al., 2011). An independent study identified additional TAF15 variants in ALS cases (Ticozzi et al., 2011). Finally, TAF15 is found aggregated in the cytoplasm and depleted from the nucleus in the degenerating neurons of some ALS (Couthouis et al., 2011) and FTLD-U (Neumann et al., 2011) patients. Interestingly, the depletion of TAF15 from the nucleus was more severe than the depletion of FUS (Neumann et al., 2011). Taken together, these data suggest that TAF15 likely contributes to ALS and FTLD-U pathogenesis. It will be important to determine whether the domain requirements for TAF15 misfolding and toxicity are similar to those defined for FUS (Couthouis et al., 2011; Sun et al., 2011). Moreover, future studies will define whether TAF15 assembles into self-templating structures. To date, TAF15 mutations have been connected to sporadic forms, but not familial forms of disease.
EWSR1 emerges in FTLD-U
The final member of the FET family, EWSR1, ranks third among human RRM-prion candidates has also recently emerged in FTLD-U pathology (Neumann et al., 2011). In FTLD-FUS cases, EWSR1 accumulates in cytoplasmic aggregates and is depleted from the nucleus (Neumann et al., 2011). The depletion of EWSR1 from the nucleus is not as severe as TAF15 (Neumann et al., 2011). EWSR1 has a prominent N-terminal prion-like domain (amino acids 1–280), which passes both the Alberti and Toombs algorithms (Figure 5, Table 1) (Alberti et al., 2009) (Toombs et al., 2010). The EWSR1 prion-like domain is more enriched for glutamine (17.5%) than asparagine (1.4%), which might render it more prone to becoming trapped in toxic oligomeric forms (Halfmann et al., 2011). EWSR1 forms cytoplasmic aggregates and is toxic in yeast, although the domain requirements remain to be identified (Couthouis et al., 2011). Efforts are now underway to identify EWSR1 mutations in neurodegenerative disease (O.D.K., A.D.G., and J.S. manuscript in preparation; Ticozzi et al., 2011) and to determine whether EWSR1 accesses prionoid forms.
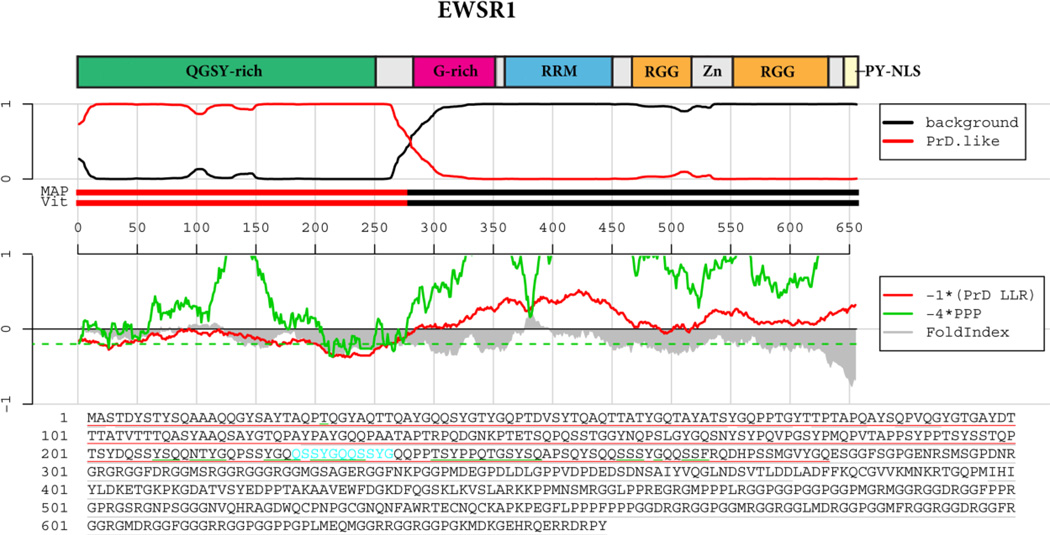
Figure 5. EWSR1 prion-like domain prediction.
The top panel shows the domain architecture of EWSR1. QGSY-rich=Glutamine, glycine, serine and tyrosine-rich domain; RRM=RNA-recognition motif; G-rich=Glycine-rich domain; RRM=RNA-recognition motif; RGG=RGG domain, a domain with repeated Gly-Gly dipeptides interspersed with Arg and aromatic residues. Zn=Zinc finger motif. Below the cartoon the probability of each residue belonging to the Hidden Markov Model state prion domain or ‘background’ is plotted; the tracks ‘MAP’ and ‘Vit’ illustrate the Maximum a Posteriori and the Viterbi parses of the protein into the prion domain or non-prion domain (Alberti et al., 2009). The plots in the middle panel show the log-likelihood ratio scores (PrD LLR) from the Alberti et al. algorithm in red (Alberti et al., 2009), the predicted prion propensity (PPP) log-odds ratio scores from the Toombs et al. algorithm in green (Toombs et al., 2010) and FoldIndex scores in grey (Prilusky et al., 2005), each averaged over sliding windows of 41 residues. Note that the curves are rescaled to give similar ranges, and so that negative scores are suggestive of both disorder and prion propensity; the rescaled cutoff corresponding to PPP > 0.05 is indicated by the dashed green line. The lower part of the panel shows the primary sequence of TDP-43. The Alberti prion domain is underlined in red (Alberti et al., 2009), the centers of windows satisfying the disorder and prion propensity criteria of Toombs are underlined in grey and green (Toombs et al., 2010), and the cyan residues represent the centers of regions that satisfy both Toombs criteria as well as the amino acid composition requirement of the Alberti algorithm.
Prion-like domains in sarcoma and leukemia
Intriguingly, all of the FET genes are directly involved in deleterious genomic rearrangements that cause sarcoma and leukemia (Tan and Manley, 2009). In all of these cases, a large portion of the prion-like domain of FUS, TAF15 or EWSR1 is translocated and appended to the N-terminal end of a transcription factor (Attwooll et al., 1999; Crozat et al., 1993; Delattre et al., 1992). Given the portable nature of yeast prion domains (Li and Lindquist, 2000; Wickner et al., 2000), it seems highly likely that appending the prion-like domain promotes misfolding, aberrant oligomerization and dysfunction of the transcription factor, which in turn leads to transformation.
Functional role of prion-like domains?
If aggregation prone RNA-binding proteins like TDP-43, FUS, and TAF15 and the others pose a major threat to neurons and contribute broadly to neurodegenerative disease pathogenesis, why are these proteins so well conserved through evolution? Perhaps the aggregation-prone nature of these proteins affords them the ability to perform essential cellular functions. One intriguing possibility is that RNA-binding proteins with prion-like domains play a role in RNA-based cellular memories or epigenetic states connected to transcriptional memory (Shorter and Lindquist, 2005). They might even be involved in long-term memory formation in a manner akin to Aplysia CPEB (Shorter and Lindquist, 2005; Si et al., 2003; Si et al., 2010). Curiously, human and other metazoan CPEB isoforms do not harbor a strong prion-like domain like the Aplysia protein. The human CPEBs pass neither the Alberti nor the Toombs prion domain algorithm. Perhaps, other RNA-binding proteins with prion-like domains have taken over the role of CPEB. Indeed, although TDP-43 and FUS are predominantly nuclear proteins, in neurons they are also involved in RNA transport to dendrites (Fujii and Takumi, 2005; Wang et al., 2008b). FUS and TDP-43 might affect mRNA transport along either actin or microtubule tracks, which could alter dendritic structure after excitation and affect long-term synaptic plasticity (Belly et al., 2005; Fujii et al., 2005; Fujii and Takumi, 2005; Liu-Yesucevitz et al., 2011; Wang et al., 2008b).
Another role of the prion-like domain could be in rapidly coalescing to form P-bodies and stress granules under situations of cellular stress. This is certainly a function of the TIA-1 prion-like domain, which ranks 11th among human RRM-bearing prion candidates (Table 1) (Gilks et al., 2004). Indeed, P-bodies and stress granules are specific types of RNA-binding protein aggregates that are used for normal biological processes (Buchan et al., 2008). However, as a consequence of having this ability, these proteins are thus poised to wreak havoc on neurons, should the quality control mechanisms regulating the assembly and disassembly of these RNA granules become corrupted. Under situations of stress, TDP-43, FUS, and other RNA-binding proteins translocate from the nucleus to the cytoplasm and associate with stress granules (Bosco et al., 2010; Dormann et al., 2010; Liu-Yesucevitz et al., 2010). When the stress dissipates, the stress granules disaggregate, and the RNA-binding proteins return to the nucleus. This repeated cycle of aggregation and disaggregation, over the course of a lifetime, perhaps has the chance to become misregulated, leading to a failure to restore one or more of these proteins to the nucleus, resulting in cytoplasmic accumulation and subsequent disease pathology. Moreover, identifying the human stress granule disaggregase machinery could yield potential therapeutic strategies. Curiously, Hsp104, a highly conserved protein disaggregase found in bacteria, fungi, plants, chromista and protozoa, is inexplicably absent from metazoa (DeSantis and Shorter, 2012; Shorter, 2008; Sweeny and Shorter, 2008; Vashist et al., 2010). Recently, however, the mammalian protein disaggregase machinery comprising Hsp110, Hsp70 and Hsp40 has been revealed (Shorter, 2011), and additional disaggregases are also likely to contribute to metazoan proteostasis (Bieschke et al., 2009; Cohen et al., 2006). It will be of great interest to determine whether these systems regulate stress granule assembly.
The concept of age-related deficits in stress granule dynamics suggests possible ways in which genetic and environmental factors might influence this process and lead to early disease onset in some cases, late onset in others, or no disease at all. For example, mutations in these RNA-binding proteins, which may accelerate their aggregation (Couthouis et al., 2011; Johnson et al., 2009), or enhanced environmental stress (for example, exposure to toxins, traumatic injury, viral infection (Chio et al., 2005; Cox et al., 2009)) could elicit exuberant cellular stress responses and increase the likelihood for RNA-binding proteins to inappropriately aggregate and accumulate in the cytoplasm of neurons. Importantly, this concept suggests that ALS and related neurodegenerative disease pathogenesis might be deeply rooted in core cell biological pathways and therefore a better understanding of the regulators of stress granule assembly and disassembly could provide new insight into disease mechanisms and suggest novel avenues for therapeutic intervention.
Genetic landscape of ALS and other RNA-binding proteinopathies
The discoveries of TDP-43 and FUS in ALS have resulted in a paradigm shift in our understanding of ALS disease mechanisms (Gitler and Shorter, 2011; Lagier-Tourenne and Cleveland, 2009). RNA-binding proteins and defects in RNA metabolism are likely central to the pathogenesis of related neurodegenerative disorders, including FTLD-U and Inclusion Body Myopathy with Paget Disease of Bone and/or Frontotemporal Dementia (IBMPFD) (Johnson et al., 2010; Neumann et al., 2006). In addition to TDP-43 and FUS, we propose that many additional RNA-binding proteins with similar properties (e.g. TAF15 and EWSR1) could also contribute to these diseases (Couthouis et al., 2011; Neumann et al., 2011; Ticozzi et al., 2011) (O.D.K., A.D.G., and J.S. manuscript in preparation). It is axiomatic that for complicated human diseases like ALS there will be both common as well as rare genetic risk factors. We envision that there may be a delicate balance in RNA processing within susceptible neuronal populations (e.g. motor neurons in ALS) such that slight perturbations from any one of several different aggregation-prone RNA-binding proteins could lead to neurodegeneration. Therefore, mutations in multiple RNA binding proteins could synergize with each other to contribute to disease. Moreover, some of these mutations will likely confer strong effects and others weaker effects. ALS-causing mutations in FUS help to illustrate this point. Certain FUS variants, like P525L and R495X, result in severe ALS clinical phenotypes and very early age of disease onset in the teenage years (Bosco et al., 2010; Huang et al., 2010). Perhaps then the accumulation of multiple weaker variants in several different aggregation-prone RNA binding proteins (e.g. the RNA-binding proteins with high-scoring prion-like domains) might be necessary to tip the balance in RNA metabolism towards ALS. Next generation sequencing approaches will empower us to test this hypothesis and to better resolve the complexities of the ALS genetic landscape.
The tip of the iceberg
More broadly, we strongly recommend that the RNA-binding prion candidates that have not yet emerged in neurodegenerative diseases (Table 1) should be investigated as potential causative agents as soon as possible. A combination of gene sequencing and histopathological examination of protein localization is warranted. We do not believe it is a coincidence that the RRM-bearing prion candidates: FUS, TAF15, EWSR1 and TDP-43, have all been connected to neurodegenerative disease. We strongly suspect that other RRM-bearing prion candidates will soon come to the fore in diverse neurodegenerative disease settings. Stay tuned.
Highlights.
- We review prion and prionoid phenomena
- We review algorithms to detect yeast prion domains
- We report list of top 29 human RRM-bearing prion candidates
- We review the function of RNA-binding proteins with prion-like domains
- We review the role of RNA-binding proteins with prion-like domains in disease
Supplementary Material
01
Acknowledgements
We thank Scott Ugras and Meredith Jackrel for thoughtful comments on the manuscript. This work was supported by NIH Director’s New Innovator Awards 1DP2OD004417-01 (A.D.G) and 1DP2OD002177-01 (J.S.), NIH R01 NS065317 (A.D.G.), NIH R21 NS067354-0110 (J.S.), a Bill and Melinda Gates Foundation Grand Challenges Explorations Award (J.S.), an Ellison Medical Foundation New Scholar in Aging Award (J.S.), and by a grant from The Robert Packard Center for ALS Research at Johns Hopkins (A.D.G. and J.S.). A.D.G. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguzzi A. Cell biology: Beyond the prion principle. Nature. 2009;459:924–925. doi: 10.1038/459924a. [DOI] [PubMed] [Google Scholar]
- Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in. C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwooll C, Tariq M, Harris M, Coyne JD, Telford N, Varley JM. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene. 1999;18:7599–7601. doi: 10.1038/sj.onc.1203156. [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D, Hilton D, Paine SM, Turner MR, Lowe J, Talbot K, Ansorge O. Juvenile ALS with basophilic inclusions is a FUS proteinopathy with FUS mutations. Neurology. 2010;75:611–618. doi: 10.1212/WNL.0b013e3181ed9cde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci U S A. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JE, Gentleman SM, Ironside JW, McCardle L, Lantos PL, Doey L, Lowe J, Fergusson J, Luthert P, McQuaid S, Allen IV. Prion protein immunocytochemistry--UK five centre consensus report. Neuropathol Appl Neurobiol. 1997;23:26–35. [PubMed] [Google Scholar]
- Belly A, Moreau-Gachelin F, Sadoul R, Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- Belzil VV, Valdmanis PN, Dion PA, Daoud H, Kabashi E, Noreau A, Gauthier J, Hince P, Desjarlais A, Bouchard JP, Lacomblez L, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–1179. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Bieschke J, Cohen E, Murray A, Dillin A, Kelly JW. A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis. Protein Sci. 2009;18:2231–2241. doi: 10.1002/pro.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, Blumbergs PC, Vucic S, Kiernan MC, Nicholson GA. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry. 2010;81:639–645. doi: 10.1136/jnnp.2009.194399. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr, Sapp P, McKenna-Yasek D, Brown RH, Jr, Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustal O, Camuzat A, Guillot-Noel L, Guy N, Millecamps S, Deffond D, Lacomblez L, Golfier V, Hannequin D, Salachas F, Camu W, Didic M, Dubois B, Meininger V, Le Ber I, Brice A. FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J Alzheimers Dis. 2010;22:765–769. [PubMed] [Google Scholar]
- Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11:301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budka H, Aguzzi A, Brown P, Brucher JM, Bugiani O, Gullotta F, Haltia M, Hauw JJ, Ironside JW, Jellinger K, et al. Neuropathological diagnostic criteria for Creutzfeldt-Jakob disease (CJD) and other human spongiform encephalopathies (prion diseases) Brain Pathol. 1995;5:459–466. doi: 10.1111/j.1750-3639.1995.tb00625.x. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 2010;7:420–429. doi: 10.4161/rna.7.4.12205. [DOI] [PubMed] [Google Scholar]
- Caccamo A, Majumder S, Oddo S. Cognitive Decline Typical of Frontotemporal Lobar Degeneration in Transgenic Mice Expressing the 25-kDa C-Terminal Fragment of TDP-43. Am J Pathol. 2012;180:293–302. doi: 10.1016/j.ajpath.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AK, Lin RY, Hsieh EZ, Tu PH, Chen RP, Liao TY, Chen W, Wang CH, Huang JJ. Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J Am Chem Soc. 2010;132:1186–1187. doi: 10.1021/ja9066207. [DOI] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- Chio A, Benzi G, Dossena M, Mutani R, Mora G. Severely increased risk of amyotrophic lateral sclerosis among Italian professional football players. Brain. 2005;128:472–476. doi: 10.1093/brain/awh373. [DOI] [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, Jucker M, Goedert M, Tolnay M. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci U S A. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, Lemus A, Cohen FE, DeArmond SJ, Prusiner SB. Protease-sensitive synthetic prions. PLoS Pathog. 2010;6:e1000736. doi: 10.1371/journal.ppat.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Prusiner SB. De novo generation of prion strains. Nat Rev Microbiol. 2011;9:771–777. doi: 10.1038/nrmicro2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, Soraru G, Carlomagno Y, Ghezzi S, Pensato V, Colombrita C, Gagliardi S, Cozzi L, Orsetti V, Mancuso M, Siciliano G, Mazzini L, Comi GP, Gellera C, Ceroni M, D'Alfonso S, Silani V. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J Med Genet. 2010;47:190–194. doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, Dejesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, Epstein J, Chiang A, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Solski JA, Williams KL, Mojsilovic-Petrovic J, Ingre C, Boylan K, Graff-Radford NR, Dickson DW, Clay-Falcone D, Elman L, McCluskey L, Greene R, Kalb RG, Lee VM, Trojanowski JQ, Ludolph A, Robberecht W, Andersen PM, Nicholson GA, Blair IP, King OD, Bonini NM, Van Deerlin V, Rademakers R, Mourelatos Z, Gitler AD. Feature Article: A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox PA, Richer R, Metcalf JS, Banack SA, Codd GA, Bradley WG. Cyanobacteria and BMAA exposure from desert dust: a possible link to sporadic ALS among Gulf War veterans. Amyotroph Lateral Scler. 2009;10(Suppl 2):109–117. doi: 10.3109/17482960903286066. [DOI] [PubMed] [Google Scholar]
- Crow ET, Du Z, Li L. A small, glutamine-free domain propagates the [SWI(+)] prion in budding yeast. Mol Cell Biol. 2011;31:3436–3444. doi: 10.1128/MCB.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011 doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Kocerha J, Finch N, Crook R, Baker M, Desaro P, Johnston A, Rutherford N, Wojtas A, Kennelly K, Wszolek ZK, Graff-Radford N, Boylan K, Rademakers R. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum Mutat. 2010;31:E1377–E1389. doi: 10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- DeSantis ME, Shorter J. The elusive middle domain of Hsp104 and ClpB: Location and function. Biochim Biophys Acta. 2012;1823:29–39. doi: 10.1016/j.bbamcr.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S A. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Koyano S, Suzuki Y, Nukina N, Kuroiwa Y. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neurosci Res. 2010;66:131–133. doi: 10.1016/j.neures.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, Neumann M, Haass C. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Haass C. TDP-43 and FUS: a nuclear affair. Trends Neurosci. 2011 doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Drepper C, Herrmann T, Wessig C, Beck M, Sendtner M. C-terminal FUS/TLS mutations in familial and sporadic ALS in Germany. Neurobiol Aging. 2011;32:548, e1–e4. doi: 10.1016/j.neurobiolaging.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Shorter J. Countering amyloid polymorphism and drug resistance with minimal drug cocktails. Prion. 2010;4:244–251. doi: 10.4161/pri.4.4.13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning CJ, Reyes JF, Steiner JA, Brundin P. Can Parkinson's disease pathology be propagated from one neuron to another? Prog Neurobiol. 2011 doi: 10.1016/j.pneurobio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele YS, Obermuller U, Heilbronner G, Baumann F, Kaeser SA, Wolburg H, Walker LC, Staufenbiel M, Heikenwalder M, Jucker M. Peripherally applied Abeta-containing inoculates induce cerebral beta-amyloidosis. Science. 2010;330:980–982. doi: 10.1126/science.1194516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink GR. A transforming principle. Cell. 2005;120:153–154. doi: 10.1016/j.cell.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba RA, Udan M, Bell S, Wegorzewska I, Shao J, Diamond MI, Weihl CC, Baloh RH. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J Biol Chem. 2010;285:26304–26314. doi: 10.1074/jbc.M110.125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem. 2011;286:18664–18672. doi: 10.1074/jbc.M111.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Long C, Jayaram N, Chen X, Li L, Wu JY. Expression of human FUS/TLS in yeast leads to protein aggregation and cytotoxicity, recapitulating key features of FUS proteinopathy. Protein Cell. 2011;2:141–149. doi: 10.1007/s13238-011-1014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendoo DM, Harrison PM. Origins and Evolution of the HET-s Prion-Forming Protein: Searching for Other Amyloid-Forming Solenoids. PLoS One. 2011;6:e27342. doi: 10.1371/journal.pone.0027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Ahn M, Lessard P, Giles K, Legname G, DeArmond SJ, Prusiner SB. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009;5:e1000673. doi: 10.1371/journal.ppat.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Baumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M, Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD. Beer and bread to brains and beyond: can yeast cells teach us about neurodegenerative disease? Neurosignals. 2008;16:52–62. doi: 10.1159/000109759. [DOI] [PubMed] [Google Scholar]
- Gitler AD, Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–187. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Clavaguera F, Tolnay M. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci. 2010;33:317–325. doi: 10.1016/j.tins.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Chen Y, Zhou X, Kar A, Ray P, Chen X, Rao EJ, Yang M, Ye H, Zhu L, Liu J, Xu M, Yang Y, Wang C, Zhang D, Bigio EH, Mesulam M, Shen Y, Xu Q, Fushimi K, Wu JY. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Eisenberg D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc Natl Acad Sci U S A. 2006;103:8042–8047. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal--unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20:125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Alberti S, Krishnan R, Lyle N, O'Donnell CW, King OD, Berger B, Pappu RV, Lindquist S. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol Cell. 2011;43:72–84. doi: 10.1016/j.molcel.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich SU, Lindquist S. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB) Proc Natl Acad Sci U S A. 2011;108:2999–3004. doi: 10.1073/pnas.1019368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt C, Kirby J, Highley JR, Hartley JA, Hibberd R, Hollinger HC, Williams TL, Ince PG, McDermott CJ, Shaw PJ. Novel FUS/TLS mutations and pathology in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2010;67:455–461. doi: 10.1001/archneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, Farazi TA, Hafner M, Borkhardt A, Sander C, Tuschl T. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18:1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Zhang J, Geser F, Trojanowski JQ, Strober JB, Dickson DW, Brown RH, Jr, Shapiro BE, Lomen-Hoerth C. Extensive FUS-Immunoreactive Pathology in Juvenile Amyotrophic Lateral Sclerosis with Basophilic Inclusions. Brain Pathol. 2010;20:1069–1076. doi: 10.1111/j.1750-3639.2010.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Bosco DA, Hayward LJ, Brown RH, Jr, Lindquist SL, Ringe D, Petsko GA. A Yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Lin L, Tradewell ML, Dion PA, Bercier V, Bourgouin P, Rochefort D, Bel Hadj S, Durham HD, Vande Velde C, Rouleau GA, Drapeau P. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- Kasyapa CS, Kunapuli P, Cowell JK. Mass spectroscopy identifies the splicing-associated proteins, PSF, hnRNP H3, hnRNP A2/B1, and TLS/FUS as interacting partners of the ZNF198 protein associated with rearrangement in myeloproliferative disease. Exp Cell Res. 2005;309:78–85. doi: 10.1016/j.yexcr.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kenan DJ, Query CC, Keene JD. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D, Wickner RB, Shewmaker F. FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP-43 in a yeast model of amyotrophic lateral sclerosis. Protein Cell. 2011;2:223–236. doi: 10.1007/s13238-011-1525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwong LK, Uryu K, Trojanowski JQ, Lee VM. TDP-43 proteinopathies: neurodegenerative protein misfolding diseases without amyloidosis. Neurosignals. 2008;16:41–51. doi: 10.1159/000109758. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EB, Lee VM, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2011;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Eisenberg D. Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process. Nat Struct Biol. 2003;10:725–730. doi: 10.1038/nsb961. [DOI] [PubMed] [Google Scholar]
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010a;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mahal SP, Demczyk CA, Weissmann C. Mutability of prions. EMBO Rep. 2011;12:1243–1250. doi: 10.1038/embor.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, 3rd, Fushimi K, Wu JY. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 2010b;107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA Translation at the Synapse and in Disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- Maclea KS, Ross ED. Strategies for identifying new prions in yeast. Prion. 2011;5 doi: 10.4161/pri.5.4.17918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Abramzon Y, Renton AE, Perry R, Bassett SS, Pletnikova O, Troncoso JC, Hardy J, Singleton AB, Traynor BJ. Repeat Expansion in C9ORF72 in Alzheimer's Disease. N Engl J Med. 2012 doi: 10.1056/NEJMc1113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Bergman A. The evolution of the evolvability properties of the yeast prion [PSI+] Evolution. 2003;57:1498–1512. doi: 10.1111/j.0014-3820.2003.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Masel J, Griswold CK. The strength of selection against the yeast prion [PSI+] Genetics. 2009;181:1057–1063. doi: 10.1534/genetics.108.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- Masison DC, Maddelein ML, Wickner RB. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci U S A. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, Vigouret JM, Paganetti P, Walsh DM, Mathews PM, Ghiso J, Staufenbiel M, Walker LC, Jucker M. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, Kuroda S, Mackenzie IR. FUS pathology in basophilic inclusion body disease. Acta Neuropathol. 2009;118:617–627. doi: 10.1007/s00401-009-0598-9. [DOI] [PubMed] [Google Scholar]
- Murray ME, Dejesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB, Rademakers R, Dickson DW. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–690. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, Rousset JP. Epigenetic control of polyamines by the prion [PSI+] Nat Cell Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, Roeber S, Kretzschmar HA, Munoz DG, Kusaka H, Yokota O, Ang LC, Bilbao J, Rademakers R, Haass C, Mackenzie IR. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara NL, Ghirlanda G, Bryson JW, Gingery M, DeGrado WF, Eisenberg D. Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc Natl Acad Sci U S A. 2001;98:1404–1409. doi: 10.1073/pnas.98.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. "Prion-proof" for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesiridis GS, Tripathy K, Tanik S, Trojanowski JQ, Lee VM. A "Two-hit" Hypothesis for Inclusion Formation by Carboxyl-terminal Fragments of TDP-43 Protein Linked to RNA Depletion and Impaired Microtubule-dependent Transport. J Biol Chem. 2011;286:18845–18855. doi: 10.1074/jbc.M111.231118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro JR, Wang F, Walsh DJ, Rees JR, Ma J, Supattapone S. Seeding specificity and ultrastructural characteristics of infectious recombinant prions. Biochemistry. 2011;50:7111–7116. doi: 10.1021/bi200786p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Cleveland DW. The Seeds of Neurodegeneration: Prion-like Spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Some speculations about prions, amyloid, and Alzheimer's disease. N Engl J Med. 1984;310:661–663. doi: 10.1056/NEJM198403083101021. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Stewart H, Dejesus-Hernandez M, Krieger C, Graff-Radford N, Fabros M, Briemberg H, Cashman N, Eisen A, Mackenzie IR. Fus gene mutations in familial and sporadic amyotrophic lateral sclerosis. Muscle Nerve. 2010;42:170–176. doi: 10.1002/mus.21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravits JM, La Spada AR. ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology. 2009;73:805–811. doi: 10.1212/WNL.0b013e3181b6bbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, Lee VM, Forman MS, Taylor JP. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BE, Duennwald ML, Wang H, Chung C, Lopreiato NP, Sweeny EA, Knight MN, Shorter J. A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat Chem Biol. 2009;5:936–946. doi: 10.1038/nchembio.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci U S A. 2010;107:10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini A, Chauhan VS. Delineation of the Core Aggregation Sequences of TDP-43 C-Terminal Fragment. Chembiochem. 2011;12:2495–2501. doi: 10.1002/cbic.201100427. [DOI] [PubMed] [Google Scholar]
- Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- Saupe SJ. A short history of small s: a prion of the fungus Podospora anserina. Prion. 2007;1:110–115. doi: 10.4161/pri.1.2.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- Shorter J. Emergence and natural selection of drug-resistant prions. Mol Biosyst. 2010;6:1115–1130. doi: 10.1039/c004550k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel KE, Gu H, Chook YM. Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 2008;6:e137. doi: 10.1371/journal.pbio.0060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2008;2:135–140. doi: 10.4161/pri.2.4.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, Sapp PC, Silani V, Bosco DA, Shaw CE, Brown RH, Jr, Landers JE. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 2010;11:823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci U S A. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan M, Baloh RH. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 2011;5:1–5. doi: 10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin H, Josephs KA, Rohrer JD, Mackenzie IR, Neumann M, Authier A, Seelaar H, Van Swieten JC, Brown JM, Johannsen P, Nielsen JE, Holm IE, Dickson DW, Rademakers R, Graff-Radford NR, Parisi JE, Petersen RC, Hatanpaa KJ, White CL, 3rd, Weiner MF, Geser F, Van Deerlin VM, Trojanowski JQ, Miller BL, Seeley WW, van der Zee J, Kumar-Singh S, Engelborghs S, De Deyn PP, Van Broeckhoven C, Bigio EH, Deng HX, Halliday GM, Kril JJ, Munoz DG, Mann DM, Pickering-Brown SM, Doodeman V, Adamson G, Ghazi-Noori S, Fisher EM, Holton JL, Revesz T, Rossor MN, Collinge J, Mead S, Isaacs AM. FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol. 2010;120:33–41. doi: 10.1007/s00401-010-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Cushman M, Shorter J. Applying Hsp104 to protein-misfolding disorders. Biochem Cell Biol. 2010;88:1–13. doi: 10.1139/o09-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Herholz D, Fiesel FC, Kaur K, Muller D, Karsten P, Weber SS, Kahle PJ, Marquardt T, Schulz JB. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LC, Levine H, 3rd, Mattson MP, Jucker M. Inducible proteopathies. Trends Neurosci. 2006;29:438–443. doi: 10.1016/j.tins.2006.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Ma J. Conversion of bacterially expressed recombinant prion protein. Methods. 2011a;53:208–213. doi: 10.1016/j.ymeth.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang Z, Wang X, Li J, Zha L, Yuan CG, Weissmann C, Ma J. Genetic informational RNA is not required for recombinant prion infectivity. J Virol. 2011b doi: 10.1128/JVI.06216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Duennwald ML, Roberts BE, Rozeboom LM, Zhang YL, Steele AD, Krishnan R, Su LJ, Griffin D, Mukhopadhyay S, Hennessy EJ, Weigele P, Blanchard BJ, King J, Deniz AA, Buchwald SL, Ingram VM, Lindquist S, Shorter J. Direct and selective elimination of specific prions and amyloids by 4,5-dianilinophthalimide and analogs. Proc Natl Acad Sci U S A. 2008a;105:7159–7164. doi: 10.1073/pnas.0801934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008b;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011c;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C, Li J, Mahal SP, Browning S. Prions on the move. EMBO Rep. 2011;12:1109–1117. doi: 10.1038/embor.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Taylor KL, Edskes HK, Maddelein ML. Prions: Portable prion domains. Curr Biol. 2000;10:R335–R337. doi: 10.1016/s0960-9822(00)00460-7. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman D, Kelly AC, Gorkovskiy A. The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion. 2011;5 doi: 10.4161/pri.5.4.17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius JJ, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woulfe J, Gray DA, Mackenzie IR. FUS-immunoreactive intranuclear inclusions in neurodegenerative disease. Brain Pathol. 2010;20:589–597. doi: 10.1111/j.1750-3639.2009.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Tan W, Whittle C, Qiu L, Cao L, Akbarian S, Xu Z. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One. 2010;5:e15878. doi: 10.1371/journal.pone.0015878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, Lin WL, Tong J, Castanedes-Casey M, Ash P, Gass J, Rangachari V, Buratti E, Baralle F, Golde TE, Dickson DW, Petrucelli L. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110(Pt 15):1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01