Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export (original) (raw)
Abstract
Export of messenger RNA (mRNA) from the nucleus to the cytoplasm is a critical step in the gene expression pathway of eukaryotic cells. Here, we report the functional and structural characterization of the mammalian TREX-2 complex and show how it links transcription/processing with nuclear mRNA export. Mammalian TREX-2 is based on a germinal-centre associated nuclear protein (GANP) scaffold to which ENY2, PCID2 and centrins bind and depletion of any of these components inhibits mRNA export. The crystal structure of the GANP:ENY2 complex shows that two ENY2 chains interact directly with GANP, but they have different orientations from those observed on yeast Sac3. GANP is required to recruit ENY2 to nuclear pore complexes (NPCs), but ENY2 is not necessary to recruit GANP, which requires both its CID and MCM3AP domains, together with nucleoporin Nup153. GANP and ENY2 associate with RNA polymerase II and inhibition of mRNA processing redistributes GANP from NPCs into nuclear foci indicating that mammalian TREX-2 is associated with transcription. Thus, we implicate TREX-2 as an integral component of the mammalian mRNA export machinery where it links transcription and nuclear export by facilitating the transfer of mature mRNPs from the nuclear interior to NPCs.
INTRODUCTION
The export of mature messenger RNA (mRNA) to the cytoplasm represents the culmination of the nuclear phase of the gene expression pathway (1–4). Transport through nuclear pore complexes (NPCs) is mediated primarily by the NXF1 export factor, but this process has to be tightly coupled to earlier steps in the pathway to ensure that they have been completed successfully (1–5). In mammalian cells, GANP (germinal-centre associated nuclear protein) integrates transcription/processing with nuclear export by facilitating the movement to nuclear pores of mRNPs generated deep in the nucleus (6). GANP binds directly to NXF1 through its N-terminal domain and is thought to shuttle between NPCs and nuclear processing centres to which it becomes relocated when transcription is inhibited (6). GANP is the mammalian orthologue of the yeast Sac3 protein that forms the scaffold of the TREX-2 transcription-export complex that also contains Thp1, Sem1, Sus1 and Cdc31 (7–12) (the corresponding mammalian proteins are PCID2, DSS1, ENY2 and centrins, respectively). The way in which GANP contributes to the gene expression pathway in metazoans is fundamentally different from the way in which its partial analogue, Sac3, contributes in budding yeast. Thus, in yeast, the Sac3-based TREX-2 complex is important in localizing a subset of actively transcribing genes to NPCs (11,13) to facilitate the export of their transcripts in a process referred to as ‘gene gating’ (14). However, in metazoa, GANP appears to function primarily in chaperoning mature mRNPs as they pass from processing centres to nuclear pores (6). Thus, in contrast to yeast, the majority of active genes in mammalian cells lie in transcription foci or ‘factories’ deep within the nucleus (15). Indeed, examination of the dynamics of single mRNPs in human U20S cells indicates that they accumulate in a particulate pattern in the nucleoplasm similar to that seen with the transcription factories (16,17). Importantly, the nuclear distribution of specific mRNPs is not influenced by the relative position of the transcription site in relation to the nuclear envelope (16,17). These fundamental differences may reflect the increased nuclear size, greater incidence of introns and genome complexity of metazoans.
In budding yeast, all the components of the TREX-2 complex are required for efficient mRNA nuclear export (7,8,12,18). We show here that although in human cells GANP forms an analogous TREX-2 complex, its contribution to the gene expression pathway and the integration of transcription/processing with nuclear export differs from that made by the analogous complex in yeast. Thus, although the human TREX-2 complex has a similar composition to its yeast analogue, there are differences in the way in which ENY2 is bound, and, importantly, the localization of the complex to NPCs requires the MCM3AP domain of GANP in addition to the presence of ENY2 and centrins. Furthermore, both GANP and ENY2 are associated with RNA polymerase II, and inhibition of mRNA processing redistributes GANP from NPCs into nuclear foci, suggesting that TREX-2 is associated with transcription.
MATERIALS AND METHODS
Protein expression and purification
Full-length ENY2, centrin-2 and centrin-3 were generated by PCR from I.M.A.G.E. cDNA clones (imaGenes) and cloned into pET30a (Novagen). Full-length PCID2 and DSS1 were cloned in to pRSFDuet-1 (Novagen). GANP fragments were generated by PCR from an I.M.A.G.E. cDNA clone and ligated into pGEXTEV (19), a modified version of pGEX-4T-1 (GE Healthcare; Chalfont St Giles, UK) in which the thrombin site has been replaced with a TEV protease site.
For crystallography, a GST-fusion with GANP residues 1162–1235 was co-expressed with untagged ENY2 in BL21 (DE3) CodonPlus RIL cells in 2xTY with conventional isopropyl-beta-d-thio-galactopyranoside (IPTG) induction overnight at 20°C. Cells were lysed by high-pressure cavitation (10–15 k psi) in 50 mM Tris–HCl (pH 8.0), 25% w/v sucrose, 1 mM ethylene glycol tetra-acetic acid (EGTA) and 1 mM phenyl methane sulphonyl fluoride. Complete EDTA-free protease inhibitor cocktail (Roche) was added to the lysed cells. Cells were clarified by centrifugation, filtered through a 0.22-µm membrane and bound to glutathione sepharose 4B resin (GE Healthcare) for 1 h at 4°C. The resin was washed with 500 ml of 50 mM Tris–HCl (pH 8.0), 250 mM NaCl, 1 mM EGTA and 2 mM DTT to remove non-specifically bound proteins, and the GANP:ENY2 complex released from the GST tag by overnight incubation with 100 µg of His-TEV protease [S219V mutant (20)]. The complex was further purified on a HiLoad Superdex 75 26/60 prep-grade column (GE Healthcare) in 20 mM Tris–HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA and 1 mM DTT. SeMet-labeled protein was expressed by IPTG induction in B834(DE3) cells as described (21) and complexes formed and purified as above, but in the presence of 5 mM DTT.
Crystallization and data collection
Crystals of the native GANP(ENY2)2 complex were grown at 19°C by hanging drop vapor diffusion in 100 mM MES pH 6.5, 11% w/v PEG8K and 0.2 M NaOAc and were briefly exposed to mother liquor containing 25% w/v glycerol before flash cooling in liquid nitrogen before data collection. Crystals of the SeMet-containing complex were grown in 100 mM MES, pH 6.5, 13% w/v PEG8K and 0.3 M NaOAc by micro-seeding using a rabbit's whisker and crystals of native complex as a source of seeds. SeMet crystals were cryo-protected in mother liquor containing 20% w/v ethylene glycol before freezing. Crystallographic data were collected either in-house using a Rigaku FR-E+ SuperBright generator equipped with Osmic mirrors and a Mar DTB camera or at beamline I03 at the Diamond Light Source (Didcot, UK) (Table 1).
Table 1.
Crystal data
| Crystals | Native | SeMet | ||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Unit cell dimensions | ||||
| a,b,c (Å) | 68.6, 76.1, 70.6 | 68.9, 75.7, 70.7 | ||
| β (°) | 117.7 | 117.7 | ||
| Data collection | ||||
| Wavelength (Å) | 0.9763 | 0.9803 | 0.9805 | 0.6989 |
| Resolution range (Å)a | 20–2.1 (2.21–2.1) | 75–2.6 (2.74–2.6) | 75–2.6 (2.74–2.6) | 75–2.6 (2.74–2.6) |
| Total observationsa | 260 408 (38 347) | 65 083 (8244) | 65 039 (8481) | 65 619 (8550) |
| Unique observationsa | 37 269 (5409) | 19 472 (2588) | 19 408 (2615) | 19 456 (2619) |
| Completeness (%)a | 99.1 (98.8) | 97.6 (89.9) | 97.8 (91.0) | 98.8 (91.3) |
| Multiplicitya | 7.0 (7.1) | 3.3 (3.2) | 3.4 (3.2) | 3.4 (3.3) |
| _R_pim (%)a | 4.3 (40.4) | 7.5 (36.8) | 6.6 (37.0) | 6.9 (38.3) |
| Mean I/σ (I)a | 13.1 (2.0) | 4.9 (2.3) | 5.0 (2.2) | 5.1 (2.2) |
| Refinement | ||||
| _R_cryst/_R_free (%) | 22.2/25.0 | |||
| Bond length rmsd (Å) | 0.002 | |||
| Bond angle rmsd (°) | 0.50 | |||
| MolProbity score/percentile | 1.22/100 | |||
| Ramachandran plot (%) | ||||
| Favoured | 99.6 | |||
| Allowed | 0.4 | |||
| Forbidden | 0 |
Structure solution and refinement
Crystals of the native GANP(ENY2)2 complex had _P_21 symmetry and diffracted to 2.1 Å using synchrotron radiation. Molecular replacement using various copies of Sus1 from the Sac3:Sus1:Cdc31 complex (18) and the Sgf11:Sus1 complex (22) in PHASER (23) did not yield a satisfactory solution. Instead, high-multiplicity MAD data collected for the Se peak, inflection and remote were processed in PHENIX (24) to determine heavy atom positions and initial phases. The two copies of the complex in the asymmetric unit were built using BUCANEER (23) and ARP/warp (23) and refined using PHENIX (24) alternating with extensive manual rebuilding in COOT (25). This preliminary model was then refined against the 2.1 Å native data to generate a final model with an _R_-factor of 25.7% (_R_free = 22.0%), excellent geometry (Table 1) and a MolProbity (26) score of 1.41 (99th percentile).
_In vitro_-binding studies
All proteins were expressed in 2xTY medium with conventional IPTG induction overnight at 20°C. Untagged ENY2, untagged centrin 2/3 and GST-GANP 686-1003 were expressed individually. His-PCID2 and DSS1 were co-expressed from pRSFDuet-1. GST-GANP CID fragments were co-expressed with either untagged ENY2 or untagged centrins. For the GANP CID domain interaction studies, binding assays were carried out by immobilizing GST fusion protein containing complexes on glutathione sepharose 4B resin from clarified bacterial lysates. Cell lysates containing GST-GANP fragments co-expressed with either untagged ENY2 or centrin (as appropriate) were mixed with cell lysate containing untagged centrin or untagged ENY2, respectively, and bound to resin at 4°C for 1 h. Resin was washed with 50 mM Tris–HCl, pH 8.0, 200 mM NaCl and 1 mM DTT and samples were analysed by SDS–PAGE. For binding studies involving PCID2, DSS1 and GANP, ternary complex was formed by mixing clarified lysates containing co-expressed His-PCID2:DSS1 and GST-GANP 686-1003 and applying to Ni-NTA resin at 4°C for 1 h. Resin was washed with 50 mM Tris–HCl, pH 8.0, 300 mM NaCl, 40 mM imidazole and 5 mM β-mercaptoethanol and samples analysed by SDS–PAGE.
siRNA-mediated depletion
For siRNA-mediated depletion, 106 HCT116 cells were transfected (Amaxa AG) with 10 µl of 20 µM siRNA. We used the following siRNAs: ENY2 siRNA4: AGAGUUGCUGAGAGCUAAA (Qiagen); ENY2 siRNA5-ON-TARGETplus SMART pool against E(Y)2: GGCACACUGUAAAGAGGUA, GAGCAGCGAUUAACCAAAA, AGAGAACGCCUCAAAGAGU, GCUGGAAGGAUCAGUUGAA, Dharmacon); Nup358 siRNA: SMART pool: GCGAAGUGAUGAUAUGUUU, CAACCACGUUAUUACUAA, CAGAACAACUUGCUAUUAG, GAAGGAAUGUUCAUCAGGA (Dharmacon); PCID2 siRNA: AUGCAGAUCAACAGUUGGUAA; centrin-2 siRNA: AAGCUCUUUGAUGAUGAUGAA and AAGCACAUGUAACUAGAUUUA (Qiagen); Nup153 siRNA: AAGCCCUUUAGCGGAUGCAAA (Qiagen); ALY siRNA: GGAACUCUUUGCUGAAUUU (Qiagen)). Cells were harvested either 48, 72 or 96 h post-transfection. Efficiency of depletion was monitored by immunoblotting with the indicated antibodies.
Immunofluorescence
Cells were fixed in 2 or 4% paraformaldehyde for 5 min at room temperature and permeabilized in PBS, 0.1% Triton X-100 (Sigma) and 0.02% SDS for 10 min at room temperature. After 30-min blocking (same buffer + 1% BSA), coverslips were incubated with the appropriate primary and secondary antibodies (Molecular Probes) and examined using a Zeiss LSM510 Meta confocal microscope. For some experiments, cells were permeabilized before fixation with PBS, 0.1% Triton X-100 for 5 min on ice. Scanning analysis of cells was performed using ImageJ software (NIH). All images used for comparative analysis were acquired using identical microscope settings. A line width of 20 was used, and pairs of cells with nuclei of same scan width as indicated by DAPI staining were used for analysis. All analyses are representative of the cell population.
Immunprecipitations and RNA FISH
RNA FISH was performed as previously described (6) using an oligo(dT) primer (Sigma). Immunoprecipitations were performed using nuclear extract as previously described (6). For immunoprecipitations with RNase-treated nuclear extract, 50 µl of a 10 mg/ml solution of RNase A (Sigma) was added to pre-cleared nuclear extract for 1 h at 4°C, after which 5 µg of antibody was added for 2 h. Samples were incubated as before with 50 µl of Protein A Sepharose (GE Healthcare) overnight with rotation, washed with lysis buffer four times and then eluted with 50 µl SDS loading buffer. Antibodies used were anti-GANP (raised against recombinant GANP 1050–1250 aa purified in E. coli) (6), anti-ENY2 (raised against full-length recombinant His-ENY2), mAb414 (Babco), β-actin (Abcam), anti-centrin, which recognizes centrin-2 but not centrin-3 (Sigma), anti-centrin-3 (Abcam), anti-RNA polymerase II (CTD4H8) (Millipore) and GFP, which recognizes YFP moiety (Clontech).
RESULTS
GANP residues 1162–1256 bind ENY2 and centrins, whereas residues 686–1003 bind PCID2 and DSS1
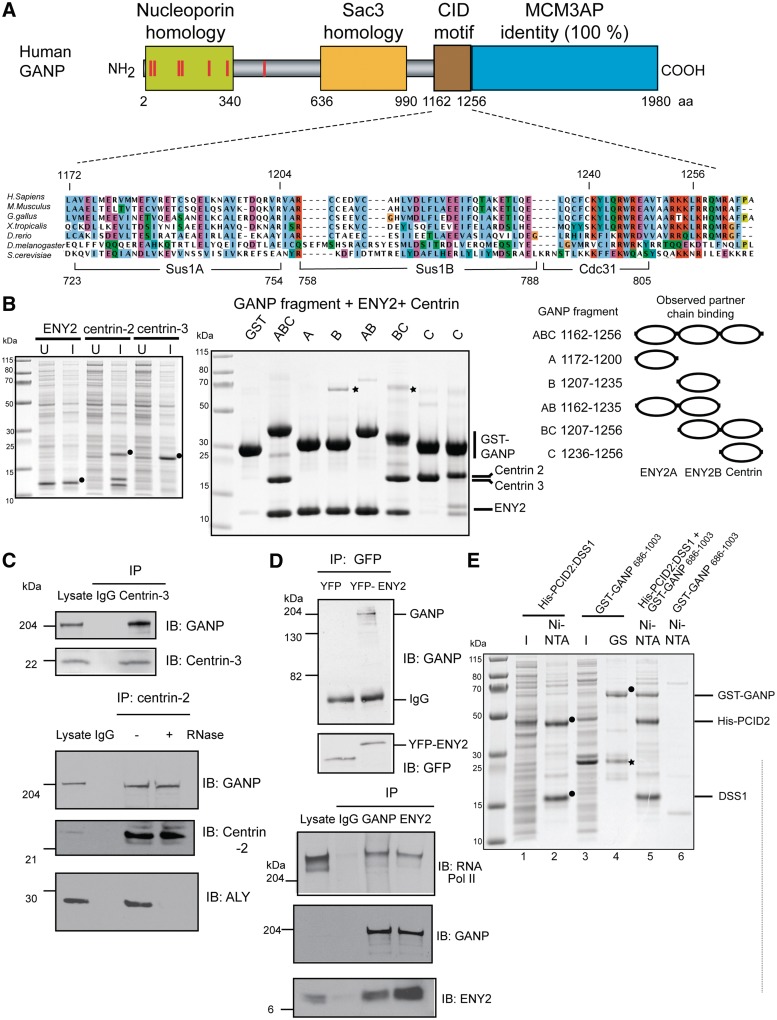
Based on the Sac3 CID (Cdc31 interaction domain) motif structure (18), secondary structure predictions and sequence conservation, we identified human GANP residues 1162–1256 as a putative region to which ENY2 and centrins [homologues of Sus1 (10,11) and Cdc31 (7), respectively] might bind (Figure 1A). In humans, multiple centrins have been identified [reviewed in Ref. (27)]. Centrin-1 has tissue-specific expression, and so we focused on centrin-2 and centrin-3 as they are expressed ubiquitously, located in the nucleus and shared 53% sequence identity (27).
Figure 1.
ENY2 and centrin-2 or centrin-3 interact with GANP in vitro and in vivo and PCID2 and DSS1 interact with GANP in vitro. (A) Sequence alignment of the Sac3 CID region showing the conservation of binding sites for Sus1A, Sus1B and Cdc31 (yeast) in orthologues from other species, including human GANP. The numbers below the alignment delimit boundaries for binding sites in yeast and those above are for GANP. (B) Clarified lysates from bacterial cells expressing untagged ENY2, centrin 2 or centrin 3 are shown with the over-expressed protein marked with a black circle. U, un-induced; I, induced. The expression vector for ENY2 shows leaky expression, and so over-expressed protein is evident in the un-induced sample. GST and GST-GANP fragments (containing predicted ENY2 and centrin-binding sites) were co-expressed with either ENY2 (in the case of fragments containing only putative ENY2-binding sites) or centrin (in the case of fragments containing the putative centrin-binding site) and their clarified lysates mixed with clarified lysate-containing centrin or ENY2, respectively. Centrin 3 was used for binding in lanes 1–7 and centrin 2 in lane 8. GST fusion protein-containing complexes were immobilized on glutathione sepharose, washed and analysed by SDS–PAGE and Coomassie staining. Impurities are marked by asterisks. A schematic representation of the GANP fragments used (black bar) depicting their residue numbers and the partner chains (ovals) they would be predicted to bind is also shown. (C) Endogenous centrin-2 and -3 were immunoprecipitated from nuclear extract of HCT116 cells and blotted for GANP, centrin-2 and centrin-3. Centrin-2 was also immunoprecipitated from RNase-treated nuclear extract and blotted for GANP, centrin-2 and ALY. The RNase treatment was effective as centrin-2 association with ALY was abolished following RNase treatment. (D) YFP or YFP-ENY2 was immunoprecipitated with GFP antibody from nuclear extracts of HCT116 cells expressing YFP or YFP-ENY2 and blotted for GFP and GANP. Endogenous ENY2 interacts with both GANP and C-terminal domain of RNA polymerase II. Endogenous GANP and ENY2 were immunoprecipitated from nuclear extract of HCT116 cells and blotted for GANP, ENY2 and RNA polymerase II. (E) Clarified lysates from bacterial cells expressing His-PCID2:DSS1 complex and GST-GANP 686-1003 were applied to Ni-NTA and glutathione sepharose (GS) resins, respectively, and showed that although protein expression levels were low such that over-expressed bands could not be observed in the lysate (I, induced), the appropriate proteins (black circles) could be enriched on the affinity matrices (lanes 1–4). When clarified lysates containing GST-GANP 686-1003 and His-PCID2:DSS1 were mixed, a ternary complex was formed and could be pulled down on Ni-NTA (lane 5). Importantly, the GST-GANP 686-1003 fusion did not have an affinity for Ni-NTA in the absence of His-PCID2:DSS1 (lane 6). Impurities are marked by asterisks.
GST pulldown experiments confirmed that GANP residues 1162–1256 bound ENY2 and either centrin-2 or centrin-3 in vitro (Figure 1B). To explore the arrangement of the ENY2 and centrin-binding sites within the GANP CID, fragments of GANP encompassing predicted binding sites were tested for binding to ENY2 and centrin-2 or 3 (Figure 1B). The results indicate that the GANP CID domain contains two different sites for ENY2-binding (GANP residues 1162–1200 and 1207–1235) together with a single centrin-binding site (GANP residues 1236–1256).
GANP also interacts with both centrins in vivo as co-immunoprecipitation studies using nuclear extracts from HCT116 colon carcinoma cells and specific antibodies against either centrin-2 or centrin-3 (Supplementary Figure S1) showed that endogenous GANP bound centrin-2 or centrin-3 in vivo (Figure 1C). Importantly, the interaction between GANP and centrin-2 was insensitive to RNase treatment (Figure 1C). The interaction of ENY2 with GANP was confirmed in vivo (Figure 1D) using a fragment generated by the fusion of YFP to full-length ENY2 that bound to endogenous GANP, whereas YFP alone did not (Figure 1D). Additionally, co-immunoprecipitation studies using specific antibodies against either ENY2 or GANP showed that endogenous ENY2 bound GANP in vivo and endogenous GANP also bound ENY2 (Figure 1D).
In yeast, the complete TREX-2 complex contains Thp1 and Sem1 in addition to Sac3, Sus1 and Cdc31 (7,8,11,12). Human TREX-2 has a similar composition and GANP (Sac3 homologue), in addition to binding ENY2 (Sus1 homologue) and centrin-2 (Cdc31 homologue), also binds the human Thp1 homologue PCID2 and Sem1 homologue Dss1 (Figure 1E). Thus, GST pulldown experiments showed that GANP residues 686–1003 (encompassing the conserved Sac3 homology domain) bound PCID2 and DSS1 (Figure 1E). Taken together, these results suggest that both the CID motif and PCID2/Thp1 and DSS1/Sem1 binding is conserved from yeast to human cells.
Crystal structure of the GANP:ENY2 complex
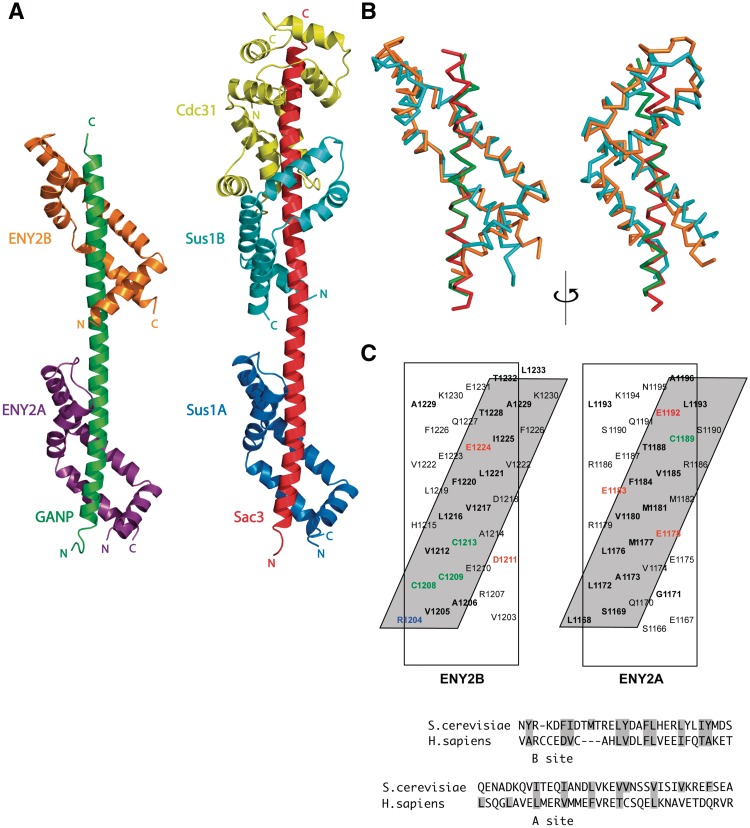
The structural basis of the interactions between GANP and ENY2 was determined using X-ray crystallography. Despite exhaustive trials, we were unable to obtain crystals of a complex containing GANP, ENY2 and centrin-2 or -3; however, we did obtain crystals of GANP residues 1162–1235 complexed with ENY2 that had P2 1 symmetry and diffracted to 2.1 Å resolution. Se MAD data was used to obtain phases, and the preliminary atomic model obtained was refined against the 2.1 Å native data to yield a final structure that had an R factor of 25.7% (_R_free = 22.0%) with excellent geometry (Table 1) and contained GANP residues 1162–1234 and two ENY2 chains in which there was electron density for residues 7–100. Figure 2A illustrates the structure in which GANP forms a 9.6-nm gently undulating α-helix to which two chains of ENY2 bind (ENY2A and ENY2B bind to GANP residues 1168–1196 and 1204–1232, respectively).
Figure 2.
Structure of the GANP:ENY2 complex. (A) Overview of the structure of the GANP:ENY2 complex (GANP residues 1162–1233 in green, ENY2A in purple and ENY2B in orange) and comparison to the yeast Sac3 CID complex (PDB 3FWC, (18), Sac3 in red, Sus1A in dark blue, Sus1B in cyan and Cdc31 in yellow). (B) Ribbon representation of ENY2B (orange) and GANP residues 1201–1233 (green) superimposed to Sus1B (cyan) and Sac3 residues 752–788 (red) [PDB 3FWB (18)]. The Cα backbones of Sac3 and GANP have a root-mean-square deviation of 0.39 Å, whereas the Cα backbones of Sus1B and ENY2B have a root-mean-square deviation of 2.10 Å. (C) The two hydrophobic stripes in the ENY2A and ENY2B-binding sites of the GANP helix. The GANP sequence is shown on a helical net, with residues that make a major contribution to the interface shown in bold. Residues forming putative salt bridges or H bonds are shown in red (for acidic) or blue (for basic). Cysteine hydrogen bonds are shown in green. The approximately four-residue repeat sequence that forms the hydrophobic stripes in GANP is broadly the same found in yeast Sac3.
The ENY2 fold resembled that of its yeast orthologue, Sus1 (18) (Figure 2A), but the higher resolution obtained in the present study enabled structural features to be defined more confidently. The ENY2 fold was based on five α-helices separated by putative hinge regions that were arranged into an anti-parallel hairpin allowing ENY2 to wrap around the GANP helix (Figure 2A–C). Both ENY2-binding sites on GANP gave rise to a hydrophobic stripe that wound around the GANP helix (Figure 2C) and contained putative salt bridge-forming residues and cysteine residues that form putative hydrogen bonds with ENY2.
Notably, the crystal structure shows that the orientation of the two ENY2 molecules relative to one another was significantly different from the orientation of the two Sus1 molecules observed in the yeast Sac3 CID complex. In the GANP complex, the two ENY2 molecules have approximately equivalent azimuthal orientations (Figure 2A and B), whereas in the Sac3 complex the two Sus1 molecules are rotated by ∼90° azimuthally (Figure 2A and B). This rotation comes about, because there is an additional residue in the alpha helix between the two ENY2 sites and GANP compared to the corresponding region in Sac3. As a result of this rotation, the exposure of the GANP helix in the complex is different from that seen for Sac3. In the Sac3 CID complex, a continuous face of the Sac3 helix is exposed (Figure 2A) as it traverses from Sus1A to Sus1B, whereas there is not a corresponding exposed face in the GANP complex. Instead, two opposing faces of the GANP helix are exposed (Figure 2A). Consequently, the Sus1/ENY2-binding regions of Sac3 and GANP have quite different surface topologies. Moreover, the different spatial positioning of the two ENY2 chains would prevent them from both binding to a single target molecule in the same way as the two Sus1 chains bound to Sac3 (Figure 2A).
Human TREX-2 components function in mRNA export
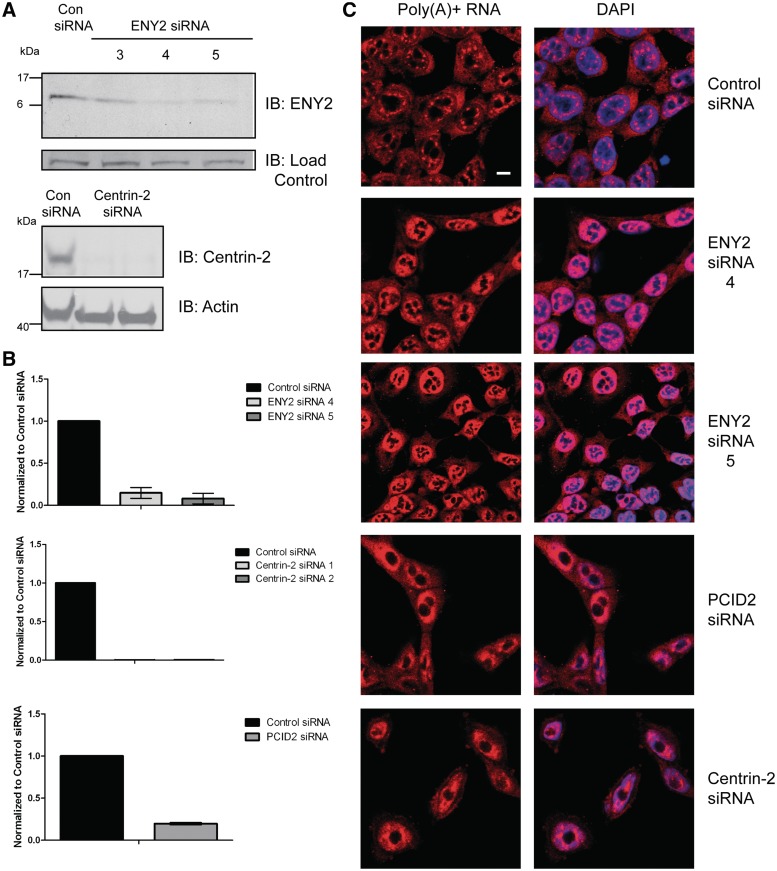
Previously, GANP was shown to function in mammalian mRNA export (6). To test if its binding partners, ENY2 and centrin-2 also, are necessary for mRNA export in human cells, we depleted both using siRNA and assayed poly(A)+ RNA localization by RNA FISH. Two individual siRNAs (siENY2-4 and -5) against ENY2 and against centrin-2 significantly decreased the levels of RNA and protein for ENY2 and centrin-2, respectively (Figure 3A and B). Whereas most poly(A) + RNA was cytoplasmic in control cells, except for a few discrete foci in nuclei, ENY2 or centrin-2 depletion resulted in nuclear accumulation of poly(A)+ RNA (Figure 3C). Depletion of the human homologue of Thp1, PCID2 (Figure 3B), an additional component of the yeast TREX-2 complex (8,28) also resulted in a nuclear accumulation of poly(A)+ RNA (Figure 3C). Therefore, these results indicate that GANP, PCID2, ENY2 and centrin-2/3 all function in mRNA export in human cells, consistent with their forming a putative TREX-2 complex analogous to that present in yeast. Indeed, expression of ENY2 in yeast cells could partially rescue mutant phenotypes resulting from a deletion of Sus1, including growth at elevated temperature and synthetic lethality when combined with mutations in yeast Mex67 (NXF1 in humans) (Supplementary Figure S3).
Figure 3.
Depletion of TREX-2 components results in nuclear accumulation of poly(A) + RNA. (A) Immunoblotting analysis of HCT116 cells depleted of ENY2 (using two independent siRNAs) or centrin-2. Samples were analysed by Western blotting with the indicated antibodies 72-h post-transfection. (B) Depletion of ENY2, centrin-2 and PCID2 was also confirmed by qRT-PCR using ENY2, centrin-2 and PCID2-specific primers 72-h post-transfection and represent the mean of triplicate readings from three independent depletion experiments ± SD. (C) FISH showing nuclear accumulation of poly(A) + RNA in either ENY2, PCID2 or centrin-2 depleted cells. ENY2-depleted cells were assayed 96-h post-transfection, whereas PCID2 and centrin-2-depleted cells were assayed 72-h post-transfection. Merged image is shown in right panels (Scale bar, 5 µm). Poly(A) + RNA was identified using a Cy3-labelled oligo(dT) probe. Nuclei were stained with DAPI.
ENY2 co-localizes with GANP but is not required for association with NPCs
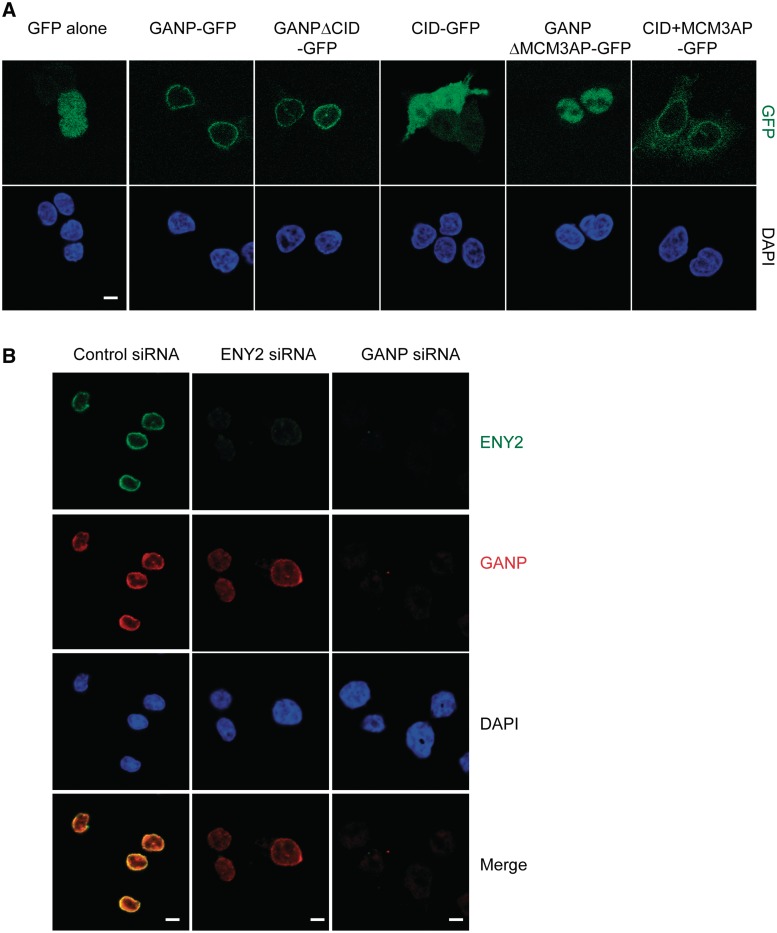
Previously, GANP was shown to localize to both the nuclear envelope and nuclear interior (6). To map the region of GANP necessary for NPC localization, we expressed GANP fragments fused to GFP in HCT116 cells and observed their cellular location by fluorescence microscopy (Figure 4A). We first analysed the location of GANPΔCID-GFP, because, in yeast, the Sac3-CID region was shown to function in the localization of TREX-2 to NPCs (7,18). Surprisingly, GANPΔCID-GFP remained localized to the nuclear envelope, suggesting that the CID domain is not necessary for the nuclear envelope localization of GANP. Indeed, a construct containing only the CID domain fused to GFP localized throughout the cell, in both the cytoplasm and nucleus, with no obvious nuclear envelope staining (Figure 4A). In GANP, the CID domain lies immediately upstream of the MCM3AP domain, which is not readily apparent in Sac3 (Supplementary Figure S1). The MCM3AP protein is an inhibitor of DNA replication initiation (29,30) and can be transcribed independently of GANP even though the nucleotide sequence for MCM3AP is completely contained within the 3′ region of the sequence of GANP (31). Interestingly, GANPΔMCM3AP-GFP did not localize to the nuclear envelope, but was instead distributed throughout the nucleoplasm (Figure 4A), suggesting that the MCM3AP domain is necessary for the NPC localization of GANP. However, overexpressed MCM3AP localizes to the cytoplasm, and overexpression of MCM3 is required for the nuclear import of MCM3AP (29). This suggests that both the CID domain and the MCM3AP domain of GANP may be contributing to its NPC localization. Consistent with this hypothesis, a GFP fusion containing both the CID region and the MCM3AP region (CID + MCM3AP-GFP) was localized primarily to the nuclear envelope with weaker cytoplasmic staining (Figure 4A). When cells were permeabilized before fixation, CID + MCM3AP-GFP displayed strong nuclear envelope staining (Supplementary Figure S4). We conclude that the CID domain by itself is not sufficient for the nuclear envelope localization of GANP, but that the CID and MCM3AP domains together are necessary and sufficient for localizing GANP to the nuclear envelope.
Figure 4.
CID and MCM3AP domains of GANP but not ENY2 are required for its NPC association. (A) CID and MCM3AP domains of GANP are required for its NPC association. Various constructs of GANP fused to GFP were transfected into HCT116 cells and localization examined by epifluorescence. GANP-GFP and GANPΔCID-GFP localize to NPCs, but GANPΔMCM3AP-GFP is nuclear. CID + MCM3AP-GFP localizes to NPCs. Nuclei were stained with DAPI (scale bar, 5 µm). (B) GANP is required for ENY2 association with NPCs. In HCT116 cells permeabilized before fixation, GANP and ENY2 co-localize at the NPC. Following depletion of ENY2, GANP NPC staining is retained, but ENY2 staining is abolished. In contrast, GANP depletion results in loss of both GANP and ENY2 staining from NPCs.
We next determined the cellular localization of ENY2 and its role in GANP localization. Immunofluorescence of permeabilized HCT116 cells showed strong nuclear envelope staining of ENY2 that was abolished following siRNA-mediated depletion of ENY2 (Figure 4B). This staining co-localized with endogenous GANP staining. However, following depletion of ENY2, GANP nuclear envelope staining was still observed (Figure 4B). In contrast, depletion of GANP abolished both GANP and ENY2 nuclear envelope staining (Figure 4B). Additionally, depletion of PCID2, a component of the human TREX-2 complex, had no effect on the localization of GANP (Figure 5G). Thus, the localization of ENY2 to NPCs is dependent on GANP, whereas the localization of GANP to NPCs is independent of ENY2 and PCID2.
Figure 5.
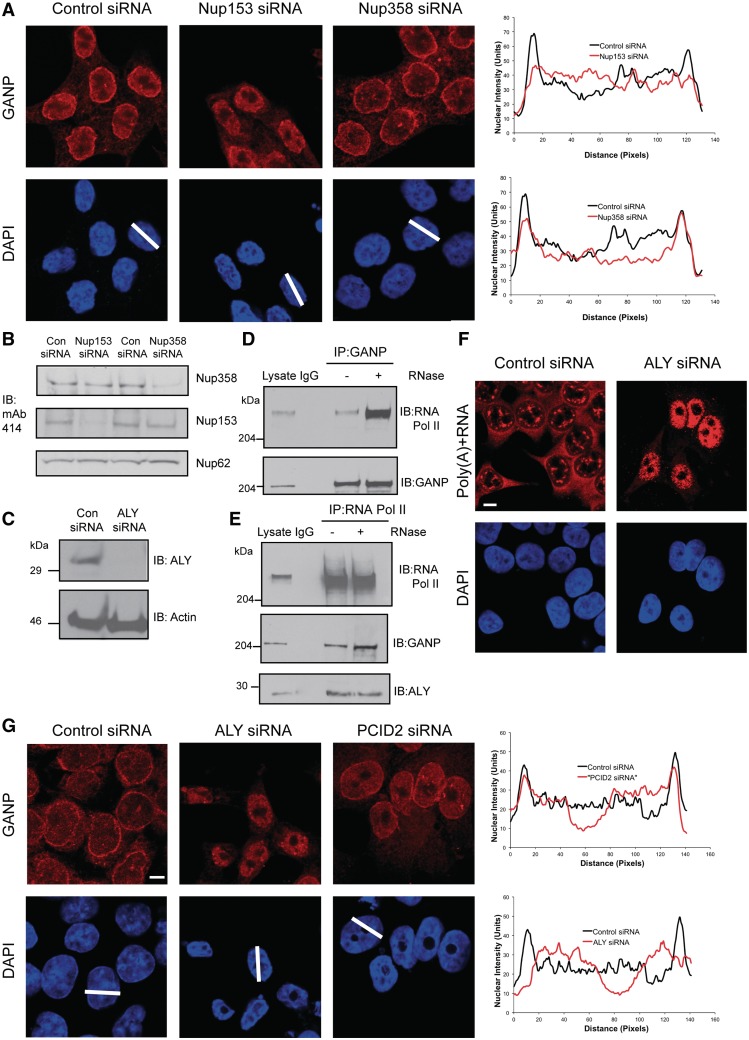
GANP localization is dependent on the early-stage mRNA export factor ALY and Nup153; GANP and ENY2 associate with RNA Pol II. (A) Depletion of Nup153, but not Nup358 redistributes GANP into nuclear foci with less at NPCs. Scanning analysis of GANP intensity in control siRNA treated or Nup153- or Nup358-depleted cells is shown. Nuclei used for scanning and the scanning axis are indicated by white lines. Pairs of nuclei of same scan width as determined by DAPI staining were used for scanning (scale bar, 5 µm). (B) Efficiency of depletion of Nup153 and Nup358 was determined by Western blotting with mAb414 antibody, which recognizes FG-repeat containing nucleoporins. (C) Efficiency of depletion of ALY was monitored by immunoblotting with the indicated antibodies. (D) The interaction between endogenous GANP and the C-terminal domain of RNA polymerase II is increased following RNase treatment. Endogenous GANP was immunoprecipitated from untreated and RNase treated nuclear extract of HCT116 cells and blotted for GANP and RNA polymerase II. (E) Endogenous RNA polymerase II was also immunoprecipitated from untreated and RNase-treated nuclear extract of HCT116 cells and blotted for GANP and RNA polymerase II. (F) Depletion of ALY results in nuclear accumulation of poly(A) + RNA. FISH showing nuclear accumulation of poly(A) + RNA in ALY-depleted cells 72-h post-transfection (scale bar, 5 µm). (G) Depletion of ALY redistributes GANP into nuclear foci with less at NPCs (scale bar, 5 µm). Scanning analysis of GANP intensity in control siRNA treated or ALY- or PCID2-depleted cells is shown and analysed as earlier.
GANP localization is dependent on the early-stage mRNA export factor ALY and nucleoporin Nup153
GANP was shown by co-immunoprecipitation to associate with Nup153 and Nup358, nucleoporins on the nuclear and cytoplasmic faces of the NPC, respectively (6). To further explore these interactions, we depleted Nup153 and Nup358 by siRNA. Interestingly, depletion of Nup153 (Figure 5B) redistributed GANP into nuclear foci, with less at the NPC (Figure 5A). In contrast, depletion of Nup358 had no effect on the localization of GANP (Figure 5A). This change in GANP localization following Nup153 depletion suggests that GANP may mediate transport of mRNPs from sites of transcription to NPCs for efficient mRNA export. In support of this hypothesis, GANP and ENY2 both co-immunoprecipitate with endogenous RNA polymerase II (Figure 1D and Supplementary Figure S5). We also used RNase digestion to demonstrate that these interactions were direct and not mediated by both proteins binding to a single mRNA chain. Thus, whereas RNase treatment resulted in ALY and centrin-2 (two proteins that do not interact directly) no longer being associated with each other (Figure 1C), instead, we observed an increase in the amount of RNA polymerase II co-immunoprecipitated with the GANP antibody from RNase-treated nuclear extract, even though an equivalent amount of GANP was immunoprecipitated from untreated nuclear extract (Figure 5D). A similar increase in the amount of GANP co-immunoprecipitated with the RNA polymerase II antibody following RNase treatment was also seen (Figure 5E). This suggests that TREX-2 may be associated with transcription. The TREX component ALY is recruited to mRNP complexes generated by splicing (32), and the TREX complex is recruited to the 5′-end of mRNA (33). Thus, recruitment of the TREX complex to sites of transcription during mRNP maturation is thought to be one of the earliest steps in mRNA export (34). Interestingly, inhibition of mRNA processing via depletion of the upstream mRNA export factor, ALY (Figure 5C), altered the localization of GANP and resulted in retention of mRNPs and GANP in punctate nuclear foci (Figure 5F and G), with a reduced level of GANP staining observed at the NPC. Taken together, our results implicate TREX-2 in the recruitment and transport of mRNPs from the nuclear interior to NPCs.
DISCUSSION
Our results have demonstrated that in mammalian cells, GANP, ENY2 and centrins form a complex that links transcription with nuclear mRNA export. Although an analogous complex is present in yeast, their function in the gene expression pathway differs in several important respects and underlies the differences that are inherent in these pathways between yeast and metazoans. Thus, although metazoans possess all the orthologous proteins corresponding to those found and participating in gene gating in yeast, this phenomenon has not been observed directly in metazoans. Instead, the majority of active genes in mammalian cells lie in transcription foci or ‘factories’ deep within the nucleus (15). Our findings implicate TREX-2 as a crucial factor linking these mRNP export complexes in the nuclear interior to NPCs.
Our immunofluorescence and co-immunoprecipitation data suggest that a proportion of TREX-2 is localized at or near sites of transcription mediated by RNA polymerase II deep in the nuclear interior of mammalian cells. Indeed, depletion of the mRNA export adaptor ALY resulted in the retention of mRNPs and GANP in punctate nuclear foci (Figure 5). Therefore, when proper mRNP assembly is inhibited, GANP's distribution is altered from predominantly NPC bound to predominantly in nuclear foci, suggesting that mammalian TREX-2 functions in a step downstream of transcription and splicing. These findings also indicate that there is a degree of co-operation between the TREX and TREX-2 complexes in mammalian cells. In mammals, TREX is recruited to mRNPs in a 5′ cap-dependent and splicing-dependent manner (3,9,33). The loss of the THO complex and UAP56 coupled with recruitment of the mRNA exporter NXF1 by ALY signals maturation of the mRNP particle to a transport competent state (3,9). We propose that it is at this stage of the gene expression pathway that the TREX-2 complex becomes attached to the mRNP particle through the interaction of the GANP component (residues 1–313; nucleoporin homology domain) with NXF1 (6) and that this attachment facilitates efficient delivery of mRNPs to NPCs.
We propose that upon reaching NPCs, GANP becomes attached via nucleoporin Nup153. In yeast, the Sac3 CID region and associated chains target Sac3 to NPCs. The different surface topologies of the Sac3 and GANP CID regions (brought about by the difference in the azimuthal orientation of the two ENY2/Sus1 chains on GANP/Sac3) suggest that these regions of GANP and Sac3 may differ in the way in which they interact with other components of the gene expression machinery. Additionally, there are likely differences in the way in which centrins bind GANP and Sac3. The crystal structure of a peptide from Xeroderma pigmentosum Group C protein bound to centrin-2 (35) suggests how centrin-2 might bind to the GANP CID (Supplementary Figure S2A). Consistent with the differences in topology of the GANP and Sac3 CID regions, we observe that the GANP CID region needs to be augmented by the MCM3AP domain to elicit NPC-binding activity (Figure 4), in contrast to yeast in which the Sac3 CID domain is sufficient for localization and in which Sac3 lacks an MCM3AP domain.
Our findings have implications for the potential role of mRNA export factors in cancer progression (36). GANP is upregulated in a variety of lymphomas (37) raising the important question of whether mRNA export derangements contribute to the development of cancer. Intriguingly, two recent studies have implicated components of TREX-2 in both embryonic stem cell specification in mammals (38) and in replication fork progression and stability during transcription in yeast (39). The structural and functional characterization of TREX-2 in mammalian cells thus provides a platform to understand its potential role in these processes in humans.
ACCESSION NUMBER
The coordinates and structure factors for the crystal structure of the GANP:ENY2 complex have been deposited in the Protein Data Bank under accession code 4DHX.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Medical Research Council (MRC) and Cancer Research UK; Wellcome Trust and Medical Research Council grant (U105178939 to M.S.) in part. Funding for open access charge: University of Cambridge, Medical Research Council, Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
V.O.W. acknowledges the support of Ashok Venkitaraman during the latter half of this study. S.L. was a recipient of an EMBO short-term fellowship to study in the lab of RAL.
REFERENCES
- 1.Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat. Rev. Mol. Cell Biol. 2011;12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Navarro S, Hurt E. Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 2011;23:302–309. doi: 10.1016/j.ceb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Stewart M. Nuclear export of mRNA. Trends Biochem. Sci. 2010;35:609–617. doi: 10.1016/j.tibs.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Carmody SR, Wente SR. mRNA nuclear export at a glance. J. Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickramasinghe VO, McMurtrie PI, Mills AD, Takei Y, Penrhyn-Lowe S, Amagase Y, Main S, Marr J, Stewart M, Laskey RA. mRNA export from mammalian cell nuclei is dependent on GANP. Curr. Biol. 2010;20:25–31. doi: 10.1016/j.cub.2009.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer T, Rodriguez-Navarro S, Pereira G, Racz A, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 8.Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 10.Pascual-Garcia P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodriguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 12.Faza MB, Kemmler S, Jimeno S, Gonzalez-Aguilera C, Aguilera A, Hurt E, Panse VG. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J. Cell Biol. 2009;184:833–846. doi: 10.1083/jcb.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 14.Blobel G. Gene gating: a hypothesis. Proc. Natl Acad. Sci. USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 16.Darzacq X, Singer RH, Shav-Tal Y. Dynamics of transcription and mRNA export. Curr. Opin. Cell Biol. 2005;17:332–339. doi: 10.1016/j.ceb.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jani D, Lutz S, Marshall NJ, Fischer T, Kohler A, Ellisdon AM, Hurt E, Stewart M. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol. Cell. 2009;33:727–737. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 20.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 21.Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 22.Ellisdon AM, Jani D, Kohler A, Hurt E, Stewart M. Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1. J. Biol. Chem. 2010;285:3850–3856. doi: 10.1074/jbc.M109.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resendes KK, Rasala BA, Forbes DJ. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol. Cell Biol. 2008;28:1755–1769. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallardo M, Luna R, Erdjument-Bromage H, Tempst P, Aguilera A. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 2003;278:24225–24232. doi: 10.1074/jbc.M302900200. [DOI] [PubMed] [Google Scholar]
- 29.Takei Y, Assenberg M, Tsujimoto G, Laskey R. The MCM3 Acetylase MCM3AP Inhibits Initiation, but Not Elongation, of DNA Replication via Interaction with MCM3. J. Biol. Chem. 2002;277:43121–43125. doi: 10.1074/jbc.C200442200. [DOI] [PubMed] [Google Scholar]
- 30.Takei Y, Swietlik M, Tanoue A, Tsujimoto G, Kouzarides T, Laskey R. MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. EMBO Rep. 2001;2:119–123. doi: 10.1093/embo-reports/kve026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickramasinghe VO, McMurtrie PI, Marr J, Amagase Y, Main S, Mills AD, Laskey RA, Takei Y. MCM3AP is transcribed from a promoter within an intron of the overlapping gene for GANP. J. Mol. Biol. 2011;406:355–361. doi: 10.1016/j.jmb.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 33.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 34.Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JR, Ryan ZC, Salisbury JL, Kumar R. The structure of the human centrin 2-xeroderma pigmentosum group C protein complex. J. Biol. Chem. 2006;281:18746–18752. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- 36.Hurt JA, Silver PA. mRNA nuclear export and human disease. Dis. Model. Mech. 2008;1:103–108. doi: 10.1242/dmm.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujimura S, Xing Y, Takeya M, Yamashita Y, Ohshima K, Kuwahara K, Sakaguchi N. Increased expression of germinal center-associated nuclear protein RNA-primase is associated with lymphomagenesis. Cancer Res. 2005;65:5925–5934. doi: 10.1158/0008-5472.CAN-04-3259. [DOI] [PubMed] [Google Scholar]
- 38.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–246. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data