Metabolic regulation of epigenetics (original) (raw)
. Author manuscript; available in PMC: 2013 Jul 3.
Abstract
How cells sense and respond to environmental cues remains a central question of biological research. Recent evidence suggests that DNA transcription is regulated by chromatin organization. However, the mechanism for relaying the cytoplasmic signaling to chromatin remodeling remains incompletely understood. Although much emphasis has been put on delineating transcriptional output of growth factor/hormonal signaling pathways, accumulated evidence from yeast and mammalian systems suggest that metabolic signals also play critical roles in determining chromatin structure. Here we summarize recent progress in understanding the molecular connection between metabolism and epigenetic modifications of chromatin implicated in a variety of diseases including cancer.
Introduction
Cells, whether as unicellular organisms or within a multicellular organism, need to make various “decisions” throughout their lifetime. Every choice – quiescence, proliferation, differentiation or migration, is made in the face of the constantly changing environment. Metazoan cells have developed elegant mechanisms to sense and integrate extracellular information into an intrinsic signaling system that regulates transcription so that variations in the form of growth factors, hormones and stromal interactions can be delivered and responded to in a timely and accurate manner. Studying these mechanisms remains a major focus of biological research and has profound implications in understanding human physiology and diseases.
One of the most exciting advances over the past 15 years is the field of epigenetics (meaning “above” genetics). We now know that in addition to primary DNA sequence information, much of the information regarding when and where to initiate transcription is stored in covalent modifications of DNA and its associated proteins. The patterns of various modifications along the chromatin, such as DNA cytosine methylation and hydroxymethylation, and acetylation, methylation, phosphorylation, ubiquitination and SUMOylation of the lysine (K) and/or arginine (R) residues of histones are thought to determine the genome accessibility to transcriptional machinery. For example, acetylation of histone lysine residues and methylation of H3K4, H3K36 and H3K79 are associated with active transcription while methylation of DNA, H3K9, H3K27 and H4K20 generally indicate silenced chromatin. In many instances, such information is heritable. The complexity and dynamics of epigenetic modifications are considered to provide a link between the extracellular environment and nuclear transcription. It is increasingly apparent that many growth factor/hormone-responsive signaling pathways such as Notch and TGFβ, often in conjunction with downstream transcription factors, can remodel the epigenome through expressing, recruiting or editing enzymes that modify chromatin (Mohammad and Baylin, 2010). One good example is the Notch effector RBP-J: depending on the presence of activated Notch, RBP-J recruits distinct histone-modifying enzymes to activate or repress target gene expression (Liefke et al., 2010).
A less studied but recently emerged concept is that information about a cell’s metabolic state is also integrated into the regulation of epigenetics and transcription (Figure 1). It is now appreciated that cells constantly adjust their metabolic state in response to extracellular signaling and/or nutrient availability (Vander Heiden et al., 2009). As a classical example, while quiescent cells fully oxidize glucose to carbon dioxide in the mitochondrial electron transport chain; proliferative cells and tumor cells consume much larger quantities of glucose, secreting excess carbon as lactate even when oxygen is abundant, a process termed “aerobic glycolysis”. Connections between metabolism and transcription are not unexpected. In unicellular organisms like yeast the major determinant of cell fate is nutrient levels. Even in metazoans where most cellular signaling events are dictated by growth factors, cytokines or hormones, metabolism still plays a significant role in transcription. This also has a potentially unifying logic, as most chromatin-modifying enzymes require substrates or co-factors that are intermediates of cell metabolism. It is not difficult to imagine that fluctuation of metabolite levels could modulate the activities of chromatin-modifying enzymes and therefore influence chromatin dynamics. As many complex diseases such as cancer and type II diabetes display abnormalities of cellular metabolism and epigenome, understanding the molecular connections between these processes may have therapeutic implications.
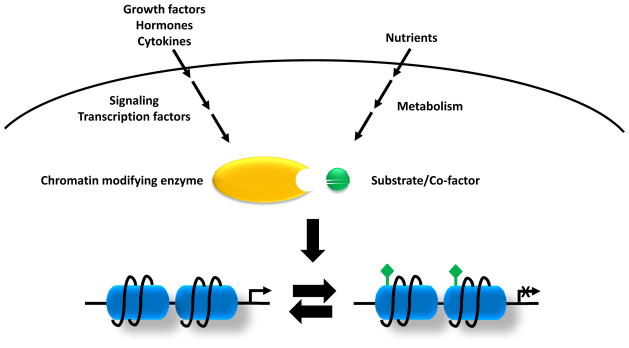
Figure 1. Signaling and metabolic inputs into epigenetics.
Growth factors, hormones and cytokines activate the classical signaling pathways and downstream transcriptional factors which will recruit chromatin-modifying enzymes to local chromatin. On the other hand, nutrient levels and cell metabolism will affect levels of the metabolites which are required substrates of chromatin-modifying enzymes that use these metabolites to post-translationally modify both histones and DNA. Variations in these two inputs will determine the epigenome remodeling and transcription.
In this perspective, we will review findings from studies that examine metabolic inputs into epigenetics and transcription in both yeast and metazoan organisms. We discuss possible mechanisms of how metabolism can influence chromatin states, and review recent efforts linking metabolic perturbation to the dysregulation of cellular differentiation.
Metabolites drive oscillatory gene expression during yeast metabolic cycle
Most laboratory strains of budding yeast Saccharomyces cerevisiae are cultured in media with abundant nutrient supply. As unicellular organisms, yeast under such conditions uptake extracellular nutrients avidly and exploit nutrient capture to fuel cell growth and proliferation at a logarithmic rate. However, under conditions where one or more nutrients are limiting, yeast tend to display oscillatory metabolic behavior with distinct phases characterized by robust changes in oxygen consumption (Chance et al., 1964). These metabolic oscillations can vary from minutes to hours depending on yeast strains and experimental conditions and are independent of cell division cycle (Slavov et al., 2011). Remarkably, when genome-wide gene expression in yeast under such culture system was analyzed, nearly 60% of the genes were expressed in a similarly cyclic fashion (Klevecz et al., 2004; Tu et al., 2005).
These data suggest that metabolite levels might be contributing to the temporal control of gene expression. This is supported by the observations that transient administration of metabolites, particularly the glycolytic intermediates, could initiate phase advancement in the oscillatory cycle (Cai et al., 2011; Tu and McKnight, 2009). Levels of acetyl-CoA, the product of glycolytic metabolism and a donor to acetylation reactions, were found to oscillate dynamically and peak during the oscillatory growth phase (Cai et al., 2011; Tu et al., 2007). Genome-wide analysis of H3K9 acetylation showed that its enrichment at genes expressed during growth phase correlated with acetyl-CoA levels and was dependent on the generation of acetyl-CoA (Cai et al., 2011). Therefore it appears that levels of acetyl-CoA can be instructive for cells to engage growth at least in part through modulating histone acetylation and transcription of growth genes. Such a mechanism may well operate at other times. For example, the exit of yeast from quiescence has been reported to require glucose catabolism but not cell cycle signaling (Laporte et al., 2011). Whether metabolites in addition to acetyl-CoA contribute to these regulatory events remains to be investigated.
Potential link between metabolism and transcriptional regulation by the circadian system
Unlike yeast, metazoan cells lack the autonomous ability to uptake and metabolize nutrients. Instead cellular nutrient uptake is under the control of a mixture of growth factor, paracrine and hormonal signaling. These factors control the positions that differentiated cells can survive in the body and dictate the share of the body’s resources a cell can utilize in maintaining itself. In addition, the metabolism of individual cells must be entrained to the activities of the organism as a whole. One way this is accomplished is through the circadian system, which has evolved to allow organisms to adapt to the 24 h day-night cycles on Earth. The circadian system regulates many aspects of animal behavior and physiology. It consists of a central clock, which is located in the hypothalamic suprachiasmatic nucleus and responsive to light; and peripheral clocks, which are present in many non-light sensitive organs. The molecular mechanism for the circadian system includes a number of transcriptional and post-translational feed-back loops. In brief, CLOCK and BMAL1, central transcriptional factors of the circadian system, regulate the expression of cryptochrome (CRY1 and CRY2) and period (PER1, PER2 and PER3). CRY and PER proteins in turn negatively regulate CLOCK-BMAL1 complex. Before a new cycle begins, CRY and PER proteins are degraded which de-represses the activities of CLOCK-BMAL1 complex (Figure 2).
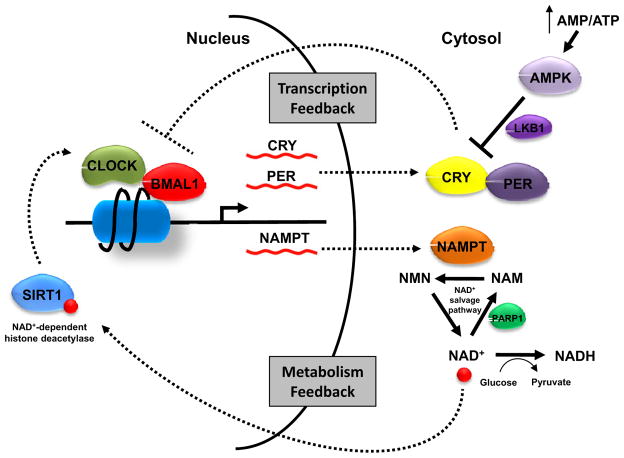
Figure 2. Metabolic feedback loop of the circadian clock.
As master regulators of the circadian clock, CLOCK and BMAL1 control the expression of CRY and PER. CRY and PER proteins in turn regulate CLOCK-BMAL1 complex activity. Recently SIRT1 was found to bind to CLOCK-BMAL1 complex and negatively regulate its target gene transcription by deacetylating histones. SIRT1 activity depends on NAD+. Increased glucose metabolism converts nuclear/cytoplasmic NAD+ to NADH and suppresses SIRT1. Another NAD+-dependent enzyme is PARP1 which catalyzes the polymerization of ADP-ribose units on proteins and generates nicotinamide. The generation of additional cytosolic NAD+ requires the rate-limiting enzyme NAMPT. The cyclic expression of NAMPT is under the control of CLOCK-BMAL1, therefore contributing to a metabolic feedback loop. The energy-responsive kinase AMPK can also affect molecular clock machinery by phosphorylating CRY1 and facilitating its degradation. AMPK also phosphorylates histones, though its implication on the circadian system remains unclear.
While light is the regulator of the central clock, there is compelling evidence linking peripheral clocks to metabolism. Bmal1−/− and Clockmut mice exhibit various abnormalities of glucose and lipid homeostasis (Oishi et al., 2006; Rudic et al., 2004; Turek et al., 2005). On the other hand, temporally restricted feeding has been shown to be a powerful entraining stimulus in several organisms (Damiola et al., 2000; Xu et al., 2011a). Moreover, high-fat diet could alter animals’ daily rhythm and the cycling of circadian clock genes (Kohsaka et al., 2007). In cell culture, glucose was demonstrated to modulate the circadian rhythm (Hirota et al., 2002). In humans, it has been shown that circadian misalignment could cause aberrant glucose response indicative of a pre-diabetic state (Scheer et al., 2009).
How peripheral clocks sense the metabolic state remains incompletely understood. Several reports suggested NAD+/NADH as a central player between metabolism and circadian system (Figure 2). Two NAD+-dependent proteins, SIRT1 deacetylase and PARP1 ADP-ribosyltransferase, were shown to bind to CLOCK-BMAL1 complex and affect its DNA binding activity and target gene expression (Asher et al., 2008; Asher et al., 2010; Nakahata et al., 2008). Part of SIRT1’s effect on circadian gene transcription could be attributed to its deacetylase activity towards histones at the promoters of circadian genes (Nakahata et al., 2008). The inhibition of nicotinamide phosphoribosyltransferase (NAMPT), a key enzyme in the NAD+ salvage pathway, promotes the oscillation of CLOCK-BMAL1 target genes, demonstrating a critical role of NAD+ in the functioning of circadian clock (Nakahata et al., 2009; Ramsey et al., 2009). Interestingly, NAD+ levels also oscillate in a circadian clock-dependent fashion and NAMPT is a direct transcriptional target of the CLOCK-BMAL1 complex, thus completing a metabolic feedback loop of the circadian system.
In addition, fluctuations of the cellular metabolic state can be sensed by signaling proteins which directly or indirectly modulate the molecular clock machinery. For example, the energy-responsive AMP-activated protein kinase (AMPK) phosphorylates and facilitates the degradation of CRY1 (Lamia et al., 2009). In mice and cells where AMPK pathway is deficient, the peripheral circadian clock is disrupted (Lamia et al., 2009; Um et al., 2011). Other nutrient sensors such as mTOR and AKT were also shown to link metabolic signals to the circadian system (Zheng and Sehgal, 2010). Both AMPK and AKT contribute to the regulation of histone phosphorylation (see below), suggesting that chromatin remodeling represents part of the underlying mechanism.
Metabolic regulation of epigenetics
Over the past 15 years, various enzymes responsible for adding or removing epigenetic modifications have been identified and functionally characterized. These enzymes include but are not limited to DNA methyltransferases (DNMTs), DNA hydroxylases (DNHDs), histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), and histone demethylases (HDMs). As discussed above, the activities of many chromatin-modifying enzymes are regulated in part by the concentrations of their required metabolic substrates or co-factors. Such substrates or co-factors are capable of diffusing through nuclear pores, therefore providing a way for the cell to deliver metabolic information to nuclear transcription (Figure. 3).
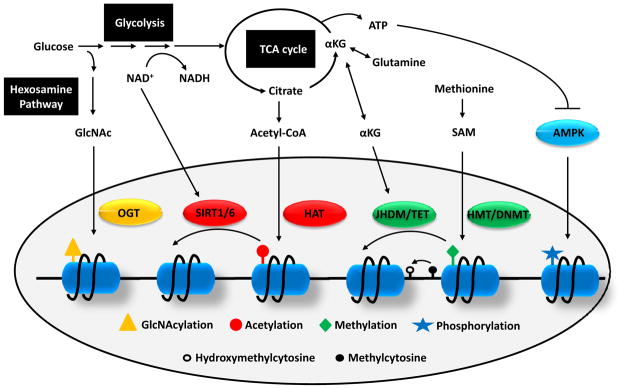
Figure 3. Crosstalk between metabolism and epigenetics.
As glucose enters the glycolytic pathway, a minor portion is branched to hexosamine biosynthetic pathway to produce GlcNAc which can be used as substrate for histone GlcNAcylation by OGT. Flux through glycolysis determines the NAD+/NADH ratio which is important for the activities of sirtuin histone deacetylases. Several TCA cycle intermediate can be exported out of mitochondria including citrate and αKG. Cytosolic citrate is converted to acetyl-CoA which is used as a donor for HAT-mediated histone acetylation. αKG is used as co-factors for histone and DNA demethylation reactions by JHDM and TET, respectively. The substrate for HMT and DNMT is SAM which is synthesized from essential amino acid methionine. Finally, a low ATP/AMP ratio can activate AMPK, a kinase that phosphorylates histones.
Histone acetylation
Histone acetylation is catalyzed by HATs. Mammalian HATs are divided into five families which share a similar enzymatic reaction: HATs transfer the acetyl group of the acetyl-CoA to the lysine residues of histones and produce CoA as an end product (Racey and Byvoet, 1971). As demonstrated in yeast, elevated levels of acetyl-CoA can be sufficient to instruct cells to enter growth by promoting histone acetylation and expression of growth-related genes (Cai et al., 2011), suggesting that the availability of acetyl-CoA is a major metabolic input into histone acetylation. Indeed, it was shown that depending on the metabolic state, intracellular acetyl-CoA concentration shows a ~10-fold variation (Cai et al., 2011; Takamura and Nomura, 1988). Since the Km of most HATs is within the range (Tanner et al., 1999), the activities of HATs are likely sensitive to the fluctuation of intracellular acetyl-CoA levels.
In mammalian cells, most cytosolic acetyl-CoA comes from citrate exported from mitochondria by a reaction catalyzed by ATP-citrate lyase (ACL). Enhanced glycolysis drives citrate synthesis and therefore the generation of cytosolic acetyl-CoA. Tracing experiments using 13C-labeled glucose in Myc-stimulated cells showed that about half of the acetyl groups on H4K16 came from glucose-derived acetyl-CoA (Morrish et al., 2010). Citrate produced from metabolism of glutamine in the TCA cycle can also contribute to the production of cytosolic acetyl-CoA (Metallo et al., 2012; Mullen et al., 2012; Wise et al., 2011). In cells where ACL was knocked down, there was a profound defect in histone acetylation and expression of genes regulating glucose metabolism (Wellen et al., 2009). In yeast, abrogation of cytosolic acetyl-CoA generating enzymes leads to global histone hypoacetylation (Takahashi et al., 2006).
Histone deacetylation
Enzymes that catalyze the removal of histone acetylation can be in principle divided into two groups based on structural and mechanistic similarities: classical HDACs and NAD+-dependent sirtuin family deacetylases. The deacetylation reaction is energetically favorable. Therefore sirtuins are intriguing as they catalyze the reaction in a seemingly wasteful way: one NAD+ molecule is hydrolyzed to yield nicotinamide and O-acetyl-ADP-ribose. The substrates of sirtuins are diverse and among seven members of the mammalian sirtuin family, SIRT1 and SIRT6 have been shown to localize to the nucleus and exhibit HDAC activities (Imai et al., 2000; Kawahara et al., 2009).
The link between sirtuins and metabolic sensing was supported by the findings that Sir2, first sirtuin gene identified in yeast, mediates the effect of caloric restriction (CR) on extending life span. It is known that glucose limitation in yeast increases life span. When Sir2 was mutated in some yeast strains, the life extension effect of CR was lost (Lin et al., 2000). Deacetylation of H4K16 appears to be essential for the anti-aging effect of Sir2 (Dang et al., 2009b). One potential mechanism is that CR in yeast increases respiration and therefore generating a more oxidized environment and a high NAD+/NADH ratio (Lin et al., 2002). Alternatively, CR was shown to decrease nicotinamide levels and increase NAD+ salvage pathway flux (Anderson et al., 2003). Further flux analysis examining the cytosolic source of NAD+ would be needed to better understand how CR activates sirtuins (Locasale and Cantley, 2011). Moreover, O-acetyl-ADP-ribose might also serve as an important metabolic signal, as it functions to assemble the SIR complex for chromatin silencing in yeast (Liou et al., 2005).
DNA and histone methylation
DNMTs and HMTs add methyl groups to DNA or lysine/arginine residues of histones, respectively. Although structurally diverse and possessing high substrate specificities, DNMTs and HMTs share a similar reaction mechanism: transferring a methyl group from S-adenosyl methionine (SAM) to the substrate with the formation of the by-product S-adenosyl homocysteine (SAH). SAM is derived from the essential amino acid methionine through methionine adenosyltransferase (MAT). It is possible to alter SAM levels through diets (Poirier et al., 2001). However, SAH is a very potent inhibitor of DNMTs and HMTs and the key metabolic determinant of methyltransferase reactions is the rate of SAH clearance (Selhub and Miller, 1992). SAH can be hydrolyzed to homocysteine. Homocysteine can be used to regenerate methionine, a step catalyzed by methionine synthetase and dependent on one-carbon metabolism. Alternately, homocysteine can enter the transulfuration pathway to generate cysteine, the precursor for glutathione synthesis.
The complexity of SAM and SAH metabolism implies that multiple metabolic inputs could affect the establishment of DNA and histone methylation. Dietary intake of methyl donors such as folate is closely linked to levels of DNA methylation, which has been excellently reviewed elsewhere (Feil and Fraga, 2012; Kim, 2005). Acute depletion of glutathione led to drainage of methionine pools and progressive DNA hypomethylation in hamsters, potentially linking the redox status to epigenetic regulation (Lertratanangkoon et al., 1997). It is expected that additional connections will unfold after further investigation of these pathways.
DNA and histone demethylation
A covalent methyl group is chemically stable. Therefore DNA and histone methylation were considered as relatively static epigenetic marks. However during embryonic development there is extensive remodeling of the cellular methylome, suggesting the existence of enzymes that actively remove methylation marks. Indeed in recent years, a variety of HDMs and DNHDs have been identified. The first identified HDM is LSD1 (Shi et al., 2004). The histone demethylation reaction catalyzed by LSD1 involves the reduction of co-factor flavin adenine dinucleotide (FAD) to FADH2 and the release of formaldehyde as a by-product. As recycling of FAD requires converting molecular oxygen to hydrogen peroxide, cellular redox status might influence the availability of FAD and thus the activity of LSD1.
Subsequently a family of HDMs named Jumonji-C domain containing HDM (JHDM) were identified and characterized (Klose and Zhang, 2007). JHDM catalyzes a distinct demethylation reaction from LSD1: it utilizes α-ketoglutarate (αKG), oxygen and Fe (II) as co-factors and releases succinate and formaldehyde as by-products. This mechanism was also adopted by the newly identified TET family enzymes that hydroxylate the 5-methylcytosine of DNA. The only difference is that in this reaction, the hydroxymethyl group is a stable product and further TET-dependent modification is undertaken with low stoichiometry (He et al., 2011; Ito et al., 2011). The function of 5-hydroxymethylcytosine is the topic of intensive investigation and thought to serve as an intermediate for subsequent active (TET-dependent) or passive (replication-dependent) demethylation (He et al., 2011; Tahiliani et al., 2009).
Studies on the metabolic regulation of JHDM- and TET-mediated demethylation are limited. However from the metabolic perspective the demethylation mechanism by JHDMs and TETs is interesting as it involves αKG and succinate, key metabolites of the TCA cycle. αKG can be derived from either glucose or glutamine and is involved in many anabolic pathways. It is estimated that in proliferating cells the intracellular αKG concentrations vary from 0.5–3 mM which is well above the Km of JHDMs (Chowdhury et al., 2011; Thirstrup et al., 2011). Nevertheless upon stimuli such as glucagon or vasopressin, the levels of αKG can rapidly drop as much as 60% (Staddon and McGivan, 1984). Moreover the transport of αKG between cytosol and mitochondria is governed by αKG-malate carrier which is part of the malate-aspartate shuttle that facilitates mitochondria-cytosol transfer of NAD+ and NADH. It has been shown that net flux of the malate-aspartate shuttle determines the rate of αKG interchange between the two compartments (Hautecler et al., 1994; Locasale and Cantley, 2011). Therefore there is a possible metabolic link between HDACs and HDMs/DNHDs. Although succinate was a weak inhibitor of JHDM in vitro (Rose et al., 2008), pathological levels of succinate can affect histone methylation (Cervera et al., 2009). These findings, though circumstantial, suggest that the rate of demethylation could be regulated at both levels of co-factor availability and feedback inhibition.
Histone phosphorylation
Multiple serine and threonine residues of histones are subject to phosphorylation by a wide array of kinases (Baek, 2011). Since the intracellular ATP concentration is well above the Km of most kinases, metabolism should not exert a direct effect on histone phosphorylation. However, some kinases that are responsive to metabolic changes can phosphorylate histones, thus providing an indirect way for metabolic control of histone phosphorylation. For example, in response to a low ATP/AMP ratio indicative of metabolic stress, AMPK can translocate to chromatin and phosphorylate histone H2B serine 36. The H2BS36 phosphorylation facilitates the expression of AMPK-dependent genes and is essential for metabolic adaptation and cell survival under energetic stress (Bungard et al., 2010). Similarly, the H3S10-specific kinase AKT is downstream of mTOR, a well-characterized amino acid sensor in cells.
Histone GlcNAcylation
Recently a new modification of histone was reported. H2B S112 can be O-linked N-acetylglucosamine (GlcNAc)-modified by the enzyme O-GlcNAc transferase (OGT) (Fujiki et al., 2011). GlcNAc comes from a minor branch of glycolytic metabolism called hexosamine biosynthetic pathway (HBP). About ~5% of glucose enters HBP and the end product GlcNAc is used as donor for various protein glycosylation reactions. GlcNAc has been reported to be a key signaling metabolite which can coordinate glucose and glutamine metabolism in cells through glycosylation of growth factor receptors (Wellen et al., 2010). Although the transcriptional implication of histone GlcNAcylation remains to be fully elucidated, it is likely that flux through HBP affects levels of this novel epigenetic mark.
Lessons from IDH mutation: metabolic reprogramming and epigenetics in cancer
It was observed more than 50 years ago by the German biochemist Otto Warburg that cancer cells utilize glucose in a fundamentally different way than normal cells (Warburg, 1956). Since then and with the recent outburst in cancer metabolism research, metabolic reprogramming as a hallmark of cancer has been increasingly appreciated (Vander Heiden et al., 2009). However, one key question remains in the field: does metabolism rewiring observed in cancer merely serve to fulfill the proliferative demand of tumor cells or play a significant role in driving cancer development? One strong argument in support of the latter would be the identification of cancer-associated mutations in metabolic enzymes. Recently the metabolic enzymes isocitrate dehydrogenase 1 (IDH1) and IDH2 were found to be frequently mutated in low-grade gliomas (Yan et al., 2009), adult de novo acute myeloid leukemias (AMLs) (Mardis et al., 2009; Ward et al., 2010) and subsets of chondrosarcomas and lymphomas (Amary et al., 2011; Cairns et al., 2012). IDH1 and IDH2 are NADP+ dependent enzymes that interconvert isocitrate and αKG in cytosol and mitochondria, respectively. The mutations observed in IDH1 and IDH2 are heterozygous and highly specific, suggesting a possible gain-of-function. Indeed, metabolomic and biochemical analysis revealed that mutant IDHs display a neomorphic activity of producing 2-hydroxyglutarate (2HG) from αKG (Dang et al., 2009a; Ward et al., 2010). In IDH mutant patient samples, 2HG levels were found to be elevated as high as 100-fold.
Since 2HG is a structural analogue of αKG, it is plausible that 2HG modulates activities of enzymes dependent on αKG. In humans there are more than 60 αKG-dependent enzymes which affect diverse aspects of cell physiology (Loenarz and Schofield, 2008). Biochemical studies suggest that 2HG can inhibit a number of αKG-dependent enzymes with varying potencies, therefore the challenge is to identify key enzymes that mediate 2HG’s oncogenic effect (Chowdhury et al., 2011; Xu et al., 2011b). We speculated that if such enzyme exists, one would expect to observe its mutations in the same malignancy where IDH mutations were identified. Genetic profiling of AML samples revealed that loss-of-function mutations of TET2, a member of the TET family DNHDs, were frequently found in AML and mutually exclusive with IDH mutations (Figueroa et al., 2010). Expression of mutant IDH in cells effectively blocked TET2-induced increase in 5-hydroxymethylcytosine (Figueroa et al., 2010). Consistently, cells stably expressing mutant IDH and AML samples with IDH mutations showed DNA hypermethylation. These findings suggest that in AML, IDH mutations and TET2 mutations share a common tumorigenic pathway which involves remodeling of the cellular epigenetic state. Interestingly, IDH mutations are similarly associated with DNA hypermethylation in gliomas where no mutations in the TET family members have been identified so far (Noushmehr et al., 2010). In vitro 2HG can competitively inhibit several αKG-dependent JHDMs (Chowdhury et al., 2011; Xu et al., 2011b). Given the close relationship between histone methylation and DNA methylation, it is tempting to speculate that aberrant histone demethylation contributes to the DNA hypermethylation phenotype in IDH mutant gliomas. Indeed, glioma samples with IDH mutations displayed higher levels of H3K9me3 and H3K27me3. In astrocyte cultures, stable expression of mutant IDH1 was sufficient to induce DNA hypermethylation, an observation that was temporally preceded by a progressive increase in H3K9me3 (Lu et al., 2012; Turcan et al., 2012). Although mutations in chromatin-modifying enzymes have not been identified in adult low-grade gliomas, in pediatric gliomas genes encoding histone variants H3.3 (H3F3A) and H3.1 (HIST1H3B) were found to be somatically mutated at specific residues including K27 which appeared to be mutually exclusive with IDH mutations. Further functional characterization of these specific histone mutations will be needed to genetically link IDH mutations to histone methylation in glioma (Schwartzentruber et al., 2012; Wu et al., 2012).
What is the consequence of epigenome aberration induced by mutant IDH? It is well known that epigenetic remodeling represents an important step in cell lineage specification and a common feature of malignancies with IDH mutations is a block to cell differentiation. When fibroblasts expressing mutant IDH were induced to differentiate to adipocytes, there was a profound defect in the expression of differentiation-related genes (Lu et al., 2012). Interestingly, such failure was associated with increased repressive histone methylation including H3K9me3 and H3K27me3 but not DNA methylation. These data, together with the findings that 1) genes showing DNA hypermethylation in cancer tend to be differentiation-related and reversibly silenced by histone methylation in stem cells (Ohm et al., 2007; Widschwendter et al., 2007), and 2) TET1 preferentially binds to the promoters of differentiation-related genes and is thought to prevent accidental DNA methylation (Williams et al., 2011; Wu and Zhang, 2011), suggest a working model (Figure 4). During differentiation of progenitor cells, JHDM inhibition by 2HG causes defective histone demethylation and blocks the accessibility of differentiation-related genes. In addition, the inhibition of TET and JHDM by 2HG leads to progressive DNA hypermethylation and permanently “locks” these genes in a silent state. The resulting differentiation arrest may facilitate cancer development through accumulation of undifferentiated cells capable of self-renewal. Future analysis of DNA and histone methylation as well as the genome-wide distribution of JHDM/TET bindings will be required to test this hypothesis.
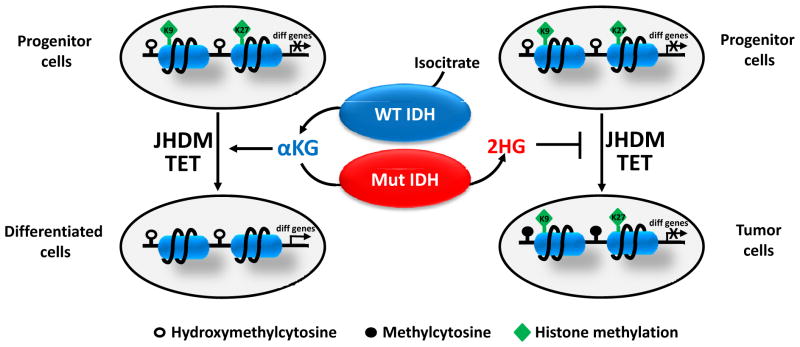
Figure 4. Proposed model for the role of 2HG in epigenetic regulation and cell differentiation.
When progenitor cells differentiate, JHDM removes repressive histone methylation marks (H3K9me3 and H3K27me3) and activates the expression of differentiation-related genes. In addition, TET acts as a failsafe mechanism to protect promoters from aberrant DNA methylation. 2HG produced by mutant IDH inhibits JHDM and TET, which leads to histone and DNA hypermethylation and permanently “locks” differentiation-related genes in a silent state. This results in a differentiation arrest and expansion of progenitor cells, thus facilitating tumor development.
The analyses of cancer-associated IDH mutations highlight the dynamics and importance of the interaction between metabolism and epigenetics. As metabolic abnormalities in cancer continue to be uncovered, it will be interesting to see if new metabolites or metabolic perturbations that affect chromatin structure are identified. For example, recently the key enzyme in serine biosynthetic pathway, phosphoglycerate dehydrogenase (PHGDH), was found to be amplified in melanoma and breast cancer (Locasale et al., 2011; Possemato et al., 2011). As the flux to serine biosynthetic pathway regulates the one-carbon metabolism which is critical for recycling of the methyl donor methionine, PHGDH amplification could potentially modulate the cellular methylome and the epigenetic state.
Concluding remarks
Research on metabolism and epigenetics are progressing at an unprecedented pace and we anticipate more exciting discoveries to arrive over the next few years. Yet technological challenges are present that must be overcome to unravel the mystery at the next level. As most metabolites are not diffusible between mitochondria and cytosol/nucleus, accurate determination of the cytoplasmic pool size is critical to understand their effects on epigenetic signaling. Although attempts have been made, so far the technology to reliably measure metabolite concentrations in different cellular compartments is lacking. Even within the same compartment, local concentration of the metabolite may vary, as substrate channeling is a common event in cellular metabolism (Huang et al., 2001). This is relevant, as metabolic enzymes are often present in transcriptional complexes, and thus could provide a local supply of substrates/co-factors to the complexes in which they localize. For example, methionine adenosyltransferase IIα (MATIIα), which generates SAM, was present in chromatin-associated complexes and thought to provide local substrate for methyltransferases reactions (Katoh et al., 2011). Alternatively metabolic enzymes may function in these complexes independently of their metabolic activity (Tristan et al., 2011; Yang et al., 2011). Finally, system biology approaches will become increasingly important to fully grasp the complexity of the connections between metabolism and chromatin dynamics. A deeper understanding of these connections may help shed light on our understanding of the etiology of a variety of complex diseases (Katada et al., 2012; Sahar and Sassone-Corsi, 2012).
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitation. We thank members of Thompson laboratory for critically reading the manuscript. C.B.T. is supported by grants from NIH and NCI. C.B.T. is a founder and consultant of Agios Pharmaceuticals and has financial interest in Agios. C.B.T. is also a member of the Board of Directors of Merck.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Baek SH. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell. 2011;42:274–284. doi: 10.1016/j.molcel.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, Parrens M, Martin A, Xerri L, Brousset P, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012 doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera AM, Bayley JP, Devilee P, McCreath KJ. Inhibition of succinate dehydrogenase dysregulates histone modification in mammalian cells. Mol Cancer. 2009;8:89. doi: 10.1186/1476-4598-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B, Estabrook RW, Ghosh A. Damped Sinusoidal Oscillations of Cytoplasmic Reduced Pyridine Nucleotide in Yeast Cells. Proc Natl Acad Sci U S A. 1964;51:1244–1251. doi: 10.1073/pnas.51.6.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009a;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009b;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautecler JJ, Sluse-Goffart CM, Evens A, Duyckaerts C, Sluse FE. Effect of aspartate and glutamate on the oxoglutarate carrier investigated in rat heart mitochondria and inverted submitochondrial vesicles. Biochim Biophys Acta. 1994;1185:153–159. doi: 10.1016/0005-2728(94)90205-4. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277:44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ito T, Ohta M, Kera Y, Noda T, Igarashi K. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011;41:554–566. doi: 10.1016/j.molcel.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI. Nutritional epigenetics: impact of folate deficiency on DNA methylation and colon cancer susceptibility. J Nutr. 2005;135:2703–2709. doi: 10.1093/jn/135.11.2703. [DOI] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci U S A. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J Cell Biol. 2011;192:949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertratanangkoon K, Wu CJ, Savaraj N, Thomas ML. Alterations of DNA methylation by glutathione depletion. Cancer Lett. 1997;120:149–156. doi: 10.1016/s0304-3835(97)00300-5. [DOI] [PubMed] [Google Scholar]
- Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, Rodriguez P, Dominguez M, Borggrefe T. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011;14:443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad HP, Baylin SB. Linking cell signaling and the epigenetic machinery. Nat Biotechnol. 2010;28:1033–1038. doi: 10.1038/nbt1010-1033. [DOI] [PubMed] [Google Scholar]
- Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, VanGilst M, Hockenbery D. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem. 2010;285:36267–36274. doi: 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Poirier LA, Wise CK, Delongchamp RR, Sinha R. Blood determinations of S-adenosylmethionine, S-adenosylhomocysteine, and homocysteine: correlations with diet. Cancer Epidemiol Biomarkers Prev. 2001;10:649–655. [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racey LA, Byvoet P. Histone Acetyltransferase in Chromatin - Evidence for in-Vitro Enzymatic Transfer of Acetate from Acetyl-Coenzyme a to Histones. Exp Cell Res. 1971;64:366. doi: 10.1016/0014-4827(71)90089-9. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Ng SS, Mecinovic J, Lienard BM, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J Med Chem. 2008;51:7053–7056. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab. 2012;23:1–8. doi: 10.1016/j.tem.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Selhub J, Miller JW. The pathogenesis of homocysteinemia: interruption of the coordinate regulation by S-adenosylmethionine of the remethylation and transsulfuration of homocysteine. Am J Clin Nutr. 1992;55:131–138. doi: 10.1093/ajcn/55.1.131. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Slavov N, Macinskas J, Caudy A, Botstein D. Metabolic cycling without cell division cycling in respiring yeast. Proc Natl Acad Sci U S A. 2011;108:19090–19095. doi: 10.1073/pnas.1116998108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staddon JM, McGivan JD. Distinct effects of glucagon and vasopressin on proline metabolism in isolated hepatocytes. The role of oxoglutarate dehydrogenase. Biochem J. 1984;217:477–483. doi: 10.1042/bj2170477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Takamura Y, Nomura G. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J Gen Microbiol. 1988;134:2249–2253. doi: 10.1099/00221287-134-8-2249. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- Thirstrup K, Christensen S, Moller HA, Ritzen A, Bergstrom AL, Sager TN, Jensen HS. Endogenous 2-oxoglutarate levels impact potencies of competitive HIF prolyl hydroxylase inhibitors. Pharmacol Res. 2011;64:268–273. doi: 10.1016/j.phrs.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. Evidence of carbon monoxide-mediated phase advancement of the yeast metabolic cycle. Proc Natl Acad Sci U S A. 2009;106:14293–14296. doi: 10.1073/pnas.0907786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Pendergast JS, Springer DA, Foretz M, Viollet B, Brown A, Kim MK, Yamazaki S, Chung JH. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS One. 2011;6:e18450. doi: 10.1371/journal.pone.0018450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Tet1 and 5-hydroxymethylation A genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011;10:2428–2436. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011a;13:639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011b;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol. 2010;20:1203–1208. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]