Complex changes in alternative pre-mRNA splicing play a central role in the Epithelial-Mesenchymal Transition (EMT) (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 1.
Abstract
The epithelial to mesenchymal transition (EMT) is an important developmental process that is also implicated in disease pathophysiology, such as cancer progression and metastasis. A wealth of literature in recent years has identified important transcriptional regulators and large-scale changes in gene expression programs that drive the phenotypic changes that occur during the EMT. However, in the past couple of years it has become apparent that extensive changes in alternative splicing also play a profound role in shaping the changes in cell behavior that characterize the EMT. While long known splicing switches in FGFR2 and p120-catenin provided hints of a larger program of EMT-associated alternative splicing, the recent identification of the epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2) began to reveal this genome-wide post-transcriptional network. Several studies have now demonstrated the truly vast extent of this alternative splicing program. The global switches in splicing associated with the EMT add an important additional layer of post-transcriptional control that works in harmony with transcriptional and epigenetic regulation to effect complex changes in cell shape, polarity, and behavior that mediate transitions between epithelial and mesenchymal cell states. Future challenges include the need to investigate the functional consequences of these splicing switches at both the individual gene as well as systems level.
Keywords: Alternative splicing, ESRP, EMT, epithelial cells, mesenchymal cells, metastasis
1. Introduction
The organ structures of all eumetazoans arise from the interactions of two basic tissue types that have fundamental differences in their physiology and behavior: the epithelium and the mesenchyme. The epithelium is composed of epithelial cells tightly bound through several types of interactions including adherens junctions, desmosomes, and tight junctions. Typically epithelial cells are polarized, with an apical surface that faces a lumen and a basolateral surface that rests on a thin extracellular matrix called the basal lamina. These properties enable the epithelium to form a semi-permeable layer that separates compartments and function in protection, directional absorption, and secretion. The mesenchyme consists of cells which have a largely undefined shape due to their relatively dynamic cytoskeleton. Mesenchymal cells, unlike epithelial cells, do not display tight cell-cell contacts, and instead interact primarily with the extracelluar matrix, enabling them to migrate in three-dimensional space.
The epithelial and mesenchymal cell phenotypes are not static as both cell types have the ability to undergo major cellular transformations that affect the morphology and behavior of the cell. These transformations are respectively known as the epithelial-to-mesenchymal transition (EMT) and the mesenchymal-to-epithelial transition (MET) and are essential for normal vertebrate development [1]. For example, during gastrulation, cells of the invaginating epithelium undergo an EMT and migrate inward to become the primary mesenchyme. Mesenchymal cells can subsequently undergo the reverse process of MET to form epithelial structures, a process that has been well documented in formation of the renal tubular epithelium during renal organogenesis [2]. While the EMT has essential roles during developmental, there is also evidence that this process can be hijacked in certain pathophysiologic conditions. For example, there is strong evidence that the EMT is one mechanism by which carcinomas spread beyond the primary tumor site and acquire invasive properties that can promote metastasis [3].
The molecular mechanisms that induce and drive the cellular conversions of EMT and MET have long been the focus of intense investigation. These studies have primarily focused on transcriptional regulation to account for changes in total transcript and protein levels in epithelial or mesenchymal cells. For example, much work has revealed important roles for the Snail, Twist, and Zeb family of transcription factors and the signaling pathways that activate them [4, 5]. However, a new avenue of investigation into the genetic program that impacts the EMT is beginning to emerge. Recent findings from our lab and several other groups has revealed that vast, coordinated changes in alternative splicing (AS) occur during the EMT and profoundly alter cellular phenotypes and behaviors. Notably, the genes that are regulated by AS during the EMT are generally not regulated transcriptionally. Thus, AS adds a post-transcriptional layer of gene regulation that functions in concert with transcriptional alterations. In this review we will briefly cover the key aspects of AS followed by a description of the recent studies performed to uncover an EMT associated splicing network and the factors that regulate it, including the recently discovered epithelial-specific splicing regulators ESRP1 and ESRP2. We will also discuss several genes showing AS changes during the EMT and speculate on the functional impact. We will conclude by placing the recent analyses in the context of the EMT and discuss some future directions for the field.
2. Alternative splicing regulation
Recent studies using high-throughput sequencing have shown that nearly all multi-exon genes within the human genome produce multiple mature mRNAs [6, 7]. Therefore, AS represents a critical mechanism for vastly expanding the protein-coding potential from a limited genome. Small changes in peptide sequence can alter a protein in many ways including localization, interactions with other proteins, enzymatic activity, and post-translational modifications as just a few examples [8]. Exemplifying its importance in regulating gene expression, many AS events are tightly regulated in a cell type or tissue specific manner, at different developmental stages, or in response to extra-cellular stimuli and activation of specific signaling pathways [9, 10].
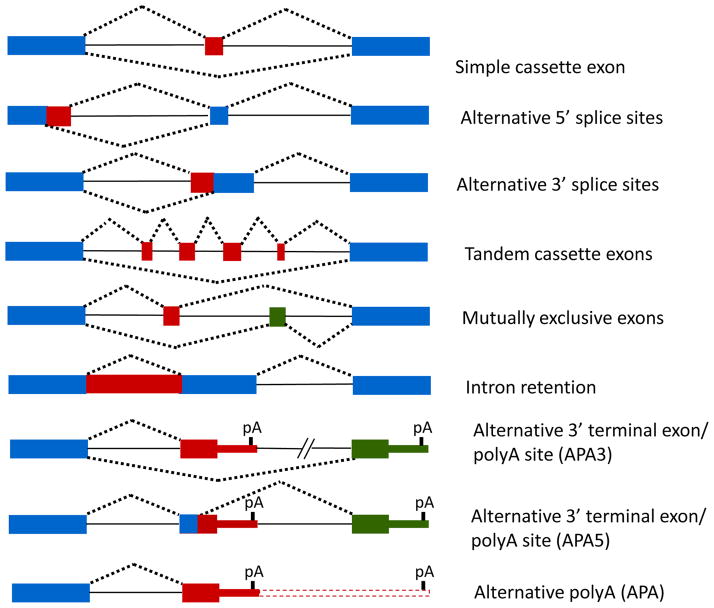
The different types of commonly observed AS events are illustrated in Fig. 1. An exon that is included or excluded from the mRNA is called a cassette exon. In addition to simple cassette exons that are singly spliced or skipped, often multiple adjacent cassette exons are spliced or skipped in tandem or spliced in a mutually exclusive manner. Exons can also be shortened or lengthened by the presence of alternative 3′ or 5′ splice sites on either end of the exon. AS can also lead to alternative polyadenylation (APA) through use of alternative 3′ splice sites (APA3) or 5′ splice sites (APA5) that are present in upstream alternative 3′ terminal exons. It is noteworthy that these events leading to use of alternative polyA sites alter the composition of the 3′ untranslated region (UTR) and thus, also subject the mRNAs to differential regulation by microRNAs and other mRNA stability or translation factors.
Figure 1.
Schematic of the different types of alternative splicing. Note that frequently there is more than one event or type of AS within a given gene transcript, further increasing the complexity of AS to expand proteomic complexity. In addition there can be overlap between the types of events, such as cassette exons that also have alternative splice sites or mutually exclusive sets of tandem exons. Blue boxes indicate constitutive exon sequences and red or green boxes are alternatively spliced exonic sequences. Solid lines indicate introns and dashed lines indicate alternative patterns with which exon sequences are ligated after splicing. The hashmarks in the APA3 and APA5 events indicate the potential for splicing of additional cassette exons between the proximal and distal 3′ terminal exons.
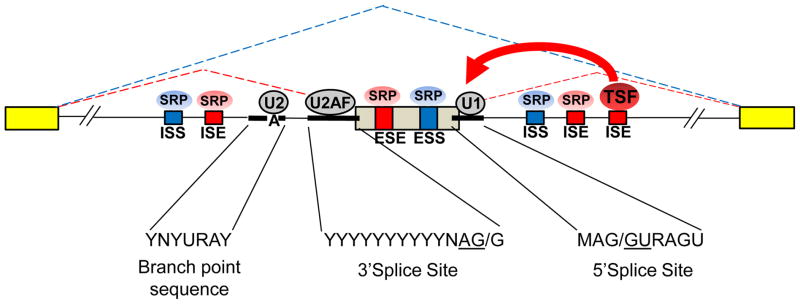
Splicing is achieved by the spliceosome, a macromolecular machine composed of five small ribonucleoproteins (snRNPs) and hundreds of additional proteins [11]. The precision of the reaction is accomplished through a coordinated series of complex RNA-protein interactions that can occur post- or co-transcriptionally. Exons are identified by the basal splicing apparatus through the recognition of conserved consensus sequences that are present at the upstream intron/exon boundary (the 3′ splice site) and the downstream exon/intron boundary (the 5′ splice site) (Fig. 2). The 3′ splice site consists of an invariant AG at the end of the intron and an upstream polyprimidine tract (PPT). In addition, the 3′ splice site is associated with a branchpoint sequence (BPS) that is usually located 20–40 nt upstream. The 5′ splice site is a 9 nt sequence of which the invariant GU is present at the start of the intron. Splice sites that are weak matches to the consensus sequences are less efficiently recognized by spliceosomal components and thus these sequence elements are one determinant of whether an exon is spliced or skipped.
Figure 2.
Regulation of cell-type or tissue-specific alternative splicing involves participation of multiple splicing regulators by combinatorial control. An example of an alternatively spliced cassette exon (gray) flanked by two constitutively spliced exons (yellow). The position of the core splice sites and branchpoint sequence are indicated by thick lines and the consensus sequences are indicated below. The 3′ splice site consists of a polypyrimidine rich sequence of variable length (the polypyrimidine tract PPT)) upstream of an invariant AG dinucleotide at the 3′ end of the intron. The 5′ splice site is a 9 nucleotide consensus sequence of which the GU at the 5′ end of the intron is invariant. The branchpoint sequence (BPS) is a loose consensus sequence that usually requires an adenosine residue as indicated. When an exon is spliced, the 5′ splice site is bound by the U1 snRNP through a base-pairing interaction, the 3′ splice site is bound by U2 auxiliary factor (U2AF), and the BPS is bound by the U2 snRNP through base pairing that excludes the branched adenosine. Note that the splice sites associated with the upstream and downstream constitutive exons are also similarly recognized and after additional spliceosomal components are recruited, the upstream and downstream introns are remove and the exons ligated. Intronic splicing enhancer (ISE) and exonic splicing enhancer (ESE) sequence elements are indicated in red associated with splicing regulatory proteins (SRP) that bind them and promote exon splicing. Intronic splicing silencer (ISS) and exonic splicing silencer (ESS) are indicated in blue along with corresponding SRPs that promote exon skipping. A general model is presented in which each auxiliary cis-element and associated factor combinatorially influences exon recognition by core spliceosomal components such as U1, U2AF, or U2, but a cell or tissue specific factor (TSF) may tip the balance towards, in this case, exon splicing.
In addition to the core splice site sequences, splicing is also influenced by _cis_-elements located within the regulated exons or flanking introns that can act to either enhance or suppress exon recognition and are therefore defined as exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), or intronic splicing silencers (ISSs). One function of the _cis_-elements is to act as binding sites for RNA binding proteins (RBPs) that bind these sequences and function either to facilitate or inhibit splicing at nearby splice sites (Fig. 2). The most well characterized splicing regulatory proteins are the generally ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNP) and the serine-arginine rich or SR proteins [12, 13]. Members of both families bind to degenerate sequence motifs within pre-mRNA transcripts to facilitate constitutive splicing as well as AS through interactions with specific cis-elements and components of the basal splicing machinery. The SR proteins have predominantly been shown to bind and mediate the functions of ESEs, whereas the hnRNP proteins have been shown to bind ESSs, ISSs, and ISEs. Additional families of splicing factors have been characterized including several with more cell type-specific expression patterns. Among these more specialized regulators are the neuronal NOVA proteins, the neural and muscle enriched Fox proteins, and the CELF and Muscleblind family of proteins that display both spatial and temporal expression patterns in muscle and nervous tissues [10]. The regulation of an AS event is largely thought to be under combinatorial control, whereby multiple RBPs bound to the pre-mRNA influence the splicing outcome and that this balance may be tipped by the presence or absence of one or more splicing factors such as those with cell-type or condition-specific expression [14].
Several studies have been undertaken to comprehensively profile AS events regulated by cell-type or developmental-stage specific splicing factors [15, 16]. Analysis of the regulated targets characterized in these studies found that many of the co-regulated splicing events occur within transcripts that encode proteins that function in biologically coherent pathways that are relevant to the tissue or cell type in which the splicing factor is expressed. For example, Nova regulates the splicing of gene transcripts that are enriched for those encoding proteins that function in processes relevant to neurons, such as synaptic transmission [15]. This work thereby established principles in which a splicing regulatory network (SRN) regulated by a specific splicing factor coordinates the expression of many protein isoforms and adds a layer of gene expression regulation independent of transcription. Such coordinated modules of post-transcriptional regulation by RNA binding proteins have been described as post-transcriptional operons [17]. An important implication of these observations is that the identification of SRNs will further define protein interaction networks and biological pathways that can impact cell morphology and function.
3. Evidence for an epithelial splicing regulatory network
Prior to the advent of high-throughput technologies, AS studies were performed on an individual gene basis and the identification of novel splice variants arose through individual cloning of mRNAs and ESTs. Nevertheless, earlier literature described several AS events in which specific splice variants showed complete or preferential expression in epithelial cells versus mesenchymal cells and/or showed changes during the EMT. These examples include the genes encoding the Fibroblast growth factor receptor 2 (FGFR2), CD44, p120-catenin (CTNND1), and Mena (ENAH) proteins. Each of these proteins has been well characterized and this section provides an overview of what is known about their isoform-specific functions. Of particular interest is that all four genes encode proteins with functional relevance to the tissue specific differences between epithelial and mesenchymal cells.
3.1. FGFR2
The fibroblast growth factor receptor 2 (FGFR2) gene encodes a transmembrane receptor tyrosine kinase that is activated by the fibroblast growth factors (FGFs) [18]. Upon FGF stimulation, a cascade of downstream signals is set in motion, ultimately influencing mitogenesis and differentiation. The second half of the third extracellular immunoglobulin-like domain is encoded by one of two mutually exclusive alternative exons: IIIb or IIIc. These exons confer distinctly different FGF ligand binding affinities to the respective receptor isoforms [19]. The expression of either isoform is highly cell-type specific with FGFR2-IIIb expression limited to epithelial cells and FGFR2-IIIc limited to mesenchymal cells [20]. Importantly, a switch in FGFR2 splicing from the FGFR2-IIIb to FGFR2-IIIc variant was the first example illustrating the role of AS during the EMT [21, 22].
The cell-type specific partitioning of these receptor isoforms establishes critical paracrine signaling pathways involved in normal cellular communication between epithelial and mesenchymal cell populations during development [23, 24]. A general model suggests that epithelial cells expressing FGFR2-IIIb specifically interact with FGFs secreted by mesenchymal cells. In contrast, FGFs secreted by epithelial cells only interact with mesenchymal FGFR2-IIIc. These directional and reciprocal growth regulatory pathways have been shown to be critical during the formation of epithelial structures in many organs [25].
3.2. p120-Catenin
p120-catenin is a master regulator of cadherin stability and a modulator of Rho GTPase activity [26, 27]. Although p120-catenin stabilizes E-cadherin at the plasma membrane and promotes cell-cell adhesion, it paradoxically can also promote cell motility and invasion in cells that have lost E-cadherin expression [28]. These seemingly contradictory functions of p120-catenin appear to be due to the different activities of the protein isoforms encoded by splice variants that predominate in epithelial versus mesenchymal cells. Mesenchymal cells express splice variants that contain alternative exons 2 and 3. These exons are predominantly skipped in epithelial cells, shifting translation from exon 3 to exon 5 and resulting in a shorter protein isoform [29]. Furthermore, expression of the mesenchymal p120-catenin isoform is induced during the EMT [30, 31]. The mesenchymal exons encode a coiled-coil domain thought to stabilize the interaction between p120-catenin and RhoA, reducing the activity of RhoA and resulting in increased cell invasiveness [28].
3.3. CD44
CD44 is a transmembrane protein that functions primarily to maintain tissue structure by mediating cell-cell and cell-matrix adhesion [32]. The N-terminal extracellular domain of the protein interacts with hyaluronic acid promoting the binding of additional extracellular ligands. This ligand complex initiates a downstream signaling cascade through the interaction of the intracellular domain with its binding partners. The CD44 transcript is composed of exons 1–5 at the 5′ end and exons 16–20 at the 3′ end that are spliced together to form the standard isoform (CD44s). Between exons 5 and 16 are ten variable exons (v1–v10) that are alternatively spliced to give rise to a plethora of isoforms. Inclusion of the variable exons lengthens the extracellular membrane-proximal region by forming a heavily glycosylated stalk-like structure that provides interaction sites for additional molecules [33].
The CD44s isoform is widely expressed and present on the surface of most cells while expression of the variant isoforms is more restricted. The CD44E isoform contains exons v8–10 and is expressed primarily in epithelial cells, is enriched in normal human breast tissue compared to metastatic carcinomas, and correlates well with the expression of E-cadherin [34–36]. When cultured cells are induced to undergo an EMT, a drastic switch in CD44 splicing from the E isoform to the standard isoform occurs, thus providing a direct relationship between CD44 splicing and the EMT [37, 38]. CD44 splicing has been shown to have a critical role in limb bud development where it is co-expressed with FGF-8 in the apical ectodermal ridge and presents this ligand to receptors on the underlying mesenchyme, likely including FGFR2-IIIc [39]. This crosstalk between epithelial and mesenchymal cells highlights the importance of appropriate isoform expression through a coordinated splicing program.
3.4. Mena (ENAH)
Mena, also known as Enabled homolog (Enah) is the mammalian ortholog of the Drosophila protein Ena. Mena is expressed in many cell types where it localizes to the leading edges of lamellipodia, the tips of filopodia, and focal adhesions and regulates the branching actin filaments [40]. Thus, Mena plays a pivotal role in controlling the motility and morpholology of a variety of cell types including fibroblasts and epithelial cells [41, 42].
An isoform of Mena that contains an exon termed 11a has shown to be expressed in primary tumor cells but not in invasive tumor cells [43]. Further analysis of tumor cells expressing Mena11a demonstrated decreased invasion, intravasation and dissemination in an in vivo invasion assay [44]. Consistent with the hypothesis that invasive tumor cells have undergone an EMT to become more mesenchymal, the Mena11a is specifically expressed in epithelial cell lines and not found in mesenchymal cell lines [45].
4. Regulators of EMT alternative splicing
4.1. Identification of the epithelial-specific splicing regulators ESRP1 and ESRP2
Given the exquisite cell-type specific expression of FGFR2 splice variants and their essential developmental roles several groups, including ours, sought to define the regulators of this important splicing choice. A number of splicing factors that influence the splicing choice between exons IIIb and IIIc in the FGFR2 transcript were identified, including PTB, Tia1/TIAR, Fox2, hnRNPM, and hnRNPA1 ([46, 47] and references therein). However, these factors are ubiquitously expressed and therefore a satisfactory model to explain the cell-type-specific regulation of FGFR2 splicing remained lacking. Data from our lab was consistent with a hypothesis that there was an epithelial cell-type specific splicing regulator that enabled splicing of the IIIb exon, whereas in non-epithelial cells, absence of this factor would lead to splicing of exon IIIc [48]. In pursuit of this hypothesis, we designed and performed a genome-wide cDNA expression screen to find novel factors that could specifically enhance exon IIIb splicing in a non-epithelial cell [49]. Among the validated hits from this screen were two protein paralogues previously known by the generic names RNA binding motif proteins 35A and 35B (RBM35A and RBM35B) whose activity depended on a previously characterized enhancer element to induce exon IIIb inclusion and repress exon IIIc splicing. Examination of mRNA levels for these two genes across a panel of cell lines as well as in mouse tissues showed that both genes were indeed highly cell-type-specific with exclusive expression in epithelial cells and absent expression in non-epithelial cell types. Thus, these genes were renamed as epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2).
Analysis of the splicing patterns of the four genes described in Section 3 (FGFR2, p120-Ctn, CD44, and ENAH) following siRNA mediated knockdown of ESRP1 and ESRP2 demonstrated a near complete switch from the epithelial to the mesenchymal splice variant. Conversely, ectopic expression of either ESRP1 or ESRP2 in mesenchymal cells switched splicing of these genes to the epithelial pattern. Together, these results demonstrate that the ESRPs are necessary and sufficient for the expression of the epithelial isoforms of these genes. An interesting note is that the loss of ESRP2 expression alone does not significantly affect splicing whereas sustained knockdown of ESRP1 expression alone is nearly sufficient to abolish epithelial splicing patterns of several transcripts. This may indicate that ESRP1 is the more essential splicing regulator, which is further supported by our observation that ESRP1 mRNA levels are generally more abundant than those of ESRP2.
The ESRPs are ~75kD RBPs consisting of three tandem RNA recognition motifs (RRMs) [50]. ESRP1 and ESRP2 share very similar amino acid sequences with the highest degree of similarity occurring within the three RRMs. The ESRPs are evolutionarily highly conserved with orthologs in both nematodes (Sym-2) and flies (Fusilli) [51–53]. Remarkably, Fusilli, also enriched in the epithelium of flies, can partially rescue the splicing of FGFR2 in mammalian epithelial cells depleted of the ESRP1 and ESRP2 [49]. The conservation of both structure and function of the ESRPs suggests that a conserved epithelial splicing program originated early during metazoan evolution and plays a profound and essential role in development and tissue homeostasis.
Our initial observations suggested that a loss of ESRP expression is likely to be a general phenomenon that occurs during the EMT. Indeed in the study that identified ESRP1 and ESRP2 we showed that they were both transcriptionally inactivated in a human mammary epithelial cell lines (HMLE) during the EMT that is induced by the mesenchymal transcription factor Twist1. Further evidence that a loss of ESRP1 and ESRP2 expression is an obligate and generalized event during the EMT is provided by studies in which the induction of EMT by TGFB1, Snail, ZEB1, ZEB2, and E-cadherin knockdown is accompanied by nearly complete abrogation of ESRP expression [54–59]. These observations suggest that just as direct and/or indirect downregulation of E-cadherin by mesenchymal transcription factors is central to the phenotypic changes during EMT, that they also direct downregulation of the ESRPs.
4.2. Additional splicing regulators associated with the EMT
To date, the ESRPs are the only known splicing factors that exhibit epithelial cell-type-specific expression and that undergo pronounced changes in expression during the EMT. As such, these proteins are likely to be key mediators of splicing differences that are observed between epithelial and non-epithelial cells. However, there is also evidence that other, more ubiquitously expressed splicing factors can also play a role in regulating the EMT. The SR protein SRSF1 (ASF/SF2) has been shown to regulate the splicing of the tyrosine kinase receptor Ron by inhibiting inclusion of exon 11, thereby promoting expression of the ΔRon isoform and inducing an EMT [60]. The ΔRon isoform is unable to undergo proteolytic cleavage rendering the protein constitutively active, a property that may account for the observation that cells overexpressing ΔRon exhibit invasive phenotypes [61]. In agreement, ΔRon expression correlates with a metastatic phenotype in tumor samples [62]. SRSF1 has also been implicated in mediating AS of the GTPase Rac1 [63]. A switch to the Rac1b isoform in response to matrix metalloproteinase-3 (MMP-3) was shown to induce EMT through generation of reactive oxygen species (ROS) and induction of Snail expression [64].
Activation of ERK1 and ERK2 leads to the phosphorylation of many substrates including the splicing regulator Sam68 (Khdrbs1). Phosphorylated Sam68 in turn upregulates SRSF1 by inhibiting the splicing of an intron from the 3′ UTR that would otherwise target the SRSF1 transcript for non-sense mediated decay [65]. In this manner, EMT is promoted through signal transduction activation of a splicing factor that then upregulates a positive EMT splicing regulator, SRSF1. A recent study also implicates RBFOX2 in contributing to the regulation of splicing changes that accompany the EMT including some combinatorial regulation of ESRP regulated events [58, 66]. Finally, Muscleblind like-1 (MBNL1) has been shown to be a negative regulator of EMT during cardiac morphogenesis, possibly by affecting the splicing of downstream effectors of the TGF-beta pathway [67].
5. Using high-throughput assays to investigate EMT associated alternative splicing
5.1. The ESRPs are master regulators of an epithelial splicing network
The advent of splicing sensitive microarrays, and more recently, high-throughput mRNA sequencing (RNA-Seq), has facilitated the ability to detect and quantify alternatively spliced mRNAs on a genome-wide basis [68, 69]. In our own studies, we sought to establish the role of the ESRPs as master regulators of an epithelial splicing program. We used microarray platforms that used probes for exons or combining probes for exons and exon junctions to identify splicing changes that occurred when we altered the expression of the ESRPs in cell lines. Two model systems were utilized: ectopic expression of Esrp1 in the human mesenchymal breast cancer cell line MDA-MB-231 and siRNA mediated knockdown of both ESRPs in the human immortalized prostrate epithelial cell line PNT2. In total, we detected AS changes in over a thousand transcripts and independently validated hundreds of these events, many undergoing a nearly complete switch from one variant to the other. Consistent with the idea that the ESRPs regulate gene products with EMT-related functions, there was enrichment for genes involved in maintenance of cell–cell adhesion, cell motility, and cell–matrix adhesion, as well as components of the MAPK pathway and regulators and effectors of the Rho GTPases [49, 70].
We also evaluated the role of ESRP expression for the maintenance of epithelial cell characteristics by observing HMLE cells during sustained knockdown of ESRP expression. HMLEs lacking ESRP1 and ESRP2 or ESRP1 alone lost cell-cell adhesion, acquired a stellate like shape, and became more motile. Our observation of changes in cell morphology and behavior through abrogation of an epithelial splicing program suggests that the ESRPs regulate transcripts that, while expressed in both epithelial and mesenchymal cells, encode protein isoforms with unique functions specific to each cell type. Together, these results strongly support a model whereby the ESRPs are master regulators of an AS program essential for the maintenance of epithelial cell function and morphology.
5.2. Alternative splicing associated with breast cancer metastasis
Breast cancers and breast cancer cell lines have been sub-divided based on their transcriptional gene expression profiles into tumor subtypes associated with different biological and prognostic properties [59, 71–73]. In contrast to luminal and basal-like tumors a core gene expression profile of a claudin-low subtype has been defined that is associated with metastasis and that reflects the EMT wherein they express mesenchymal markers such as Vimentin and N-cadherin and a reduction in epithelial cell markers such as the prototypical E-cadherin [73]. Breast cancer cell lines with a claudin-low gene expression profile (also known as basal B) similarly show increased invasiveness and metastatic potential when compared with luminal cell line subtypes that have a gene expression signature reminiscent of epithelial cells [74, 75]. These characteristics indicate that claudin-low cells arose through an EMT. Recently it has been shown that these cells can be faithfully sub-divided in the same manner based on patterns of AS as assessed by splicing sensitive microarrays [66]. Importantly, we found a large and significant overlap of genes between this data set and the set of ESRP targets found in our studies. This observation was expected as the claudin-low cells express very low levels of the ESRPs compared to the luminal cells [75]. Interestingly, when gene pathway enrichment was compared between genes that were differentially expressed to those with changes in AS, there were distinct differences in function between the two sets of genes. These results provide further evidence that coordinated changes of AS is an important mechanism of gene regulation during the EMT that adds an additional layer beyond transcriptional regulation. It is important to point out, however, that while the role of EMT in cancer metastasis has been most clearly defined in breast cancer, there is ample evidence that it is a more general feature in the dissemination of carcinomas as well as their response to therapies (reviewed in [1]). Thus, for example, a role for the EMT in metastasis of colorectal, pancreatic, ovarian, prostate, cervical, thyroid, and other tumors has also been defined suggesting that a switch in an AS signature is likely to be more generally associated with cancer metastasis.
5.3. Profiling alternative splicing upon Twist induced EMT
Another approach taken was to use RNA-Seq to identify changes in AS in HMLEs that were induced to undergo an EMT by expression of Twist [58]. This experiment offered an opportunity to observe changes in splicing that were a direct consequence of the EMT. We had previously established that the ESRPs are down-regulated in this experimental system and indeed many of the more robust changes in AS detected were also identified in our own experiments. However, the use of different cell types and detection methodologies complicated a more direct comparison of these results with ESRP target events. Notably, when the authors of this study investigated changes in expression of RBPs during the EMT, they found that the ESRPs showed the most discordance in expression between control cells and those expressing Twist. In experiments parallel to knocking down ESRP expression to partially induce an EMT, this study and another demonstrated that forced expression of ESRP1 could attenuate the induction of an EMT [57, 58]. The collective evidence strongly suggests that the ESRPs, in combination with more ubiquitously expressed splicing factors, are the primary drivers of splicing changes that occur during the EMT.
6. New examples of EMT induced changes in AS that are strongly suspected to be functionally relevant
We will first highlight examples of genes that function in cellular processes that underlie the EMT that have been shown to exhibit isoform specific functions or localization. However, we emphasize that even in most of these cases our understanding of how such differential activities of these isoforms impact cell behavior is wanting. In addition, there remains a second intriguing and significantly longer list of splicing changes in genes encoding proteins with highly relevant functions in the EMT, but for which isoform-dependent functions have been neither entertained nor studied. It is worth pointing out that in these cases there are examples where previous studies have uncovered paradoxes or inconsistent observations on the functions of these genes that might be accounted for by isoform specific functions (see 3.2. p120-Catenin). Several examples of each class of gene are summarized in Table 1. We touch upon the details of a few AS events with large changes in splicing in response to ESRP expression and/or EMT inducers that merit discussion.
Table 1.
| Gene (Other names) | Function | Epithelial splicing | Domain affected/functional difference | Ref. |
|---|---|---|---|---|
| FGFR2 | Transmembrane receptor tyrosine kinase | Exon IIIb | Confers ligand binding specificity | [19, 20] |
| CTNND1 (p120-Catenin) | Delta-catenin; regulator of cell adhesion and signaling | Skipped | Coiled-coil domain; stabilizes interaction with RalA | [28, 29] |
| CD44 | Cell-surface glycoprotein involved in cell adhesion and migration | Exons v8–v10 | Extra-cellular membrane proximal region; creates a heavily glycosylated stalk | [33] |
| ENAH (Mena) | Regulator of actin dynamics | Included | Ena/Vasp homology domain; contains a phosphorylation site that may disrupt actin binding | [100] |
| NUMB | Complex protein implicated in many roles including cell migration and adhesion | Included | Phosphotyrosine binding domain; the encoded peptide confers localization to the plasma membrane | [82] |
| FLNB | F-actin cross-linking protein | Included | Contributes to the hinge domain; allows for more rigid actin branching | [101] |
| DNM2 | GTPase that binds cytosketal proteins | Included | Pleckstrin homology domain; affects subcellular localization | [85, 86] |
| TCF7L2 (Tcf4) | Transcription factor involved in Wnt signaling pathway | Included | Differential activation of Wnt/β-catenin target genes | [84] |
| BAIAP2 (Irsp53) | Cdc42 effector protein involved in lamellipodia and filopodia formation | Included | Pentultimate exon with stop codon; differentially phosphorylated in response to IGF-1 | [78] |
| MAP3K7 (Tak1) | Kinase that mediates TGF-β and BMP signal transduction | Included; Skipped | Peptide encoded by downstream exon is required for interaction with Tab2/3 | [102] |
| ARHGAP17 (Rich1) | GTPase-activating protein involved in maintenance of the tight junction | Skipped | Part of proline rich domain | [88] |
| MAGI1 (Baiap1) | Scaffolding protein associated with complexes at the inner plasma membrane | Skipped | Encodes peptide between the two WW domains | [89] |
| LRRFIP2 | Involved in activation of Wnt signaling | Skipped | Predicted coiled coil domain; encoded peptide may enhance interaction with Dvl3 | [90] |
| SCRIB | Scaffolding protein associated with tight junctions and cell polarity | Skipped | Encodes a peptide proximal to the first PDZ domain | [91, 93] |
| EPB41L5 (Ymo1) | A FERM protein that interacts with Crumbs complex to regulate cell architecture | Short isoform | Paxillin-binding domain; enhances focal adhesion complexes | [95, 98] |
| RALGPS2 | A guanine nucleotide exchange factor involved in cytoskeleton reorganization | Included | Between a PxxP motif and a pleckstrin homology domain; may influence GEF activity | [103] |
| ITGA6 | Alpha subunit of integrin, a laminin receptor | Included | Light chain and cytoplasmic domain; changes C-terminus sequence | [104] |
| SLK | STE20-like kinase with a role in promoting cell motility | Included | Predicted coiled-coil domain; may specify interaction partners | [105] |
| ARHGEF11 (PDZ-RhoGEF) | RhoA-specific guanine nucleotide exchange factor | Skipped | C-terminus; may influence homodimerization or interaction with PAK4 and LARG | [106, 107] |
6.1. EMT associated AS events in which isoform specific functions have been demonstrated
6.1.1. BAIAP2 (IRSp53)
BAIAP2, better known as insulin receptor substrate p53 (IRSp53), is a CDC42 effector protein involved in stress fiber formation as well as filopodia and lamellopodia formation in motile cells [76]. ESRP enhances splicing of a penultimate exon that contains a stop codon 18 nt from the 3′ end of the exon and thereby changes the C-terminal coding sequence from that of mesenchymal isoforms that skip the exon. Interestingly, BAIAP2 interacts with Mena and both act synergistically to promote filopodia formation [77]. Differential, isoform-specific phosphorylation of tyrosine residues in the common portion of the protein was observed in response to insulin or IGF-1 stimulation. However, the functional consequences of this observation remain unclear [78].
6.1.2. NUMB
Recent studies have shown diverse roles for Numb in regulating cell-cell adhesion, polarity and migration during EMT. Numb has been shown to interact with E-cadherin in the maintenance of adherens junctions and also directly interacts with p120-catenin [79–81]. The interaction of Numb with E-cadherin involves an N-terminal phosphotyrosine-binding (PTB) domain and abrogation of the Numb-E-cadherin interaction was proposed to contribute to loss of cell polarity and cell-cell adhesion in early EMT events. The 33 nt epithelial-specific exon in Numb encodes an 11 amino acid insert in this PTB domain and it is therefore predicted that loss of this insert and consequent changes within the PTB binding domain during the EMT may also abrogate the Numb-E-cadherin interaction and thereby promote loss of cell-cell-adhesion and promote cell motility. Consistent with this possibility, isoforms that contain this insert in the PTB domain are predominantly associated with the plasma membrane, whereas isoforms that exclude it are diffusely cytoplasmic [82].
6.1.3. TCF7L2 (Tcf-4)
Wnt signaling has been implicated in the EMT and TCF/LEF transcription factors together with beta-catenin are downstream effectors that induce transcriptional changes that are associated with the EMT [83]. In addition to a previously demonstrated epithelial specific exon in TCF7L2 (TCF-4), our more recent studies have shown additional complex regulation of TCF7L2 exons. The complex AS of TCF7L2 has been shown to yield various isoforms that differentially activate several Wnt/β-catenin target genes [84]. It will therefore be of interest to determine whether some of the transcriptional changes that occur during the EMT might reflect changes in splicing of TCF7L2 and potentially other transcription factors.
6.1.4. DNM2
Dynamin 2 (DNM2) is a GTPase that interacts with numerous actin binding proteins to regulate assembly and turnover of the actin cytoskeleton during membrane turnover, endocytosis, and cell migration. A 12 nt epithelial exon encodes a short four amino acid sequence (GEIL) that was shown to mediate localization to the GOLGI apparatus, whereas the forms without it localized to punctuate spots throughout the plasma membrane [85]. However, another study suggested that the primary difference was that the insert containing isoforms had reduced ability to rescue transport of p75 (an apical plasma cell membrane marker) from the TGN to the plasma membrane [86]. How either of these activities differentially affects endocytic and other pathways in the EMT requires further investigation.
6.2. EMT associated AS events with uncharacterized isoform specific functions
6.2.1. MAP3K7 (TAK1)
TGF-beta activated protein kinase (TAK1; official gene symbol MAP3K7) contains two highly conserved alternatively spliced exons of which one is enhanced by ESRP expression and the other is silenced. TGF-beta is a well-described inducer of the EMT through both SMAD-dependent and SMAD-independent pathways. Recent studies have shown that TAK1 is required for TGF-beta induced EMT through participation in a TRAF6-TAK1-JNK/p38 pathway [87]. Given the central role of TAK1 in cell signaling events that regulate the EMT it will thus be of great interest to determine whether epithelial versus mesenchymal TAK1 isoforms have distinct signaling properties and protein interactions that impact cell behaviors. It is notable that a peptide sequence at the C-terminus of TAK1 that was shown to mediate interactions with TAK1-binding proteins 2 and 3 (TAB2 and TAB3) is missing from the epithelial isoform, though whether this exon switch indeed abrogates this binding and whether such a change might impact the EMT has not been shown.
6.2.2. ARHGAP17 and MAGI1
Epithelial tight junctions are essential for maintenance of epithelial cell polarity and epithelial barrier functions. The CDC42 associated guanine nucleotide exchange protein ARHGAP17 and the membrane-associated guanylate kinase (MAGUK) protein MAGI1 play roles in tight junction maintenance through CDC effector and scaffolding functions, respectively [88, 89]. It therefore might be predicted that these functions are mediated primarily by the epithelial isoforms and that the switch towards the mesenchymal forms might promote tight junction dissolution during the EMT.
6.2.3. LRRFIP2
Leucine rich repeat (in FLII) interacting protein (LRRFIP2) is an activator of canonical Wnt signaling that functions upstream of beta catenin though interactions with disheveled 3 (Dvl3) [90]. The exon in LRRFIP2 that is silenced by ESRP is within or near the domain that mediates the interaction with Dvl3, suggesting that skipping of this exon might induce downregulation of Wnt targets in response to ESRP induced splicing changes, although this remains to be determined. Together with the case of TCF7L2, these changes in splicing of Wnt pathway components suggest that differences in splicing between epithelial and mesenchymal cell types impact the transcriptional programs that are downstream of Wnt signaling at multiple steps.
6.2.4. SCRIB
SCRIB is a scaffolding protein associated with cell-cell junctions in epithelial cells and in epithelial MDCK cells a loss of SCRIB was associated with increased cell migration and phenotypic changes consistent with EMT [91]. In contrast, in different contexts SCRIB knockdown inhibited cell migration and invasion and induced downregulation of mesenchymal markers [92, 93]. In addition it was shown that the localization of SCRIB to lateral junctions was determined by E-cadherin expression and in cells that did not express this epithelial marker, SCRIB displayed differential localization. It is notable that the mesenchymal SCRIB isoform contains an exon that is skipped in epithelial cells and this exon overlaps with a domain shown to mediate interaction with beta-PIX. Thus, it may also be the case that these context dependent functions may reflect isoform-specific functions, but this has not been examined.
6.2.5 EPB41L5
Mouse knockout studies demonstrated that EPB41L5 is required for the EMT at areas of enhanced cell-cell adhesion during gastrulation [94, 95]. This was further shown to involve interactions with p120-catenin and E-cadherin that disrupted cell-cell adhesions, as well as a C-terminal paxillin-binding domain that enhanced focal adhesion formation. However, it was noted that EPB41L5 was present in all germ layers and embryonic epithelia begging the question as to why it did not promote EMT in those contexts [94]. In addition, EPB41L5 and its orthologs in zebrafish and Drosophila were also shown to regulate epithelial polarity [96–98]. Of interest is that the extended C-terminus that includes the paxillin-binding domain is not present in the shorter epithelial EPB41L5 isoform, suggesting that this may account for some of these context dependent differences, although here again this has not been explored.
7. Conclusion and future directions
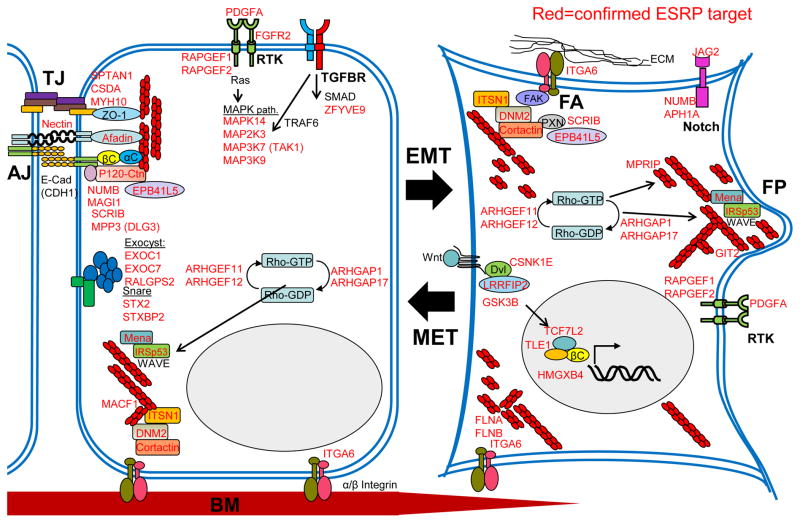
The current view, supported by the work reviewed here, is that numerous AS events are coordinately regulated in epithelial cells through the actions of ESRP1 and ESRP2. Central to the maintenance of epithelial cell polarity are the tight and adherens junctions. Coordinated changes in the organization of the actin cytoskeleton during the EMT involve contributions from key signaling pathways that dissolve these structures. At the same time, these cellular changes induce the formation of processes that mediate cell motility through the formation of focal adhesions and other structures. In Figure 3 we highlight some of these key protein complexes and pathways and illustrate how validated ESRP-regulated targets function at each these nodes. This epithelial SRN thus controls the expression of specific protein isoforms that function to maintain the morphology, behavior, and cellular characteristics of epithelial cells. When an EMT is elicited, a coordinated switch in splicing occurs in hundreds of genes through the repression of ESRP expression to give rise to a proteome that confers mesenchymal-like properties in the cell. Three independent studies aiming to comprehensively identify splicing changes that occur during the EMT have produced largely overlapping data sets. This is a significant finding given the variety of cell lines used in each of the studies and suggests that a repertoire of genes common to many epithelial and mesenchymal cells have different functions in these cell types via AS. This interpretation thus provides an impetus for many labs to consider and investigate the possibility that such apparently divergent functions that have been assigned to the same gene/protein may reflect the consequence of a switch in AS.
Figure 3.
Gene transcripts that switch splicing during the EMT function in complexes or pathways that define epithelial and mesenchymal cell behaviors. Gene names or symbols indicated in red type represent validated ESRP target genes whereas those in black type are nodes shown for orientation. A representative epithelial cell is shown at left and a mesenchymal cell after EMT is shown at right. Complexes important for epithelial cell polarity including tight junctions (TJ) and adherens junctions (AJ) are shown along with proteins associated with each complex. Additional signaling pathways and protein-protein interactions with relevance to the EMT are also shown. The red chain-like structures represent filamentous actin and sites of actin-cytoskeletal organization. FA- focal adhesion; FP- filopodia or other dynamic cell extensions; RTK- receptor tyrosine kinase; BM- basement membrane.
The establishment of this novel genetic program is a significant development for the EMT field but has also given rise to new interesting questions and challenges. Thus far all of the evidence for an epithelial SRN and the role of the ESRPs in the EMT has been obtained through experiments performed in cell lines. To truly understand this splicing network in the context of development will require the generation of knockout mice to delineate the extent to which the ESRPs are involved in the EMT. Interestingly, a recent study demonstrated that reprogramming of mouse embryonic fibroblasts to create induced pluripotent stem cells (iPSCs) is characterized by an MET during the initiation phase [99]. Among the most highly upregulated genes during this process is Esrp1. It is tempting to speculate that Esrp1 could be used as an additional reprogramming factor to induce the MET and thereby boost the efficiency of iPSC production. Another issue to be addressed is the utility of the splicing signature whereby analysis of a core set of AS events can determine if a cell type or tumor is epithelial, mesenchymal, or undergoing a dynamic transition between these cell types [58, 70]. Such an assay may prove to be a valuable diagnostic tool in the clinic as cancer cells undergoing an EMT display a more aggressive tumor phenotype.
The most challenging goal going forward, yet one of great importance, will be to elucidate the specific functions for the protein isoforms produced from the various splice variants. While it may be possible to use cell-based assays to systematically investigate these genes in some of these processes, a thorough analysis will require a more gene-by-gene focused analysis that necessarily will require contributions by multiple investigators. Nevertheless, in order to gain a full understanding of the events that occur at the molecular level which give rise to the EMT or cancer metastasis, this is an objective that must be pursued.
Acknowledgments
The authors gratefully acknowledge members of the Carstens lab for helpful discussion. We also thank Wei Guo, Peter Stoilov, and Yi Xing for critical review of the manuscript. We apologize to those authors whose work could not be cited due to space limitations. Work in the Carstens lab was supported by NIH grants R01 CA093769 and R01 GM088809.
Abbreviations
AS
alternative splicing
ESRP
epithelial splicing regulatory protein
RBP
RNA binding protein
RRM
RNA recognition motif
SRN
splicing regulatory network
Footnotes
Conflict of interest statement
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–29. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 5.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 6.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–54. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–47. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 13.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 14.Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–5. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 15.Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–52. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 16.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–8. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 18.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carstens RP, Eaton JV, Krigman HR, Walther PJ, Garcia-Blanco MA. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene. 1997;15:3059–65. doi: 10.1038/sj.onc.1201498. [DOI] [PubMed] [Google Scholar]
- 21.Savagner P, Valles AM, Jouanneau J, Yamada KM, Thiery JP. Alternative splicing in fibroblast growth factor receptor 2 is associated with induced epithelial-mesenchymal transition in rat bladder carcinoma cells. Mol Biol Cell. 1994;5:851–62. doi: 10.1091/mbc.5.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Medina SG, Popov Z, Chopin DK, Southgate J, Tucker GC, Delouvee A, et al. Relationship between E-cadherin and fibroblast growth factor receptor 2b expression in bladder carcinomas. Oncogene. 1999;18:5722–6. doi: 10.1038/sj.onc.1202958. [DOI] [PubMed] [Google Scholar]
- 23.Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- 24.De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–92. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 25.Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P. Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc Natl Acad Sci U S A. 1999;96:11895–9. doi: 10.1073/pnas.96.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–34. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosheva I, Shtutman M, Elbaum M, Bershadsky AD. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J Cell Sci. 2001;114:695–707. doi: 10.1242/jcs.114.4.695. [DOI] [PubMed] [Google Scholar]
- 28.Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, et al. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem. 2008;283:18344–54. doi: 10.1074/jbc.M801192200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keirsebilck A, Bonne S, Staes K, van Hengel J, Nollet F, Reynolds A, et al. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–46. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- 30.Ohkubo T, Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci. 2004;117:1675–85. doi: 10.1242/jcs.01004. [DOI] [PubMed] [Google Scholar]
- 31.Sarrio D, Perez-Mies B, Hardisson D, Moreno-Bueno G, Suarez A, Cano A, et al. Cytoplasmic localization of p120ctn and E-cadherin loss characterize lobular breast carcinoma from preinvasive to metastatic lesions. Oncogene. 2004;23:3272–83. doi: 10.1038/sj.onc.1207439. [DOI] [PubMed] [Google Scholar]
- 32.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–67. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 33.Bennett KL, Modrell B, Greenfield B, Bartolazzi A, Stamenkovic I, Peach R, et al. Regulation of CD44 binding to hyaluronan by glycosylation of variably spliced exons. J Cell Biol. 1995;131:1623–33. doi: 10.1083/jcb.131.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terpe HJ, Stark H, Prehm P, Gunthert U. CD44 variant isoforms are preferentially expressed in basal epithelial of non-malignant human fetal and adult tissues. Histochemistry. 1994;101:79–89. doi: 10.1007/BF00269353. [DOI] [PubMed] [Google Scholar]
- 35.Hong RL, Pu YS, Chu JS, Lee WJ, Chen YC, Wu CW. Correlation of expression of CD44 isoforms and E-cadherin with differentiation in human urothelial cell lines and transitional cell carcinoma. Cancer Lett. 1995;89:81–7. doi: 10.1016/0304-3835(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 36.Iida N, Bourguignon LY. Coexpression of CD44 variant (v10/ex14) and CD44S in human mammary epithelial cells promotes tumorigenesis. J Cell Physiol. 1997;171:152–60. doi: 10.1002/(SICI)1097-4652(199705)171:2<152::AID-JCP5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 37.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–74. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman L, Wainwright D, Ponta H, Herrlich P. A splice variant of CD44 expressed in the apical ectodermal ridge presents fibroblast growth factors to limb mesenchyme and is required for limb outgrowth. Genes Dev. 1998;12:1058–71. doi: 10.1101/gad.12.7.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–64. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 41.Gates J, Mahaffey JP, Rogers SL, Emerson M, Rogers EM, Sottile SL, et al. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–39. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- 42.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–21. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 43.Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W, et al. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis. 2009;26:153–9. doi: 10.1007/s10585-008-9225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roussos ET, Balsamo M, Alford SK, Wyckoff JB, Gligorijevic B, Wang Y, et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci. 2011;124:2120–31. doi: 10.1242/jcs.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pino MS, Balsamo M, Di Modugno F, Mottolese M, Alessio M, Melucci E, et al. Human Mena+11a isoform serves as a marker of epithelial phenotype and sensitivity to epidermal growth factor receptor inhibition in human pancreatic cancer cell lines. Clin Cancer Res. 2008;14:4943–50. doi: 10.1158/1078-0432.CCR-08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hovhannisyan RH, Carstens RP. Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem. 2007;282:36265–74. doi: 10.1074/jbc.M704188200. [DOI] [PubMed] [Google Scholar]
- 47.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–22. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hovhannisyan RH, Carstens RP. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol Cell Biol. 2005;25:250–63. doi: 10.1128/MCB.25.1.250-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–62. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dominguez C, Fisette JF, Chabot B, Allain FH. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol. 2010;17:853–61. doi: 10.1038/nsmb.1814. [DOI] [PubMed] [Google Scholar]
- 51.Barberan-Soler S, Zahler AM. Alternative splicing regulation during C. elegans development: splicing factors as regulated targets. PLoS Genet. 2008;4:e1000001. doi: 10.1371/journal.pgen.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yochem J, Bell LR, Herman RK. The identities of sym-2, sym-3 and sym-4, three genes that are synthetically lethal with mec-8 in Caenorhabditis elegans. Genetics. 2004;168:1293–306. doi: 10.1534/genetics.104.029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wakabayashi-Ito N, Belvin MP, Bluestein DA, Anderson KV. fusilli, an essential gene with a maternal role in Drosophila embryonic dorsal-ventral patterning. Dev Biol. 2001;229:44–54. doi: 10.1006/dbio.2000.9954. [DOI] [PubMed] [Google Scholar]
- 54.Huang Y, Fernandez SV, Goodwin S, Russo PA, Russo IH, Sutter TR, et al. Epithelial to mesenchymal transition in human breast epithelial cells transformed by 17beta-estradiol. Cancer Res. 2007;67:11147–57. doi: 10.1158/0008-5472.CAN-07-1371. [DOI] [PubMed] [Google Scholar]
- 55.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–54. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 56.Thomson S, Petti F, Sujka-Kwok I, Mercado P, Bean J, Monaghan M, et al. A systems view of epithelial-mesenchymal transition signaling states. Clin Exp Metastasis. 2011;28:137–55. doi: 10.1007/s10585-010-9367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2011 doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–90. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 61.Collesi C, Santoro MM, Gaudino G, Comoglio PM. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol Cell Biol. 1996;16:5518–26. doi: 10.1128/mcb.16.10.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou YQ, He C, Chen YQ, Wang D, Wang MH. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: generation of different splicing RON variants and their oncogenic potential. Oncogene. 2003;22:186–97. doi: 10.1038/sj.onc.1206075. [DOI] [PubMed] [Google Scholar]
- 63.Goncalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet. 2009;18:3696–707. doi: 10.1093/hmg/ddp317. [DOI] [PubMed] [Google Scholar]
- 64.Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–7. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, et al. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol. 2010;191:87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res. 2010;8:961–74. doi: 10.1158/1541-7786.MCR-09-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vajda NA, Brimacombe KR, LeMasters KE, Ladd AN. Muscleblind-like 1 is a negative regulator of TGF-beta-dependent epithelial-mesenchymal transition of atrioventricular canal endocardial cells. Dev Dyn. 2009;238:3266–72. doi: 10.1002/dvdy.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore MJ, Silver PA. Global analysis of mRNA splicing. RNA. 2008;14:197–203. doi: 10.1261/rna.868008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–24. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–97. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 74.Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–42. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 75.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol. 2001;11:1645–55. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- 77.Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. 2008;283:20454–72. doi: 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- 78.Okamura-Oho Y, Miyashita T, Yamada M. Distinctive tissue distribution and phosphorylation of IRSp53 isoforms. Biochem Biophys Res Commun. 2001;289:957–60. doi: 10.1006/bbrc.2001.6102. [DOI] [PubMed] [Google Scholar]
- 79.Sato K, Watanabe T, Wang S, Kakeno M, Matsuzawa K, Matsui T, et al. Numb controls E-cadherin endocytosis through p120 catenin with aPKC. Mol Biol Cell. 2011;22:3103–19. doi: 10.1091/mbc.E11-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, Li SS. Numb: A new player in EMT. Cell Adh Migr. 2010;4:176–9. doi: 10.4161/cam.4.2.10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Sandiford S, Wu C, Li SS. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. Embo J. 2009;28:2360–73. doi: 10.1038/emboj.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dho SE, French MB, Woods SA, McGlade CJ. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem. 1999;274:33097–104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 83.Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell. 2006;17:1871–9. doi: 10.1091/mbc.E05-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wohrle S, et al. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucleic Acids Res. 2010;38:1964–81. doi: 10.1093/nar/gkp1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao H, Garcia F, McNiven MA. Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell. 1998;9:2595–609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells. Mol Biol Cell. 2008;19:5347–59. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–39. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, et al. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125:535–48. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 89.Laura RP, Ross S, Koeppen H, Lasky LA. MAGI-1: a widely expressed, alternatively spliced tight junction protein. Exp Cell Res. 2002;275:155–70. doi: 10.1006/excr.2002.5475. [DOI] [PubMed] [Google Scholar]
- 90.Liu J, Bang AG, Kintner C, Orth AP, Chanda SK, Ding S, et al. Identification of the Wnt signaling activator leucine-rich repeat in Flightless interaction protein 2 by a genome-wide functional analysis. Proc Natl Acad Sci U S A. 2005;102:1927–32. doi: 10.1073/pnas.0409472102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J Cell Biol. 2005;171:1061–71. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vaira V, Faversani A, Dohi T, Maggioni M, Nosotti M, Tosi D, et al. Aberrant overexpression of the cell polarity module scribble in human cancer. Am J Pathol. 2011;178:2478–83. doi: 10.1016/j.ajpath.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dow LE, Kauffman JS, Caddy J, Zarbalis K, Peterson AS, Jane SM, et al. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene. 2007;26:2272–82. doi: 10.1038/sj.onc.1210016. [DOI] [PubMed] [Google Scholar]
- 94.Hirano M, Hashimoto S, Yonemura S, Sabe H, Aizawa S. EPB41L5 functions to post-transcriptionally regulate cadherin and integrin during epithelial-mesenchymal transition. J Cell Biol. 2008;182:1217–30. doi: 10.1083/jcb.200712086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee JD, Silva-Gagliardi NF, Tepass U, McGlade CJ, Anderson KV. The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial-mesenchymal transition at the primitive streak of the mouse embryo. Development. 2007;134:2007–16. doi: 10.1242/dev.000885. [DOI] [PubMed] [Google Scholar]
- 96.Gosens I, Sessa A, den Hollander AI, Letteboer SJ, Belloni V, Arends ML, et al. FERM protein EPB41L5 is a novel member of the mammalian CRB-MPP5 polarity complex. Exp Cell Res. 2007;313:3959–70. doi: 10.1016/j.yexcr.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 97.Laprise P, Beronja S, Silva-Gagliardi NF, Pellikka M, Jensen AM, McGlade CJ, et al. The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev Cell. 2006;11:363–74. doi: 10.1016/j.devcel.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Christensen AK, Jensen AM. Tissue-specific requirements for specific domains in the FERM protein Moe/Epb4.1l5 during early zebrafish development. BMC Dev Biol. 2008;8:3. doi: 10.1186/1471-213X-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 100.Di Modugno F, DeMonte L, Balsamo M, Bronzi G, Nicotra MR, Alessio M, et al. Molecular cloning of hMena (ENAH) and its splice variant hMena+11a: epidermal growth factor increases their expression and stimulates hMena+11a phosphorylation in breast cancer cell lines. Cancer Res. 2007;67:2657–65. doi: 10.1158/0008-5472.CAN-06-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van der Flier A, Kuikman I, Kramer D, Geerts D, Kreft M, Takafuta T, et al. Different splice variants of filamin-B affect myogenesis, subcellular distribution, and determine binding to integrin [beta] subunits. J Cell Biol. 2002;156:361–76. doi: 10.1083/jcb.200103037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Besse A, Lamothe B, Campos AD, Webster WK, Maddineni U, Lin SC, et al. TAK1-dependent signaling requires functional interaction with TAB2/TAB3. J Biol Chem. 2007;282:3918–28. doi: 10.1074/jbc.M608867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ceriani M, Scandiuzzi C, Amigoni L, Tisi R, Berruti G, Martegani E. Functional analysis of RalGPS2, a murine guanine nucleotide exchange factor for RalA GTPase. Exp Cell Res. 2007;313:2293–307. doi: 10.1016/j.yexcr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 104.Hogervorst F, Admiraal LG, Niessen C, Kuikman I, Janssen H, Daams H, et al. Biochemical characterization and tissue distribution of the A and B variants of the integrin alpha 6 subunit. J Cell Biol. 1993;121:179–91. doi: 10.1083/jcb.121.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roovers K, Wagner S, Storbeck CJ, O’Reilly P, Lo V, Northey JJ, et al. The Ste20-like kinase SLK is required for ErbB2-driven breast cancer cell motility. Oncogene. 2009;28:2839–48. doi: 10.1038/onc.2009.146. [DOI] [PubMed] [Google Scholar]
- 106.Barac A, Basile J, Vazquez-Prado J, Gao Y, Zheng Y, Gutkind JS. Direct interaction of p21-activated kinase 4 with PDZ-RhoGEF, a G protein-linked Rho guanine exchange factor. J Biol Chem. 2004;279:6182–9. doi: 10.1074/jbc.M309579200. [DOI] [PubMed] [Google Scholar]
- 107.Chikumi H, Barac A, Behbahani B, Gao Y, Teramoto H, Zheng Y, et al. Homo- and hetero-oligomerization of PDZ-RhoGEF, LARG and p115RhoGEF by their C-terminal region regulates their in vivo Rho GEF activity and transforming potential. Oncogene. 2004;23:233–40. doi: 10.1038/sj.onc.1207012. [DOI] [PubMed] [Google Scholar]