Light-Controlled Synthetic Gene Circuits (original) (raw)
. Author manuscript; available in PMC: 2013 Aug 1.
Published in final edited form as: Curr Opin Chem Biol. 2012 May 25;16(3-4):292–299. doi: 10.1016/j.cbpa.2012.04.010
Abstract
Highly complex synthetic gene circuits have been engineered in living organisms to develop systems with new biological properties. A precise trigger to activate or deactivate these complex systems is desired in order to tightly control different parts of a synthetic or natural network. Light represents an excellent tool to achieve this goal as it can be regulated in timing, location, intensity, and wavelength, which allows for precise spatiotemporal control over genetic circuits. Recently, light has been used as a trigger to control the biological function of small molecules, oligonucleotides, and proteins involved as parts in gene circuits. Light activation has enabled the construction of unique systems in living organisms such as band-pass filters and edge-detectors in bacterial cells. Additionally, light also allows for the regulation of intermediate steps of complex dynamic pathways in mammalian cells such as those involved in kinase networks. Herein we describe recent advancements in the area of light-controlled synthetic networks.
Introduction
Gene expression is regulated by precisely orchestrated genetic networks that operate with a high degree of spatiotemporal control. These genetic circuits represent building blocks for synthetic biology, a discipline that strives to apply electrical engineering principles to the design and understanding of biological processes and to create new biological systems with useful properties [1-3].
In order to program synthetic gene circuits to perform their various functions, precise external control over their activity needs to be achieved. Any input for an engineered system under study must be accurate, precise, and tunable to ensure stable output generation. Light serves as an excellent trigger to achieve precise control over synthetic systems (Figure 1A) as it can be regulated in wavelength, timing, intensity, and location [4-6]. In this review, we present select recent advances in the development of light-controlled synthetic gene networks and their applications.
Figure 1.
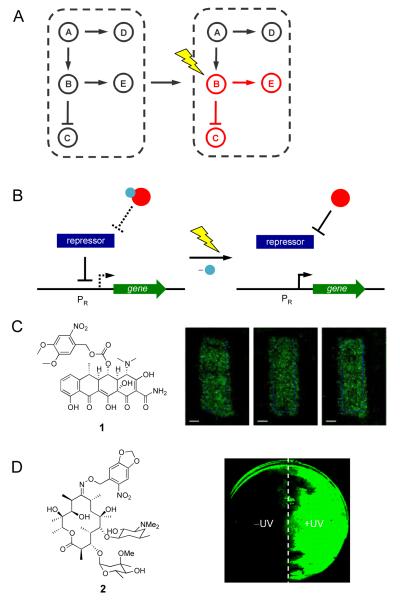
(A) Light enables precise spatial and temporal activation of genetic circuits, even the activation of specific nodes in a natural or synthetic network. (B) Control over gene expression with photocaged small molecule inducers of transcription. The photocaging group is represented by a blue sphere and the small molecule inducer is represented by a red sphere. When the small molecule is caged, the repressor protein will bind the promoter PR. After UV irradiation the caging group is removed and the small molecule will bind the repressor, which releases PR, and allows for transcription to occur. (C) NvOC-Dox (1) was used to create photolithographic images onto an NIH 3T3 monolayer expressing GFP. (D) Photocaged erythromycin (2) was used in the spatial control of a light activated logic gate; cells treated with 2 where one half of the plate was exposed to UV light. Adapted with permission from the American Chemical Society, © 2010, from [17] and the Royal Society of Chemistry, © 2011, from [18].
Photochemical regulation of gene circuits with caged small molecules
Small molecule inducible gene expression systems are a fundamental method of gene regulation in pro- and eukaryotic organisms [7]. These gene switches are comprised of a repressor protein that binds to a promoter region upstream of the gene of interest inhibiting gene expression. Binding of a small molecule induces a conformational change and releases the repressor from the DNA thereby inducing gene expression. These switches exhibit high specificity for the small molecule and its cognate repressor protein, tight regulation between ‘on’ and ‘off’ states, and low basal levels of gene expression [7]. Small molecule activated gene switches have long been used in traditional molecular biology to control expression of recombinant proteins. These features, as well as the ease of manipulation of small organic molecules, make small molecule inducers targets for photochemical control of gene networks. Light-inducible small molecule systems are often generated through the installation of a light-cleavable photo-protecting (photocaging) group on the small molecule inducer of gene expression, rendering the small molecule inactive until the caging group is removed through irradiation (Figure 1B).
The first instance of using a photocaged small molecule inducer to control a genetic circuit was the application of a photocaged estradiol for control of transcriptional activator activity in eukaryotic cells [8]. This light-inducible system demonstrated the modular nature of small molecule inducible systems, as the necessary components of transcriptional regulation can be separated from their endogenous genes and recombined to create new synthetic circuits. Since then, various other photocaged small molecules have been developed to created light-activatable molecular switches including a caged ecdysone [9], a caged IPTG for use with the lac operon [10], caged toyocamycin for ribozyme-mediated gene expression [11], caged doxycycline for use with the Tet-on system [12], and caged rapamycin for light-mediated FKBP/FRB dimerization [13-15].
Various photocaged analogues of doxycycline have been shown to control gene expression in eukaryotic systems, including transgenic mice [16]. Recently, Koh et. al, reported a new photocaged doxycycline, NvOC-Dox 1 (Figure 1C), to create photolithographic patterns of gene expression in mammalian cells [17].
Discrete patterning was achieved in NIH 3T3 cells that grew into defined monolayers and contained a tetracycline-inducible GFP reporter (HRSp-GFP). Photolithography experiments were performed by treating cells harboring HRSp-GFP with 1, followed by irradiation with UV light through a photomask applied to the bottom of a cell culture. Patterns of GFP expression with clearly defined edges were obtained using masks of different shapes (Figure 1C). This system was also used to control cell adhesion of human embryonic kidney (HEK293T) cells to a mouse fibroblast (NIH 3T3) cellular monolayer in a spatially directed fashion. Light-inducible expression of ephrin A5, a membrane bound ligand that mediates cell-cell interactions by interacting with EphA7 receptors, allowed for photochemical control of cell attachment.
Photocontrolled synthetic gene networks have also been reported for prokaryotic systems. The light-activated erythromycin 2 (Figure 1D) was recently developed for UV-inducible gene expression and was used to create a light activated AND gate in bacterial cells [18]. A gene cassette, which confers erythromycin resistance to E. coli, was used to construct a synthetic erythromycin-inducible gene network that regulates expression of the reporter gene EGFP. This synthetic gene cassette, when paired with the photocaged analogue of erythromycin, functions as a light-activated logic gate where both inputs and the corresponding output are specific wavelengths of light. The molecular AND gate performed its logic operation in a spatially restricted fashion, as shown by localized EGFP expression in a bacterial lawn (Figure 1D). Tunable and modular building blocks of synthetic biology, such as this light-triggered AND gate in bacterial cells and light-triggered promoter systems in mammalian cells, will allow for the construction of advanced photochemically controlled gene networks.
Photochemical regulation of gene circuits with caged oligonucleotides
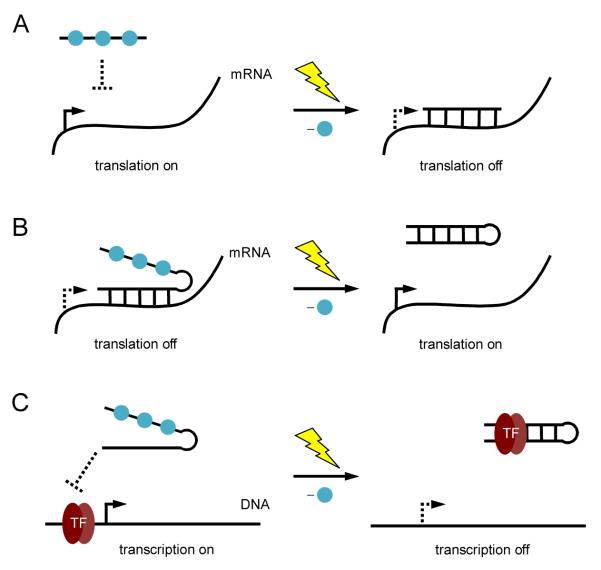
Oligonucleotides are another module of gene regulatory tools whose function can be precisely controlled with light. This has been explored previously in mammalian cells by using photocaged antisense agents to control gene silencing [19-26]. By placing photocaging groups on nucleotide bases, duplex formation between the antisense agent the target mRNA can be controlled in a spatiotemporal fashion and either deactivation (Figure 2A) or activation of gene function (Figure 2B). In addition to light-induced deactivation of gene expression, this technology was further developed to achieve photochemical activation of gene expression through control of antisense activity [27]. Caged oligonucleotides have also been used to control gene expression at the transcriptional level. A photocaged DNA decoy for the transcription factor NF-κB was used to regulate NF-κB activity with light [28]. Photocaging groups were installed to disrupt hairpin formation of the DNA decoy, and prevent binding of the DNA decoy with NF-κB. After irradiation the DNA decoy is able to form a hairpin and sequester NF-κB, which turns off gene transcription (Figure 2C).
Figure 2.
(A) Photochemical activation of gene silencing using a caged antisense agent. Photocaging groups (blue spheres) were placed on the antisense agent to prevent hybridization to its target mRNA. UV irradiation induces decaging and gene silencing. (B) Photochemical deactivation of an antisense agent. Hairpin formation of the antisense agent is disrupted through nucleobase caging, allowing the antisense agent to bind its target mRNA and downregulate translation. After UV irradiation, hairpin formation occurs rendering the antisense agent inactive and activating gene expression. (C) Photochemical control of transcription using a caged DNA decoy. The caged decoy is unable to form a hairpin and does not bind the transcription factor NF-κB. After UV irradiation, the DNA decoy forms a hairpin and is able to bind the transcription factor.
Recent advances in building DNA circuits [29] that are able to perform complex computational algorithms were combined with photochemical oligonucleotide activation in the construction of a light-activated DNA logic gate [30]. This lays the foundation toward using light as an interface between biological, DNA-based computation and electrical, silicon-based circuitry.
Photochemical regulation of gene circuits with caged proteins
Proteins represent excellent targets for the engineering of light-activated cellular networks as site-specific modification of active residues can be achieved through the in vivo incorporation of photocaged amino acids. Re-engineering of the genetic code to develop caged proteins has seen application in prokaryotes and eukaryotes as regulators of gene circuits [31,32]. This was achieved through engineering of cells with an orthogonal biosynthetic machinery comprised of evolved tRNA synthetases and their cognate tRNAs. Various photocaged amino acids are accessible for incorporation, which in turn makes a wide variety of protein gene-regulators accessible for modification. The light-regulation of enzymatic function was achieved through incorporation of photocaged analogues of tyrosine [33], cysteine [34], serine [35], and lysine [36].
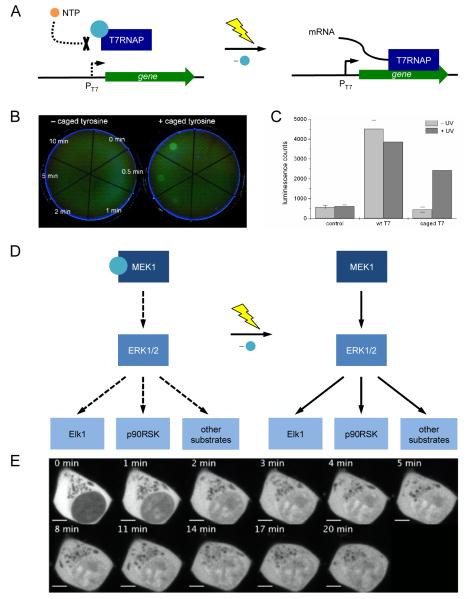
The temporary blocking of an active site with a caging group was recently used in the photochemical control of T7RNA polymerase activity and, therefore gene expression, in bacterial and mammalian cells [37]. Transcription of EGFP (under control of a T7 promoter), was photochemically controlled by caging the essential tyrosine 639 in the active site of T7RNAP. The caged polymerase was completely inactive; however, after a brief UV exposure, bacterial cells harboring plasmids expressing photocaged T7RNAP and T7 promoter-driven EGFP showed fully activated gene function (Figure 3A). This enabled the engineering of bacterial cells that respond to light exposure with the activation of a specific gene circuit and allowed for spatial control of reporter gene expression (Figure 3B). Since the T7 RNA polymerase and promoter system is orthogonal to endogenous gene circuits in E. coli, this system can be used to regulate the activity of any gene of interest with UV light.
Figure 3.
(A) Schematic representation of the photocaged T7RNAP system. The photocaging group is represented by a blue sphere. NTPs are blocked from entering the active site of T7RNAP when the enzyme is caged. After UV irradiation the caging group is removed, NTPs are free to enter the active site and transcription is activated. (B) Photoactivation of EGFP expression in E. coli through light-activation of a photocaged T7RNAP; EGFP expression is restored after 10 minutes exposure to localized 365 nm irradiation. (C) Light-activation of T7RNAP activity and gene expression in vivo. (D) Light-activated MAP kinase signaling pathway. The photocaging group is represented by a blue sphere. (E) Light-activated translocation of EGFP-tagged ERK into the nucleus of HEK293 cells. Adapted with permission from Wiley-VCH Verlag GmbH & Co, © 2010, from [37] and from the American Chemical Society, © 2011, from [36].
This system was also functional in mammalian cells as shown in the expression of a T7 promoter-driven firefly luciferase gene (Figure 3C). Co-transfection of a firefly luciferase reporter plasmid with purified caged or wild-type T7RNAP into human embryonic kidney (HEK293T) cells enabled photochemical activation of gene function. Photocaging of protein residues involved in post-translational modifications has also been used to create a light-responsive genetic circuit. Specifically, photocaged serine was applied to photochemically control phosphorylation of the yeast transcription factor Pho4, which is involved in various signaling pathways of phosphorylation responsive genes [35].
Photocaged unnatural amino acids have also allowed for the regulation of complex signaling networks such as mammalian kinase pathways [36]. The complexity and adaptivity of kinase signaling networks makes their investigation through traditional genetic approaches difficult. Chin and co-workers site-specifically incorporated a photocaged lysine into the active site of MEK1 to control the MAP kinase signaling pathway. The MAP kinase network (Figure 3D) is activated through phosphorylation of MEK1, which in turn activates ERK1 and ERK2, two kinases that translocate to the nucleus to phosphorylate transcription factors and other targets. The photocaged MEK1 was used to induce translocation of EGFP-tagged ERK2 after a brief UV irradiation (Figure 3E). When compared to stimulation with epidermal growth factor, photoactivation of the pathway led to a quicker translocation of ERK2. This revealed that factors acting upstream of MEK1 activation in the pathway are responsible for a slower ERK2 translocation rate. Photocaging of MEK1 allowed for precise control of a single node in the MAP kinase pathway.
Photochemical regulation of gene circuits with light-responsive protein domains
Proteins that are naturally light responsive can be found in organisms involved in photosynthesis or phototropism [38]. The particular protein domains responsible for the photoactivity have been used to create synthetic systems that are triggered by light [39-41].
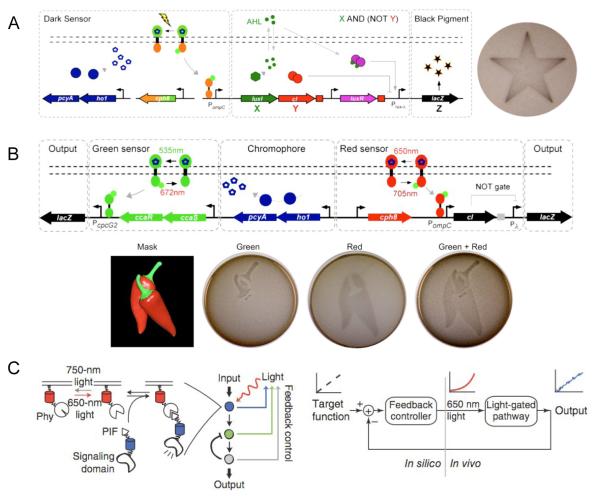
Ellington and co-workers applied the light-responsive protein Cph1 to the construction of a synthetic edge detection program in E. coli [42]. This edge detector functions by distinguishing between inputs of either light or dark (NOT light), and generating a diffusible signal in response to “NOT light”. The diffusion of the signal from cells that sense dark to neighboring cells that sense light creates the edge image by producing an output of black pigment as a result of β-galactosidase expression. Genetically, the “NOT light” sensor is comprised of a fusion of the photoreceptor domain of Cph1 (from a photosynthetic cyanobacteria) and the kinase domain of EnvZ, termed Cph8 (Figure 4A). In the absence of light, Cph8 is activated and promotes the transcription of an enzyme which synthesizes the diffusable cell signal X (which activates LacZ expression) and the non-diffusable signal Y (which dominantly represses LacZ expression). Cells in the dark therefore do not produce a black pigment as Y represses LacZ. However, when cells are exposed to red light, Cph8 activity is blocked, X and Y are not expressed, and thus LacZ is not expressed. The cell diffusion signal X produced by cells in the dark can reach neighboring cells. If the neighboring cells are also in the dark, LacZ is not expressed due to Y production. However, if those neighboring cells are exposed to light, and therefore at the light-dark boundary, transcription of LacZ will be activated, turning on pigment production. Accordingly, only cells that exist in light exposed areas and are juxtaposed to cells in the dark produce the black pigment. Due to diffusion, the darkest black pigment is found in cells immediately at the edge of light exposure and gradually decreases from there.
Figure 4.
(A) Synthetic edge detector for spatiotemporal control over gene expression in E coli. Exposure to light inhibits the diffusion signal X and β-galactosidase expression Z. When cells are in the dark (NOT light), X and Y are turned on. Y functions as an inverter, thus X AND (NOT Y) generates the signal Z. (B) A multichromic light-inducible gene expression system. The green sensor turns on one β-galactosidase reporter to generate Z in response to green light. When exposed to red light, the green sensor is turned off, and the red sensor is turned on to depress Y and activate the second β-galactosidase reporter Z. (C) Light-regulated network that uses feedback to adjust input levels to create stable output patterns. The PIF/PhyB system responds to 650 nm light to produce an output from a signaling domain. This output is then fed back through a controller which will alter light input levels to maintain a desired cellular output. Adapted with permission from Elsevier, © 2009, from [42] and © 2010, from [43], and from Nature Publishing Group, © 2011, from [40].
This chimeric Cph8 light sensor was subsequently applied in conjunction with another photoresponsive kinase CcaS and its response regulator CcaR by Voigt and co-workers in order to construct a synthetic gene circuit that can sense two different colors of light [43]. The CcaS/R system is a photoswitchable sensor. CcaS responds to green light (535 nm) to phosphorylate CcaR and activate transcription, while red light (672 nm) leads to the opposite response. Discrete patterns of gene expression in E. coli were achieved using the green light inducible CcaS/R circuit and the red light inducible Cph8 circuit (Figure 4B).
The PIF-PhyB interaction found in Arabidopsis thaliana is another light-controlled system that can be reversibly activated upon exposure to different wavelengths of light. This interaction has been applied to the development of synthetic systems to photochemically control cell signaling [44,45]. Recently, the PIF-PhyB interaction was used to create a light-based cellular network that uses a feedback loop to generate stable output signals [40]. As protein expression levels differ between individual cells and the varying presence of endogenous components can perturb a linear relationship between input signal and activity, even a precisely controlled photochemical input signal can result in variability of intracellular activity. Toettcher et al. designed a system based on PIF/PhyB that directly responds to variances in output signal and uses this information to adjust the intensity of the light input in order to maintain a desired correlation between input and output (Figure 4C). A membrane bound PhyB recruits a PIF-signaling domain fusion in response to 650 nm light and the process is reversed with 750 nm light. The signaling domain output, e.g., fluorescence of a reporter protein detected by cell imaging, is then computationally recalculated to adjust the light intensity in response to the signal. This system was used to produce a defined, constant activity of phosphoinositide 3-kinase in live cells.
The light-responsive LOV domain from Avena sativa has also been implemented in light activated systems, including the construction of light-activated GTPases [39,46]. Moreover, various opsin proteins have been used to elegantly engineer light-sensitive ion channels in mammalian cells and model organisms [41,47].
Conclusion
Many advances in the construction of synthetic light-activated genetic and cell signaling circuits have been made in recent years. These technologies provide an excellent source of control over delicate biological networks while being minimally invasive. The development of light-triggered gene networks involves the application of many biological parts engineered to be light-responsive, such as small molecules, oligonucleotides, and proteins. The technologies discussed here provide many general methods that can be applied to a wide range of genetic systems, even highly complex and dynamic systems such as signal transduction, while retaining precise temporal and spatial control over the engineered networks.
Highlights.
- ➢
photochemical control of gene function - ➢
photochemical control of cell signaling - ➢
caged oligonucleotides and caged proteins - ➢
light-responsive proteins domains
Acknowledgements
The authors thank the NIH (R01GM79114) and NSF (1200160) for financial support and Dr. Gavin Williams for helpful comments on the manuscript. LG acknowledges a GAANN Molecular Biotechnology graduate fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• of outstanding interest
- 1.Agapakis CM, Silver PA. Synthetic biology: exploring and exploiting genetic modularity through the design of novel biological networks. Mol Biosyst. 2009;5:704–713. doi: 10.1039/b901484e. [DOI] [PubMed] [Google Scholar]
- 2.Tanouchi Y, Pai A, You L. Decoding biological principles using gene circuits. Mol Biosyst. 2009;5:695–703. doi: 10.1039/b901584c. [DOI] [PubMed] [Google Scholar]
- 3.Weber W, Fussenegger M. Molecular diversity--the toolbox for synthetic gene switches and networks. Curr Opin Chem Biol. 2011;15:414–420. doi: 10.1016/j.cbpa.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Deiters A. Light activation as a method of regulating and studying gene expression. Curr Opin Chem Biol. 2009;13:678–686. doi: 10.1016/j.cbpa.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deiters A. Principles and applications of the photochemical control of cellular processes. Chembiochem. 2010;11:47–53. doi: 10.1002/cbic.200900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riggsbee CW, Deiters A. Recent advances in the photochemical control of protein function. Trends Biotechnol. 2010;28:468–475. doi: 10.1016/j.tibtech.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buskirk AR, Liu DR. Creating small-molecule-dependent switches to modulate biological functions. Chem Biol. 2005;12:151–161. doi: 10.1016/j.chembiol.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Cruz F, Koh J, Link K. Light-activated gene expression. Journal of the American Chemical Society. 2000;122:8777–8778. [Google Scholar]
- 9.Lin W, Albanese C, Pestell RG, Lawrence DS. Spatially discrete, light-driven protein expression. Chem Biol. 2002;9:1347–1353. doi: 10.1016/s1074-5521(02)00288-0. [DOI] [PubMed] [Google Scholar]
- 10.Young DD, Deiters A. Photochemical activation of protein expression in bacterial cells. Angew Chem Int Ed Engl. 2007;46:4290–4292. doi: 10.1002/anie.200700057. [DOI] [PubMed] [Google Scholar]
- 11.Young DD, Garner RA, Yoder JA, Deiters A. Light-activation of gene function in mammalian cells via ribozymes. Chem Commun (Camb) 2009:568–570. doi: 10.1039/b819375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cambridge SB, Geissler D, Keller S, Cürten B. A caged doxycycline analogue for photoactivated gene expression. Angew Chem Int Ed Engl. 2006;45:2229–2231. doi: 10.1002/anie.200503339. [DOI] [PubMed] [Google Scholar]
- 13.Sadovski O, Jaikaran AS, Samanta S, Fabian MR, Dowling RJ, Sonenberg N, Woolley GA. A collection of caged compounds for probing roles of local translation in neurobiology. Bioorg Med Chem. 2010;18:7746–7752. doi: 10.1016/j.bmc.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karginov A, Zou Y, Shirvanyants D, Kota P, Dokholyan N, Young D, Hahn K, Deiters A. Light Regulation of Protein Dimerization and Kinase Activity in Living Cells Using Photocaged Rapamycin and Engineered FKBP. Journal of the American Chemical Society. 2011;133:420–423. doi: 10.1021/ja109630v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umeda N, Ueno T, Pohlmeyer C, Nagano T, Inoue T. A photocleavable rapamycin conjugate for spatiotemporal control of small GTPase activity. J Am Chem Soc. 2011;133:12–14. doi: 10.1021/ja108258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cambridge SB, Geissler D, Calegari F, Anastassiadis K, Hasan MT, Stewart AF, Huttner WB, Hagen V, Bonhoeffer T. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat Methods. 2009;6:527–531. doi: 10.1038/nmeth.1340. [DOI] [PubMed] [Google Scholar]
- 17.Sauers DJ, Temburni MK, Biggins JB, Ceo LM, Galileo DS, Koh JT. Light-activated gene expression directs segregation of co-cultured cells in vitro. ACS Chem Biol. 2010;5:313–320. doi: 10.1021/cb9002305. [DOI] [PubMed] [Google Scholar]
- •18.Gardner L, Zou Y, Mara A, Cropp TA, Deiters A. Photochemical control of bacterial signal processing using a light-activated erythromycin. Mol Biosyst. 2011;7:2554–2557. doi: 10.1039/c1mb05166k. This article demonstrates the construction of a light-activated AND gate as well as a bandpass filter in bacterial cells.
- 19.Mikat V, Heckel A. Light-dependent RNA interference with nucleobase-caged siRNAs. RNA. 2007;13:2341–2347. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang X, Maegawa S, Weinberg ES, Dmochowski IJ. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J Am Chem Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 21.Blidner RA, Svoboda KR, Hammer RP, Monroe WT. Photoinduced RNA interference using DMNPE-caged 2′-deoxy-2′-fluoro substituted nucleic acids in vitro and in vivo. Mol Biosyst. 2008;4:431–440. doi: 10.1039/b801532e. [DOI] [PubMed] [Google Scholar]
- 22.Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Regulating gene expression in human leukemia cells using light-activated oligodeoxynucleotides. Nucleic Acids Res. 2008;36:559–569. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young DD, Lusic H, Lively MO, Yoder JA, Deiters A. Gene silencing in mammalian cells with light-activated antisense agents. Chembiochem. 2008;9:2937–2940. doi: 10.1002/cbic.200800627. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, Olson AJ, Chen JK. Versatile synthesis and rational design of caged morpholinos. J Am Chem Soc. 2009;131:13255–13269. doi: 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain PK, Shah S, Friedman SH. Patterning of Gene Expression Using New Photolabile Groups Applied to Light Activated RNAi. J Am Chem Soc. 2010 doi: 10.1021/ja107226e. [DOI] [PubMed] [Google Scholar]
- 26.Tang X, Su M, Yu L, Lv C, Wang J, Li Z. Photomodulating RNA cleavage using photolabile circular antisense oligodeoxynucleotides. Nucleic Acids Res. 2010;38:3848–3855. doi: 10.1093/nar/gkq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •27.Young DD, Lively MO, Deiters A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J Am Chem Soc. 2010;132:6183–6193. doi: 10.1021/ja100710j. By installing caging groups on various positions of DNAzyme molecules and antisense agents, both activation and deactivation of gene expression was achieved.
- 28.Govan JM, Lively MO, Deiters A. Photochemical control of DNA decoy function enables precise regulation of nuclear factor κB activity. J Am Chem Soc. 2011;133:13176–13182. doi: 10.1021/ja204980v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 30.Prokup A, Hemphill J, Deiters A. DNA Computation: A Photochemically Controlled AND Gate. J Am Chem Soc. 2012 doi: 10.1021/ja210050s. [DOI] [PubMed] [Google Scholar]
- 31.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 32.Chin JW. Reprogramming the genetic code. EMBO J. 2011;30:2312–2324. doi: 10.1038/emboj.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards WF, Young DD, Deiters A. Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem Biol. 2009;4:441–445. doi: 10.1021/cb900041s. [DOI] [PubMed] [Google Scholar]
- 34.Wu N, Deiters A, Cropp TA, King D, Schultz PG. A genetically encoded photocaged amino acid. J Am Chem Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- 35.Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat Chem Biol. 2007;3:769–772. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- ••36.Gautier A, Deiters A, Chin JW. Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. A good example of how using a small molecule (caging group) can offer a unique avenue of control over a very comlpex pathway.
- 37.Chou C, Young DD, Deiters A. Photocaged t7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. Chembiochem. 2010;11:972–977. doi: 10.1002/cbic.201000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahn KM, Kuhlman B. Hold me tightly LOV. Nat Methods. 2010;7:595, 597. doi: 10.1038/nmeth0810-595. [DOI] [PubMed] [Google Scholar]
- •40.Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. This work cleverly uses the system’s own ouput signals to adjust input intensity to maintain stable output patterns.
- 41.Fenno L, Yizhar O, Deisseroth K, Hyman S, Jessell T, Shatz C, Stevens C, Zoghbi H. The Development and Application of Optogenetics. Annual Review of Neuroscience, Vol 34. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••42.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. This work elegantly describes the manipulation of various genetic circuits to create a highly precise method to create edge detectors in E. coli.
- •43.Tabor JJ, Levskaya A, Voigt CA. Multichromatic control of gene expression in Escherichia coli. J Mol Biol. 2011;405:315–324. doi: 10.1016/j.jmb.2010.10.038. Another great demonstration of utilizing various cellular circtuits to create highly sensitive patterns of gene expression in E. coli.
- 44.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Georgianna WE, Deiters A. Reversible light switching of cell signalling by genetically encoded protein dimerization. Chembiochem. 2010;11:301–303. doi: 10.1002/cbic.200900754. [DOI] [PubMed] [Google Scholar]
- 46.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, Prigge M, Berndt A, Cushman J, Polle J, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]