Discovery of hundreds of mirtrons in mouse and human small RNA data (original) (raw)
Abstract
Atypical miRNA substrates do not fit criteria often used to annotate canonical miRNAs, and can escape the notice of miRNA genefinders. Recent analyses expanded the catalogs of invertebrate splicing-derived miRNAs (“mirtrons”), but only a few tens of mammalian mirtrons have been recognized to date. We performed meta-analysis of 737 mouse and human small RNA data sets comprising 2.83 billion raw reads. Using strict and conservative criteria, we provide confident annotation for 237 mouse and 240 human splicing-derived miRNAs, the vast majority of which are novel genes. These comprise three classes of splicing-derived miRNAs in mammals: conventional mirtrons, 5′-tailed mirtrons, and 3′-tailed mirtrons. In addition, we segregated several hundred additional human and mouse loci with candidate (and often compelling) evidence. Most of these loci arose relatively recently in their respective lineages. Nevertheless, some members in each of the three mirtron classes are conserved, indicating their incorporation into beneficial regulatory networks. We also provide the first Northern validation for mammalian mirtrons, and demonstrate Dicer-dependent association of mature miRNAs from all three classes of mirtrons with Ago2. The recognition of hundreds of mammalian mirtrons provides a new foundation for understanding the scope and evolutionary dynamics of Dicer substrates in mammals.
Diverse pathways of conserved post-transcriptional gene regulation are mediated by Argonaute proteins and their guide, short RNAs. Among Argonaute-mediated small RNA pathways, the best-studied are the microRNAs (miRNAs). Generally speaking, miRNAs are ∼21 to 24-nucleotide (nt) RNAs whose termini are precisely defined, and derive from precursor transcripts bearing one or more inverted repeats or hairpins (Axtell et al. 2011). The first miRNAs emerged from genetic studies of Caenorhabditis elegans developmental mutants (Lee et al. 1993; Reinhart et al. 2000), and were only recognized as noncoding loci upon their cloning. This set the stage for the directed identification of miRNA genes from cloned short RNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). In animals, most miRNAs are generated by stepwise cleavage of primary miRNA transcripts (Kim et al. 2009). These are processed in the nucleus by the Drosha RNase III enzyme to release an ∼50- to 80-nt pre-miRNA hairpin, and again in the cytoplasm by a Dicer-class RNase III enzyme to yield a small RNA duplex. One of the strands is preferentially stably incorporated as a single-stranded RNA in an Argonaute (Ago) complex, and guides it to target transcripts (Czech and Hannon 2010).
Although bioinformatic strategies have been used to identify miRNA genes (Lai et al. 2003; Lim et al. 2003; Huang et al. 2007; van der Burgt et al. 2009), these have mostly been superseded by deep sequencing. This is in large part due to the fact that effective computational methods rely on comparative genomics and are ill-suited to identify species-specific miRNAs with reasonable specificity and sensitivity. Another limitation of the forward computational approach is the need to set substrate parameters based on current conceptions of biogenesis pathways. Indeed, careful perusal of sequenced short RNAs has revealed diverse classes of noncanonical miRNA substrates that deviate from criteria used in forward computational approaches (Yang and Lai 2011).
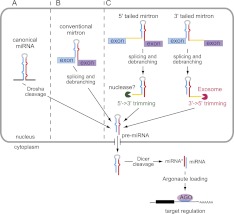
The most prevalent alternative pathway involves short hairpin introns known as mirtrons (Fig. 1) that serve as pre-miRNA mimics (Westholm and Lai 2011). The biochemical and functional properties of mirtrons have been studied most carefully in Drosophila (Okamura et al. 2007; Ruby et al. 2007a), but they also exist in C. elegans (Ruby et al. 2007a; Chung et al. 2011; Jan et al. 2011) and vertebrates (Berezikov et al. 2007; Babiarz et al. 2008, 2011; Glazov et al. 2008; Chiang et al. 2010). In addition to conventional mirtrons, where splicing defines both ends of the pre-miRNA hairpin, so-called “tailed” mirtrons contain an unstructured region 5′ or 3′ to a splicing-derived terminal intron hairpin. A number of 3′-tailed mirtrons exist in Drosophila, and where studied, the RNA exosome is responsible for removal of the 3′ tail (Flynt et al. 2010). In contrast, tailed mirtrons in vertebrates are essentially only of the 5′-tailed class (Babiarz et al. 2008, 2011; Glazov et al. 2008; Chiang et al. 2010; Valen et al. 2011); the relevant nuclease involved in their biogenesis is not known.
Figure 1.
Summary of canonical and splicing-mediated miRNA pathways. (A) Canonical miRNA hairpins are cleaved by Drosha to release a pre-miRNA hairpin, which is exported to the cytoplasm and cleaved by Dicer to release a miRNA/star duplex. One strand is preferentially loaded into an Argonaute (AGO) protein and guides it to targets. (B) Conventional mirtrons are short hairpin introns that are spliced and debranched to form a pre-miRNA hairpin, which then enters the canonical pathway. (C) With 5′- and 3′-tailed mirtrons, splicing generates only one end of the hairpin, with an unstructured “tail” that extends to the 3′ splice acceptor or 5′ splice donor, respectively. These require additional ribonucleolytic processing to generate the pre-miRNA hairpin.
Following the initial recognition of mirtrons in mammals (Berezikov et al. 2007), efforts to discover additional splicing-derived miRNAs have mostly been incidental, and adjunct to efforts to annotate canonical miRNAs. However, algorithms that identify miRNAs from deep sequence data do not necessarily identify mirtrons, owing to their distinct properties. We recently compiled >100–200 small RNA data sets from Drosophila melanogaster and C. elegans and identified a plethora of new mirtrons (Chung et al. 2011). This motivated us to perform similar analyses of the large amount of available mouse and human small RNA data. While only a few tens of mirtrons have been reported in these species, compared to 700–1000 canonical miRNA loci, our dedicated inspection revealed hundreds of novel 5′-tailed mirtrons supported by stringent small RNA evidence, and hundreds more with candidate evidence. We also identified a smaller number of confident novel mirtrons, as well as a previously unrecognized class of mammalian 3′-tailed mirtrons. We provide first evidence for incorporation of mammalian mirtron-derived miRNAs into Ago complexes using Northern assays, validating this association for members of all three mirtron biogenesis classes. These findings substantially revise the scope of endogenous Dicer substrates in mammals.
Results
Curation of mouse and human small RNA data sets
We obtained available raw data for small RNA libraries of mouse or human origin from the NCBI GEO or SRA archives (Supplemental Tables S1, S2) and analyzed these using a standard pipeline (see Methods); only in the small minority of cases where the raw data were lacking did we use preprocessed data. In total, we collected 501 human data sets and 236 mouse data sets comprising 1,358,711,357 and 1,480,375,413 raw reads, respectively (Supplemental Table S3). From these, we could clip ∼1 billion reads in each species that were ≥17 nt, of which >650 million mapped perfectly to the reference human (hg19) and mouse (mm9) genomes. A majority of these reads derived from miRNA loci (as defined by miRBase; http://www.mirbase.org/), with 494 and 431 million reads mapping perfectly to known human and mouse miRNAs, respectively (Supplemental Table S3).
Most miRNAs annotated in miRBase are generated by the canonical miRNA pathway, but some derive from a variety of alternative biogenesis routes (Yang and Lai 2011). To follow our interests in splicing-derived miRNAs, we analyzed in detail those reads mapping in the vicinity of intron termini (defined by the UCSC Genome Browser; http://genome.ucsc.edu). We mapped 133,254,416 human and 92,872,757 mouse reads to the terminal 5′ or 3′ 250 nt of annotated introns. In addition to heterogeneous read mapping patterns and intron-terminal reads of heterogeneous sizes, these included distinctive patterns derived from snoRNAs, tRNAs, canonical miRNAs, and mirtrons located within this distance to intron termini. The 3′ ends of canonical miRNAs are often subject to untemplated additions (Burroughs et al. 2010; Chiang et al. 2010; Berezikov et al. 2011), especially the 3′ ends of mirtron-3p species (Westholm et al. 2012). We therefore paid attention to reads that mapped to intron termini following trimming of 3′ nucleotides, yielding an additional 15,169,877 human and 10,600,599 mouse mapped reads (Supplemental Table S3).
We built visual interfaces that displayed all of the different reads aligned to each locus, which sometimes numbered in the hundreds of variants mapping to an individual intron terminus. These data were divided up by library of origin and included other information such as the read density across the intron and flanking exonic sequence, the predicted secondary structure, and so forth. Those interfaces that are informative with respect to mirtrons are publicly accessible (Supplemental HTML documents).
Reevaluation of miRBase annotations reveals three classes of mammalian mirtrons
Many annotators of miRNAs do not distinguish between canonical loci and those produced from alternative pathways, and neither does the miRBase repository at present. We examined whether any presumed canonical miRBase loci might have a splicing-dependent origin. While this work was in preparation, Jensen and colleagues noted that five human miRNA annotations were actually 5′-tailed mirtrons (Valen et al. 2011). Our analysis largely concurred with their evaluation but revealed the scope of unrecognized 5′-tailed mirtrons in miRBase to be much larger. We find that 14 human and 10 mouse annotations can be recognized as 5′-tailed mirtrons (Supplemental Tables S4, S5, “reclassified” loci).
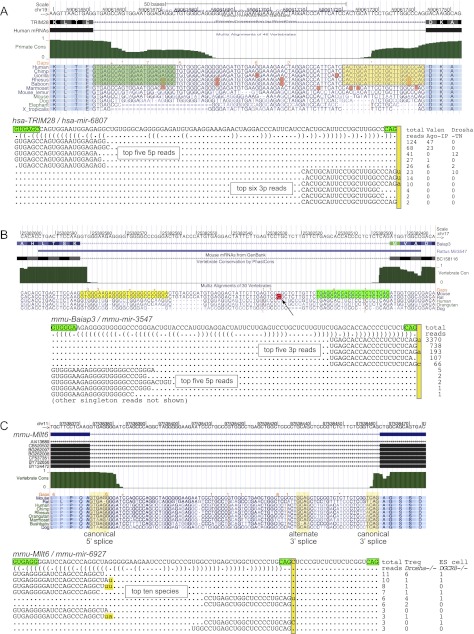
Of particular note was our recognition of 3′-tailed mirtrons, i.e., loci for which splicing directly generates only the 5′ end of the pre-miRNA hairpin. To date, these have only been experimentally characterized in Drosophila, with the RNA exosome performing 3′ trimming following intron splicing (Flynt et al. 2010). We were intrigued to realize that the 5p reads of the conserved mammalian locus Mir668 began with the canonical splice donor “GUAAGU” (Fig. 2A). Mir668 is located in a supercluster of many tens of miRNAs (Seitz et al. 2004), many of which are associated with a maternally imprinted primary transcript cloned from mouse termed Mirg (“miRNA containing gene”). Indeed, mmu-miR-668-5p phases precisely with a highly conserved Mirg splice donor site, and its hairpin is followed by a tailing region of 26 nt extending to a highly conserved splice acceptor (Fig. 2A). We also identified conserved polypyrimidine tract and branchpoint sequences, providing further evidence that it resides in a functional intron. Notably, the flanking exons on either side are much more poorly conserved than the intron. There is no alternate “AG” acceptor in the tailing region, suggesting that biogenesis of the pre-mir-668 hairpin involves 3′ resection following splicing. Consistent with this notion, the 3′ ends of miR-668-3p species exhibit greater heterogeneity than is typical for conserved miRNAs, analogous to the Drosophila 3′-tailed mirtron miR-1017-3p (Flynt et al. 2010).
Figure 2.
Evidence for conserved and nonconserved 3′-tailed mirtrons in mammals. (A) The Dlk imprinted region contains a large cluster of miRNAs, at least some of which are associated with the Mirg transcription unit, presumed to be a noncoding RNA. Closeup of mir-668 alignments reveals a conserved hairpin that resides at the 5′ splice donor of Mirg, with a 3′ tail extending to the 3′ splice acceptor; this intron also encodes a branchpoint consensus and a typical 3′ poly-polypyrimidine tract. The top expressed 3p (miRNA) and 5p (star) reads from mmu-mir-668 support this locus as being derived by 5′ splicing and 3′ trimming. (B) Example of a 3′-tailed mirtron in human PTBP1 (hsa-mir-4745), for which the dominant species derives from the 5p arm. Reads were also recovered in two independent Ago-IP data sets, supporting their classification as bona fide miRNAs. Although most 5p reads began with uridine, less abundant reads presumed to be the primary spliced products initiate with the “GU” splice donor. Note that an “aberrant” 3p species produced from a different register was not associated with Ago-IP evidence (arrow). Density plot to the right highlights the specificity of these reads from the intronic hairpin. (C) Example of 3′-tailed mirtron in human FKBP1A (hsa-mir-6869), for which reads were recovered in independent Ago-IP data sets. The secondary structure illustrates that the dominant small RNAs arrange as a duplex with 3′ overhangs.
Two human 3′-tailed mirtrons were recently reported independently by Jensen and colleagues (Valen et al. 2011). Our further inspection of miRBase loci actually revealed five additional 3′-tailed mirtrons in human (Table 1; Supplemental Table S4, “reclassified” tab). One illustrative example was located in human PTBP1 (hsa-mir-4745) and generated several hundred reads. Both 5p (mature) and 3p (star) reads were recorded in independent Ago1/2-IP and Ago2-IP data sets, providing strong confirmation that they constitute genuine miRNAs (Fig. 2B). A tail of ∼14 nt is resected from the 3′ end following splicing, and again there is no alternate “AG” acceptor. Therefore, the repertoire of mouse and human miRNAs includes the 3′-tailed mechanism, and some (e.g., miR-668) are deeply conserved among mammals.
Table 1.
Numbers of mammalian mirtron loci
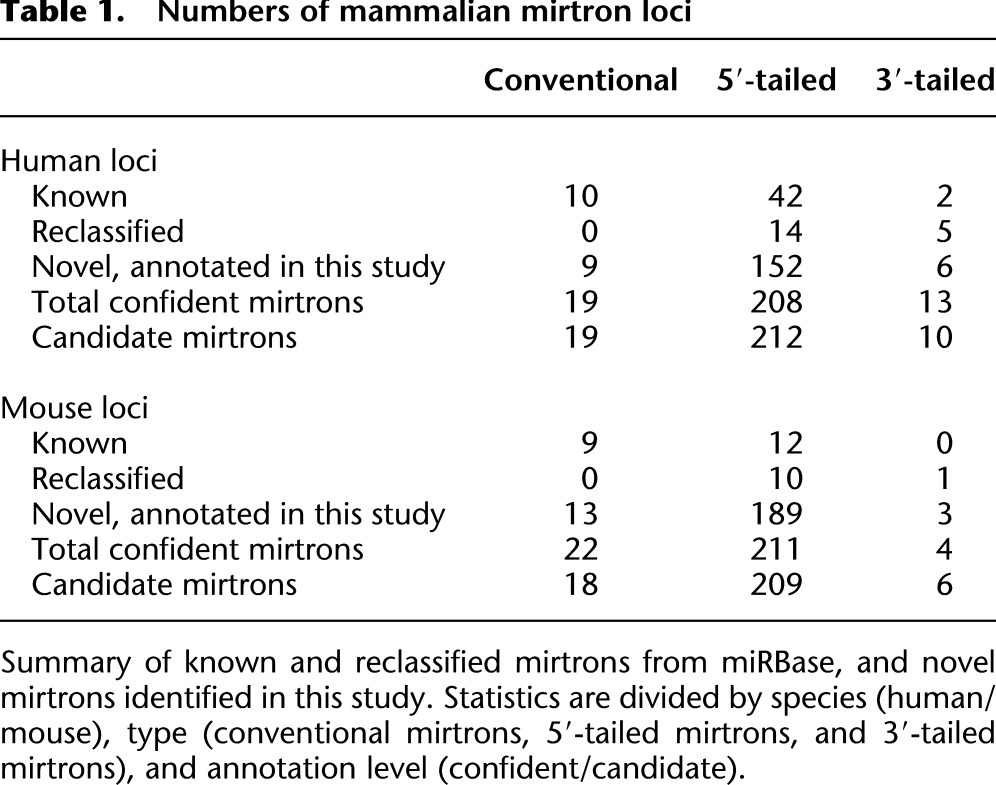
Altogether, >40 mammalian loci in miRBase (Release 17), annotated in literature reporting canonical miRNA loci, actually derive from splicing. These comprise three subpathways of miRNA biogenesis, which in order of their locus abundance are 5′-tailed mirtrons, conventional mirtrons, and 3′-tailed mirtrons (Table 1, “reclassified” loci). We provide summaries of the read mappings to known and reannotated human and mouse mirtrons in Supplemental Tables S4 and S5, and comprehensive read data in Supplemental Figures S1 and S2.
Annotation of novel human and mouse mirtrons
With this new perspective on the diversity of splicing-derived miRNAs in mammals, we sought to identify novel mirtrons. To do so, we used all the reads mapping in the vicinity of intron termini in mouse and human, and annotated hairpins coinciding with one or both splice sites, for which relatively specific sRNA reads formed clear miRNA/star duplexes with a 3′ overhang on the inferred Dicer-cleaved end. In brief, we searched for straight hairpins that generated miRNA/star duplexes with relatively specific 5′ ends, appropriate 3′ overhangs to the inferred Dicer-cut site, and a minimum of at least 50 total miRNA/star reads with at least three star reads. Further details of the criteria are discussed in the Methods and in the following sections, and statistics of the winnowing process of human and mouse introns are provided in Supplemental Table S6.
We emphasize the stringency of our annotation criteria to highlight the very conservative nature of our efforts, which exceed the levels of small RNA evidence associated with hundreds of miRNA annotations in miRBase (Supplemental Table S7). Given this, it was striking that we could confidently classify nearly 500 novel 5′-tailed, 3′-tailed, and conventional mirtrons in the mammalian small RNA data (Supplemental Tables S4, S5; Supplemental Figs. S1, S2). We segregated an additional 235 mouse and 241 human introns with candidate miRNA read patterns. Many of these have compelling evidence that might have led to miRBase annotations in other studies, such as paired miRNA/star reads and/or reads in Ago-IP libraries. We presume that some of these may be genuine Dicer products, but we felt it prudent at this point to reserve them as candidates.
We note that several miRBase mirtrons may be genuine but would have fallen into our “candidates” category based on paucity of reads. For example, hsa-mir-1178 is a 5′-tailed mirtron supported by only four duplex reads, insufficient to assess the nature of its biogenesis. On the other hand, the conventional mirtron hsa-mir-1233-1 and the 5′-tailed mirtron hsa-mir-4701 each fell short of our 50-read cutoff by a handful of reads, and would therefore not have qualified for de novo “confident” annotations in this study. However, because these straight hairpins generate highly specific miRNA/star duplex reads, even with some AGO-IP reads, these are very likely genuine splicing-derived miRNAs. The 19 human and two mouse miRBase mirtrons (both recognized loci and ones that we reclassified as splicing-derived) whose small numbers of star reads or total reads would have disqualified them in our annotation are noted in Supplemental Tables S4 and S5. Finally, two loci we earlier reported as mirtrons based on more limited small RNA sequencing (Berezikov et al. 2007) were not associated with read patterns indicative of Dicer cleavage in these deep data sets, even though terminal small RNA reads were found (hsa-mir-1231 and hsa-mir-1225); these should be flagged as questionable.
In addition to the mapping summaries provided in Supplemental Figures S1 and S2, comprehensive data for all of the mouse and human mirtron annotations are provided in the Supplemental HTML websites (http://ericlailab.com/mammalian_mirtrons/hg19/ and http://ericlailab.com/mammalian_mirtrons/mm9/). The pages are subdivided according to biogenesis class (conventional, 5′-tailed, and 3′-tailed) and certainty of annotation (confident or candidate), and they collect a broad set of information on individual genome mapping reads, reads that match following 3′ trimming, the library origin of each of the sequences, their lengths, the distribution of reads in the intron and in the flanking exons, and the predicted secondary structure of the miRNA-generating hairpin, along with links to the loci on the UCSC Genome Browser. In the following sections, we highlight some notable novel examples in each of the mirtron categories.
Novel conventional mirtrons
We identified 22 confident novel conventional mirtrons, that is, pre-miRNAs for which both hairpin ends are defined by splicing. A compelling human example was located in TRIM28 (hsa-mir-6807), whose abundant reads included data from Drosha-TN and hAgo1/2-IP libraries, providing support for their identity as microprocessor-independent, functional miRNAs (Fig. 3A). Interestingly, this intron exhibited evidence of purifying selection among a set of primate genomes as an miRNA-type locus, in that more positions of divergence were found in the terminal loop region than the hairpin arms. Therefore, even though this intron gained miRNA capacity recently, its evolutionary pattern was consistent with the acquisition of endogenously beneficial regulatory function. We also identified mirtrons that emerged recently in the rodent lineage, including mouse Baiap3, whose mirtron (mmu-mir-3547) generated >4700 reads, mostly from its 3p end (Fig. 3B). This mouse intron is nearly identical with its rat ortholog, save for the presence of two diverged loop nucleotides. Interestingly, rno-mir-3547 was also annotated from rat (Linsen et al. 2010), which we can now reassign as a conserved rodent mirtron.
Figure 3.
Examples of novel mammalian conventional mirtrons annotated in this study. (A) Human TRIM28 contains a mirtron (hsa-mir-6807) that is selectively conserved among primates. The dominant 5p and 3p reads form a typical miRNA/star duplex with 3′ overhang on the Dicer-cleaved end, and mature species were recovered from independent Ago-IP data sets. (B) Murine Baiap3 contains a mirtron (mmu-mir-3547) with relatively high expression. The orthologous intron was annotated as the canonical miRNA Mir3547 in rat; our observations indicate this as a conserved rodent mirtron. The red box and arrow highlight a terminal loop nucleotide that has diverged between the mouse and rat orthologs. Note that the 3p species of Trim28 and Baiap3 mirtrons accumulate high levels of untemplated uridine, with a lower level of untemplated adenine (yellow highlight). (C) An intron in mouse Mllt6 generates small RNAs that are reminiscent of a 3′-tailed layout. However, the 3′ end of its dominant 3p species end in CAG and are strongly uridylated, and the CAG is conserved across mammals. These features suggest it to be a conventional mirtron (mmu-mir-6927) in an alternatively spliced intron.
Baiap3/miR-3547-3p species were notable for their extraordinary accumulation of 3′ uridylated reads. Although terminally modified miRNAs are usually considered to represent a few percent of base reads, there were 3370 “AGu” Baiap3/miR-3547-3p reads compared with 738 “AG” reads; smaller numbers of A-tailed and C-tailed reads were noted. In fact, Trim28/miR-6807-3p reads, which represent a star species and were comparatively rare, were overwhelmingly 3′ modified (37 “AGu” and 10 “AGa” reads, compared with only two “AG” reads). This is consistent with our recent report that the 3p species of conventional mirtrons are strongly uridylated in both invertebrates and vertebrates (Westholm et al. 2012). In total, our new annotations comprise 13 confident conventional mirtrons in mouse (and 18 additional candidates), and nine confident conventional mirtrons (and 19 candidates) in human (Table 1). The confident loci alone more than double the set of known mammalian conventional mirtrons.
Novel 3′-tailed mirtrons
As mentioned, our reannotation of the conserved vertebrate locus Mir668 and other loci provided evidence for 3′-tailed mirtrons in vertebrates. We newly recognized three mouse and seven human 3′-tailed mirtrons that pass strict criteria, with additional candidate members of this biogenesis class (Table 1). An example of a novel locus in human FKBP1A (hsa-mir-6869) is shown in Figure 2C. Although modestly expressed, reads were recovered in independent Ago-IP libraries, and the dominant reads form a duplex with 3′ overhangs. Moreover, this intron appears to be under selective constraint for miRNA potential within primates, since it exhibits accelerated divergence within the terminal and stable hairpin arms (http://genome.ucsc.edu/). Overall, while the number of 3′-tailed mirtrons is much smaller than the number of conventional or 5′-tailed mirtrons, their confident annotation in both mouse and human data along with the evolutionary selection of some of them together support the notion of their maturation via a specific pathway in mammals.
Although most annotated mirtrons involve constitutively spliced introns, Drosophila CG17560 contains a mirtron in an alternatively spliced intron (Chung et al. 2011). A mirtron in mouse Mllt6 (mmu-mir-6927) was noteworthy in this regard. Both miRNA and star species of a hairpin anchored at the 5′ end of its intron were recovered, with an apparent 3′-tailing region (Fig. 3C). Although not abundantly expressed, reads were present in libraries from Drosha knockout regulatory T cells (Tregs) and Dgcr8 knockout ES cells, providing evidence for microprocessor-independent accumulation. However, the 3′ termini of its 3p reads caught our attention, since the dominant species ended in GCAG and GCAGu. These might be interpreted as mirtron-3p reads ending in an optimal splice acceptor site (YCAG), and its 3′-uridylated species. While this mirtron hairpin exists only in mouse, the GCAG sequence was specifically conserved among diverse mammals (Fig. 3C). Therefore, this apparent 3′-tailed mirtron may actually represent a conventional mirtron using an unannotated splice site.
Novel 5′-tailed mirtrons
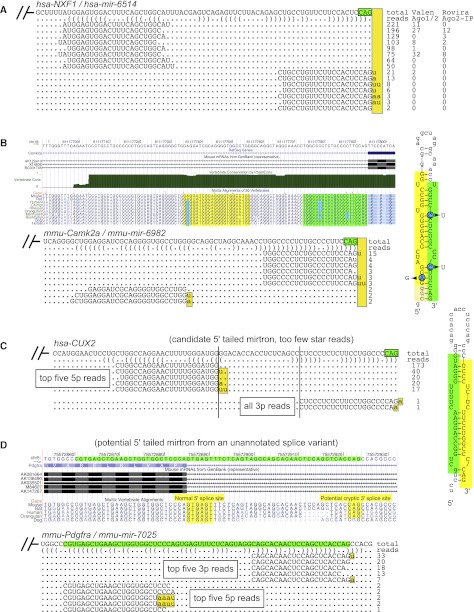
Our efforts yielded a plethora of novel 5′-tailed mirtrons: 189 mouse and 152 human loci passed strict criteria, respectively (Table 1; Supplemental Tables S4, S5; the Supplemental websites). These represent a tremendous increase in the numbers of recognized 5′-tailed mirtrons in mammals. Figure 4A shows an example of a novel human 5′-tailed mirtron located in NXF1 (hsa-mir-6514). Additional evidence that it generates bona fide miRNAs came from its recovery in independent Ago1/2-IP and Ago-IP libraries, as well as highly uridylated 3p species as seen with conventional mirtrons.
Figure 4.
Examples of novel mammalian 5′-tailed mirtrons annotated in this study. (A) Human NXF1 (hsa-mir-6514) contains a 5′-tailed mirtron, whose mature species were recovered in independent Ago-IP data sets. (B) Small RNAs mapped to murine Camk2a (mmu-mir-6982) define a 5′-tailed mirtron that, although modestly expressed, is very well-conserved among mammals. Curiously, the presumed seed sequences of both 5p and 3p species have diverged, but all of these changes are structurally conservative. Such evolutionary patterns suggest that the structure, rather than the precise sequence, of this mirtron hairpin is under selection. (C) A candidate 5′-tailed mirtron in the intron of human CUX2. While this hairpin generates a typical miRNA/star duplex with high 5′ specificity, the existence of only two star reads was below our requirement for confident annotation. (D) A locus that emerged from the control analysis of “shifted” intron coordinates. This hairpin spans an exon–intron junction of mmu-Pdgfra and generates a typical miRNA/star duplex, for which the dominant 3p species terminate in CAG or exhibit untemplated uridylation. This layout strongly suggests that this region of Pdgfra harbors a 5′-tailed mirtron (mmu-mir-7025) that bypasses an annotated 5′ splice acceptor and uses an unannotated 3′ splice acceptor; note that the CAG acceptor is conserved in many mammals. Overall, note that the 3p species of 5′-tailed mirtrons generally accumulate high levels of untemplated uridine (yellow highlight), with a lower level of untemplated adenine.
A 5′-tailed mirtron that was strikingly conserved across the mammals was found in Camk2a (Fig. 4B). Although the locus is clearly present in human, confident read patterns were only found in mouse (mmu-mir-6982), and a modest number of reads at that. However, its dominant 3p species exhibited an invariant 5′ end and were mostly uridylated, providing confidence that it generates a genuine miRNA. The small number of reads recovered from this conserved locus is a reminder that even with catalogs of a billion reads, there may remain highly conserved, uncloned miRNA hairpins. Notably, the Camk2a hairpin has incurred divergence on both of its hairpin arms, which is unusual among conserved miRNA loci (Fig. 4B). Interestingly, all three of the major positions bearing nucleotide changes were structurally compensatory, either changing GC pairs to GU pairs, or AU pairs to GU pairs. This tailed mirtron hairpin may potentially serve some structural function in cis.
Many loci in the 5′-tailed category did not fully meet all of our annotation criteria, but were compelling nonetheless. These candidates included 212 human and 211 mouse loci. The human CUX2 locus in Figure 4C illustrates the stringency of our annotation criteria. This straight hairpin generated hundreds of reads with high precision of 5′ identities on both putative miRNA and star strands, and appropriate geometry of 3′ duplex overhangs. However, we only recorded two star reads, which was below our minimum to confidently assess Dicer cleavage; moreover, the two reads were 3′ modified so there, in fact, were no perfect genome-matched star reads. This level of annotation stringency clearly exceeds that of many other miRNA loci that are deposited in the miRBase registry, and thus it is certainly the case that many of the >400 5′-tailed mirtron candidates that we report will eventually be deserving of “genic” status. As with the confidently annotated loci, the basic details of their sequence, structure, and mature species are presented in Supplemental Tables S2 and S3, and fuller accountings are presented in Supplemental Figures S1 and S2 and the Supplemental HTML documents.
Assessment of false negatives in mirtron annotation
To test the specificity of our annotation pipeline, we re-ran the analysis on control loci generated by shifting the coordinates of all annotated mouse and human introns 30 nt upstream. Of 453,367 “shifted” human intron termini, only one locus passed criteria. This resides in an intron of PILRB (hsa-mir-6840) and generates clear miRNA/star duplex reads from a straight hairpin (Supplemental Fig. S3). This locus might simply represent an intronic canonical miRNA, but its lack of “lower stem” base-pairing may potentially suggest it as an endo-shRNA. In mouse, we evaluated 358,228 “shifted” intron termini and found only two loci that passed criteria (Supplemental Fig. S3). One of these resides in Pdgfra (mmu-mir-7025), for which the annotated splice site is highly conserved across vertebrates (Fig. 4D). Notably, the 3′ end of the dominant 3p reads end in CAGu, where the terminal nucleotide is untemplated; otherwise CAG terminating reads are the next most abundant. Because the CAG sequence is well-conserved among diverse mammals (Supplemental Fig. S3), we speculate that these shifted coordinates fortuitously uncover a cryptic intron that happens to generate a mirtron (Fig. 4D). Therefore, as with the case of mmu-Mllt6/mmu-mir-6927 (Fig. 3C), alternative splicing may generate miRNAs in a nonconstitutive fashion.
The other hit from the shifted mouse intron analysis is located in Ighg2a (mmu-mir-7094a). We picked this up from an intron annotation in this region available from the UCSC Genome Browser (uc011yvj.1, also known as “_abParts_”) for which the annotated splice acceptor is noncanonical (TGG). Curiously, the shifted locus corresponds to a straight hairpin that generates precise miRNA/star reads. Although lowly expressed, the 3′ end of certain 3p species terminates in a CAG sequence, for which a spliced EST (CN661462) supports its precise usage as a splice acceptor site (Supplemental Fig. S3). Taken together, these control analyses demonstrate the specificity of the mirtron production and suggest a negligible (∼0.5%) false discovery rate of mirtron annotation when compared with the hundreds of highly confident loci observed with genuine introns. In fact, the control shifted intron analysis proved effective at identifying mirtrons from unannotated splice sites.
Numbers and expression of canonical miRNA and mirtron loci in mammals
Deeply conserved miRNA loci typically generate more reads than recently evolved loci in small RNA libraries (Ruby et al. 2007b; Chiang et al. 2010). This trend is the case despite the uncertainty of interpreting whether low numbers of reads from given loci are due to low expression, poor processing, or highly cell-specific or state-specific expression. While certain mirtron loci are broadly conserved across mammals, the vast majority of mirtrons appear to have gained miRNA-generating potential within the primate and rodent lineages. This contrasts with the hundreds of canonical miRNAs well-conserved across mammals. Consistent with this, the cumulative number of reads assigned to mirtrons is substantially less than map to canonical miRNAs, on the order of 1000- to 5000-fold less (Supplemental Table S7). This comparison must be taken with a grain of salt, because there is no way to normalize the diverse representation of small RNA libraries in a meaningful way. For example, the miRNA with the highest number of reads across the analyzed human libraries is not a broadly expressed locus, as one might expect. Instead, the liver-specific locus MIR122 topped the aggregate tallies of miRNA reads; this appears to be due to its extraordinary representation in a set of liver libraries (Hou et al. 2011). Still, it is clear that all of the highest-expressed miRNAs derive from canonical biogenesis (Supplemental Table S7).
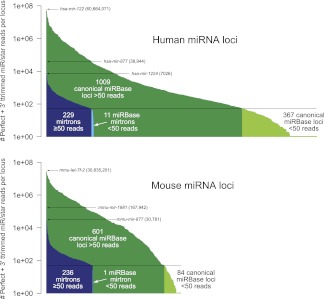
Nevertheless, it is worth pointing out that the expression range of most mirtrons, while clearly far less than the best-studied conserved miRNAs, is still within the range of hundreds of annotated mouse and human miRNAs (Fig. 5; Supplemental Table S7). Moreover, the cutoff of 50 total miRNA/star reads that we used to segregate “confident” mirtron calls exceeds the number of reads that we could map to nearly 450 human/mouse loci available in miRBase. While some of these miRBase annotations may be suspect, this comparison indicates that mirtrons are within an expression range of a substantial fraction of known miRNA loci.
Figure 5.
Comparison of read abundance from canonical miRNA loci and mirtrons. Plotted are the total miRNA/star reads, including 3′-trimmed reads, collected from the 501 human and 236 mouse small RNA data sets; each bar represents an individual locus. Read numbers were normalized for number of genomic matches. The highest expressed canonical miRNAs and mirtrons are indicated. Our annotation of confident mirtrons required at least 50 miRNA/star reads; note that hundreds of loci deposited in miRBase do not meet this minimum. The underlying data used to generate this table are presented in Supplemental Table S7.
This can also be examined with respect to the number of loci. When considering those miRNA loci that generated more than 50 reads, the cutoff we used to annotate novel loci, then mirtrons comprise about one-fourth of murine miRNA loci and one-sixth of human miRNA loci (Fig. 5). Thus, while mirtrons are overall modestly expressed, splicing-derived miRNAs contribute substantially to the pool of distinct, confident, endogenous Dicer substrates transcribed from the mouse and human genomes. Given that most of these are recently evolved loci, this may imply that mirtrons may have a comparable contribution to canonical loci with regard to regulatory networks involving species-specific miRNAs.
Mechanistically diverse, Dicer-dependent, mirtron-derived miRNAs associate with Ago
We wished to obtain evidence that mature small RNAs from mirtrons are incorporated into Ago effector complexes. Analysis of extensive published human Ago1-IP and Ago2-IP data sets (Persson et al. 2011; Valen et al. 2011) identified 212 confident mirtrons with reads in human Ago-IP data; moreover, 146 candidate mirtrons had two or more reads in such Ago-IP data (Supplemental Fig. S4; Supplemental Table S8). The available mouse Ago-IP data we analyzed are much more modest (Schwamborn et al. 2009), but also contained reads from 27 mirtrons. The abundance of Ago-associated mirtron reads was overall modest, but as with the total RNA data, compared favorably with hundreds of miRBase canonical miRNA loci.
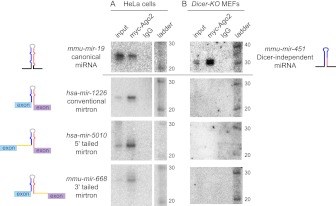
We sought further experimental evidence for the association of mirtron-derived miRNAs with Ago2 beyond library sequencing. The most direct method would be to detect these species using Northern blot assays. While mirtrons have been extensively analyzed this way in invertebrates (Okamura et al. 2007; Ruby et al. 2007a; Chung et al. 2011), to date we are not aware that this has been done in mammalian cells; the only studies thus far used indirect methods (Havens et al. 2012; Sibley et al. 2012). With this in mind, we selected members of all three mirtron biogenesis classes for functional study: conventional (hsa-mir-1226), 5′-tailed (hsa-mir-5010), and 3′-tailed (mmu-mir-668).
We cloned their host introns and flanking exonic sequence downstream of CMV>GFP, and transfected these constructs into HeLa cells together with a myc-Ago2 construct. We were encouraged to observe accumulation of mature species derived from all of these constructs in total RNA, although these were more difficult to detect compared with the canonical endogenous miRNA miR-19 (Fig. 6A). We then immunoprecipitated Ago2 from these cells and analyzed their associated RNAs. We now observed more robust Northern signals for mature species from each of the constructs, indicating their presence in Ago2 complexes (Fig. 6A). Parallel control immunoprecipitations using IgG showed no signals, demonstrating the specificity of these assays. These assays validate the association of mature miRNAs from all three mirtron biogenesis classes in Ago2 complexes.
Figure 6.
Northern validation of the incorporation of mirtron-derived miRNAs into Ago2 complexes. The structures of small RNA precursors of various biogenesis classes are shown at far left and far right. (A) Mirtron expression constructs were transfected into HeLa cells along with myc-tagged Ago2 vector. Endogenous miR-19 was detected in input samples and Ago2 immunoprecipitate, but not IgG immunoprecipitate. We detected mature mirtron-derived small RNAs from each of the three biogenesis classes in input samples. These were strongly enriched in Ago2 immunoprecipitates but absent from control IgG immunoprecipitates, demonstrating their specific residence in effector complexes. (White bar) Designates that the mirtron lanes were not adjacent to the ladder lanes, although they were taken from the same blots in all cases. The data for miR-1226 and miR-19 were generated by sequential stripping and reprobing of the same blot as an internal control; the other blots also showed mature miR-19 (data not shown). (B) Mirtron expression constructs were transfected into Dicer-KO MEFs along with a Mir144/451 expression construct and myc-tagged Ago2 vector. While the Dicer-independent species miR-451 was successfully processed in Dicer-KO cells, none of the three mirtrons generated mature small RNAs in either total RNA or Ago2 complexes. The data for miR-1226 and miR-451 were generated by sequential stripping and reprobing of the same blot as an internal control; the other blots also showed mature miR-451 (data not shown).
To further validate that mirtrons are matured via Dicer, we performed stringent assays in mouse embryonic fibroblasts stably knocked out for Dicer (Dicer-KO MEFs). We previously used these cells to analyze Dicer-independent processing of the Ago2-dependent locus Mir451 (Yang et al. 2010). This permitted a robust internal control to our experiments in which we co-transfected an Mir144/451 expression plasmid along with GFP-mirtron and myc-Ago2 expression plasmids into Dicer-KO MEFs. These tests validated that miR-451 was detected in total RNA and in Ago2 complexes, whereas co-transfected mirtrons failed to generate mature small RNAs in either condition (Fig. 6B). Therefore, mammalian mirtrons of all three classes are completely dependent on Dicer for their maturation. These data are consistent with our strict annotation criteria for which we demanded the presence of small RNA duplexes bearing a 3′ overhang on the terminal hairpin loop side, corresponding to the inferred Dicer cleavage site.
Discussion
Unexpected diversity and prevalence of mammalian mirtrons
Animal transcriptomes exhibit surprising flexibility in endogenous substrates of miRNA biogenesis pathways. Although the bulk of animal miRNAs are generated by the canonical Drosha–Dicer pathway, a cornucopia of Drosha-independent and even Dicer-independent miRNA pathways has been described (Yang and Lai 2011). These include splicing-mediated mirtrons (Berezikov et al. 2007; Okamura et al. 2007; Ruby et al. 2007a), tailed mirtrons (Babiarz et al. 2008; Flynt et al. 2010), snoRNA- and tRNA-derived miRNAs (Babiarz et al. 2008; Ender et al. 2008), tRNaseZ-derived miRNAs (Bogerd et al. 2010), Integrator-mediated miRNA biogenesis (Cazalla et al. 2011), and Ago2-mediated miRNA biogenesis (Cheloufi et al. 2010; Cifuentes et al. 2010; Yang and Lai 2010).
Nevertheless, by far the bulk of animal miRNA reads derives from canonical biogenesis. Since miRNA activity is concentration-dependent, this might be taken as evidence that the alternative biogenesis pathways are collectively of minor consequence. However, such a view is challenged by increasing reports of phenotypic discrepancies among mutants in core miRNA pathway machinery (Yang and Lai 2011). Furthermore, the embedding of noncanonical miRNA genes into conserved regulatory networks provides a teleological view that they have acquired essential functions. For example, the 3′-tailed mirtron pathway in Drosophila seems to be used to generate but a single well-conserved miRNA in flies, miR-1017 (Flynt et al. 2010), and the Ago2-mediated pathway in vertebrates seems to be used to manufacture only a single well-conserved miRNA, miR-451 (Yang and Lai 2010). Even though the endogenous repertoire of these pathways appears limited, their rigid preservation across flies and vertebrates indicates that mir-1017 and Mir451 have been evolutionarily selected for reasons that are apparently not satisfied by the canonical miRNA pathway.
Although 3′-tailed mirtrons had only been noted in Drosophila and 5′-tailed mirtrons in vertebrates (Westholm and Lai 2011), we now recognize that mammals have the capacity to generate 3′-tailed mirtrons, extending very recent observations from Jensen and colleagues (Valen et al. 2011). Moreover, while a subset of conventional mirtrons is well-conserved among mammals (Berezikov et al. 2007), we can now extend this to include atypical mirtrons of both the 5′-tailed and 3′-tailed classes. Thus, each of these alternative pathways has acquired a niche in the miRNA universe that is detectably “useful” to vertebrate gene regulation. Equally notable is the fact that mirtron pathways are no longer a minor pathway, at least in terms of numbers of mammalian loci. While mirtrons provide a small fraction of total mature miRNA reads, we now recognize that the various mirtron pathways act upon a number of loci that is about one-third to one-fourth those of annotated canonical miRNAs. We note that we have been quite strict in our annotation policies and that there are several hundred additional “candidate” mirtrons with compelling expression evidence (e.g., Fig. 4C), at least some of which are undoubtedly genuine (Supplemental Figs. S1, S2). Thus, splicing-derived hairpins contribute substantially to the diversity of endogenous Dicer substrates in mammals.
Regulatory possibilities for mirtrons
The mirtron pathway is best characterized as an alternate source of miRNA-class regulatory RNAs. For those mirtrons that are conserved (e.g., Figs. 2–4), their targets may be predicted using comparative genomics as for conserved canonical miRNAs. For the bulk of mirtrons, which are neither well-conserved nor abundantly expressed, their quantitative impact as _trans_-regulatory RNAs is likely subtle. Nevertheless, hundreds of them are detected in Ago-IP data sets (Supplemental Fig. S4; Supplemental Table S8), and to that extent may be inferred to exert at least some function as regulatory RNAs. As we now appreciate mirtrons to comprise a substantial fraction of confident, newly emerged miRNAs, they may conceivably mediate substantial aspects of species-specific miRNA regulatory networks, compared with canonical miRNAs. Indeed, we observed cases of primate- and rodent-specific mirtrons with loop-preferred divergence (Fig. 3), an evolutionary pattern consistent with positive selection for miRNA regulatory activity (Lai 2003; Lai et al. 2003).
However, it is also worth considering whether mirtronic hairpins may have any direct effects on their host mRNAs (Westholm and Lai 2011). In the scenario of _cis_-regulation, a paucity of small RNA products of these hairpins may or may not be meaningful. For example, a small amount of mature miRNAs are cleaved from the 5′ region of Dgcr8 (Friedlander et al. 2008), but it is not yet evident that they serve a substantial _trans_-regulatory function since the pre-miRNAs are mostly retained in the nucleus (Han et al. 2009). Instead, Drosha-mediated cleavage of these hairpins functions in cis to limit the production of this Drosha cofactor (Han et al. 2009; Kadener et al. 2009; Triboulet et al. 2009; Smibert et al. 2011).
In Drosophila, select mirtrons are generated by alternative splicing, so that production of the mRNA and the miRNA are exclusive, as opposed to coordinated, events (Chung et al. 2011). We similarly identified potential cases of mammalian mirtrons that reside in alternatively spliced introns (Fig. 3C; Supplemental Fig. S3). More generally, RNA structures have been reported to influence mRNA splicing (Graveley 2005; Warf and Berglund 2010). A close look at the conserved 5′-tailed mirtron Camk2a shows that it has accumulated several compensatory changes within the hairpin (Fig. 4B). Such a pattern is rare among conserved canonical miRNAs and mirtrons (Lai et al. 2003; Okamura et al. 2007; Flynt et al. 2010), which by and large exhibit greater nucleotide divergence in the terminal loop. In contrast, compensatory evolution is common for _cis_-regulatory structured RNAs. We should therefore consider whether mirtronic hairpins might influence splicing, either coordinately or independently of their capacity to be diced into miRNAs. In general, the mechanistic basis for the extraordinary bias for mammalian mirtrons to emerge in the 5′-tailed format begs for explanation, since conventional mirtrons and 3′-tailed mirtrons can all generate mature miRNAs that load Ago complexes in mammals (Fig. 6). Future studies should begin to address the regulatory influence of mirtrons, both in trans and in cis.
Methods
Primary data collection and processing
We downloaded 236 mouse and 501 human small RNA data sets from NCBI GEO/SRA, as summarized in Supplemental Tables S1 and S2; relevant ancillary information was captured from GEO and SRA to help organize the data in local databases. Our preference was to use raw data in the form of fastq files when available, and processed data when this was the only option. Those reads in which the adaptor was successfully clipped were then filtered by size >16 nt and <36 nt and collapsed by total read count to minimize the amount of redundant information. Bowtie was then used for genome mapping to either Mus musculus or Homo sapiens (mm9 or hg19 builds, respectively). Bowtie's parameters were to allow only perfect matches to the genome -v0. Unmapped reads were saved to a separate file for later processing at the end of the pipeline.
Intron analysis
Intron annotations were obtained from the UCSC Genome Browser (http://genome.ucsc.edu/) via the table browser. We de-duplicated these annotations by comparing the chromosome, start, end, and strand information. Next, we created a Bowtie index for each species' introns. Depending on its size, the intron was either retained as is, or split into two 250-nt portions, at the splice donor and the splice acceptor, adding 50-nt upstream and downstream flanking portions to assess exonic reads. The perfect genome mapping reads were then used as the input set of reads for mapping to introns. Reads mapping perfectly to introns were normalized by total genome hit count. The unmapped genome reads are then used for intron mapping by serial trimming of up to four 3′ nt. The trimmed nucleotides were recorded for each read, and the process was reiterated until a successful mapping to the genome was obtained. In this way we retain perfect mapping and terminally modified reads.
Mirtron annotation
To determine and analyze the set of “known” mirtrons, including loci that have been published but were not recognized as deriving from splicing, we reanalyzed all loci in miRBase release 17 (http://www.mirbase.org). We mapped all of our aggregate human and mouse small RNA data to the respective entries in these species and searched for loci with evidence of reads that mapped to splice acceptor and/or donor sites. We also incorporated loci reported in the literature that are not presently available in miRBase, namely, from Blelloch and colleagues (Babiarz et al. 2011) and Jensen and colleagues (Valen et al. 2011).
To identify novel loci, we examined all other introns whose termini were not associated with an miRNA annotation. Intron mappings were read into an interface processing program that generates html outputs for visual inspection. Custom scripts were used to automatically check a series of read properties that could indicate potential mirtrons (summarized in Supplemental Table S6).
- (1) A putative mature and star strand was predicted that involved the 5′-intron end, the 3′-intron end, or both (at most 13 nt from the splice site).
- (2) The mature strand was between 18 and 25 nt and the star strand between 18 and 26 nt; the relatively long length was necessary to capture a few mirtrons that generate reads of atypical size (Westholm et al 2012).
- (3) Neither the mature nor star strands were found to map more than nine times elsewhere in the genome; however, the vast majority of mirtrons had only uniquely mapping reads.
- (4) A straight hairpin that involved a duplex pairing from the mature and star strand of at least 13 bases, assessed using RNAshapes (Steffen et al. 2006).
- (5) The total reads from the mature and star strand must be at least 50 reads with the star strand containing a minimum of 3 reads. These had to exhibit 3′ overhangs on the Dicer cut site.
- (6) At least 15 reads mapped to the dominant arm, of which 85% displayed homogenous 5′ ends in an 11-nt window. To measure 5′ heterogeneity, we extended an 11-nt window centered at the 5′ end of the mature read. Those reads with 5′ ends beginning within this window were collected by position. A ratio of 85% of more of the top three starting positions among all starting positions within the window was necessary to consider homogeneous start positions.
These criteria alone were not sufficient to assess miRNAs confidently. All loci that passed were subject to detailed manual inspection in an effort to be very conservative in mirtron annotation. For example, we carefully considered the existence of specific miRNA/star duplexes bearing a 3′ overhang at the Dicer-cleaved site (on the terminal loop side). However, some Ago-loaded small RNAs are subject to extensive 3′ trimming, resulting in an apparent 5′ overhang (Westholm et al. 2012), so the inferred Dicer-cleaved duplex did not necessarily correspond to the most abundant reads. Furthermore, the 5′ end of many pre-miRNA hairpins from 5′-tailed mirtrons was somewhat heterogeneous, creating heterogeneity on the Dicer cleavage site. Therefore, duplex specificity had to be evaluated carefully, and many loci were downgraded to candidate or negative status if their status as a Dicer substrate was deemed at all ambiguous. Additional loci that failed but passed a majority of criteria were further inspected for compelling evidence and were kept as candidates for future analysis. Summaries of the known, novel, and candidate loci, comprising mirtrons, 5′-tailed mirtrons, and 3′-tailed mirtrons, in both human and mouse, are provided in Supplemental Tables S4 and S5, and Supplemental Figures S1 and S2. Much more comprehensive information on all these loci can be browsed and sorted at the following online resources: human (http://ericlailab.com/mammalian_mirtrons/hg19/) and mouse (http://ericlailab.com/mammalian_mirtrons/mm9/).
Validation of mirtron processing and Ago2 association
Mirtron expression constructs contained the intron and flanking exonic sequence; the primers used for amplification are listed below. HeLa cells and Dicer-KO MEFs were grown to confluency in six-well plates (9.5 cm2). HeLa cells were transfected with Lipofectamine LTX using 750 ng of pcDNA-Myc-Ago2 and 500 ng of pcDNA-GFP-mir-668, pcDNA-GFP-mir-5010, or pcDNA-GFP-mir-1226 per well. Dcr-KO MEFs were transfected with Lipofectamine 2000 using 2 μg of pcDNA-Myc-Ago2 and 1 μg of pcDNA-GFP-mir-144/451 (Yang et al. 2010), along with 1 μg of pcDNA-GFP-mir-668, pcDNA-GFP-mir-5010 intron, or pcDNA-GFP-mir-1226 per well. Cells were harvested 24 h later and lysed on ice in passive buffer. Cell lysates from three wells were pooled for IPs using anti-Myc (A-14, Santa Cruz) and IgG (Jackson Labs) antibodies. Northern blotting was performed with DNA oligos.
- hsa-mir-5010 F: GACTTTGGGGACACCATGGTCC
- hsa-mir-5010 R: CAGTGTAAGCGCAGTGCGTG
- hsa-miR-5010 probe: AATTTTGCTCTGCCATCCCCC
- mmu-mir-668 F: TGTGAGATGATACCAACCTCTAGAAGC
- mmu-mir-668 R: TGGGAACAGTAGTGGCTTGAGAAG
- mmu-miR-668 probe: GGTAGTGGGCCGAGCCGAGTGACA
- hsa-mir-1226 F: AGCCCAACAGCGTCACATA
- hsa-mir-1226 R: GTCAGCAGCAGCACAGCTA
- hsa-miR-1226 probe: CCCCATCCAGGCCTGCATGCCCTCAC
Acknowledgments
We are indebted to the many researchers that made this meta-analysis possible by depositing their small RNA data in public databases. J.O.W. was supported by a fellowship from the Swedish Research Council. K.O. was supported by the Japan Society for the Promotion of Science. Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Starr Cancer Consortium (I3-A139), and the NIH (R01-GM083300, U01-HG004261, and RC2-HG005639).
Footnotes
[Supplemental material is available for this article.]
References
- Axtell MJ, Westholm JO, Lai EC 2011. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12: 221 doi: 10.1186/gb-2011-12-4-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Hsu R, Melton C, Thomas M, Ullian EM, Blelloch R 2011. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA 17: 1489–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC 2007. Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. 2011. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21: 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol Cell 37: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Xie M, Steitz JA 2011. A primate herpesvirus uses the Integrator complex to generate viral microRNAs. Mol Cell 43: 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ 2010. A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC 2011. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res 21: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson N, et al. 2010. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328: 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ 2010. Small RNA sorting: Matchmaking for Argonautes. Nat Rev Genet 12: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Flynt AS, Chung WJ, Greimann JC, Lima CD, Lai EC 2010. microRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell 38: 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML 2008. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res 18: 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR 2005. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123: 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens MA, Reich AA, Duelli DM, Hastings ML 2012. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 40: 4626–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. 2011. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 19: 232–243 [DOI] [PubMed] [Google Scholar]
- Huang TH, Fan B, Rothschild MF, Hu ZL, Li K, Zhao SH 2007. MiRFinder: An improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinformatics 8: 341 doi: 10.1186/1471-2105-8-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT II, Nelson S, Rosbash M 2009. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA 15: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lai EC 2003. microRNAs: Runts of the genome assert themselves. Curr Biol 13: R925–R936 [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM 2003. Computational identification of Drosophila microRNA genes. Genome Biol 4: R42 doi: 10.1186/gb-2003-4-7-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N, Lim L, Weinstein E, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP 2003. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, de Bruijn E, Cuppen E 2010. Small RNA expression and strain specificity in the rat. BMC Genomics 11: 249 doi: 10.1186/1471-2164-11-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, Loman N, Jonsson G, Naya H, Hoglund M, et al. 2011. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res 71: 78–86 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007a. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007b. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J 2004. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 14: 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ 2012. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res 40: 438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Bejarano F, Wang D, Garaulet DL, Yang JS, Martin R, Bortolamiol-Becet D, Robine N, Hiesinger PR, Lai EC 2011. A Drosophila genetic screen yields allelic series of core microRNA biogenesis factors and reveals post-developmental roles for microRNAs. RNA 17: 1997–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R 2006. RNAshapes: An integrated RNA analysis package based on abstract shapes. Bioinformatics 22: 500–503 [DOI] [PubMed] [Google Scholar]
- Triboulet R, Chang HM, Lapierre RJ, Gregory RI 2009. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 15: 1005–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH 2011. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol 18: 1075–1082 [DOI] [PubMed] [Google Scholar]
- van der Burgt A, Fiers MW, Nap JP, van Ham RC 2009. In silico miRNA prediction in metazoan genomes: Balancing between sensitivity and specificity. BMC Genomics 10: 204 doi: 10.1186/1471-2164-10-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA 2010. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci 35: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Lai EC 2011. Mirtrons: microRNA biogenesis via splicing. Biochimie 93: 1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Ladewig E, Okamura K, Robine N, Lai EC 2012. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA 18: 177–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC 2010. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle 9: 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC 2011. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 43: 892–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC 2010. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci 107: 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]