Chaperoning histones during DNA replication and repair (original) (raw)
. Author manuscript; available in PMC: 2012 Sep 5.
Abstract
Nuclear DNA is tightly packaged into chromatin, which profoundly influences DNA replication, transcription, repair, and recombination. The extensive interactions between the basic histones proteins and acidic DNA make the nucleosomal unit of chromatin a highly stable entity. For the cellular machinery to access the DNA, the chromatin must be unwound and the DNA cleared of histone proteins. Conversely, the DNA has to be repackaged into chromatin afterwards. This review focuses on the roles of the histone chaperones in assembling and disassembling chromatin during the processes of DNA replication and repair.
Introduction
The term “molecular chaperone” was first coined by Ron Laskey (Laskey et al., 1978) to describe nuclear proteins in extracts of frog oocytes that prevent incorrect interactions between histones and DNA that would otherwise lead to the formation of an insoluble precipitate rather than nucleosomes. Subsequently, the term has been more broadly adopted to describe proteins that prevent or reverse incorrect interactions that occur when interactive surfaces are exposed to the environment. Histone chaperones shield non-specific interactions between the negatively charged DNA and the positively charged histones, to allow the ordered formation of the nucleosome structure. An additional important requisite of histone chaperones is that they are not a permanent component of the final product, the nucleosome.
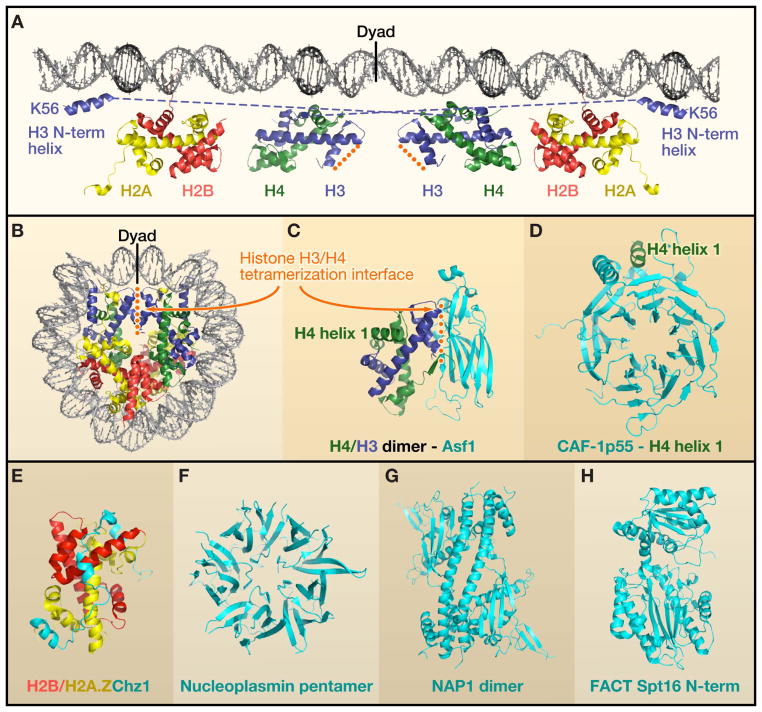
To better understand the function of histone chaperones, it is important to appreciate that the nucleosome is a modular assembly of stable heterodimers of histone 2A/2B (H2A/H2B) and H3/H4 associated with 146bp of DNA. The central ~80 bp of the nucleosomal DNA is organized by a heterotetramer of H3/H4, whereas the peripheral 40 bp of DNA on each side are bound more loosely by the H2A/H2B dimers (Figure 1A). The penultimate 10 bp are also organized by the N-terminal alpha helix of histone H3. Because the two H2A/H2B dimers occupy peripheral locations flanking the central H3/H4 heterotetramer within the nucleosome (Figure 1B), H2A/H2B dimers are not assembled onto the DNA until after H3/H4 have been deposited. Polynucleosome arrays further condense into higher order chromatin structures that are stabilized by association of additional proteins including linker histones. Conversely, during chromatin disassembly, H2A/H2B dimers are removed prior to removal of H3/H4 from the DNA. Perhaps due to the stepwise nature of the chromatin assembly and disassembly reactions, different histone chaperones exist to chaperone H2A/H2B and H3/H4.
Figure 1. Anatomy of the nucleosome and histone chaperones.
A. Unraveling of the nucleosomal DNA to indicate which regions of the 147 bp of DNA are organized by which histone proteins. The surface of the histones closest to the DNA is the one that binds to the DNA. The interactions within the stable dimer units have been maintained, but all other histone-histone interactions have been dissolved. The orange dotted line indicates the H3/H4 tetramerization interface. The N-terminal alpha helix of H3 interacts with a different DNA gyre from that which the remainder of the H3/H4 dimer interacts. PDB 1KX5. B. The nucleosome core particle (Luger et al., 1997), Protein Data Bank(PDB): 1KX5. C. The H3/H4 dimer binds to the Asf1 histone chaperone via the H3/H4 tetramerization interface (English et al., 2006), PDB: ID2Hue. D. CAF-1 p55 bound to the N-terminal alpha helix of histone H4 (Song et al., 2008), PDB 3C99. E. Chz1 bound to H2AZ-H2B (Zhou et al., 2008), PDB: 2JSS. F. Nucleoplasmin pentamer that binds H2A/H2B (Dutta et al., 2001), PDB: 1K5J. G. NAP1 that binds H2A/H2B (Park and Luger, 2006), PDB: 2AYU. H. N-terminus of FACT Spt16 that binds to H3/H4 (Stuwe et al., 2008), PDB: 3CB5.
Many of the histone chaperones that function during repair and replication were discovered via biochemical fractionation of chromatin assembly activities assayed in cell-free systems for chromatin assembly coupled to SV40 DNA replication (Laskey et al., 1977; Stillman, 1986). This review will detail the molecular insight that has since been gained into the function of histone chaperones during repair and replication. Although not discussed further here, the function of histone chaperones is intimately coupled to the action of ATP-dependent chromatin remodeling machines that use the energy provided by ATP hydrolysis to break histone-DNA contacts during chromatin disassembly and to reposition histones during chromatin assembly (Haushalter and Kadonaga, 2003).
How Do Histone Chaperones Bind Histones?
Other than having the common feature of being acidic, histone chaperones are a diverse group of proteins with little sequence similarity. Structurally, many histone chaperones include a hydrophobic beta sheet motif (Figure 1C,D,F–H). In the case of the ubiquitous histone chaperone Asf1, we know that the edge of the hydrophobic beta sheet motif mediates the interaction with histones (English et al., 2006) (Figure 1C). It will be interesting to determine whether this is also the case for the other histone chaperones bearing this motif. Certain other histone chaperones lack this motif. For example, Chz1 bears the irregular chain and alpha helices that mediate the interaction with dimers of H2B and the variant histone H2A.Z (Zhou et al., 2008) (Figure 1E).
Histones H3/H4 exist as a dimer when bound to some histone chaperones (Tagami et al., 2004), yet they exist as H3/H4 heterotetramers within the nucleosome (Figure 1B). The explanation for this apparent discrepancy, shown by the structure of Asf1 bound to an H3/H4 dimer (English et al., 2006; Natsume et al., 2007), is that Asf1 physically occludes the H3/H4 tetramerization interface (Figure 1C). The fact that Asf1 binds to newly-synthesized dimers of H3/H4 (Tyler et al., 1999) indicates that the process of chromatin assembly involves the formation of an H3/H4 heterotetramer from two dimers of H3/H4. Future studies should reveal whether histone chaperones downstream of Asf1 in the chromatin assembly pathways bind to dimers or heterotetramers of H3/H4, which will help determine whether the H3/H4 heterotetramer is formed on the DNA or prior to being incorporated onto the DNA. One histone chaperone that is downstream of Asf1 is chromatin assembly factor 1 (CAF-1). Recent crystal structures of the p55 subunit of Drosophila CAF-1 and the related protein human RbAp46 (Murzina et al., 2008; Song et al., 2008) reveal that they interact with alpha helix 1 of histone H4 (Figure 1D). This is a region of histone H4 that is far from the H3/H4 tetramerization interface (Figure 1C), raising the possibility that CAF-1 may be able to bind to and deposit tetramers of H3/H4 onto DNA.
Nucleosome Disassembly Ahead of the Replication Fork
Duplication of the genome requires that numerous replisome proteins intimately contact the DNA. The steps involved include unwinding of the DNA duplex by the MCM2-7 (minichromosome maintenance) helicase complex, followed by passing of the single-stranded DNA through various polymerases. The processivity of DNA replication is maintained by PCNA (proliferating cell nuclear antigen), a ring-like trimer that encircles the DNA and tethers DNA polymerase to prevent its dissociation. Conceptually, it is difficult to imagine that the physical process of threading the DNA through the MCM helicase, PCNA, and the polymerases, as well as separating the DNA strands, can occur on nucleosomal DNA. As such, DNA must be nucleosome free or “naked” while it is replicated. The best evidence for the removal of histones during DNA replication has come from electron micrographs of replicating SV40 minichromosomes that had been psoralen-crosslinked to preserve the location of where nucleosomes were positioned. These studies revealed that 250 bp or more of naked DNA resides behind the replication fork (Sogo et al., 1986), while a stretch of approximately 300 bp of naked DNA lies ahead of the replication fork (Gasser et al., 1996). Whether this destabilization of nucleosomes ahead of the replication fork is due to the passage of the replication machinery itself, or is mediated by histone chaperones or ATP-dependent chromatin remodelers, is not yet known.
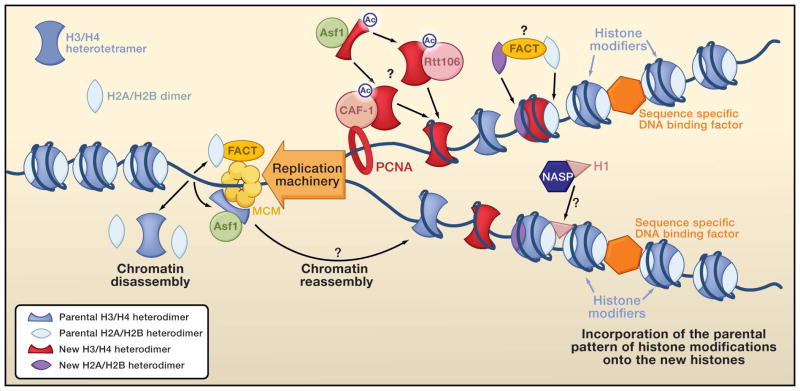
The loss of the histone octamer from the parental DNA during DNA replication is accompanied by the dissociation of H3/H4 from H2A/H2B (Figure 2) (Jackson, 1988). There are candidate histone chaperones for both H2A/H2B and H3/H4 that may disassemble chromatin at the DNA replication fork. However, because chromatin assembly and chromatin disassembly are so tightly coupled to DNA replication, it is often difficult to distinguish between a role for histone chaperones in chromatin disassembly versus a role in chromatin assembly during replication in vivo. To add to the confusion, inhibition of chromatin assembly after DNA replication negatively feeds back to inhibit DNA replication. Because inhibition of DNA replication would also be expected to occur as a consequence of inhibition of chromatin disassembly ahead of the replication fork, it is difficult to differentiate between whether a factor is solely required for chromatin assembly or is required for both chromatin disassembly and assembly during DNA replication in vivo.
Figure 2. Replication coupled chromatin assembly and disassembly.
The histone chaperones, but not the ATP-dependent chromatin remodelers involved in these processes are shown. Once the new histones have been post-translationally modified to adopt the pattern carried by the parental histones, they are considered parental histones and are colored accordingly. Question marks indicate steps that are somewhat speculative.
Circumstantial evidence supports a role for FACT (facilitates chromatin transcription) as an H2A/H2B chaperone during DNA replication (Table 1, Figure 2). FACT is a two subunit complex containing Spt16 (suppressor of Ty) and Pob3 (polymerase one binding) in yeast and SPT16 and SSRP1 (structure specific recognition protein) in humans. FACT has received considerable attention for its function in disassembling and assembling chromatin during transcriptional elongation. FACT is unusual for a histone chaperone in that it binds to both H2A/H2B and H3/H4, seemingly via multiple functionally overlapping interaction surfaces (Stuwe et al., 2008; VanDemark et al., 2008). FACT facilitates transcription dependent nucleosome alterations that can result in the displacement of a dimer of histones H2A/H2B from nucleosomes (Belotserkovskaya et al., 2003). However, it appears that H2A/H2B displacement may be an indirect consequence of the nucleosomal reorganization that is induced by FACT (Xin et al., 2009).
Table 1.
Histone chaperones implicated in DNA replication and repair.
| Histone Cargo | Histone Chaperone | Interactions and Function during Replication | Interactions and Function during Repair |
|---|---|---|---|
| H3/H4 | CAF-1 (p150, p60, p48) | PCNA, Asf1Chromatin assemblyHeterochromatin silencing | PCNA, BLM, WRN, Asf1Chromatin assemblyPromotes checkpoint recovery |
| Asf1 | RFC, MCM, CAF-1Promotes H3 K56 acetylation leading to chromatin assemblyChromatin disassembly*Histone buffer | CAF-1Promotes H3 K56 acetylation leading to chromatin assemblyPromotes checkpoint recovery | |
| Rtt106 | Chromatin assemblyHeterochromatin silencing | ||
| NASP | Histone buffer* | Histone buffer* | |
| H2A/H2B | FACT (Spt16 and Pob3) also binds H3/H4 | MCM, RPA, DNA pol IChromatin assembly*Chromatin disassembly* | γH2AX/H2B exchange for H2A/H2B |
| NAP1 | Histone shuttlingChromatin assembly**Chromatin disassembly** | ||
| Chz1 | H2AZ.H2B exchange for H2A/H2BPromotes DNA resection | ||
| H1 | NASP | Chromatin assembly* | |
| CenH3/H4 | Scm3 (S.c and S.p)HJURP (H.s.) | Mis18, RbAp48, NucleophosminCentromere identity |
Regardless of whether the role of FACT in histone H2A/H2B displacement is direct or indirect, FACT clearly is involved in altering chromatin structure during DNA replication. Indeed, the Pob3 subunit of budding yeast FACT was first discovered as a protein bound to DNA polymerase I (Wittmeyer and Formosa, 1997). FACT is required for DNA replication in Xenopus egg extracts (Okuhara et al., 1999). A role for FACT during DNA replication is further suggested by the synthetic phenotypes of yeast double mutants between FACT and other replication components, as well as the sensitivity of yeast FACT mutants to hydroxyurea, a drug that interferes with DNA replication by causing replication forks to stall (Schlesinger and Formosa, 2000). Human and mouse FACT localize to replication origins (Hertel et al., 1999; Tan et al., 2006) whereas budding yeast FACT interacts with the single-stranded DNA binding protein replication protein A (RPA) (VanDemark et al., 2006) and human FACT interacts with the MCM complex (Tan et al., 2006). Yeast FACT also co-purified as part of the replication fork progression complex (Gambus et al., 2006). Ultimately, the most convincing evidence for a role for human FACT in chromatin disassembly during DNA replication comes from its ability to promote replication initiation in vivo and to promote DNA unwinding by the MCM helicase on nucleosomal templates in vitro (Tan et al., 2006).
Another histone chaperone that could be involved in removing H2A/H2B from DNA is Nap1. Nap1 interacts with H2A/H2B in co-immunoprecipitation studies and can assemble chromatin in vitro (Ito et al., 1996). Conversely, Nap1 can also disassemble histone octamers from chromatin in concert with the ATP-dependent chromatin remodeling factor RSC in vitro (Lorch et al., 2006). However, great care has to be taken when inferring physiological relevance from biochemical analyses of histone chaperone function. This is because in vitro chromatin disassembly assays (in the absence of ongoing DNA repair or replication) require a negatively charged molecule to capture the histones when they are loosened from the DNA by the action of an ATP-dependent remodeler or during the process of nucleosome breathing (spontaneous partial unwinding). Similarly, in vitro chromatin assembly assays (in the absence of ongoing DNA repair or replication) also require a negatively charged molecule to hold onto the free histones long enough to slow down the intrinsic electrostatic attraction between DNA and histones. By empirical optimization, assays can be created to achieve both chromatin assembly and chromatin disassembly by the same negatively charged molecule, as is the case for Nap1. Consequently, it is important to confirm the findings of biochemical analyses of chromatin assembly and disassembly with in vivo studies. However, the interpretation of in vivo studies becomes complicated when the histone chaperones are involved in multiple pathways, when they are essential, or when they are functionally redundant.
Regardless of how H2A/H2B are removed, it is clear that once they have exited from the DNA, the more stable H3/H4 dimers can then be removed. One histone chaperone that has the potential to remove H3/H4 from replicating chromatin is Asf1 (anti-silencing function) (Fig. 2). Asf1 localizes to replication foci in Drosophila S2 cells, but rapidly dissociates when replication is halted in response to hydroxyurea treatment (Schulz and Tyler, 2006). In yeast, Asf1 interacts with the PCNA loader, RFC (replication factor C) and Asf1 is required for the integrity of the replisome in the face of replicational stress (Franco et al., 2005). Like FACT, human Asf1 has been shown to bind to the MCM helicase complex, through histones H3 and H4 (Groth et al., 2007) (Fig. 2). As such, Asf1 is at the right place at the right time to influence chromatin structure during replication. The strongest evidence for a role for Asf1 in histone disassembly during replication is that the histones in the Asf1-H3/H4-MCM complex display a parental distribution of post-translational modifications, suggesting that the histones bound to Asf1 have been removed from the parental chromatin (Groth et al., 2007). Furthermore, in the absence of human Asf1, DNA unwinding by the MCM helicase is greatly reduced in vivo, potentially due to the failure to remove the nucleosomes ahead of the replication fork (Groth et al., 2007).
The definitive proof of whether any histone chaperone is required for chromatin disassembly during DNA replication awaits the development of in vivo assays for chromatin disassembly during replication. Alternatively, biochemically manipulable in vitro systems, such as the Xenopus egg extract chromatin replication system, hold the potential to provide molecular insight into chromatin disassembly during replication and the understanding of how the parental pattern of histone variants and histone post-translational modifications are re-established on the daughter DNA strands to facilitate epigenetic inheritance.
Nucleosome Reassembly Following Replication Fork Passage
Chromatin is rapidly reassembled onto newly-replicated DNA. This occurs almost as soon as the length of DNA needed to wrap around the core histone octamer has passed through the replication machinery (Sogo et al., 1986). In the reassembled chromatin, half of the histones are recycled from the parental chromatin while the other half are newly-synthesized. The mechanism used to transfer the parental histones to the newly-replicated DNA appears to be somewhat distinct from the mechanism whereby newly-synthesized histones are deposited, as discussed below. Nevertheless, the assembly of both parental and newly-synthesized histones occurs without particular preference for either the leading or lagging DNA strand, that is it occurs in a semi-conservative manner (Jackson, 1988; Sogo et al., 1986). The first step in chromatin assembly is the deposition of H3/H4 onto the DNA, followed by deposition of the two H2A/H2B dimers and later by incorporation of linker histones such as H1, if appropriate.
De novo incorporation of newly-synthesized H3/H4 after DNA replication is mediated by the CAF-1 histone chaperone (Figure 2). CAF-1 (chromatin assembly factor) is a highly conserved three subunit complex that favors assembly of chromatin onto newly-replicated DNA over bulk DNA in vitro (Smith and Stillman, 1989). Based on their post-translational modification pattern, CAF-1 binds to newly-synthesized molecules of histones H3 and H4 (Verreault et al., 1996). A role for CAF-1 in chromatin assembly during replication in vivo is suggested by the fact that it binds to the replication-specific histone H3.1 variant but not to the replication-independent histone H3.3 variant (Tagami et al., 2004). CAF-1 is found at sites of replication, presumably through its interaction with PCNA (Krude, 1995; Shibahara and Stillman, 1999). Consistent with a role for CAF-1 in assembling chromatin following DNA replication in vivo, both RNA interference (RNAi)-mediated depletion of CAF-1 and expression of a dominant negative CAF-1 mutant profoundly decrease the assembly of newly-replicated DNA into chromatin (Hoek and Stillman, 2003; Nabatiyan and Krude, 2004; Ye et al., 2003). Interestingly, depletion of Human CAF-1 also activates the DNA damage checkpoint and stalls DNA replication itself (Nabatiyan and Krude, 2004; Ye et al., 2003). This result suggests that rapid chromatin assembly behind the replication fork is required, either directly or indirectly, for the efficient progression of the replication fork. Although the nature of this negative feedback mechanism is unclear, it further highlights the tight coupling between chromatin assembly and DNA replication.
The histone chaperone Asf1 appears to deliver the newly-synthesized histones to CAF-1 for deposition onto the newly-replicated DNA (Figure 2). Accordingly, chromatin assembly onto replicating DNA in eukaryotes larger than budding yeast requires Asf1 in vitro (Tyler et al., 1999) and in vivo (Galvani et al., 2008; Groth et al., 2005; Sanematsu et al., 2006). The one exception to this is that chromatin assembly by CAF-1 after DNA synthesis in Xenopus egg extracts does not require Asf1, presumably because CAF-1 can obtain its histones from the large storage pools of histones present in this unique system (Ray-Gallet et al., 2007). In adult cells, the transfer of newly-synthesized histones from Asf1 to CAF-1 is likely to be mediated via a physical interaction between these two histone chaperones (Tyler et al., 2001). The role of Asf1 at the replication fork is clearly important because depletion of human, chicken, or Drosophila Asf1 slows down DNA replication (Groth et al., 2005; Sanematsu et al., 2006; Schulz and Tyler, 2006) and slows down DNA unwinding by the MCM helicase in human cells (Groth et al., 2007). Given that Asf1 binds to histone H3/H4 dimers (Figure 1C), it is still unclear exactly how the H3/H4 tetramers form on DNA. Clearly, Asf1 has to be removed from the tetramerization interface of H3 to enable formation of the H3/H4 heterotetramer (Figure 1D). But exactly how Asf1 is removed from the H3/H4 dimer and how the histones are transferred to CAF-1 remains unclear (Figure 2). Depending on whether CAF-1 binds to H3/H4 dimers or H3/H4 tetramers, it is possible that the H3/H4 tetramer is first assembled on CAF-1 followed by its deposition onto DNA. Alternatively, CAF-1 may transfer two H3/H4 dimers to the DNA to form the H3/H4 tetramer on DNA. Yet another H3/H4 histone chaperone, Rtt106, is also implicated in the assembly of chromatin after DNA replication, because it interacts physically and functionally with CAF-1 (Huang et al., 2005; Huang et al., 2007) (Figure 2). Exactly how Rtt106 and CAF-1 coordinate to assemble the newly-synthesized H3/H4 tetramer remains to be determined.
The other half of the histones that are assembled onto the newly-replicated DNA are derived from the parental chromatin. It is far from clear which histone chaperones, if any, are involved in the transfer of parental histones from the parental DNA to the newly-replicated DNA. However, the question of whether old H3/H4 is transferred from the parental chromatin to the newly-replicated DNA as a dimer or a heterotetramer has recently become a matter of debate (Corpet and Almouzni, 2009; Tagami et al., 2004). The answer to this seemingly trivial question may have profound consequences for understanding the mechanistic basis for epigenetic inheritance of histone modifications. If the parental H3/H4 tetramer were split into two H3/H4 dimers, then one H3/H4 dimer could be deposited onto each of the two newly formed daughter-parental DNA duplexes. In this manner, the parental H3/H4 dimer could provide a template for the copying of the parental pattern of post-translational modifications onto the newly synthesized and incorporated H3/H4 dimer thereby recreating the parental pattern of histone modifications present on the parental chromatin over the same DNA sequence on the new DNA duplexes. However, if the original stimuli for the histone modifications and the epigenetic initiators (such as transcriptional activators or repressors) are still present, splitting the parental H3/H4 tetramer would not be necessary for the reestablishment of the parental pattern of histone modifications (Figure 2). In this scenario, once the association of the sequence-specific DNA binding factors has been re-established after DNA replication, they would re-recruit the histone modifying machinery to the chromatin and subsequently reset the pattern of post-translational histone modifications on the daughter DNA duplexes back to that of the parental chromatin (Figure 2).
There is strong evidence for the parental H3/H4 tetramer either being transferred to or reassembled onto the newly-replicated DNA as a whole, as opposed to formation of a mixed heterotetramer consisting of a parental H3/H4 dimer and a new H3/H4 dimer. In a set of classical experiments from Vaughn Jackson’s lab, proteins were radiolabeled with heavy amino acids by a pulse-chase technique followed by nuclear isolation, formaldehyde cross-linking and separation of new labeled histones from old unlabeled histones on density gradients. From this analysis, it was determined that although new H2A/H2B dimers could freely associate with parental H3/H4 tetramers, new H3/H4 dimers were undetected in complexes with parental H3/H4 dimers following DNA replication (Jackson, 1988). This result indicates that the majority of parental H3/H4 heterotetramers do not dissociate into H3/H4 dimers to reassort with new H3/H4 dimers during DNA replication. This interpretation is supported by experiments in slime molds that were fed fluorescent H3. Fluorescent H3 emits light at a different wavelength when paired with unmodified H3. If H3 dissociated from the parental tetramer and reformed with the new H3, a change in fluorescence could be measured. However, such a change in emission could not be detected, indicating that either the vast majority of parental H3/H4 heterotetramers remain intact, or the two parental H3/H4 dimers preferentially reassociate with each other after DNA replication (Prior et al., 1980).
Even though tetramers of parental H3/H4 are clearly passed to the daughter genomes, it is unclear whether the parental H3/H4 tetramer is transiently dissociated into two H3/H4 dimers or whether it always remains intact. Given that the histone chaperone Asf1 binds to dimers of H3/H4 and not to tetramers of H3/H4 (Figure 1C) (English et al., 2006; Natsume et al., 2007), it is intriguing that human Asf1 binds to histones carrying the parental pattern of histone post-translational modifications in the context of the MCM-Asf1-histone complex (Groth et al., 2007). Assuming that the histones within the MCM-Asf1-histone complex are directly binding to Asf1, this would suggest that H3/H4 tetramers may at least be transiently split into dimers during their transfer, via Asf1, from the parental chromatin to the newly-replicated DNA (Figure 2). Currently, it is not clear whether Asf1 is actively or passively involved in the displacement of the H3/H4 dimers from the parental chromatin.
After the H3/H4 tetramer has been assembled onto the DNA, a dimer of H2A/H2B is incorporated on either side of the tetramer. As discussed previously, both FACT and Nap1 interact with H2A/H2B; however, it is still unclear whether one or both of them donate H2A/H2B dimers to DNA following replication. Notably, Drosophila Nap1 relocates to the nucleus during S-phase of the cell cycle, potentially functioning to bring newly-synthesized histones H2A/H2B to the replication fork (Ito et al., 1996).
Histone H1 is the last histone to be assembled onto the chromatin following replication (Figure 2). Although the emphasis has traditionally been on the core histone proteins, there is data that suggests the proper incorporation of H1 is also required for S phase progression. The NASP (nuclear autoantigenic sperm protein) histone chaperone binds to H1 in vitro and in vivo, and can incorporate H1 onto nucleosome arrays (Finn et al., 2008). Unexpectedly, human NASP exists in multi-chaperone complexes with H3/H4 and the H3/H4 chaperones (Tagami et al., 2004) and is found as the only histone chaperone within cytoplasmic H3/H4 complexes (Eric Campos and Danny Reinberg, personal communication). NASP, like the other chaperones involved in replication, is required for efficient replication and cell cycle progression (Richardson et al., 2006).
A Histone Code for Chromatin Assembly
Newly-synthesized histones H3 and H4 have a specific pattern of post-translational modifications, and nearly all newly-synthesized histone H3 is acetylated on lysine 56 (H3K27ac; Masumoto et al., 2005). H3 lysine 56 resides within the N-terminal alpha helix of H3, which interacts with the DNA as it enters and exits the nucleosome (Figure 1A). Specifically, lysine 56 contacts DNA via a water molecule and acetylation of lysine 56 breaks this histone-DNA contact and increases the rate of unwrapping or “breathing” of the ends of the nucleosomal DNA from the histone octamer seven fold over the unacetylated state (Neumann et al., 2009). Interestingly, the histone chaperone Asf1 is required to achieve acetylation of H3 on K56 via its role in presenting H3/H4 dimers to the Rtt109 acetyltransferase (Han et al., 2007; Recht et al., 2006; Tsubota et al., 2007). Strikingly, H3K56 acetylation greatly increases the affinity of CAF-1 and Rtt106 for H3, which in turn results in efficient chromatin assembly following replication (Li et al., 2008). Basically, H3K56ac acts to recruit H3/H4 dimers to the histone chaperones that are poised at the sites of DNA synthesis, in order to promote their assembly onto DNA.
It is intriguing that the specific histone modification (H3K56ac) that targets histones to the sites of DNA synthesis for chromatin assembly is the same histone modification that loosens chromatin structure to promote chromatin disassembly from transcribed genes (Williams et al., 2008). Conveniently for the cell, acetylated H3K56 blocks the formation of two distinct histone-DNA contacts causing the newly-formed nucleosomes to be slightly looser. The available evidence indicates that histone chaperones deposit histones onto DNA in a random manner to form irregularly spaced or closely packed arrays of nucleosomes. The subsequent action of ATP-dependent chromatin remodelers enables the histone octamer to move along the DNA to find its optimal position. This spot is likely to be determined by a combination of DNA sequence, nearby sequence-specific DNA binding proteins and electrostatic repulsion from adjacent histone octamers. Having K56 acetylated on the newly-deposited histone octamers would facilitate the nucleosome repositioning process during chromatin maturation. Once the nucleosomes have achieved their optimal position after DNA replication, H3K56ac is deacetylated (Masumoto et al., 2005). This deacetylation step may help stabilize the nucleosomes once the octamers are appropriately positioned on the DNA.
In addition to H3 being acetylated on lysine 56, newly-synthesized histones also carry other specific acetylation marks that may be important for their assembly into chromatin. These include acetylation of the N-terminal tails on H4K5 and K12 residues and H3 on K9/14 (depending on the species being examined). Indeed, the N-terminal tails of either H3 or H4 are essential for chromatin assembly in vivo and in vitro (Ling et al., 1996). Although we still do not understand how the H4 N-terminal tail acetylations contribute to chromatin assembly, it appears that the acetylation of the H3 N-terminal tails by Gcn5 (general control nonderepressable 1) is required for efficient incorporation of new histones following DNA replication by CAF-1, via increasing the affinity of the histones for CAF-1 (Zhigou Zhang, personal communication). Furthermore, different histone acetylations appear to be targeted or read by different histone chaperones. For example, while H3K56ac promotes binding to both CAF-1 and Rtt106, the N-terminal acetylations of H3 promote binding to CAF-1, yet only have a minor effect on Rtt106 binding (Zhigou Zhang, personal communication). Therefore, acetylation of H3 at the N-terminus and at lysine 56 is important for targeting histones to the histone chaperones in order to drive chromatin assembly after DNA synthesis.
Whereas acetylation of histone H3 K56 regulates the interaction between the DNA and the histone components of the nucleosome, other histone modifications regulate the interaction between H2A/H2B dimers and H3/H4 tetramers during chromatin assembly. For example, newly-synthesized histone H4 is acetylated on lysine 91, which is a residue that lies at the interface of the histone H2A/H2B dimer and the H3/H4 tetramer (Ye et al., 2005). Indeed, this acetylation event destabilizes the histone octamer (Ye et al., 2005). It is intriguing to consider that the cell may use this and other histone modifications to coordinate the process of chromatin assembly, perhaps to prevent premature interactions between the dimer and tetramer subunits of the nucleosome chromatin assembly, whereas deacetylation of H4 K91 would promote formation of the histone octamer.
Chaperones for Centromeric Histone H3 Variants
The centromere is a unique heterochromatic domain that mediates spindle microtubule attachment and is required for correct chromosome segregation. Centromere function depends on a unique H3 histone variant called CENP-A in humans, Cid in D. melanogaster, Cse4 in S. cerevisiae and Cnp1 in S. pombe. These centromere specific histone H3 variants, collectively termed CenH3, define the identity of the chromatin region as a centromere and potentially act as an epigenetic mark to identify centromeres. As such, it is critically important that CenH3 is efficiently and specifically incorporated onto only centromeric DNA.
In contrast to the highly conserved amino acid sequences of H3 proteins among species, CenH3 is highly diverged. Nonetheless, members of this diverse family are conserved in function. For example, human CENP-A can functionally replace Cse4 in yeast (Wieland et al., 2004). The amino acid differences between CenH3 and canonical H3 confer structural rigidity to the CenH3 nucleosome and this in turn is essential for specifying the centromere location (Black et al., 2007). In larger eukaryotes, the CenH3 containing nucleosomes are interspersed between canonical nucleosomes promoting the folding of centromeric chromatin into unique higher order structures during metaphase. At this time, all the CenH3 containing nucleosomes face outwards on one surface (Figure 3) (Black and Bassett, 2008). The CenH3 surface forms the foundation for assembly of the kinetechore and microtubule attachment to enable faithful segregation of sister chromatids to each daughter cell. Budding yeast, by contrast, have a single CenH3 nucleosome on each chromosome. Rather unexpectedly, it has recently been reported that the yeast CenH3 nucleosome has the DNA wrapped in a right-handed manner inducing positive DNA supercoiling (Furuyama and Henikoff, 2009). This is the opposite of the left-handed wrapping of the DNA around the canonical histone octamer, which induces negative DNA supercoiling and could play an important role in the epigenetic maintenance or function of the yeast centromere.
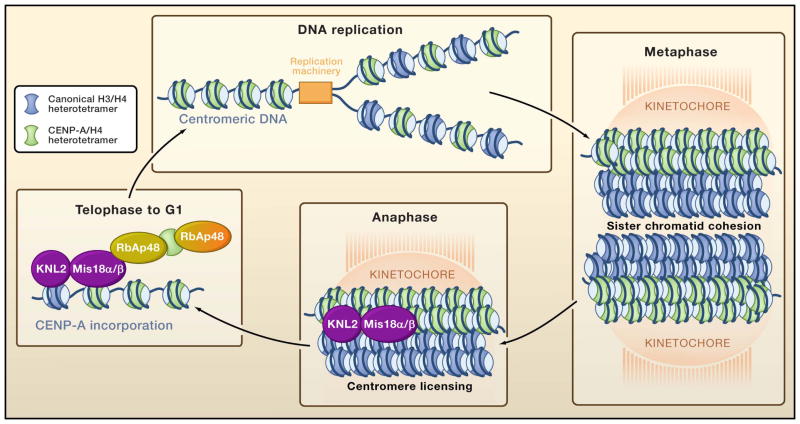
Figure 3. Incorporation of the centromere specific histone variant CENP-A.
Parental CENP-A is passed to the newly-replicated DNA through DNA replication, but de novo incorporation of CENP-A, mediated by the histone chaperones HJURP and RbAp48 does not occur until after chromosome segregation to the daughter cells.
Recently, new insight has been gained into how the centromeric specific histone H3 variants are assembled into centromeric DNA. Unlike canonical histone H3, newly-synthesized CenH3 is not incorporated onto the DNA following DNA replication. Indeed, animal cells proceed through chromosome segregation having filled only half of the available sites for CenH3 assembly because CenH3 is incorporated into chromatin only during telophase to G1 phase of the cell cycle (Figure 3) (Jansen et al., 2007; Schuh et al., 2007). This temporal control of loading of CenH3 is dependent on the prior incorporation of Mis18α/β and KLN2, which occurs transiently in anaphase and mediates a priming or licensing event that is necessary to maintain centromere identity (Fujita et al., 2007) (Figure 3).
In an effort to isolate potential CENP-A chaperones, epitope-tagged human CENP-A proteins were recently used to identify interacting proteins in prenucleosomal complexes (Dunleavy et al., 2009; Foltz et al., 2009). Several proteins have identified in this fashionm including HJURP (Holliday junction recognition protein), also known as hFLEG1 (human fetal liver expressing gene 1), and the two histone chaperones retinoblastoma binding accessory protein 48 (RbAp48) and nucleophosmin (Npm1) (Figure 3). When compared to other H3 prenucleosomal complexes, only HJURP and Npm1 are specifically enriched in CENP-A complexes, whereas RbAp48 is present in many different chromatin assembly and chromatin modifying complexes. Consistent with a role in CENP-A incorporation into chromatin, HJURP localizes to the centromere during the short window of CENP-A incorporation during late telophase/early G1 phase of the cell cycle. Using RNAi to knockdown the expression of the putative CENP-A histone chaperones, it has been shown that HJURP and RbAp48 are both required for CENP-A localization (Dunleavy et al., 2009; Foltz et al., 2009). Providing a possible link between the timing of CENP-A recruitment in late mitosis, Mis18α/β interacts with RbAp48 (Fujita et al., 2007). As such, one could imagine how CENP-A may be recruited to the centromere via interactions between RbAp48 and Mis18α/β, while HJURP incorporates the CENP-A into the adjacent centromeric chromatin (Figure 3).
Although it lacks sequence similarity with HJURP, Scm3 (suppressor of chromosome misegregation) has been proposed as the histone chaperone that mediates the incorporation of CenH3 into centromeric chromatin in both budding and fission yeast. Scm3 interacts with the yeast CenH3 both in vivo and in vitro, and is required both for the incorporation of CenH3 into centromeric chromatin and for proper chromosome segregation (Mizuguchi et al., 2007; Pidoux et al., 2009; Stoler et al., 2007; Williams et al., 2009). Despite the recent progress in discovering the histone chaperones that incorporate CenH3 into centromeric chromatin, many questions remain regarding how centromeric nucleosomes are formed, the exact structural nature of the centromeric nucleosome and how they interact with centromere specific proteins.
The access, repair, restore model for DNA repair
The steps and histone chaperones involved in chromatin assembly after DNA repair are closely related to those during DNA replication, which is not too surprising given the similarity of the machinery involved in copying the DNA template during both repair and replication. DNA is continually damaged by intrinsic and extrinsic sources such as free radicals, ultraviolet (UV) light, gamma rays, and mutagenic chemicals. This damage must be recognized and repaired rapidly and efficiently to prevent the propagation of mutations. The tightly packaged chromatin structure impedes DNA repair and the current model for DNA repair on chromatin is the “access, repair, restore” model (Smerdon, 1991). The first step in the repair process is recognition of the damage. Once the DNA damage has been recognized, the histones are shifted or removed to allow the DNA repair proteins to access the DNA lesion. Chromatin alteration is followed by the repair of the DNA lesion and the restoration of the chromatin structure. Although the study of chromatin dynamics during DNA repair is in its infancy compared to the understanding of the relationship between chromatin and either replication or transcription, many similar themes are emerging. Here, we will highlight the chromatin dynamics that are unique to DNA repair as opposed to DNA replication. Post-translational histone modifications are important for the DNA damage response and have been thoroughly reviewed elsewhere (Dinant et al., 2008; Escargueil et al., 2008), but will not be discussed here unless they pertain directly to the processes of chromatin assembly and disassembly.
Chromatin disassembly during DNA repair
Just as there are many types of DNA lesions, there are multiple distinct DNA repair pathways. The four main pathways are nucleotide excision repair, base excision repair, homologous recombination and non-homologous end joining (NHEJ). Nucleotide excision repair removes bulky DNA lesions that distort the DNA helix such as those caused by ultraviolet (UV) light. Small chemical alterations in the bases and single strand breaks are targeted by base excision repair. In both nucleotide and base excision repair, the DNA lesion is only on one strand of DNA. The lesion is excised and the resulting gap is filled in by DNA synthesis using the intact DNA strand as a template. The more problematic double-strand breaks are repaired by either homologous recombination or NHEJ. Homologous recombination is used when a homologous copy of the sequence is available, for example on the sister chromatid, while the more error-prone NHEJ pathway is used to “stick” the DNA ends back together. Despite the different machineries mediating repair, where studied, all pathways require opening chromatin to enable access of the repair machinery to the DNA.
During nucleotide excision repair approximately 30 base pairs of DNA are replaced. The pioneering work of Michael Smerdon over 30 years ago demonstrated that nucleotide excision repair is accompanied by nucleosome rearrangements (Smerdon and Lieberman, 1978) and is promoted by histone acetylation (Smerdon et al., 1982) which may serve to increase access to the DNA. Subsequent studies have lead to a consensus that chromatin is remodeled by ATP-dependent chromatin remodelers (such as SWI/SNF) at sites of UV damage in order to promote nucleotide excision repair (Dinant et al., 2008). In contrast to DNA replication, both nucleotide excision repiar and double-strand break repair are preceded by localized increases in histone acetylation (Dinant et al., 2008; Tamburini and Tyler, 2005). The end result of the chromatin changes is to promote nucleotide excision repair. Whether this is achieved by sliding the histone octamers along the DNA or removing them from DNA with histone chaperones is still not clear.
The study of chromatin dynamics during double-strand DNA break repair has been greatly facilitated by the introduction of inducible site-specific endonucleases that cleave the DNA at one or a limited number of defined positions within the genome. Using such systems, the chromatin structure flanking the induced DSB can be assayed by chromatin immunoprecipitation (ChIP) while simultaneously following the DNA damage and repair processes by polymerase chain reaction (PCR) or southern blotting analyses. Both pathways for double-strand break repair require that the DNA ends be resected by a combination of the action of the exonucleases Exo1, Dna2 and MRX (Mre11/Rad50/Xrs2) complex and the Sgs1 helicase complex (Raynard et al., 2008) to yield single-stranded DNA that then either anneals (in the case of NHEJ) or invades (in the case of homologous recombination) homologous DNA sequences. The resection of the DNA during double-strand break repair is accompanied by at least some degree of chromatin disassembly, as revealed by histone ChIP analyses and increased accessibility to micrococcal nuclease digestion (Tsukuda et al., 2005; van Attikum et al., 2007). Further confirmation for histone eviction from the single stranded DNA following DNA resection comes from the fact that they are replaced by the single stranded DNA binding proteins RPA and Rad51 (Dubrana et al., 2007; Tsukuda et al., 2005). To date, it has not been possible to kinetically separate the processes of DNA resection and chromatin disassembly. For example, mutants that slow down DNA resection also slow down chromatin disassembly (Chen et al., 2008; Tsukuda et al., 2005; van Attikum et al., 2007).
The degree of chromatin disassembly flanking a double strand break is a matter of debate. The extent of histone loss by ChIP analysis is not striking when one normalizes to the input DNA signal (Shroff et al., 2004; Tamburini and Tyler, 2005). However, in mutants that are not able to repair the DNA break, such as rad52 mutants, the degree of histone loss near the DNA break is more profound (Chen et al., 2008). Furthermore, although yeast strains lacking the histone chaperone Asf1 do repair their DNA damage, they do not reassemble chromatin following DNA repair, indicating that some histones flanking the double-strand break must have been lost (Chen et al., 2008). The requirement for the INO80 ATP-dependent chromatin remodeler for both the nucleosome disassembly flanking a double-strand break and for DNA resection (van Attikum et al., 2007), suggests that although these two processes are tightly coupled, histone removal is likely to facilitate DNA resection rather than being a consequence of DNA resection. Because the rate of DNA resection determines the rate of chromatin disassembly flanking a double strand break (Chen et al., 2008; Tsukuda et al., 2005; van Attikum et al., 2007), one might predict that the resection machinery and the chromatin disassembly machinery are physically coupled.
To date, there is no evidence for histone chaperones playing an active role in chromatin disassembly flanking a double strand break or a UV lesion. Neither Asf1 nor CAF-1 are required for DNA resection or for nucleotide excision repair. This lack of a requirement for histone chaperones in chromatin disassembly during DNA repair contrasts with DNA replication, where the absence of histone chaperones inhibits DNA replication. However, this does not rule out a potential role for histone chaperones acting as histone acceptors during the chromatin disassembly step that is coupled to DNA repair.
Chromatin reassembly after DNA repair
Following nucleotide excision repair, the chromatin is reassembled over the site of repair by the human H3/H4 histone chaperones Asf1 and CAF-1 (Gaillard et al., 1996; Mello et al., 2002). Current evidence indicates that this process is very similar to that during replication: PCNA recruits CAF-1 to the site of DNA repair and Asf1 hands the histones to CAF-1 for deposition onto the DNA (Green and Almouzni, 2003; Moggs et al., 2000). In vivo, CAF-1 localizes to sites of UV damage and is required for the incorporation of newly-synthesized, replication-specific, H3.1 onto the repaired DNA (Green and Almouzni, 2003; Polo et al., 2006).
CAF-1 knockdown does not inhibit nucleotide excision repair in mammalian cells, suggesting that this H3.1 deposition is part of a chromatin restoration step after repair with no, or limited, influence on the repair rate itself (Polo et al., 2006). To unequivocally rule out any interference between the role of CAF-1 during DNA replication and during DNA repair, the response to DNA damage has been investigated in quiescent cells (Nabatiyan et al., 2006). CAF-1 is highly induced and recruited to sites of nucleotide excision repair and double-strand break repair in quiescent cells following UV irradiation, gamma irradiation or treatment with a radiomimetic (Nabatiyan et al., 2006).
All available data indicate that during double-strand break repair, like nucleotide excision repair, CAF-1 and Asf1 mediate chromatin assembly over the repaired DNA. In yeast, neither CAF-1 nor Asf1 mutants show any clear defects in repair of the double strand breaks themselves (Linger and Tyler, 2005; Ramey et al., 2004), consistent with a role after rather than before DNA repair. Asf1’s role in promoting chromatin assembly after double-strand break repair is an indirect consequence of its requirement to acetylate H3K56 (Chen et al., 2008). As is the case during chromatin assembly after DNA replication, acetylation of H3 on K56 on the free histones plays a critical role in targeting the histones to the sites of double-strand break repair in order to achieve chromatin assembly (Chen et al., 2008). In parallel to the situation during replication where H3K56ac targets the histones to CAF-1 for chromatin assembly (Li et al., 2008), CAF-1 also contributes to H3K56ac-mediated chromatin assembly after double-strand break repair (C.-C. Chen and J.K. Tyler, unpublished data).
The function of CAF-1 during double-strand break repair is likely to be subject to tight regulation. For example, CAF-1 interacts with the RecQ family DNA helicase BLM that is mutated in the genetic instability disorder Bloom’s syndrome (Jiao et al., 2004). BLM is the human counterpart of the Sgs1 helicase and facilitates the separation of the two strands of the DNA helix to allow extensive resection during double-strand break repair (Gravel et al., 2008). BLM inhibits the ability of CAF-1 to assemble chromatin after DNA repair in vitro and BLM is required for a normal CAF-1 response to hydroxyurea treatment in vivo. These results raise the possibility that BLM impedes CAF-1-mediated chromatin assembly at sites of stalled replication, presumably to allow more efficient lesion processing prior to chromatin reassembly. Additionally, the interaction between CAF-1 and the Werner syndrome protein (WRN), another RecQ family DNA helicase involved in DNA synthesis, is necessary for the localization of CAF-1 to sites of DNA damage (Jiao et al., 2007). So far, no evidence has been found for the involvement of an H2A/H2B histone chaperone in chromatin assembly following DNA repair, although possible candidates for this function are Nap1 and FACT.
Given that the extent of chromatin disassembly around a double-strand break is still unclear, it remains possible that the reassembly of chromatin and incorporation of new histones that is observed during DNA repair occurs during specific events. For example, during the later stages of the single-strand annealing repair, there is extensive filling in of the gaps left over from the resection reactions. It is possible that this step is uniquely accompanied by histone loss and histone reassembly in a DNA synthesis-coupled manner during repair.
In contrast to the situation with DNA replication, DNA repair is usually accompanied by activation of the DNA damage checkpoint. This checkpoint confers cell cycle arrest which provides time for DNA repair. After DNA repair is complete, it is essential to turn off the DNA damage checkpoint in order for the cells to reenter the cell cycle, a process called “checkpoint recovery”. Somewhat surprisingly, the repair of DNA damage itself is not sufficient to signal for checkpoint recovery: the DNA damage checkpoint remains active when chromatin reassembly after DNA repair is blocked by inactivation of the Asf1 and CAF-1 histone chaperones (Chen et al., 2008; Kim and Haber, 2009). Consequently, it appears that chromatin assembly after DNA repair is the elusive signal that DNA repair is complete. Future studies should reveal the mechanism whereby chromatin reassembly signals to turn off the DNA damage checkpoint.
H2A histone variants during DNA repair
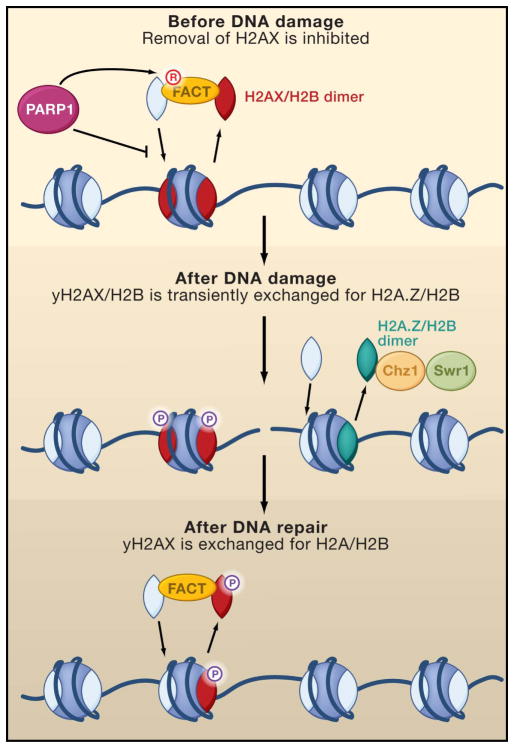
Histone variants play unique roles during DNA repair. The best known example of this is the phosphorylation of the histone H2AX variant. Phosphorylation occurs in a large domain flanking the DNA lesion, which although not required for DNA repair, does facilitate the signaling of DNA damage. Accordingly, one aspect of turning off the DNA damage checkpoint after DNA repair involves the removal of phosphorylated H2A.X (termed γH2AX) from the chromatin, followed by its replacement with canonical H2A. This exchange of γH2AX for H2A is mediated by the human FACT histone chaperone and is stimulated by phosphorylation of H2AX, which itself disrupts the nucleosome structure (Heo et al., 2008) (Figure 4). In mammals, poly (ADP)-ribosylation of proteins by PARP1 (poly ADP ribose polymerase) is a rapid response to DNA lesions and is important for preventing genomic instability. In vitro, the ability of FACT to remove unphosphorylated H2AX from chromatin is inhibited by human PARP1-mediated poly-ADP ribosylation of FACT (Heo et al., 2008). Therefore, an important role of PARP1 may be to maintain unphosphorylated H2AX on the chromatin in order to poise it for a rapid response to DNA breaks (Figure 4). In Drosophila, the removal of the fly version of γH2AX (H2Av) from chromatin is stimulated by Tip60-mediated acetylation of phosphorylated H2Av, which promotes the subsequent removal of phosphorylated H2Av from the chromatin by the ATP-dependent chromatin remodeler Domino/p400 (Kusch et al., 2004).
Figure 4. H2A histone variants play roles unique to DNA repair.
The loss of human histone H2A variant H2AX from chromatin is prevented by ADP-ribosylation of the FACT histone chaperone by PARP1. Rapidly after DNA damage, yeast Chz1 and SWR1 incorporate dimers of H2A.Z/H2B to promote DNA resection of the DNA ends. Phosphorylated H2AX is removed from DNA by the human FACT histone chaperone, and H2A is inserted in its place.
Another histone variant, H2A.Z, appears to promote the DNA repair process itself. Exchange of H2A.Z/H2B dimers for H2A/H2B dimers is mediated by the Chz1 histone chaperone in concert with the Swr1 ATP-dependent chromatin remodeler (Luk et al., 2007; Mizuguchi et al., 2003). Current data are consistent with the exchange of H2A.Z into the regions flanking a double strand break being important for DNA repair (Fig. 4). Swr1 and H2A.Z are recruited to double strand breaks very early in the repair process, albeit transiently (Kalocsay et al., 2009; van Attikum et al., 2007). Consistent with a role for incorporation of H2A.Z around a DNA break site during DNA repair, DNA resection is markedly delayed in yeast lacking H2A.Z (Kalocsay et al., 2009). Consequently, recruitment of RPA, which binds to single stranded DNA, is also delayed in yeast lacking H2A.Z, as is activation of the DNA damage checkpoint (Kalocsay et al., 2009). Swr1 mutants also show delayed recruitment of the Ku80 NHEJ component, which presumably leads to the inefficient error prone NHEJ that occurs in Swr1 mutants (van Attikum et al., 2007). In agreement with a role for H2A.Z incorporation at sites of double-strand break repair, inactivation of SWR1, Chz1, or H2A.Z leads to DNA damage sensitivity. Although more studies are required, one possible explanation for these results is that H2A.Z incorporation flanking the site of DNA damage may promote chromatin disassembly from around double strand breaks to enable efficient DNA resection.
Perspectives
In eukaryotic cells the assembly and disassembly of chromatin that accompanies DNA synthesis is a complex process involving many steps and proteins of various functions. Here we have emphasized the roles that histone chaperones play in regulating these processes during DNA replication and repair. Although many of the histone chaperones involved have been discovered over the last 20 years of study, there are still many important questions left to address regarding the mechanisms of chromatin assembly and disassembly. For example, is chromatin actively disassembled or is it a passive consequence of physical displacement of histones by the DNA replication and repair machineries? Are histone chaperones recruited to disassemble chromatin or do they simply act as receptors to store the histones ready for reassembly? Is the parental H3/H4 tetramer disassembled into H3/H4 dimers, and if so, how does this occur? How are the parental histones transferred to the daughter DNA strands? Are there mechanisms at play to ensure the epigenetic inheritance of the parental pattern of histone post-translational modifications and histone variants in the absence of the original stimulus or “epigenators” for the mark or histone variants? Do post-translational histone modifications regulate the formation of histone-histone contacts and histone-DNA contacts during chromatin assembly and disassembly? How does post-translational modification of the histone chaperones regulate their function? Is FACT the H2A/H2B chaperone during replication and repair or is another chaperone responsible? Hopefully, the development of new assays and new technologies will reveal the answers to these currently technically challenging questions within the next 20 years.
Acknowledgments
This work was supported by NIH funding to JKT from GM and NCI. We are extremely grateful for the kind assistance of Siddhartha Roy with the generation of figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Dinant C, Houtsmuller AB, Vermeulen W. Chromatin structure and DNA damage repair. Epigenetics Chromatin. 2008;1:9. doi: 10.1186/1756-8935-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrana K, van Attikum H, Hediger F, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci. 2007;120:4209–4220. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escargueil AE, Soares DG, Salvador M, Larsen AK, Henriques JA. What histone code for DNA repair? Mutat Res. 2008;658:259–270. doi: 10.1016/j.mrrev.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Finn RM, Browne K, Hodgson KC, Ausio J. sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys J. 2008;95:1314–1325. doi: 10.1529/biophysj.108.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AA, Lam WM, Burgers PM, Kaufman PD. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- Galvani A, Courbeyrette R, Agez M, Ochsenbein F, Mann C, Thuret JY. In vivo study of the nucleosome assembly functions of ASF1 histone chaperones in human cells. Mol Cell Biol. 2008;28:3672–3685. doi: 10.1128/MCB.00510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gasser R, Koller T, Sogo JM. The stability of nucleosomes at the replication fork. J Mol Biol. 1996;258:224–239. doi: 10.1006/jmbi.1996.0245. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. Embo J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 Regulates the Flow of S Phase Histones during Replicational Stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Haushalter KA, Kadonaga JT. Chromatin assembly by DNA-translocating motors. Nat Rev Mol Cell Biol. 2003;4:613–620. doi: 10.1038/nrm1177. [DOI] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Hertel L, De Andrea M, Bellomo G, Santoro P, Landolfo S, Gariglio M. The HMG protein T160 colocalizes with DNA replication foci and is down-regulated during cell differentiation. Exp Cell Res. 1999;250:313–328. doi: 10.1006/excr.1999.4495. [DOI] [PubMed] [Google Scholar]
- Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc Natl Acad Sci U S A. 2005;102:13410–13415. doi: 10.1073/pnas.0506176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP- facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 1988;27:2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao R, Bachrati CZ, Pedrazzi G, Kuster P, Petkovic M, Li JL, Egli D, Hickson ID, Stagljar I. Physical and functional interaction between the Bloom’s syndrome gene product and the largest subunit of chromatin assembly factor 1. Mol Cell Biol. 2004;24:4710–4719. doi: 10.1128/MCB.24.11.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao R, Harrigan JA, Shevelev I, Dietschy T, Selak N, Indig FE, Piotrowski J, Janscak P, Bohr VA, Stagljar I. The Werner syndrome protein is required for recruitment of chromatin assembly factor 1 following DNA damage. Oncogene. 2007;26:3811–3822. doi: 10.1038/sj.onc.1210150. [DOI] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci U S A. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Mills AD, Morris NR. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X, Harkness TA, Schultz MC, Fisher-Adams G, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- Linger J, Tyler JK. The Yeast Histone Chaperone Chromatin Assembly Factor 1 Protects Against Double-Strand DNA-Damaging Agents. Genetics. 2005;171:1513–1522. doi: 10.1534/genetics.105.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, et al. Chz1, a nuclear chaperone for histone H2AZ. Mol Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-Driven Exchange of Histone H2AZ Variant Catalyzed by SWR1 Chromatin Remodeling Complex. Science. 2003 doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatiyan A, Krude T. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol Cell Biol. 2004;24:2853–2862. doi: 10.1128/MCB.24.7.2853-2862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatiyan A, Szuts D, Krude T. Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells. Mol Cell Biol. 2006;26:1839–1849. doi: 10.1128/MCB.26.5.1839-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol. 1999;9:341–350. doi: 10.1016/s0960-9822(99)80160-2. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Prior CP, Cantor CR, Johnson EM, Allfrey VG. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980;20:597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol Cell Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Sillje HW, Nigg EA, Almouzni G. The histone chaperone Asf1 is dispensable for direct de novo histone deposition in Xenopus egg extracts. Chromosoma. 2007;116:487–496. doi: 10.1007/s00412-007-0112-x. [DOI] [PubMed] [Google Scholar]
- Raynard S, Niu H, Sung P. DNA double-strand break processing: the beginning of the end. Genes Dev. 2008;22:2903–2907. doi: 10.1101/gad.1742408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RT, Alekseev OM, Grossman G, Widgren EE, Thresher R, Wagner EJ, Sullivan KD, Marzluff WF, O’Rand MG. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem. 2006;281:21526–21534. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- Sanematsu F, Takami Y, Barman HK, Fukagawa T, Ono T, Shibahara KI, Nakayama T. Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J Biol Chem. 2006 doi: 10.1074/jbc.M511590200. [DOI] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics. 2000;155:1593–1606. doi: 10.1093/genetics/155.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Schulz LL, Tyler JK. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. Faseb J. 2006 doi: 10.1096/fj.05-5020fje. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon MJ. DNA repair and the role of chromatin structure. Curr Opin Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- Smerdon MJ, Lan SY, Calza RE, Reeves R. Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J Biol Chem. 1982;257:13441–13447. [PubMed] [Google Scholar]
- Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- reapir synthesis. Proc Natl Acad Sci U S A. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Hothorn M, Lejeune E, Rybin V, Bortfeld M, Scheffzek K, Ladurner AG. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc Natl Acad Sci U S A. 2008;105:8884–8889. doi: 10.1073/pnas.0712293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Chien CT, Hirose S, Lee SC. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006;25:3975–3985. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 Chromatin Assembly Factors. Mol Biol Cell. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Xin H, McCullough L, Rawlins R, Bentley S, Heroux A, Stillman DJ, Hill CP, Formosa T. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J Biol Chem. 2008;283:5058–5068. doi: 10.1074/jbc.M708682200. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat Struct Mol Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]