Genome-Wide Expression Profiling Identifies Type 1 Interferon Response Pathways in Active Tuberculosis (original) (raw)
Abstract
Tuberculosis (TB), caused by Mycobacterium tuberculosis (M.tb), remains the leading cause of mortality from a single infectious agent. Each year around 9 million individuals newly develop active TB disease, and over 2 billion individuals are latently infected with M.tb worldwide, thus being at risk of developing TB reactivation disease later in life. The underlying mechanisms and pathways of protection against TB in humans, as well as the dynamics of the host response to M.tb infection, are incompletely understood. We carried out whole-genome expression profiling on a cohort of TB patients longitudinally sampled along 3 time-points: during active infection, during treatment, and after completion of curative treatment. We identified molecular signatures involving the upregulation of type-1 interferon (α/β) mediated signaling and chronic inflammation during active TB disease in an Indonesian population, in line with results from two recent studies in ethnically and epidemiologically different populations in Europe and South Africa. Expression profiles were captured in neutrophil-depleted blood samples, indicating a major contribution of lymphocytes and myeloid cells. Expression of type-1 interferon (α/β) genes mediated was also upregulated in the lungs of M.tb infected mice and in infected human macrophages. In patients, the regulated gene expression-signature normalized during treatment, including the type-1 interferon mediated signaling and a concurrent opposite regulation of interferon-gamma. Further analysis revealed IL15RA, UBE2L6 and GBP4 as molecules involved in the type-I interferon response in all three experimental models. Our data is highly suggestive that the innate immune type-I interferon signaling cascade could be used as a quantitative tool for monitoring active TB disease, and provide evidence that components of the patient’s blood gene expression signature bear similarities to the pulmonary and macrophage response to mycobacterial infection.
Introduction
Approximately one third of the world’s population is estimated to be latently infected with Mycobacterium tuberculosis (M.tb), the etiological agent of tuberculosis (TB), which is responsible for about 8–10 million active new cases of TB each year [1], [2]. A bacteriologically confirmed diagnosis of TB is challenging, as direct microscopy has low sensitivity, bacterial culture takes 2 to 4 weeks, and sputum samples can be microbiologically false negative. Effective treatment of active TB is crucial in containing spread of the disease, and better diagnostics would greatly impact on TB control by facilitating early treatment. Little is known about the precise mechanisms and pathways that control protective versus pathogenic immunity during TB [3], [4]. The IL-12/IFN-γ and the TNF/TNFR axis are both crucial in protection during M.tb infection, as genetic (e.g. defects in the IL-12/IL-23/IFN-γ/STAT1 axis) or acquired (HIV; immune-suppressants; anti-cytokine auto-antibodies) defects strongly increase host-susceptibility to mycobacterial disease [5]–[7]. IFN-γ is produced mostly by CD4+ T cells and assists macrophages in controlling intracellular M.tb infection through various anti-microbial immunity pathways, which include phagosomal maturation; anti-microbial peptides; oxidative stress; apoptosis and autophagy [8]. Although IFN-γ is essential to protection against M.tb, it is not in itself a correlate of protection, however.
Several studies have profiled gene expression patterns in peripheral blood from TB patients [9]–[15]. In a most comprehensive recent study, Berry et al. [15] reported a previously unappreciated role of IFN-αβ signaling in TB pathogenesis. Blood transcriptional profiles of active TB patients were dominated by a neutrophil driven IFN-αβ inducible gene expression profile [15]. A more recent study compared gene expression patterns in the blood of tuberculosis and sarcoidosis patients (of European descent), a chronic inflammatory lung disease with granulomatous inflammatory pathology similar to TB, and observed a very similar pattern in gene-expression, miRNA expression and serum cytokine levels between the two diseases [16]. These data strongly suggest the presence of a major core pro-inflammatory pathway in TB and sarcoidosis, although also disease specific patterns could be distinguished marking TB and sarcoidosis. Thus there has been substantial success in identifying biomarkers associated with active TB.
Notwithstanding the value of these two genome wide gene expression studies reported recently, a major outstanding question is whether these signatures, captured mostly in EU and South African populations, also translate to populations and regions with a very different host genetic and TB epidemiological background. Our study addresses this issue directly by assessing unbiased whole genome expression profiles in Indonesia, an Asian country with the third highest TB prevalence worldwide. Specifically, we have examined gene expression patterns in PBMC from a longitudinal patient cohort during active TB, TB treatment and TB convalescence. Moreover, we replicated the findings in two independent model systems, a mouse _M.tb_-infection model and a mycobacterium-infected human macrophage cell line model. Although mouse models typically do not develop caseous necrotic lesions, the hallmark of human pulmonary TB [17], both mice and humans develop TB granulomas in the lung with activation of similar pro-inflammatory and anti-inflammatory responses [18], and develop highly parallel innate and adaptive immune responses during M.tb infection [8], [19], [20]. Our results validate and significantly extend the results available in the current literature, extend these to different ethnic and TB epidemiological settings, and further refine the pro-inflammatory biomarker signature associated with active TB.
Results
Gene Expression Profiles of Active TB Patients Versus Healthy Controls in a Longitudinal Patient Cohort from Indonesia
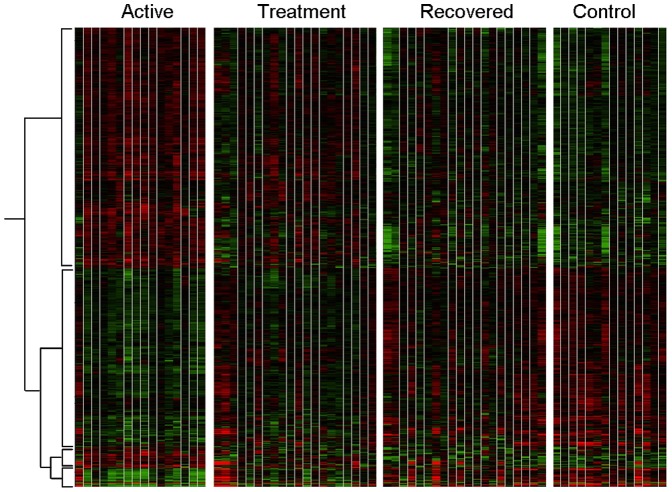
To identify molecular signatures involved in active TB, we applied microarray analysis to compare global gene expression in 23 patients with active TB (t = 0 weeks), whilst they were undergoing treatment (t = 8 weeks), and after they had completed the entire course of treatment (t = 28 weeks). The same microarray analysis was also applied to 23 matched healthy controls from the same population from the same area (see Methods). A total of 875 transcripts were significantly differentially expressed with an additional filter of a fold change greater than 2 compared to controls. The heatmap generated by these transcripts is shown ( Figure 1 ). Looking at the overall gene-expression profile patterns as a continuum from active disease (t = 0 weeks) to treatment (t = 8 weeks) and cure (t = 28 weeks), we observed that global gene expression profile patterns were more similar for patients in the active disease stage (t = 0 weeks) and at the treatment stage (t = 8 weeks), whilst the gene expression profile patterns for patients after 28 weeks of curative treatment were more similar to those of the healthy controls.
Figure 1. Differential gene expression in blood from TB patients and controls determined by microarrays.
Heat map of 875 transcripts which were statistically different in one condition from the other condition, using ANOVA and were hierarchically clustered. active = patients with active disease, treatment = patients after 8 weeks of treatment, recovered = patients after 28 weeks of treatment and controls = healthy controls. Red represents an increase in gene expression and green represents a decrease in gene expression.
Active TB Disease Involves the Expression of Genes Related to Several Branches of Immunity
Gene ontology clustering was performed on the 875 differentially expressed transcripts described above using the Panther analysis software, comparing them to the entire reference list (N ≥24,000) of genes. Of the 875 transcripts 460 fall within eight broad groupings which were most statistically significant: amongst others immunity and defense, interferon-mediated and macrophage-mediated immunity, and the chemokine and cytokine pathways ( Table 1 , Bonferroni-corrected _P_-values <0.05 for all the groups).
Table 1. The distinct pathways in which gene ontology clusters 460 of the 875 differentially expressed genes.
| Biological process | Total numberof genes | Number of genes differentially expressed between active TB patients and controls | Expected numberof genes | Bonferroni correctedp-value |
|---|---|---|---|---|
| Immunity and defense | 1393 | 126 | 53.85 | 3.57×10−17 |
| Interferon mediated immunity | 69 | 17 | 2.67 | 5.15×10−7 |
| Macrophage-mediated immunity | 150 | 24 | 5.8 | 1.57×10−6 |
| Cytokine/chemokine mediated signaling pathway | 272 | 31 | 10.51 | 3.53×10−5 |
| Natural Killer Cell mediated immunity | 77 | 14 | 2.98 | 4.21×10−4 |
| Apoptosis | 606 | 44 | 23.42 | 2.2×10−3 |
| T-cell mediated immunity | 210 | 21 | 8.12 | 1.5×10−2 |
| Signal transduction | 3791 | 183 | 146.54 | 2.1×10−2 |
Differential Gene Regulation in Humans During Active TB Disease and Upon Treatment
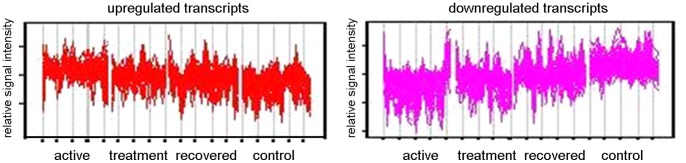
Further analysis was undertaken by hierarchically clustering the 875-transcript list using the Self Organizing Map (SOM) algorithm in Genespring GX10, taking into account the patients’ disease stage at the time of sampling (untreated active disease (t = 0), undergoing treatment (t = 8 weeks), during convalescence after curative treatment (t = 28 weeks)) and for comparison, healthy controls. In order to distinguish between different expression profiles across these four conditions, ANOVA was used and the 875 statistically significant transcripts discovered from gene ontology clustering were initially subjected to a 3 row by 4 column SOM matrix (model free). Transcripts which exhibited the same behavior were clustered together, and two groups of transcripts ( Figure 2 ) can be observed to have the clearest evidence of increased ( Figure 2, left panel) or decreased expression ( Figure 2, right panel) when comparing with active TB (t = 0 weeks), with time points during treatment (t = 8, 28 weeks), as well as comparing with controls. Both clusters were investigated for gene ontology clustering using Panther analysis. Analysis with the group of transcripts showing significantly increased expression revealed 56 genes which were significantly involved in discrete biological pathways. The type 1 interferon mediated immunity (a subgroup of immunity and defense), underlined by interferon α/β-mediated signaling, was highlighted as a very significant biological sub-process, (P = 5.21×10−8; Table 2 ). Conversely, analysis with the group of transcripts showing significantly decreased expression revealed 88 genes involved in significant biological pathways. Apart from immunity and defense (P = 5.84×10−11), other highly significant biological processes highlighted include chemokine and chemokine-mediated signaling (P = 4.53×10−5 Table 3 ). Interestingly, a concurrent down-regulation of interferon-gamma was observed.
Figure 2. Cluster analysis using self-organizing map with further Panther analysis.
Genespring GX10 was used to analyse transcripts which were significantly different between the four conditions (Active disease, Treatment, Recovered and Controls) using one-way ANOVA and subsequently subjected to self-organizing map clustering analysis. Upregulated transcripts (left panel) and downregulated transcripts (right panel) were subjected to further analysis through Panther.
Table 2. The distinct pathways in which gene ontology clusters 56 of the genes with increased expression normalizing during treatment.
| Biological process | Total numberof genes | Number of genes with significantlyincreased expression in active TB patientsversus controls, normalizing duringTB treatment | Expected numberof genes | Bonferroni correctedp-value |
|---|---|---|---|---|
| Immunity and defense | 1393 | 39 | 11.20 | 1.83×10−10 |
| Type-I interferon mediated immunity | 69 | 10 | 0.55 | 5.21×10−8 |
| Macrophage mediated immunity | 150 | 7 | 1.21 | 0.034 |
Table 3. The distinct pathways in which gene ontology clusters 88 of the genes with decreased expression normalizing during treatment.
| Biological process | Total numberof genes | Number of genes with significantlydecreased expression in active TBpatients versus controls normalizingduring TB treatment | Expected numberof genes | Bonferroni correctedp-value |
|---|---|---|---|---|
| Immunity and defense | 1393 | 35 | 8.93 | 5.84×10−11 |
| Cytokine and chemokine mediated signaling pathway | 272 | 12 | 1.74 | 4.53×10−5 |
| Ligand mediated signaling | 451 | 14 | 2.89 | 2.83×10−4 |
| Cytokine/chemokine mediated immunity | 123 | 7 | 0.79 | 2.42×10−3 |
| Apoptosis | 606 | 13 | 3.88 | 4.76×10−3 |
| Macrophage mediated immunity | 150 | 7 | 0.96 | 8.42×10−3 |
Comparison of Gene Expression Signatures from the Longitudinal TB Cohort Versus Those Obtained in a Human in vitro Macrophage Infection Model
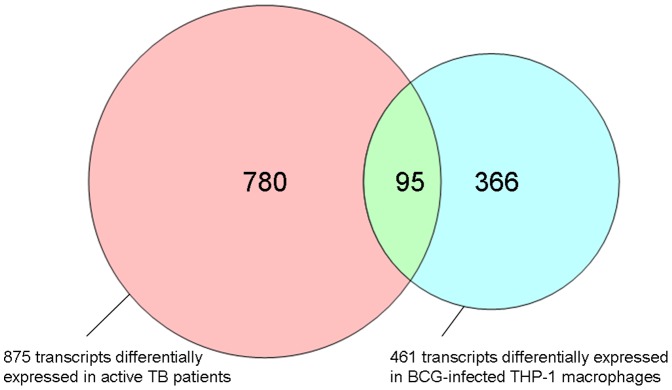
In order to examine whether the findings using patient blood mRNA responses could be confirmed in a well defined in vitro human macrophage (THP-1) cell line infection model, THP-1 cells were infected with the TB vaccine strain, M.bovis BCG (for 20 hours). A total of 461 transcripts were found by microarray analysis to be significantly differentially expressed in THP-1 macrophages following infection with BCG compared to uninfected control cells. A significant proportion, 95 out of these 461 transcripts, was found to be shared with the significantly differentially in vivo expressed transcripts in the above patient cohort ( Figure 3 ). These 95 transcripts were investigated for functional clustering using Panther analysis, of which 77 out of the 95 were found to be significantly involved within functional clusters. The most significant biological process was again found to be in interferon mediated immunity (P = 1.14×10−17; Table 4 ).
Figure 3. Comparison of differential gene expression between M.tb infected patients and BCG infected THP-1 cells.
Venn diagram representing the 875 significantly differentially expressed in TB patients and 461 transcripts significantly differentially expressed in THP-1 BCG in vitro model. A total of 95 transcripts were found to be in common between these two systems. Red – transcripts for active TB patients only; Green – Common between TB patients and BCG-infected THP-1 cell line; Blue – transcripts for BCG-infected THP-1 only.
Table 4. Differentially expressed genes in common between active TB patients versus controls and BCG-infected versus uninfected THP-1 cells.
| Biological process | Total numberof genes | Number of genes in common betweenactive TB patients vs controls andBCG-infected vs uninfected THP-1 cells | Expected numberof genes | Bonferroni correctedp-value |
|---|---|---|---|---|
| Interferon-mediated immunity | 69 | 14 | 0.29 | 1.14×10−17 |
| Immunity and defense | 1393 | 31 | 5.8 | 9.22×10−14 |
| Cytokine/chemokine mediated signaling pathway | 272 | 10 | 1.13 | 4.43×10−5 |
| Cytokine/chemokine mediated immunity | 123 | 7 | 0.51 | 1.37×10−4 |
| Macrophage mediated immunity | 150 | 6 | 0.62 | 6.08×10−3 |
| Ligand-mediated signaling | 451 | 9 | 1.88 | 2.26×10−2 |
Human Host Responses to M.tb Infection Common with an in vivo Mouse Infection Model
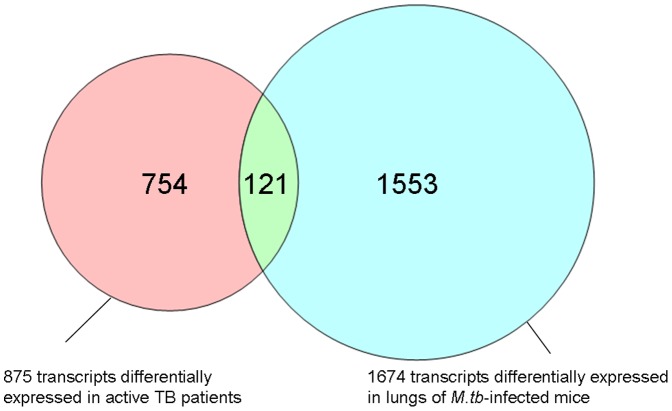
In a third independent approach, we used an in vivo mouse TB infection model to examine whether the findings in the longitudinal patient cohorts blood samples could be confirmed in the infected mouse lung, the major organ affected by TB. A total of 1,674 transcripts were found by microarray analysis to be significantly differentially expressed in C57BL/6 mice infected with M.tb compared to non-infected controls. Of these transcripts, 121 were common to the significantly differentially expressed transcripts in the human cohort ( Figure 4 ). These common transcripts were subjected to Panther gene ontology analysis, and 85 out of the 121 transcripts clustered into significant biological pathways. The most significant biological pathway was once again type1 interferon-mediated immunity ( Table 5 ).
Figure 4. Comparison of differential gene expression between patients with active TB and mice infected M.tb in vivo model.
Venn diagram representing the 875 significantly differentially expressed in TB patients and 1674 transcripts significantly differentially expressed in TB actively infected mice. A total of 121 transcripts were found to be in common between these two systems. Red – transcripts for TB patient only; Green – Common between TB patients and mice infected with TB; Blue – transcripts for mice infected with TB only.
Table 5. Differentially expressed genes in common between active TB patients versus controls and TB-infected versus uninfected mice.
| Biological process | Total numberof genes | Number of genes in common betweenactive TB patients vs controls andTB-infected vs uninfected mice | Expected numberof genes | Bonferroni correctedp-value |
|---|---|---|---|---|
| Immunity and defense | 1393 | 39 | 7.46 | 6.71×10−17 |
| Interferon-mediated immunity | 69 | 12 | 0.37 | 7.90×10−13 |
| Macrophage mediated immunity | 150 | 11 | 0.8 | 1.02×10−7 |
| Cytokine/chemokine mediated signaling pathway | 272 | 11 | 1.46 | 5.63×10−5 |
| Cytokine/chemokine mediated immunity | 123 | 7 | 0.66 | 7.48×10−4 |
| Natural Killer cell mediated immunity | 77 | 5 | 0.41 | 9.51×10−3 |
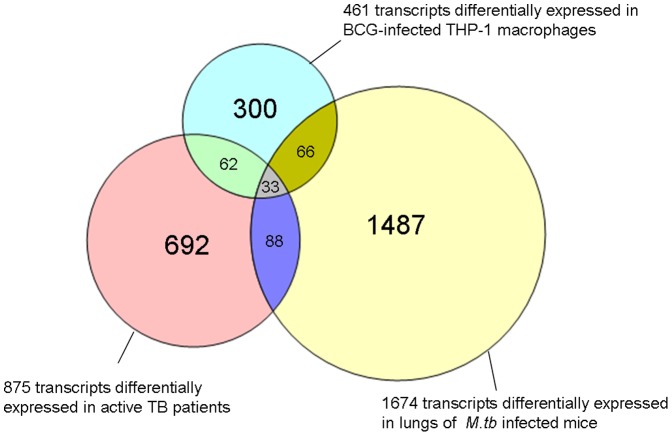
Genes Found in Common in All 3 Models of TB
A total of 26 genes (or 33 entities due to the presence of multiple transcripts for several genes) were found to be significantly differentially expressed in all 3 models (longitudinal human TB cohort, human macrophage cell line, and in vivo mouse model; Table 6 , Figure 5 ). These 26 genes were then subjected to Ingenuity Pathway Analysis (IPA). The most significant network (P<10−32) includes 18 focus molecules showing a high level of interaction between these molecules which involves both NFκB activation and the type I interferon response.
Table 6. Validation by qRT-PCR of 26 genes differentially expressed in all three models based on microarray analyses.
| gene | Validated in longitudinal cohort TB patients | Validated in BCG-infected human macrophages | Validated in _M.tb_-infectedlungs of mice | Direction of expression changea |
|---|---|---|---|---|
| BATF2 | yes | yes | no | up |
| C1QC | yes | no | yes | up |
| CCL2 | no | no | no | down |
| CCL7 | no | no | no | down |
| CD40 | yes | yes | no | up |
| CX3CR1 | yes | no | yes | up |
| CXCL10 | no | no | no | up |
| DDX58 | yes | yes | no | up |
| GBP4 | yes | yes | yes | up |
| IFIT2 | no | no | no | up |
| IFIT3 | no | no | no | up |
| IL15RA | yes | yes | yes | up |
| IL1A | no | no | no | Down |
| IMPA2 | yes | yes | no | Up |
| MEFV | yes | yes | no | Up |
| OAS1 | yes | yes | no | Up |
| OAS2 | yes | yes | no | Up |
| PBEF1 | no | no | no | Down |
| PIM1 | yes | yes | no | Up |
| SLAMF7 | no | no | no | Up |
| TLR5 | yes | yes | no | Up |
| TNFAIP2 | yes | yes | no | Up |
| TRAFD1 | yes | yes | no | Up |
| TRIM5 | yes | yes | no | Up |
| UBE2L6 | yes | yes | yes | Up |
| WARS | no | no | no | Up |
| 17 genes | 15 genes | 5 genes |
Figure 5. Comparison of differential gene expression from all three models.
Venn diagram representing the 875 significantly differentially expressed in TB patients, 461 significantly differentially expressed in BCG-infected THP-1 cells and 1674 transcripts significantly differentially expressed in TB actively infected mice. A total of 33 transcripts (representing 26 genes) were found to be in common between all three systems. Red – transcripts for patients with active TB; Blue – transcripts for BCG-infected THP-1 cells; yYellow – transcripts for mice infected with active TB.
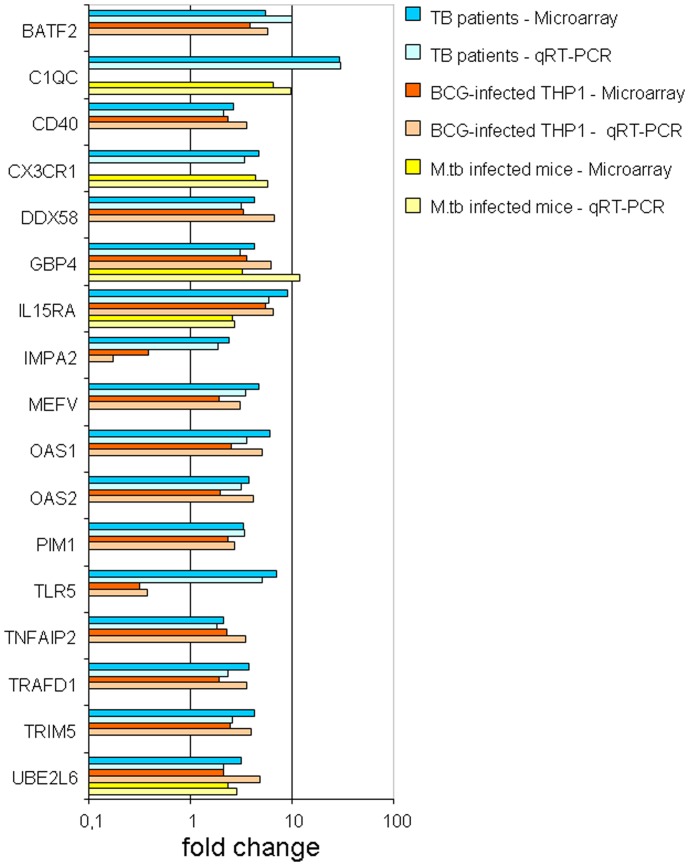
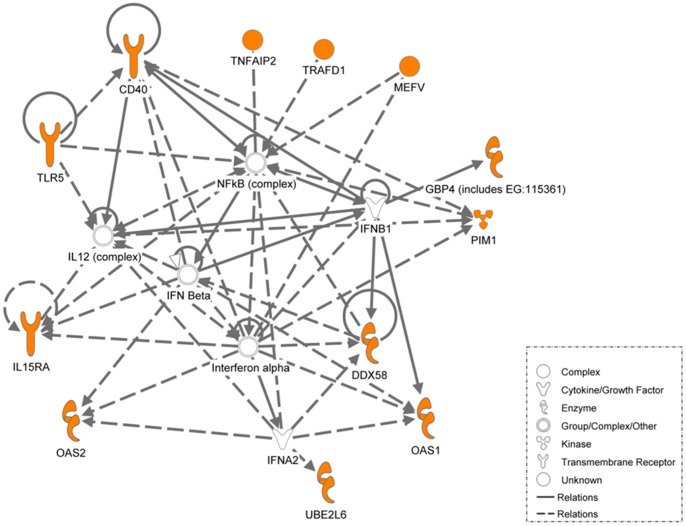
We then proceeded to validate the microarray findings for these 26 genes using quantitative RT-PCR (qRT-PCR); for successful validation, the direction for the fold-change in gene expression using qRT-PCR should be consistent with the micro-array findings. A total of 17 out of the 26 genes were successfully validated via qRT-PCR in the longitudinal patient cohort and 15 out of the 17 genes were validated in the human macrophage model (BCG-infected THP-1 cells) ( Table 6 ). These 15 genes were common to both the human models (longitudinal patient cohort and BCG-infected THP-1 cells). However, only 5 out of the 17 genes could be validated in the in vivo mouse model ( Table 6 ). Considering all validation data, 3 genes (IL15RA, UBE2L6 and GBP4) out of the 26 genes common to all 3 models were found to be consistently and reproducibly upregulated across 3 model systems ( Figure 6 ). It is clear from the network analysis ( Figure 7 ) that IL15RA, UBE2L6 and GBP4 are involved in the type-I interferon signaling response.
Figure 6. Validating microarray data by qRT-PCR.
Microarray results were validated by qRT-PCR. 17 genes from the 26 genes were found to be significantly different between TB patients with active disease and healthy controls, 15 genes from the 26 genes were found to be significantly different between live BCG-infected THP-1 cells at 20 hours compared to uninfected controls, and 5 genes from 26 genes were found to be significantly different between mice infected with active TB and mock infected mice, using student’s T test (P<0.05). Three genes which were common to all 3 models show similar levels of expression with both techniques applied. The fold increase for microarray technique (blue, orange and yellow) and qRT-PCR technique (light blue, light orange and light yellow) for patients, THP-1 cells and mice respectively. Microarray data (and their corresponding qRT-PCR data) that were not validated by qRT-PCR are not shown.
Figure 7. Ingenuity pathway analysis of the key genes identified.
Ingenuity pathway analysis of the genes differentially regulated and common to the three systems described above (in orange). Network of genes in the interferon 1 signaling pathways that were found to be common in active TB patients, BCG-infected THP-1 cells at 20 hours and mice infected with active TB are illustrated. The lines in between genes represent known interactions, with solid lines representing direct interactions and dashed lines representing indirect interactions.
Discussion
Comparing gene expression profiles in 3 models of TB, a longitudinal human TB cohort study comprising of newly-diagnosed TB patients followed up through treatment, a BCG-infected human macrophage cell line and an in vivo mouse lung TB infection model, we identified in all three models a highly prominent type-I interferon molecular signature. This result significantly extends those of Berry et al [15], and Maertzdorf et al [16]. who found highly similar signatures in the blood of TB patients in Europe and South Africa; our results now confirm and extend this to a TB endemic setting on a third continent in a different host genetic as well as TB epidemiological setting, namely Indonesia. In addition, our results confirm and validate part of the findings in two independent models of TB infection, thus connecting the TB patient blood expression profiles to pulmonary and macrophage expression signatures.
Our un-biased whole genome microarray analysis revealed that the most prominent host responses in all three models of infection were immune related. The most significant pathways were interferon mediated immunity, macrophage mediated immunity and cytokine and chemokine mediated signaling pathways. Further analysis of the differentially expressed genes in the TB patient study revealed that the type-1 interferon signaling activity was the predominant pathway up-regulated during active disease, and that there was concurrent down-regulation of interferon-gamma. This is consistent with findings that key protective immune mechanisms are repressed by type-1 interferon signaling during active TB infection [21], [22] as a possible immune evasion response.
The network analysis of genes found to be commonly regulated in all 3 independent TB disease models showed that the most significant network was mainly involved in IL-12, IFN-α and IFN-β signaling. This is indeed noteworthy as evidence implicating the involvement of type 1 interferon (IFN-α and IFN-β) in TB have only begun to emerge very recently [15]. In the present study, we have shown an up regulation in type 1 interferon (α/β) and the genes involved. Kuchtey et al., showed that during early infection (2–3 weeks post infection) there was an enhanced growth of M. bovis BCG in the lungs of mice deficient in IFN-αβ receptors, and during late infection the burden of BCG was similar in lungs of IFN-αβ−/− and wild type mice [23]. Desvignes et al. recently reported a similar non redundant protective role for type-1 interferons during early M.tb infection in a mouse model [24]. In a large human study using whole blood from active TB patients vs. uninfected controls, Berry et al., found an increase in type 1 IFN which was attributed predominantly to neutrophils [15]. Here, however, we present highly similar findings in TB patients’ blood leucocytes, which were depleted of neutrophils through Ficoll density centrifugation, but still include monocyte/macrophages. The fact that the latter are the most likely source of this signature was supported by the finding of a similar signature in an independent, human macrophage cell line model of mycobacterial infection. Also in another study using whole blood to compare gene expression profiles in TB patients with active disease vs. individuals with latent M.tb infection several interferon inducible genes were found to be profoundly upregulated, including IFN-α protein 6 and 27 and ISG15 [12]. Furthermore, exogenously added IFN-α was found to inhibit the priming effect of IFN-γ on key cytokine release by monocytes [22]. IFN-β was found to have a similar effect [25]. This is compatible with a model in which enhanced type I interferon signaling inhibits type II interferon mediated protective immunity in TB.
It is unsurprising that of all immune cells and corresponding signaling systems (T-cells, B-cells, monocytes/dendritic cells/macrophages, natural killer cells), the macrophage-mediated immunity process was consistently found to play a dominant role ( Tables 1 – 4 ) in the transcriptional response in each of the three different study models. This highlights the key role that M.tb/macrophage interactions play during infection. Alveolar macrophages phagocytose inhaled M.tb by a wide range of receptors such as complement receptors CR1, 3 and 4; mannose receptors; Toll-like receptors; C-type-2 lectin receptors and scavenger receptors [26]. The further influx of macrophages following the formation of a primary lesion further highlights the key role of these professional phagocytes in pulmonary host defence in TB [12]. M.tb however inhibits macrophage activity by inhibiting autophagy, an important cellular mechanism to control intracellular M.tb [27], [28]. In addition, M.tb blocks the maturation of phagosomes such that they fail to traffic further along the endosomal-lysosomal pathway and fail to fuse with lysosomes. One newly identified receptor which may be important in this process is human TLR8. A polymorphism resulting in a missense mutation in TLR8 (Met1Val) was recently shown to be strongly associated with human susceptibility to TB [29]. Increased credence to the possible involvement of TLR8 in upstream M.tb recognition is lent by findings from the expression array in the longitudinal patient cohort, which highlighted the significance of the immunity and defense pathway in active disease, including TLRs. It may be of interest to note that TLR8 is absent from mice, which could account for part of the differences in TB infection biology between humans and mice.
Further validation of our findings using qRT-PCR revealed that IL15RA, UBE2L6 and GBP4 were involved in this molecular signature in all three models ( Figure 6 ). IL15RA encodes for interleukin-15 (IL-15) receptor alpha (IL-15Rα), a common cytokine receptor expressed by macrophages that plays an important role in the development, survival, and proliferation of various immune cells (e.g. natural killer and CD8+ T cells). IL-15, which is co expressed with IL-15Rα by antigen presenting cells allowing trans-production of cytokines to immune effector cells [30] is generally considered to be a regulator of T cell homeostasis as it cooperates with other cytokines like IL-2 and IL-7 to maintain pools of naive and memory T cell populations. Mortier et al., has described that IL-15Rα expression on macrophages supports the early transition of antigen specific effector CD8+ T cells to memory cells [31]. As these processes are critical in the early immune response against microbial infections, and as data on the potential protective effects of IL-15 on murine experimental tuberculosis is starting to emerge, further studies are necessary in order to document the precise mechanism of action of IL-15-mediated type-I interferon signaling on M.tb survival and proliferation.
The second gene with consistent validation across the three TB models studied here, UBE2L6, is a member of the ubiquitin family of proteins. The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation, and recent work linking the ubiquitin family of proteins to type-I interferon mediated host immune responses is now beginning to emerge [32]–[35].
The third gene, GBP4, encodes Guanylate binding protein 4. With IFN-α being a main hypothesis in our study, it is reassuring that GBP4 was commonly found in all 3 models, and indeed GBP4 displays a direct interaction with IFN-α ( Figure 7 ). Type 1 and 2 IFNs have been shown to differentially regulate the GBP family of genes, and GBP4 is exclusively regulated by IFN-α production through macrophages [36].
In our study a substantial number of overlapping genes were identified when comparing BCG infected human macrophage cells and circulating leucocytes from TB patients with active disease ( Figure 3 ). The majority of these genes was present in the immune-mediated and host defense pathways. It needs to be highlighted that these genes were also common when the mouse TB infection model was compared to the data from the human models ( Figure 4 ). The genes overlapping in all three systems were further validated with qRT-PCR. The 15 genes that were successfully validated in the THP-1 cell line model by qRT-PCR completely overlapped with the 17 genes validated by qRT-PCR in the longitudinal patient cohort. ( Figure 6 ). This is not surprising since both models share a common human host background. The fact that only 3 genes were common to all three models may reflect important differences between the human and mouse models of TB. The PCR validation of these genes may represent an underestimate of the of the true array findings as the detection probes did not directly overlap with the array probes and so may not be able to confirm the specific RNA transcript identified by the array, particularly in the mouse to human comparisons. Although the use of BCG rather than M.tb in the THP-1 infection model may also have contributed to this relatively low number, it should be emphasized that the analysis focused only on those genes found in all three models, and that the variation between the M.tb infected human patients and the M.tb infected mice was significantly larger that that between the M.tb infected human patients and the BCG infected THP-1 cells. Our approach thus excludes the identification of genes based on only one individual model, and identifies genes only when they have been validated in three models, which is a significant advance compared to previous studies. In our study the mice were infected with M.tb intravenously, and it will be of interest to see whether infection via the aerosolic route, which is reflective of natural TB transmission, would have yielded similar response patterns. A cardinal feature of human TB is of central caseous necrosis in pulmonary granulomatous lesions, while such necrotic lesions are not seen in the mouse TB infected lung. Caseous necrosis is considered an essential step towards developing cavernous TB, in which phase M.tb bacilli are released into the open airways prior to transmission. Thus, there are clear differences in the pathological manifestations of TB in human and mouse systems and this difference in disease pathology has been central to the argument that the mouse is an imperfect model for TB. However, notwithstanding these differences, we were able to observe consistent molecular signatures of active TB across the distinct models, thus supporting the general conclusion that mouse models are appropriate for studying certain aspects of the immune molecular host response to infection.
The use of TB patients cohorts, in vitro human macrophage cell lines and inbred mouse models of TB have provided new information which is in line with, but also significantly extends recent work from other groups [15], [16]. Collectively, these studies demonstrate the major role of dysregulated inflammation in general, and of strongly enhanced type 1 IFN-αβ signaling in active TB disease. Importantly whereas this latter finding was originally attributed to the activity of neutrophils, we here find these same gene expression patterns also in cells devoid of neutrophils, such as PBMCs and macrophages. Finally, our study discovered genes commonly expressed in all three TB models we have studied, for example IL15RA, UBE2L6 and GBP4 which are all common to the IFN-α network. Future studies should extend this also to protein expression profiles to determine whether these differences are also present at the protein level. This will allow further insights into their possible function during TB disease and M.tb infection, and help to better delineate new TB biomarker signatures.
Materials and Methods
Ethical Statement
This study was approved by the Ethical Committee of the Faculty of Medicine, University of Indonesia, Jakarta, and by the Eijkman Institute Research Ethics Committee, Jakarta, and written consent was voluntarily signed by all patients and control subjects [37].
Patient Recruitment
After informed consent, 23 new pulmonary tuberculosis patients above 15 years of age were recruited from an outpatient tuberculosis clinic in central Jakarta (Indonesia) [37]. Diagnosis of TB was performed according to the World Health Organization criteria, on the basis of the clinical presentation and a chest X-ray radiograph (CXR) and confirmed by sputum microscopy positive for mycobacteria using Ziehl-Nielsen stain. Free anti-TB drug treatment was provided to all patients and consisted of a standard regimen of isoniazid, rifampin, pyrazinamide and ethambutol (2HRZE, 4H3R3) for a total of 6 months according to the national TB program. Treatment was supervised once weekly by a directly observed treatment program.
Human immunodeficiency virus (HIV)-seropositive patients, diabetes mellitus affected patients, patients with heart diseases and patients with incomplete data records were excluded. 23 randomly selected control subjects with the same sex and age (+/−10%) were recruited from neighboring households, with first degree relatives excluded. Controls with signs, symptoms and CXR results suggestive of active TB, a history of prior anti-TB treatment, diabetes mellitus or incomplete data entry were excluded. HIV status was not tested in control group. Indonesia is classified as a country with a low HIV prevalence of ≤0.1% during the sample period [38]. Patients’ self reported ethnicities were recorded upon recruitment. A Javanese origin characterized three groups – the Jawa, Betawi and Sunda – and altogether comprised more than 80% of the total sample. The non-Javanese category included individuals born on other Indonesian islands.
RNA Extraction
Blood samples at three time points of disease spectrum (active phase: 0 weeks, treatment phase: 8 weeks and convalescent phase: 28 weeks) were obtained and collected in heparinized tubes. Peripheral blood mononuclear cells (PBMCs) were separated using a Ficoll density gradient, and kept frozen until use. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions, and subsequently subjected to DNAse treatment using RNAse free DNAse Set (Qiagen, Germany). RNA integrity was assessed by 1% agarose gel electrophoresis and samples of poor quality were excluded from analysis.
Mycobacterium bovis Bacillus Calmette Guérin (BCG) Infection of Differentiated THP-1 Macrophages
The human monocytic cell line THP-1 (ATCC, Rockville, MD) cells were cultured in RPMI 1640 supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 µg/ml) (Invitrogen). Cells were plated at a density of 106 in 6-well tissue culture plates (Nunclon). Monocytes were allowed to adhere and differentiate into macrophages for 48 hours with 5 nM PMA (Sigma Aldrich) at 37°C in a humidified atmosphere of 5% CO2. Differentiated macrophages were infected with BCG at an MOI of 5∶1 and incubated at 37°C, 5% CO2. Infected cells at 4 hr post infection were washed twice with RPMI without antibiotics to remove un-ingested and un-adhered bacteria to minimize cell death and re-incubated for a further 16 hours. The cells were washed once in 1× distilled PBS before 350 µL RTL buffer was added to lyse the cells and RNA extraction was then carried out using the RNeasy Mini Kit (Qiagen, Germany), according to the manufacturer’s instructions.
C57BL/6 Mice Infected with Acute TB
C57BL/6Jbrc female mice (18–19 weeks old) were purchased from the Medical Research Centre (Biopolis, Singapore) from the specific pathogen free (SPF) facility. Mice were kept at the Novartis Institute for Tropical Diseases (NITD, Singapore) ABSL-3 facility for a week before the experiment. The experiment was approved by the Institute Animal Care and Use Committee of NITD. A total of eleven mice were used. Five mice were infected intravenously with 0.2 ml (approximately 106 CFU) of M.tb Erdman strain. Six mice served as mock infected controls.
Briefly, M.tb Erdman strain was grown in Dubos broth to the logarithmic stage. Bacteria were diluted in saline containing 0.05% Tween 80 for infection in mice. Mock infected mice were intravenously administered PBS containing 0.05% Tween 80. After four weeks, infected and control mice were sacrificed. Half of the lungs was immersed in saline containing 0.05% Tween 80 for homogenizing to determine the CFU (data not shown) and the other half was immersed in the RNA later® stabilizer solution (Ambion®, USA) prior to preparing tissue lysate for extracting total RNA.
RNA Extraction from Mice Lungs
For preparation of tissue lysate, lung tissue immersed in the RNA later® stabilizer solution (Ambion®, USA) was transferred to the lysing Matrix D tube of FastPrep system Pro Green Kit (Plants and Animals) (Qbiogen, Germany). Tissue lysate was then prepared according to the manufacturer’s instructions.
Total RNA was extracted from tissue lysate using the RNA Easy kit (Qiagen, Germany). Extracted RNA was subjected to DNAse treatment using RNAse free DNAse Set (Qiagen, Germany). Integrity of the RNA was assessed by 1% agarose gel electrophoresis and samples of poor quality were excluded from analysis.
Microarrays
Total RNA (250 ng) was amplified in a single round of in vitro transcription amplification that allowed incorporation of biotin-labelled nucleotides using the Illumina TotalPrep RNA amplification kit (Ambion, Austin, TX), according to the manufacturer’s instructions. cRNA (750 ng) of each sample was hybridized to an Illumina HumanRef-8 V3 BeadChip (containing probes to 24526 RefSeq gene sequences) at 58°C for 16 hours according to the manufacturer’s instructions (Illumina, Inc., San Diego, CA). This was followed by washing, blocking and streptavidin-Cy3 staining steps, followed by scanning with a high resolution Illumina bead array reader confocal scanner, according to the manufacturer’s instructions. For all arrays at the different time points, a rigorous quality check was carried out to ensure good performance before the array data was extracted. The data extraction was performed by using Illumina BeadStudio v3 software and uploaded into Genespring GX10 (Silicon Genetics, Redwood City, CA) software for downstream analysis.
Array Normalization
The standard normalization procedures recommended by Genespring software for one-colour array were followed. Data transformation was corrected for a low signal, with values recorded at <0.01 increased to the minimum (0.01). Chip variability was accounted for using per-chip normalization by dividing all of the measurements on each chip by a 75th percentile value. Probe set variability for different genes were accounted for using per-gene normalization by dividing all genes by the median of all genes. Analysis was restricted to probe sets for which a detection level of 0.9899 was obtained in at least 50% of arrays in at least one condition stated. Following quality control of probes, statistical analysis, hierarchical clustering and functional classification were performed.
Selection of Differentially Expressed Genes from Microarray Data
Differentially expressed genes were selected from the normalized data by using a procedure known as significance analysis of microarrays (SAM) [39] as installed on Genespring GX10. The statistic in SAM is given as d = (u1−u2)/(s−s0), where the numerator is the group mean difference, s is the standard error and s0 is a regularizing constant. Setting s0 = 0 will yield a t-statistic. This value, called the fudge constant, is found by removing the trend in d as a function of s in moving windows across the data to reduce false positive results. Since the statistic is not t-distributed, significance is computed using a permutation test. Genes with computed statistic larger than the threshold are called significant. 1000 permutations were conducted and a threshold false discovery rate (FDR) of 0.05 set to determine the differentially expressed genes which were significant, and subsequently subjected to filtering through fold change of 2 or greater.
Clustering using Self Organizing Map (SOM) function was used to group genes with different gene expression profiles, with Euclidean distance metric, a maximum iterations of 50, grid rows of 3 by 4, initial learning rate of 0.03, initial neighborhood radius of 5, neighborhood bubble type, with a hexagonal grid topology.
Gene Ontology Clustering
The PANTHER (www.pantherdb.org) online gene expression analysis system was used to group significantly overrepresented genes into functional relationships. Similarly, the Ingenuity Pathway Analysis (IPA; www.ingenuity.com) was also used to classify differentially expressed genes into functional relationships and further show canonical pathways and networks involving these genes, with nodes defining potentially critical host mediators of infection. The canonical pathways are described in the library of the Ingenuity Pathways knowledge base and are based on well-known established textbook literature where molecular links are known. The networks are built from a compilation of literature searches. Connecting lines between nodes represent one or more publications which imply a direct or indirect connection between the genes. The Ingenuity program is updated monthly to reflect new publications.
For canonical pathways, the significance of the association was measured in two ways: (i) by the ratio of the number of genes from the data set that map to the pathway divided by the total number of genes in that pathway and (ii) by using the Benjamin-Hochberg multiple corrected p-value to obtain a p-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone, correcting for multiple testing of the same genes against different canonical pathways.
Network-wise, IPA computes a score for each network related to the fit of the input set of differentially expressed genes. The score is obtained from the likelihood that the focus genes in a network are together due to chance alone, with a score of 2 being equivalent to a p-value of 0.01.
Quantitative RT-PCR (qRT-PCR) by Fluidigm System
Using the extracted RNA from the three systems above, 100 ng of total RNA was reverse transcribed using the High-Capacity cDNA Kit for 1000 reactions (ABI, USA), and processed for the 48.48 Fluidigm Microfluidic Chips, according to the manufacturer’s instructions, together with data analysis using the Biomark qRT-PCR analysis and Biomark Melting Curve analysis software (Fluidigm, USA). We normalized the qRT-PCR data in relation to a reference gene expression quantitation using the gene 18S, according to well established procedures. The RNA samples used for PCR were the same as those used for the microarray.
Microarray Statistical Analysis
For microarray analysis, the Illumina HumanRef-8 V3 BeadChip was utilized, containing more than 24,000 genes. For the initial analysis, the T-test with Benjamin-Hochberg procedure was used to determine differentially expressed genes from the normalized data in any one condition from the controls (see Materials and Methods). Analysis of variance (ANOVA) was used to determine differentially expressed genes across patient groups.
Acknowledgments
We would like to thank Dr Sabai Phyu and Dr Mark Schreiber from the Novartis Institute of Tropical disease, Singapore for facilitating the mouse infections and useful discussions on the manuscript.
Funding Statement
The authors gratefully acknowledge the support of the following organizations: the Netherlands’ Organization of Scientific Research (NWO); the Royal Netherlands’ Academy of Arts and Sciences (KNAW); WOTRO/NACCAP; the Bill and Melinda Gates Foundation Grand Challenges in Global Health (grant GC6#74); the Netherlands Leprosy Relief Foundation; the Q.M. Gastmann Wichers Foundation. The research also received funding from the European Union's Seventh Framework Programme [FP7/2007–2013] for projects NEWTBVAC; IDEA; and ADITEC (No: 280873). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Raviglione MC (2003) The TB epidemic from 1992 to 2002. Tuberculosis 83: 4–14. [DOI] [PubMed] [Google Scholar]
- 2.Schuck SD, Mueller H, Kunitz F, Neher A, Hoffmann H, et al. (2009) Identification of T-Cell Antigens Specific for Latent Mycobacterium Tuberculosis Infection. PLoS ONE 4: e5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ottenhoff TH, Kaufmann SH (2012) Vaccines against tuberculosis: where are we and where do we need to go? PLoS Pathog 8: e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottenhoff TH, Ellner JJ, Kaufmann SH (2012) Ten challenges for TB biomarkers. Tuberculosis (Edinb ) 92 Suppl 1S17–S20. [DOI] [PubMed] [Google Scholar]
- 5.van de Vosse E, van Dissel JT, Ottenhoff TH (2009) Genetic deficiencies of innate immune signaling in human infectious disease. Lancet Infect Dis 9: 688–698. [DOI] [PubMed] [Google Scholar]
- 6.de Jong R, Altare F, Haagen IA, Elferink DG, Boer T, et al. (1998) Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 280: 1435–1438. [DOI] [PubMed] [Google Scholar]
- 7.Levin M, Newport M (2000) Inherited predisposition to mycobacterial infection: historical considerations. Microbes Infect 2: 1549–1552. [DOI] [PubMed] [Google Scholar]
- 8.Ottenhoff THM (2012) New pathways of protective and pathological host defense to mycobacteria. Trends Microbiol 2012 (9 July Epub ahead of print). [DOI] [PubMed]
- 9.Mistry R, Cliff JM, Clayton CL, Beyers N, Mohamed YS, et al. (2007) Gene-Expression Patterns in Whole Blood Identify Subjects at Risk for Recurrent Tuberculosis. J Infect Dis 195: 357–365. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, et al. (2007) Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med 85: 613–621. [DOI] [PubMed] [Google Scholar]
- 11.Joosten SA, Goeman JJ, Sutherland JS, Opmeer L, de Boer KG, et al. (2011) Identification of biomarkers for tuberculosis disease using a novel dual-color RT-MLPA assay. Genes Immun 13: 71–82. [DOI] [PubMed] [Google Scholar]
- 12.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, et al. (2011) Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun 12: 15–22. [DOI] [PubMed] [Google Scholar]
- 13.Maertzdorf J, Ota M, Repsilber D, Mollenkopf HJ, Weiner J, et al. (2011) Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS ONE 6: e26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesho E, Forestiero FJ, Hirata MH, Hirata RD, Cecon L, et al. (2011) Transcriptional responses of host peripheral blood cells to tuberculosis infection. Tuberculosis (Edinb ) 91: 390–399. [DOI] [PubMed] [Google Scholar]
- 15.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, et al. (2010) An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maertzdorf J, Weiner J, Mollenkopf HJ, TBornotTB Network, Bauer T, et al. (2012) Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A 109: 7853–7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abebe M, Doherty TM, Wassie L, Aseffa A, Bobosha K, et al. (2010) Expression of apoptosis-related genes in an Ethiopian cohort study correlates with tuberculosis clinical status. Eur J Immunol 40: 291–301. [DOI] [PubMed] [Google Scholar]
- 18.Schafer G, Guler R, Murray G, Brombacher F, Brown GD (2009) The Role of Scavenger Receptor B1 in Infection with Mycobacterium tuberculosis in a Murine Model. PLoS ONE 4: e8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dharmadhikari AS, Nardell EA (2008) What Animal Models Teach Humans about Tuberculosis. Am J Respir Cell Mol Biol 39: 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ottenhoff TH, Verreck FA, Lichtenauer-Kaligis EG, Hoeve MA, Sanal O, et al. (2002) Genetics, cytokines and human infectious disease: lessons from weakly pathogenic mycobacteria and salmonellae. Nat Genet 32: 97–105. [DOI] [PubMed] [Google Scholar]
- 21.Korbel DS, Schneider BE, Schaible UE (2008) Innate immunity in tuberculosis: myths and truth. Microbes Infect 10: 995–1004. [DOI] [PubMed] [Google Scholar]
- 22.van de Wetering D, van Wengen A, Savage ND, van de Vosse E, van Dissel JT (2011) IFN-α cannot substitute lack of IFN-γ responsiveness in cells of an IFN-γR1 deficient patient. Clin Immunol 138: 282–290. [DOI] [PubMed] [Google Scholar]
- 23.Kuchtey J, Fulton SA, Reba SM, Harding CV, Boom WH (2006) Interferon-αβ mediates partial control of early pulmonary Mycobacterium bovis bacillus Calmette-Guérin infection. Immunology 118: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desvignes L, Wolf AJ, Ernst JD (2012) Dynamic roles of type I and type II IFNs in early infection with Mycobacterium tuberculosis. J Immunol 188: 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Paus RA, van Wengen A, Schmidt I, Visser M, Verdegaal EME, et al.. (2012) Inhibition of the type I immune responses of human monocytes by IFN-α and IFN-β. submitted. [DOI] [PubMed]
- 26.Sinai AP, Joiner KA (1997) Safe Haven: The Cell Biology of Nonfusogenic Pathogen Vacuoles. Annu Rev Microbiol 51: 415–462. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, et al. (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753–766. [DOI] [PubMed] [Google Scholar]
- 28.Petruccioli E, Romagnoli A, Corazzari M, Coccia EM, Butera O, et al. (2012) Specific T cells restore the autophagic flux inhibited by Mycobacterium tuberculosis in human primary macrophages. J Infect Dis 205: 1425–1435. [DOI] [PubMed] [Google Scholar]
- 29.Davila S, Hibberd ML, Hari Dass R, Wong HE, Sahiratmadja E, et al. (2008) Genetic association and expression studies indicate a role of Toll-Like Receptor 8 in pulmonary tuberculosis. PLoS Genet 4: e1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong RL, Liu B, Zhu X, You L, Kong L, et al. (2011) Interleukin-15:Interleukin-15 receptor α scaffold for creation of multivalent targeted immune molecules. Protein Eng Des Sel 24: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortier E, Advincula R, Kim L, Chmura S, Barrera J, et al. (2009) Macrophage- and Dendritic-Cell-Derived Interleukin-15 Receptor Alpha Supports Homeostasis of Distinct CD8+ T Cell Subsets. Immunity 31: 811–822. [DOI] [PubMed] [Google Scholar]
- 32.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, et al. (2010) The Ubiquitin Ligase Riplet Is Essential for RIG-I-Dependent Innate Immune Responses to RNA Virus Infection. Cell Host Microbe 8: 496–509. [DOI] [PubMed] [Google Scholar]
- 33.Arimoto Ki, Takahashi H, Hishiki T, Konishi H, Fujita T, et al. (2007) Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA 104: 7500–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, et al. (2004) The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN- α/β-induced ubiquitin-like protein. Proc Natl Acad Sci USA 101: 7578–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, et al. (2009) ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep 10: 374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decker T, Lew DJ, Darnell JE (1991) Two distinct alpha-interferon-dependent signal transduction pathways may contribute to activation of transcription of the guanylate-binding protein gene. Mol Cell Biol 11: 5147–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahiratmadja E, Alisjahbana B, de Boer T, Adnan I, Maya A, et al. (2007) Dynamic changes in pro- and anti-inflammatory cytokine profiles (IFN-γ, TNF-α, IL-12/23, and IL-10) and IFN-γ receptor signalling integrity correlate with tuberculosis disease activity and response to curative treatment. Infect Immun 75: 820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.WHO (2012) Global Health Observatory Data Repository. Available at: http://apps.who.int/ghodata/(last accessed August 2012).
- 39.Leyten EM, Lin MY, Franken KL, Friggen AH, Prins C, et al. (2006) Human T-cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis. Microbes Infect 8: 2052–2060. [DOI] [PubMed] [Google Scholar]