Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso (original) (raw)
Abstract
Methylation of CpG dinucleotides in DNA is a common epigenetic modification in eukaryotes that plays a central role in maintenance of genome stability, gene silencing, genomic imprinting, development, and disease. Kaiso, a bifunctional Cys2His2 zinc finger protein implicated in tumor-cell proliferation, binds to both methylated CpG (mCpG) sites and a specific nonmethylated DNA motif (TCCTGCNA) and represses transcription by recruiting chromatin remodeling corepression machinery to target genes. Here we report structures of the Kaiso zinc finger DNA-binding domain in complex with its nonmethylated, sequence-specific DNA target (KBS) and with a symmetrically methylated DNA sequence derived from the promoter region of E-cadherin. Recognition of specific bases in the major groove of the core KBS and mCpG sites is accomplished through both classical and methyl CH···O hydrogen-bonding interactions with residues in the first two zinc fingers, whereas residues in the C-terminal extension following the third zinc finger bind in the opposing minor groove and are required for high-affinity binding. The C-terminal region is disordered in the free protein and adopts an ordered structure upon binding to DNA. The structures of these Kaiso complexes provide insights into the mechanism by which a zinc finger protein can recognize mCpG sites as well as a specific, nonmethylated regulatory DNA sequence.
Keywords: protein–DNA interaction, NMR spectroscopy, X-ray crystallography, folding upon binding, intrinsic disorder
In eukaryotes, DNA methylation is a common epigenetic modification that is central to the maintenance of genome stability, gene silencing, genomic imprinting, development, and disease (1, 2). Methyl-CpG–binding proteins (MBPs) mediate these processes by binding to methylated DNA signals and recruiting chromatin remodeling corepressor complexes, resulting in compaction of chromatin into its transcriptionally inactive state (3). To date, three classes of MBPs that recognize 5-methyl cytosine (5mC) have been identified. The SRA domain family has specificity for hemimethylated sites and is required for maintenance methylation during DNA replication (4). In contrast, the methyl CpG-binding domain (MBD) and Kaiso families of MBPs function as essential mediators of epigenetically controlled gene silencing by recognizing symmetrically methylated CpG sites (5). Kaiso is a MBP belonging to the BTB/POZ (broad complex, tramtrak, bric à brac/pox virus and zinc finger) subfamily of transcription factors that recognize cognate DNA sequences through a C-terminal zinc finger domain; the N-terminal BTB/POZ domain mediates protein–protein interactions (6). Kaiso is a POZ protein that participates in both methyl-dependent and sequence-specific transcriptional repression, using its three Cys2His2 zinc fingers to recognize either two consecutive symmetrically methylated CpG dinucleotides (mCpG) (7, 8) or a TCCTGCNA consensus (termed “KBS”) (9). Although originally it was reported that Kaiso only requires zinc fingers 2 and 3 for DNA recognition (9), we determined that Kaiso in fact requires all three zinc fingers plus adjacent regions for structural stability and high-affinity binding to both sequence classes (10).
Transcriptional repression by Kaiso has implications in both development and cancer (11–13). Kaiso binds to and silences aberrantly methylated tumor-suppressor and DNA-repair genes in cancer cells (14). Specific Kaiso-binding sites are found in the promoters of several target genes (matrilysin, cyclin-D1, siamois, c-myc, and Wnt11) regulated by the Wnt signaling pathway, which plays a critical role in early development and tumor progression (9, 15, 16). The cytoplasmic armadillo repeat protein p120 catenin (p120ctn) binds Kaiso and relieves Kaiso-mediated transcriptional repression (6, 17, 18). The interaction of Kaiso with p120ctn has been implicated in regulation of canonical and noncanonical Wnt signaling pathways through a mechanism that involves KBS recognition (15, 16). Additionally, there is increasing evidence that Kaiso expression levels are elevated in tumorigenesis, indicating that Kaiso may play a direct role in modulating cancer through transcriptional control of aberrantly methylated and KBS-containing genes (14, 19, 20). It still is unclear whether the transcriptional regulation of the two distinctly different DNA sequences is synergistic in nature or mutually exclusive.
Although structures of the SRA (UHRF1) (21–23) and MBD (MBD1, MeCP2, and MBD2) (24–26) domains in complex with methylated DNA have been determined, the structural basis for recognition of both mCpG and KBS sites by the Kaiso family is unknown. Here we report crystal and solution structures of the Kaiso zinc finger domain in complex with its nonmethylated, sequence-specific DNA site (KBS) (Fig. 1_A_) and the crystal structure of the complex with an oligonucleotide derived from the promoter region of E-cadherin, a known Kaiso-binding site (7, 27) that contains two sequential, symmetrically methylated CpG sites (MeECad) (Fig. 1_B_). In normal cells, E-cadherin is an important mediator of cell–cell adhesion, but hypermethylation of the promoter in several cancers results in down-regulation of E-cadherin expression and has been correlated with increasing invasion and metastasis (28). The current studies provide insights into the mechanism by which Kaiso recognizes both methylated DNA and the unmethylated KBS DNA sequence. Remarkably, the molecular interactions that mediate binding to these distinct DNA target sites are very similar.
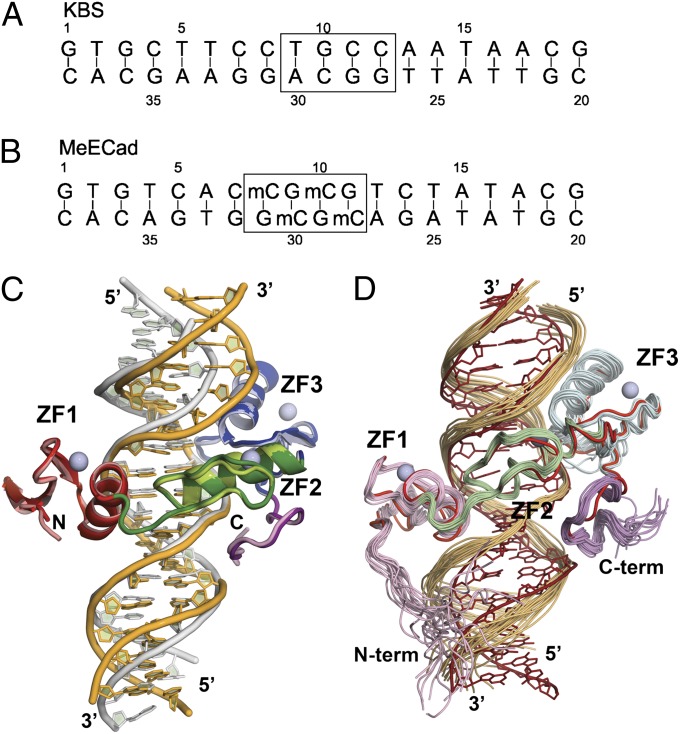
Fig. 1.
(A) Sequence of oligonucleotide containing the KBS motif (TCCTGCNA). The box shows the core sequence equivalent to the methylated DNA site shown in B. (B) Sequence of the MeECad oligonucleotide containing two symmetrically methylated CpG sites (boxed). (C) Overlay of the Kaiso:KBS (dark colors, yellow DNA) and Kaiso:MeECad (light colors, gray DNA) X-ray crystal structures, superimposed on the protein backbone. The three Cys2His2 zinc finger DNA-binding domains are colored red (ZF1), green (ZF2), and blue (ZF3), and the zinc atoms are shown as gray spheres. The C-terminal extension is colored purple. In the crystal structure of the KBS complex, the protein backbone is ordered between residues 482–597, whereas the MeECad complex is ordered between residues 481–600. (D) Superposition of the crystal structure (red protein and DNA) and the 20 lowest-energy structures in the NMR ensemble of the Kaiso:KBS complex. The structures are superimposed on their protein backbones. The zinc finger domains in the NMR structures are colored pink (N-terminal extension and ZF1), pale green (ZF2), pale blue (ZF3), and magenta (C-terminal extension). This figure was prepared using PYMol.
Results and Discussion
Overall Structure of the Kaiso:DNA Complexes.
The crystal structure of the Kaiso:KBS complex was determined at 2.4-Å resolution using single-wavelength anomalous dispersion from the zinc, and the Kaiso:MeECad complex then was solved by molecular replacement at 2.8-Å resolution as detailed in SI Methods. The diffraction and refinement statistics are summarized in Table S1. The two crystal structures are superimposed in Fig. 1_C_. The solution structure of the Kaiso:KBS complex (Fig. 1_D_) was determined from experimentally derived distance and dihedral angle restraints (SI Methods and Table S2). The overall arrangement of the three zinc fingers (ZF1–3) of the Kaiso DNA-binding domain is very similar in the KBS and MeECad complexes (backbone rmsd between the crystal structures is 0.50 Å). ZF2 adopts the canonical ββα zinc finger fold, whereas ZF1 and ZF3 contain three-stranded β-sheets with topology βββα and ββαβ, respectively (Fig. S1). The three Kaiso zinc fingers wrap around the DNA with their α-helices in the major groove, contacting only five or six base pairs in total (Fig. 1); this interaction contrasts markedly with canonical Cys2His2 zinc finger proteins, which typically contact three base pairs per finger (29).
DNA Binding Induces Structure in the C Terminus.
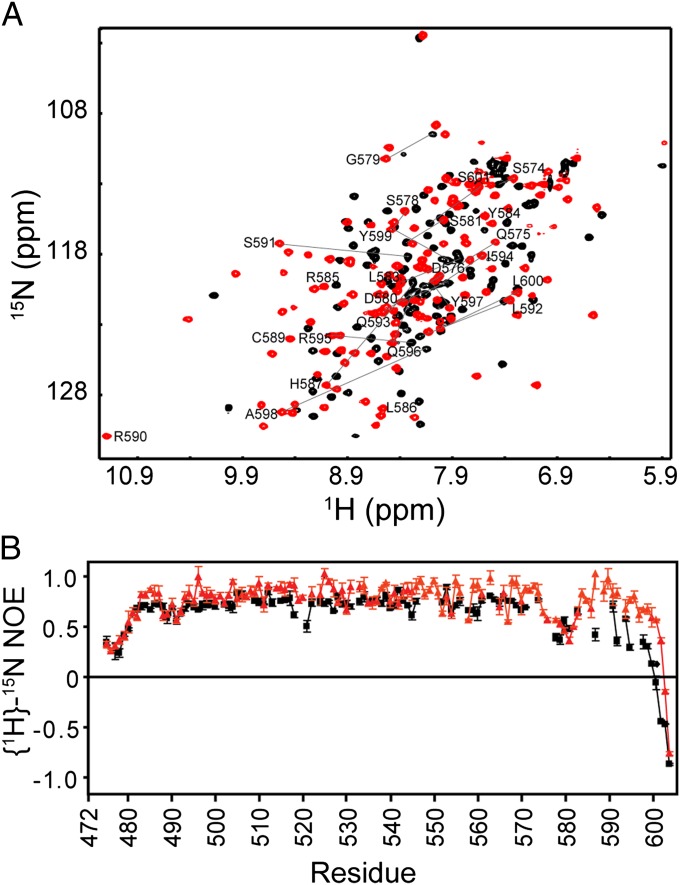
The ββαβ topology of ZF3 appears to be unique; residues 583–586, C-terminal to the ZF3 helix, fold back along the DNA phosphate backbone to form the third β-strand of ZF3 (Fig. S1). NMR experiments indicate that the C-terminal region, residues 575–604, is intrinsically disordered in the free protein and undergoes a conformational change to a more ordered state upon binding to DNA. The cross peaks of many of these residues lie in the random coil region of the 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of free Kaiso but undergo large shifts in the spectrum of the KBS complex, indicating formation of folded structure in the presence of DNA (Fig. 2_A_). Backbone {1H}-15N heteronuclear nuclear Overhauser effect (NOE) measurements (Fig. 2_B_) confirm that this region is highly flexible in the free protein but becomes ordered, with NOE values similar to those of the rest of the protein, in the presence of KBS. The structure induced in the C-terminal region is stabilized by incorporation into the ZF3 β-sheet, by hydrophobic packing and hydrogen-bonding interactions with ZF2, and by interactions with the phosphate backbone (Figs. S1 and S3_A_).
Fig. 2.
(A) 1H-15N HSQC spectra of free (black) and KBS-bound (red) Kaiso. Selected cross peaks are labeled to indicate the large changes in chemical shift that occur as the C-terminal extension becomes structured upon binding to DNA. (B) {1H}-15N heteronuclear NOEs for free Kaiso (black squares) and Kaiso in complex with KBS (red triangles). The small values of the heteronuclear NOE and the nearly random coil chemical shifts for residues 575–604 in the free protein indicate that this region is disordered in the absence of DNA.
Molecular Determinants of KBS and MeECad DNA Recognition.
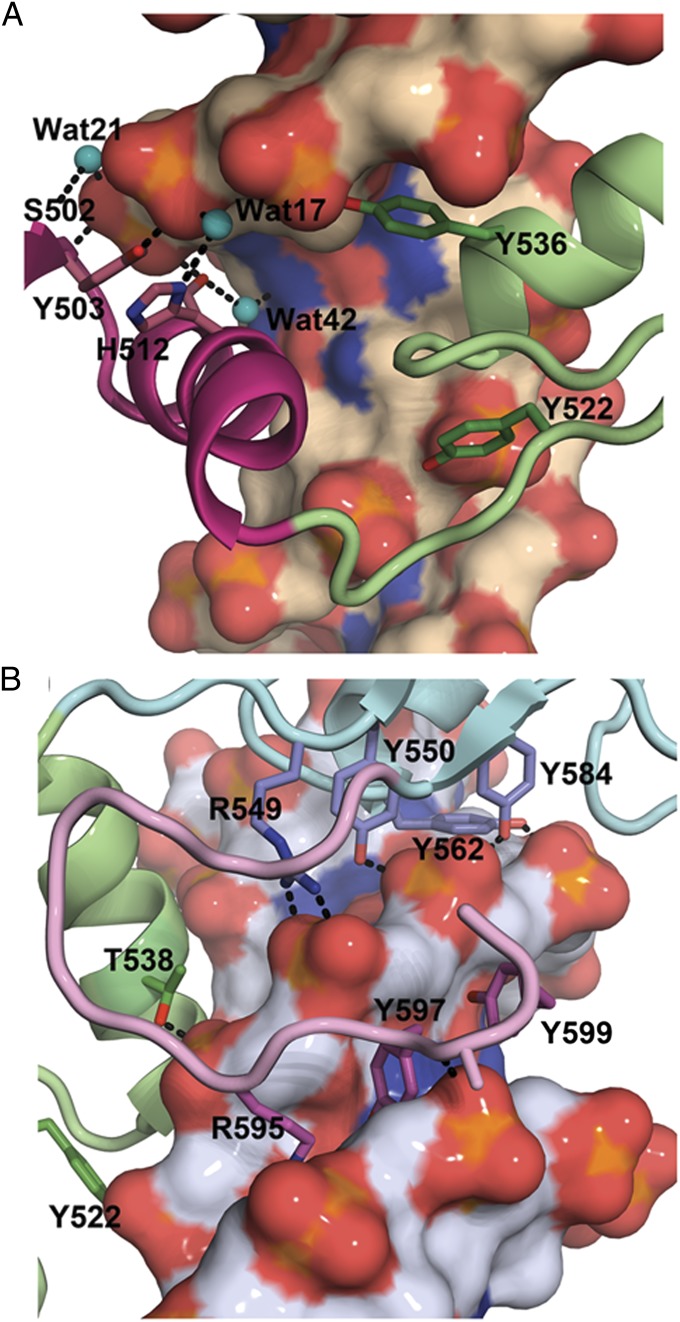
In both the KBS and MeECad complexes, the zinc fingers are anchored to the DNA by an extensive network of direct and water-mediated hydrogen bonds to the phosphate backbone as well as by van der Waals interactions with the sugar rings (Fig. 3_A_). Subtle differences are observed in the phosphate backbone interactions for the KBS and MeECad complexes (Fig. S2). ZF3 is anchored firmly to the DNA backbone by a network of hydrogen bonds from a cluster of tyrosine residues (Fig. 3_B_). Residues 590–594 form a structured loop, which is anchored to the side chain of Glu-547 in the ZF2–ZF3 linker by hydrogen bonds involving Arg-590 and Ser-591 backbone amides, and packs, by way of hydrophobic interactions, against the helix of ZF2, thereby positioning the C-terminal region of the polypeptide chain to enter the minor groove (Fig. 3_B_ and Fig. S3_A_). A notable difference between the X-ray and NMR structures is observed at the N terminus. There is no electron density for residues preceding the N-terminal β-strand of ZF1 in either crystal structure. The conformation of this region also is disordered in the NMR structural ensemble, but weak intermolecular NOEs from the Arg-475 and Lys-477 side chains indicate transient interactions with the DNA backbone across the minor groove (Fig. 1_D_).
Fig. 3.
(A) DNA backbone contacts between ZF1 (magenta) and ZF2 (green) and the coding strand of the KBS complex. Cyan spheres indicate water molecules that mediate hydrogen bonding to the phosphate backbone. (B) Interactions of ZF2 (green) and ZF3 (blue) with the backbone of the noncoding strand in the MeECad complex. ZF3 is anchored to the DNA backbone by a network of hydrogen bonds from a cluster of tyrosines (Tyr-550, Tyr-562, and Tyr-584) and from the Arg-549 guanidinium group. The side chains of Arg-595, Tyr-597, and Tyr-599 in the C-terminal extension (pink) project deeply into the narrowed minor groove. This figure was prepared using PyMol.
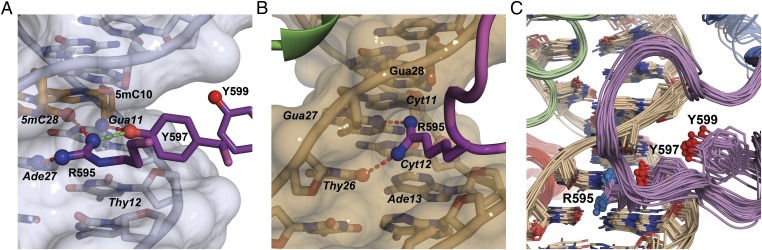
The C-terminal extension to ZF3, which is intrinsically disordered in the absence of DNA, is absolutely essential for high-affinity DNA binding. Truncation of the zinc finger construct at Gly-579, following the ZF3 helix, severely impairs KBS binding (10). The structures show that this impairment results from the elimination of DNA backbone contacts (from Tyr-584 and Leu-586) and the disruption of an extensive network of minor groove interactions. In the crystal structures of both complexes, the Arg-595 side chain is inserted deeply into the minor groove opposite the core-recognition site, where it forms hydrogen bonds with the O2 of 5mC28 and the N3 of A27 in MeECad (Fig. 4_A_) and with the O2 of T26 and the N3 of G27 in KBS (Fig. 4_B_). In the MeECad complex, the aromatic rings of Tyr-597 and Tyr-599 are inserted deeply into the narrow minor groove at the core mCpG site, where they pack tightly against the sugars of T12, C13, and G29. The OηH atom of the Tyr-597 side chain forms a hydrogen bond to the N2 amino group of G11, an interaction that is buttressed by an additional hydrogen bond to the Arg-595 guanidinium group. The Ala-598 amide hydrogen bonds to the phosphate backbone of C13, helping anchor the neighboring Tyr residues in the minor groove. In the crystal structure of the KBS complex, the location of the two Tyr side chains is indeterminate, because no electron density is observed for residues beyond position 597, and no side-chain density is observed for Gln-596 or Tyr-597. In solution, however, intense intermolecular NOEs are observed between the ribose protons of A13/G28 and the aromatic protons of Tyr-597/Tyr-599, showing that these side chains do in fact bind in the minor groove of the KBS DNA (Fig. 4_C_ and Fig. S3_B_). We conclude that the C-terminal extension assumes essentially the same conformation in the two complexes, making extensive minor groove contacts that are essential for high-affinity binding to both the mCpG and the KBS sites.
Fig. 4.
Minor groove interactions. (A) View of minor groove protein:DNA contacts for the MeECad complex, showing hydrogen bonding (dashed red lines) between the Arg-595 and Tyr-597 side chains and the bases of A27, 5mC28 (orange), and G11. A buttressing hydrogen bond between Tyr-597 and Arg-595 is shown in green. The protein backbone is cut away for clarity. (B) View of minor groove contacts in the KBS complex showing hydrogen bonding of the Arg-595 side chain to T26 and G27. (C) DNA contacts made by C-terminal extension [purple backbone with nitrogen (red) and oxygen (blue) molecules] in NMR structures of the Kaiso:KBS complex, showing the insertion of Arg-595, Tyr-597, and Tyr-599 side chains into the minor groove (DNA in beige with nitrogen in blue and oxygen in red). This figure was prepared using PyMol.
Base-specific interactions are dominated by side chains in the N-terminal regions of the ZF1 and ZF2 helices, whereas ZF3 interacts primarily with the phosphate backbone and makes no (MeECad) or few (KBS) base-specific contacts. In both the crystal and NMR structures of the KBS complex (Fig. 1), deviations from regular B-form geometry in the core-recognition site (6TCCTGCCA13) allow the ZF3 helix to penetrate the major groove so that the side chain of Gln-563 can form direct and water-mediated hydrogen bonds to the bases of G32 and C7 (Table S3). In contrast, because of differences in DNA conformation, the ZF3 helix is unable to extend deeply enough into the major groove of the MeECad DNA to form base-specific contacts (Fig. 1_A_). Instead, Nε2 of the Gln-563 side chain forms a hydrogen bond to the phosphate backbone of C5 (Table S3). Overall, the area of the major groove contacted by ZF3 is smaller in the MeECad complex than in the complex with KBS.
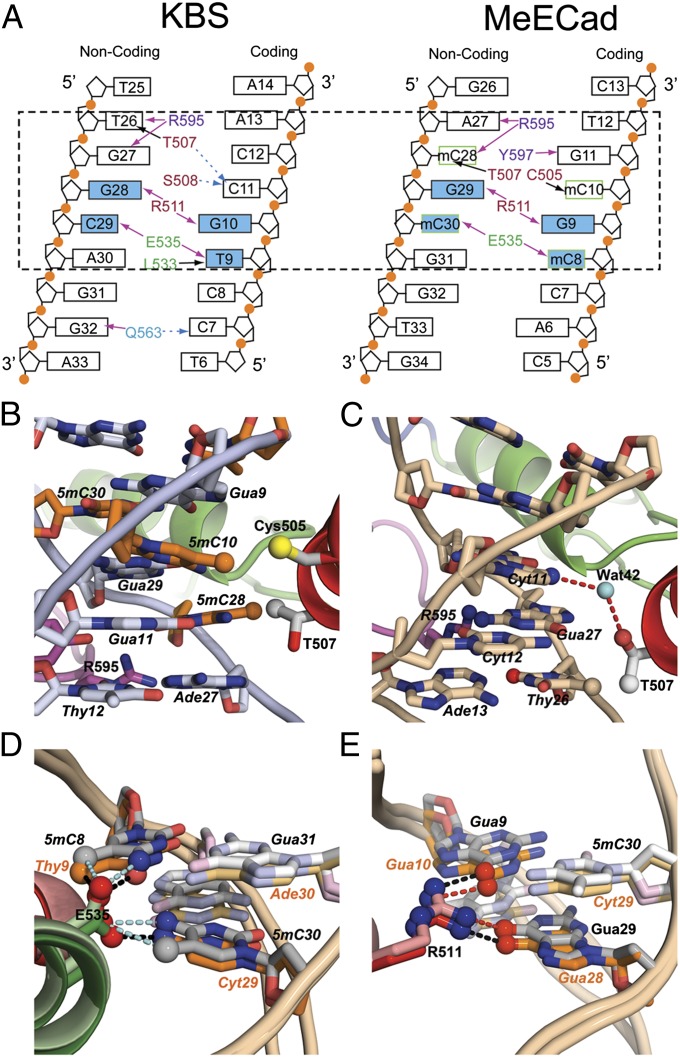
Recognition of the KBS and mCpG sites by Kaiso is accomplished through base-specific hydrogen bonding and hydrophobic interactions with the C5-methyl groups of thymines (KBS) or methylated cytosines (MeECad) within the major groove (Fig. 5_A_). In the MeECad complex, residues in the N-terminal regions of the ZF1 and ZF2 helices provide hydrophobic environments for the C5-methyls of 5mC10/5mC28 and 5mC8/5mC30, respectively. The Cys-505 side chain and the Cγ-methyl of Thr-507 form a hydrophobic pocket that accommodates the C5-methyl group of 5mC10 (Fig. 5_B_); if the cytosine were unmethylated, the distance between Cys-505 and the base edge would be greater, and the effectiveness of these hydrophobic interactions would be diminished. The Thr-507 Cγ-methyl also packs against the C5-methyl of 5mC28 (Fig. 5_B_). Similar interactions are observed in the KBS complex, through a change in the side-chain rotamer that places the Thr-507 Cγ-methyl in contact with the T26 C5-methyl and leads to the formation of a water-mediated hydrogen bond between Thr-507 Oγ and the N4-amino of C11 (Fig. 5_C_). A further contribution to specificity comes from hydrophobic interactions with the Cδ2-methyl of Leu-533, which lies close to the C5-methyl groups of 5mC8 and T9 in the MeECad and KBS complexes, respectively.
Fig. 5.
Kaiso:KBS and Kaiso:MeECad base-specific interactions. (A) Summary of major groove interactions. Recognition of MeECad is mediated by major groove interactions with only the four 5mC:G base pairs; additional minor groove hydrogen bonds extend the binding site to A27. The contact surface for KBS is larger because of the bending of the DNA around the core recognition sequence. The dashed box highlights the core contact site in the two DNA sequences. Residues are colored red, green, blue, and purple to denote their location in ZF1, ZF2, ZF3, and the C-terminal extension, respectively. Residues in the C-terminal extension (purple) make minor groove contacts. Black arrows denote hydrophobic interactions, pink arrows indicated classical hydrogen bonds, and dashed blue arrows indicate water-mediated hydrogen bonds. A complete summary of all DNA base and backbone contacts is shown in Fig. S2 and Table S3. (B) Cys-505 and Thr-507 form a hydrophobic pocket that accommodates the methyl groups (orange spheres) of 5mC10 and 5mC28. The 5-methylcytosines are highlighted in orange. (C) Base-specific interactions of Thr-507 in the KBS complex. The red lines show water-mediated hydrogen bonds to the N4-amino of C11. The water is shown as a cyan sphere. (D) Hydrogen-bonding interactions involving Glu-535 in the core mCpG- (gray) and KBS- (orange) binding sites. (E) Hydrogen-bonding interactions involving Arg-511 in the core mCpG- (gray) and KBS- (orange) binding sites. The Kaiso color scheme is as in Fig. 1_C_. This figure was prepared using PyMol.
There are marked similarities in the hydrogen-bonding interactions with bases in the major groove of the core MeECad and KBS sites, despite differences in sequence and cytosine methylation. These interactions are mediated almost entirely by the side chains of Glu-535 (ZF2) and Arg-511 (ZF1). These residues are invariant in Kaiso sequences from different species and in the Kaiso-like zinc finger proteins ZBTB4 and ZBTB38 (30), suggesting that these proteins all share a common mechanism of DNA recognition. The Glu-535 side chain plays a critical role in the recognition of the symmetrically methylated mCpG site in MeECad by forming CH···O hydrogen bonds with the C5-methyls of these bases (Fig. 5_D_ and Fig. S4). The marked decrease in binding affinity observed when all of the 5mC bases are replaced with nonmethylated cytosines (7, 10) confirms the importance of the methyl CH···O hydrogen bonds in specific recognition of mCpG sites by Kaiso. Methyl CH···O hydrogen bonds appear to be common in 5-mC recognition, because they have been observed in complexes of MeCP2 and UHRF1 with methylated or hemimethylated DNA, respectively (22, 23, 25). Interestingly, the position of the Glu-535 side chain is virtually identical in the KBS complex of Kaiso, where it forms a hydrogen bond with the N4-amino group of C29 as well as a methyl CH···O hydrogen bond with the C5-methyl of T9 (Fig. 5_D_ and Fig. S4). The Glu-535 Oε2 atom is surprisingly close (2.7 Å) to the O4 atom of T9, suggesting formation of a hydrogen bond through protonation of the carboxylic side chain. A comparison of the NMR spectrum of Kaiso in complex with the two different DNA sequences (Fig. S5) shows a large difference in the 15N amide chemical shifts of Glu-535 and Tyr-536, likely reflecting a change in the Glu-535 protonation state between the two complexes. The hydrogen-bonding interactions of Arg-511 and Glu-535 are poorly defined in the NMR ensemble because of a lack of intermolecular NOEs between their side chains and the DNA.
Base-specific interactions involving the guanidinium of Arg-511 also are similar in the two complexes (Fig. 5_E_). In the MeECad complex, Arg-511 forms asymmetrical hydrogen-bonding contacts across the major groove with the guanines of the central two mC:G base pairs (O6 atoms of G9 and G29). This observation provides insights into why Kaiso prefers two consecutive mCpG base pairs at the center of the recognition site or, at the very least, requires that a guanine occupy both positions, irrespective of the methylation status of the base-paired cytosine. Although Arg-511 cannot distinguish 5mC from C, it clearly discriminates against ApT or TpA steps and thus plays a key role in recognition of the central G9:mC30/mC10:G29 base pairs. In the KBS complex, Arg-511 forms similar side-chain hydrogen bonds to the O6/N7 of G10 and the O6 of G28 (Fig. 5_E_); substitution of either guanine by thymine greatly impairs binding (9). The importance of these hydrogen bonds for all DNA-binding interactions is demonstrated by mutagenesis (to alanine) of the equivalent Arg residue (Arg-326) in the Kaiso homolog ZBTB4, which abrogates binding to both methylated and unmethylated DNA (31). Comparison of the KBS and MeECad complexes shows that as long as the Arg-511/guanine hydrogen bonds are maintained, Kaiso can tolerate thymine substitutions at the 5mC8 position (equivalent to T9 in KBS) and likely at 5mC28, although substitution of either 5mC10 or 5mC30 with a thymine would greatly impair binding (9).
Recognition of Hemimethylated DNA.
To assess the extent to which each 5mC site contributes to recognition, we examined the binding of Kaiso to hemimethylated and single-site symmetrically methylated DNA. Kaiso binds equally well to MeECad and to a probe methylated only on the coding strand (5mC8/5mC10), whereas methylation on only the noncoding strand (5mC28/5mC30) reduces the affinity slightly (Fig. S6 A and B). Additionally, symmetric methylation at a single site decreases the binding affinity only slightly, by ∼1.5-fold for the 5′-site (5mC8/5mC30) and ∼twofold for the 5mC10/5mC28 site (Fig. S6_C_). Thus, Kaiso binds with significant affinity to hemimethylated DNA and to a single mCpG site within the context of the ECad sequence. Binding to hemimethylated DNA appears to be asymmetric, with a slight preference for methylation on the coding strand. Similar behavior has been observed for the Kaiso-like protein ZBTB4 (24, 31), suggesting a common mechanism by which Kaiso family members read and interpret the methylation signal.
Conclusions
Our work has shown how the three Cys2His2 zinc fingers and flanking regions of Kaiso are uniquely adapted for recognition of both mCpG and specific nonmethylated DNA sequences. Although the mode of DNA binding, with three zinc fingers in the major groove, is generally similar to other Cys2His2 zinc finger proteins, Kaiso differs markedly in the length of its core-recognition site. Canonical three-zinc finger proteins “read” the sequence in the major groove through hydrogen bonding and hydrophobic contacts with nine or 10 base pairs (29). In contrast, Kaiso has evolved to recognize a highly localized site, containing only four or five base pairs, through hydrogen-bonding interactions in both the major and minor grooves. Like Kaiso, the MBD family proteins MBD1, MeCP2, and MBD2 bind to a highly localized site, making minimal major groove contacts in the immediate vicinity of a single symmetrical mCpG site (24–26). Kaiso is anchored to the DNA by a plethora of phosphate backbone contacts that undoubtedly contribute to binding affinity. Major groove contacts with the four or five base pairs in the core DNA sequences are mediated entirely by side chains within ZF1 and ZF2. The sequences of these two zinc fingers are strongly conserved between Kaiso and its ZBTB4 and ZBTB38 homologs, but the sequence of ZF3 has diverged considerably (30), suggesting that the third zinc finger of the various family members may interact differently with DNA or with other proteins. The minor groove interactions, made by residues in the C-terminal extension following ZF3, are required for high-affinity binding to both KBS and methylated DNA (10). The present structural analysis reveals the mechanism of DNA recognition by the Kaiso zinc finger domain and represents a first step toward a molecular-level description of the complex pathways involved in methylation-dependent silencing of tumor-suppressor genes and in the regulation of the Wnt signaling pathway by Kaiso (14, 32). Once bound to DNA, Kaiso represses the target gene by recruiting an _N_-CoR/histone deacetylase complex through its N-terminal BTB/POZ domain (33). Further studies will be required to elucidate the complex network of downstream protein–protein interactions through which Kaiso regulates gene expression in development and carcinogenesis.
Methods
Sample Preparation.
Kaiso-ZF123(472-604) uniformly labeled with 15N, 15N/13C, or 15N/13C/2H was expressed and purified, and protein–DNA complexes were prepared as described previously (10). Methylated and unmethylated oligonucleotides were prepared synthetically (IDT Inc.) and dissolved in 10 mM Tris (pH 7.0). Equimolar amounts of complementary oligonucleotides were heated to 90 °C and annealed by slow cooling to room temperature to form duplex DNA. All samples of the complex were prepared in argon-saturated 10 mM d11-Tris buffer (pH 7.0) containing 1 mM Tris(2-carboxyethyl)phosphine, 0.005% (wt/vol) NaN3, and 5% (vol/vol) D2O (10). Complete formation of the 1:1 Kaiso:MeECad and Kaiso:KBS complexes used for NMR and crystallography was verified from 1D NMR spectra of the DNA imino proton resonances and 2D 15_N_-1H HSQC spectra of protein amide resonances.
EMSA.
EMSA experiments were run on 5% nondenaturing acrylamide gels using 5′-end 32P-labeled duplex DNA sequences. Experimental details are given in SI Methods.
X-Ray Structure Determination.
Crystals of the Kaiso:DNA complexes were obtained using sitting-drop vapor diffusion. Crystallographic data were collected on cryo-cooled crystals at the Advanced Photon Source (APS) beamline 23ID-D and the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9-2 for the KBS and MeECad complexes, respectively. The Kaiso:KBS complex crystal structure was solved by zinc single-wavelength anomalous dispersion and refined to 2.4-Å resolution. The crystal structure of the Kaiso:MeECad complex was solved by molecular replacement using the Kaiso from the Kaiso:KBS crystal structure as a search model and was refined to 2.8 Å. A more comprehensive description of the methods and refinement procedure can be found in SI Methods. The crystallographic and refinement statistics are summarized in Table S1.
NMR Spectroscopy.
NMR spectra of free Kaiso and of the KBS and MeECad complexes were recorded at 298K on Bruker 600, 750, 800, and 900 MHz spectrometers. Assignments of backbone and side-chain resonances were made using standard multidimensional NMR experiments. Structures were determined using experimentally derived distance, dihedral angle, and residual dipolar coupling restraints. Initial structures were generated using the program CYANA (34) and were refined using restrained molecular dynamics simulated annealing using the Amber 10 software package (35). The experimental restraints and refinement statistics for the NMR structure ensemble are summarized in Table S2. Details of the assignment procedure and structure calculations are given in the SI Methods.
Supplementary Material
Supporting Information
Acknowledgments
We thank Joel Gottesfeld for assistance with EMSA experiments, Gerard Kroon for assistance with NMR experiments, and Brian M. Lee, Brendan Borin, Chul Won Lee, and Christine Beuck for helpful discussions. X-ray data for 4F6M were collected at the Advanced Photon Source General Medicine/Cancer (GM/CA) beamline 23ID-D. The GM/CA Collaborative Access Team has been funded in whole or in part with federal funds through National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Science Grant Y1-GM-1104. Use of the Advanced Photon Source was supported by the US Department of Energy (DOE), Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. X-ray data for 4F6N were collected at Beamline 9-2 of the Stanford Synchrotron Radiation Lightsource (SSRL), a Directorate of the Stanford Linear Accelerator Center National Accelerator Laboratory and an Office of Science User Facility operated for the DOE Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, by the National Institutes of Health (NIH), National Center for Research Resources, Biomedical Technology Program Grant P41RR001209, and by the National Institute of General Medical Sciences. This work was supported by the NIH Grant GM036643 and the Skaggs Institute of Chemical Biology. B.A.B.-K. was supported by Grant PF-07-124-01-GMC from the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: Atomic coordinates and structure factors have been deposited in Protein Data Bank, www.pdb.org [PDB ID codes 4F6M (crystal structure of the KBS complex), 4F6N (crystal structure of the MeECad complex), and 2LT7 (NMR structures)] and resonance assignments in the Biological Magnetic Resonance Bank, http://www.bmrb.wisc.edu [accession no. 18462 (NMR assignments)].
References
- 1.Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 2.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Clouaire T, Stancheva I. Methyl-CpG binding proteins: Specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci. 2008;65:1509–1522. doi: 10.1007/s00018-008-7324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 5.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prokhortchouk A, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prokhortchouk AV, Aitkhozhina DS, Sablina AA, Ruzov AS, Prokhortchouk EB. Kaiso, a new protein of the BTB/POZ family, specifically binds to methylated DNA sequences. Russ J Genet. 2001;37(6):603–609. [PubMed] [Google Scholar]
- 9.Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck-Koehntop BA, Martinez-Yamout MA, Dyson HJ, Wright PE. Kaiso uses all three zinc fingers and adjacent sequence motifs for high affinity binding to sequence-specific and methyl-CpG DNA targets. FEBS Lett. 2012;586:734–739. doi: 10.1016/j.febslet.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel JM. Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim Biophys Acta. 2007;1773:59–68. doi: 10.1016/j.bbamcr.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 12.Kelly KF, Daniel JM. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 13.van Roy FM, McCrea PD. A role for Kaiso-p120ctn complexes in cancer? Nat Rev Cancer. 2005;5:956–964. doi: 10.1038/nrc1752. [DOI] [PubMed] [Google Scholar]
- 14.Lopes EC, et al. Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res. 2008;68:7258–7263. doi: 10.1158/0008-5472.CAN-08-0344. [DOI] [PubMed] [Google Scholar]
- 15.Kim SW, et al. Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat Cell Biol. 2004;6:1212–1220. doi: 10.1038/ncb1191. [DOI] [PubMed] [Google Scholar]
- 16.Park JI, et al. Kaiso/p120-catenin and TCF/β-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Kelly KF, Spring CM, Otchere AA, Daniel JM. NLS-dependent nuclear localization of p120ctn is necessary to relieve Kaiso-mediated transcriptional repression. J Cell Sci. 2004;117:2675–2686. doi: 10.1242/jcs.01101. [DOI] [PubMed] [Google Scholar]
- 18.Spring CM, et al. The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp Cell Res. 2005;305:253–265. doi: 10.1016/j.yexcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Prokhortchouk A, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208. doi: 10.1128/MCB.26.1.199-208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai SD, et al. Upregulation of δ-catenin is associated with poor prognosis and enhances transcriptional activity through Kaiso in non-small-cell lung cancer. Cancer Sci. 2011;102:95–103. doi: 10.1111/j.1349-7006.2010.01766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto H, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avvakumov GV, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 23.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 24.Ohki I, et al. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 25.Ho KL, et al. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 26.Scarsdale JN, Webb HD, Ginder GD, Williams DC., Jr Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Res. 2011;39:6741–6752. doi: 10.1093/nar/gkr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang BZ, Gu LK, Deng DJ. [Methylation specific binding activity of zinc finger protein Kaiso] Zhonghua Yu Fang Yi Xue Za Zhi. 2007;41(Suppl):43–46. Chinese. [PubMed] [Google Scholar]
- 28.Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 2003;94:575–581. doi: 10.1111/j.1349-7006.2003.tb01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 30.Filion GJP, et al. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 2010;38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Del Valle-Pérez B, et al. Wnt controls the transcriptional activity of Kaiso through CK1ε-dependent phosphorylation of p120-catenin. J Cell Sci. 2011;124:2298–2309. doi: 10.1242/jcs.082693. [DOI] [PubMed] [Google Scholar]
- 33.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 35.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information