Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation (original) (raw)
At the same time that BubR1 acts as a pseudosubstrate inhibitor in the spindle assembly checkpoint complex to inhibit Cdc20, Mad2 binds to Cdc20 to prevent activation of APC/C and subsequent mitotic exit when chromosome attachment is not complete.
Abstract
The spindle assembly checkpoint (SAC) is essential to ensure proper chromosome segregation and thereby maintain genomic stability. The SAC monitors chromosome attachment, and any unattached chromosomes generate a “wait anaphase” signal that blocks chromosome segregation. The target of the SAC is Cdc20, which activates the anaphase-promoting complex/cyclosome (APC/C) that triggers anaphase and mitotic exit by ubiquitylating securin and cyclin B1. The inhibitory complex formed by the SAC has recently been shown to inhibit Cdc20 by acting as a pseudosubstrate inhibitor, but in this paper, we show that Mad2 also inhibits Cdc20 by binding directly to a site required to bind the APC/C. Mad2 and the APC/C competed for Cdc20 in vitro, and a Cdc20 mutant that does not bind stably to Mad2 abrogated the SAC in vivo. Thus, we provide insights into how Cdc20 binds the APC/C and uncover a second mechanism by which the SAC inhibits the APC/C.
Introduction
The spindle assembly checkpoint (SAC) is essential for mitosis in mammalian cells: in its absence, cells rapidly become aneuploid, and mouse embryos die early in development (Dobles et al., 2000; Wang et al., 2004). The SAC monitors the attachment of spindle microtubules to kinetochores and delays mitosis until all the chromosomes have attached to the spindle (Musacchio and Salmon, 2007; Khodjakov and Pines, 2010). The SAC inhibits the anaphase-promoting complex/cyclosome (APC/C), the crucial ubiquitin ligase in mitosis (Pines, 2011). By preventing the destruction of two key APC/C substrates, securin and Cyclin B1, while any chromosomes remain unattached, the SAC ensures that an identical set of chromosomes is inherited by each of the two daughter cells.
Genetic evidence identified the target of the SAC as Cdc20 (Hwang et al., 1998; Kim et al., 1998), a coactivator of the APC/C. Cdc20 is thought to form part of a bipartite receptor for APC/C substrates (by analogy with another coactivator, Cdh1; Buschhorn et al., 2011; da Fonseca et al., 2011), and recent structure data show how the SAC effector proteins Mad2 and BubR1 (Mad3 in yeast) bind Cdc20 (Chao et al., 2012). Mad2 and BubR1 are essential to establish the SAC (Hoyt et al., 1991; Li and Murray, 1991; Meraldi et al., 2004). In mammalian cells, depleting the levels of these proteins accelerates mitosis (Meraldi et al., 2004) because the destruction of Cyclin B1 and securin is advanced to begin at nuclear envelope breakdown (NEBD; Mansfeld et al., 2011). Unattached kinetochores are the primary signal for the SAC and are thought to catalyze the conversion of Mad2 from its inactive “O” (open or N1) to its active “C” (closed or N2) conformation, which binds to Cdc20 (Luo et al., 2000; Sironi et al., 2002) and to BubR1 (Tipton et al., 2011; Chao et al., 2012). Mad2 and BubR1 synergize to inhibit the APC/C (Tang et al., 2001; Fang, 2002; Morrow et al., 2005; Davenport et al., 2006; Kulukian et al., 2009) by binding to Cdc20 to form the mitotic checkpoint complex (MCC; Sudakin et al., 2001; Kops et al., 2010), although we, and others, find that Mad2 is a substoichiometric component of the MCC (Nilsson et al., 2008; Maciejowski et al., 2010; Westhorpe et al., 2011). The structure of fission yeast MCC (Chao et al., 2012) shows that the N-terminal KEN box in Mad3 blocks the putative substrate binding site for KEN box degrons on the top face of the β-propeller domain of Cdc20. This supports biochemical evidence that Mad3/BubR1 acts as a pseudosubstrate inhibitor of Cdc20 (Burton and Solomon, 2007; Sczaniecka et al., 2008; Rahmani et al., 2009; Elowe et al., 2010). Modeling this structure onto the pseudoatomic structure of the APC/C reveals that the MCC will displace Cdc20 away from the site that it should occupy to form a bipartite degron receptor with APC10 (Chao et al., 2012). Thus, the MCC should block substrate recognition as a pseudosubstrate inhibitor for KEN box degrons and prevent the formation of the putative bipartite Destruction box receptor.
Here, we provide a second mechanism by which the SAC can inhibit Cdc20 through the Mad2 protein. We show that Mad2 binds to a motif on Cdc20 that is itself required for Cdc20 to bind to and activate the APC/C. Thus, Mad2 competes directly for Cdc20 with the APC/C, which would contribute to the rapid and potent inhibition of Cdc20.
Results and discussion
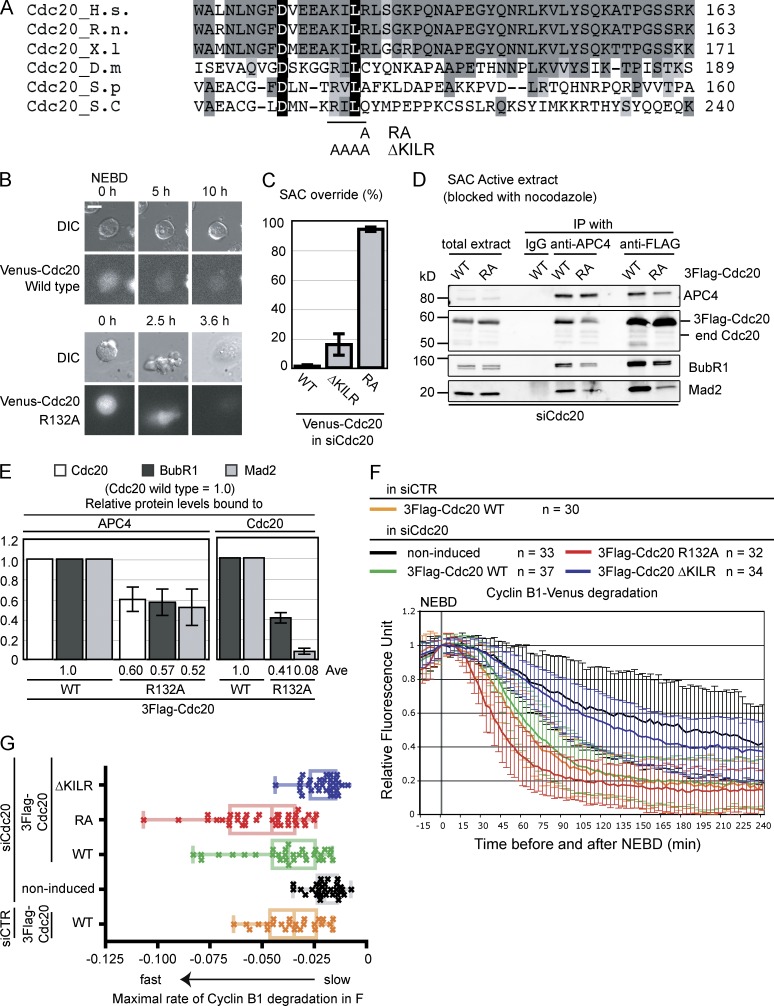
Cdc20 binds to Mad2 through a motif that is conserved through evolution (Fig. 1 A; Hwang et al., 1998; Luo et al., 2000; Zhang and Lees, 2001; Sironi et al., 2002). A previously described point mutation in this motif (Cdc20R132A; Fig. 1 A; Zhang and Lees, 2001; Nilsson et al., 2008; Ge et al., 2009) overrides the SAC, such that cells go through mitosis even in the presence of unattached kinetochores (Fig. 1, B and C). Consistent with this, the Cdc20R132A mutant binds much less Mad2 than wild-type Cdc20 and consequently binds less BubR1 and APC/C (Fig. 1, D and E, quantification). Cdc20 with a more extensive mutation in the motif (K129ILR to AAAA termed ΔKILR), however, could not override the SAC (Fig. 1 C), and most cells remained in mitosis. We were puzzled by this result, therefore, we compared the properties of wild-type Cdc20 and the ΔKILR mutant by generating cell lines expressing inducible siRNA-resistant 3×Flag-tagged wild-type Cdc20 (Cdc20wt) or Cdc20ΔKILR or Cdc20R132A at similar levels (Fig. S1 A). Cells depleted of endogenous Cdc20 slowed the kinetics of Cyclin B1–Venus destruction (Fig. 1, F [noninduced] and G [quantified]), and normal kinetics were restored by inducing Cdc20wt (Fig. 1, F and G). The initiation of Cyclin B1 destruction was much earlier than normal in cells expressing the Cdc20R132A mutant, consistent with its SAC-deficient phenotype (Fig. 1 F). In contrast, Cdc20ΔKILR did not restore the degradation of Cyclin B1; indeed, the rate of Cyclin B1 destruction was very similar to that in Cdc20-depleted control cells (P = 0.98). We excluded the trivial explanation that Cdc20ΔKILR was misfolded because it migrated correctly on size-exclusion chromatography (see Fig. 3 E) and was able to bind to Cyclin A (Fig. S1 B; Wolthuis et al., 2008; Di Fiore and Pines, 2010).
Figure 1.
The Mad2 binding motif is required for Cdc20 activity. (A) Alignment of the Mad2 binding motif in Cdc20 from different organisms. Black shows residues present in all species; dark gray shows residues present in three or more species; gray shows similar residues. The R132A and ΔKILR mutations (129KILR132 is substituted by four alanines) are also shown. H.s., Homo sapiens; R.n., Rattus norvegicus; X.l, Xenopus; D.m, Drosophila melanogaster; S.p, Schizosaccharomyces pombe; S.c, Saccharomyces cerevisiae. (B and C) The RA, but not the KILR mutant, overrides the SAC. Plasmids expressing siRNA-resistant Venus-tagged wild type, RA, or ΔKILR mutant of Cdc20 were transfected into HeLa cells with siRNA against human Cdc20. Cells were analyzed by time-lapse differential interference contrast (DIC) and fluorescence microscopy at 10-min intervals in the presence of 100 ng/ml nocodazole. (B) Representative images of cells. Bar, 10 µM. (C) Percentage of cells exiting from mitosis within 10 h. 100 cells were analyzed in each from three independent experiments. (D and E) The Cdc20R132A mutant partially forms the MCC but weakly binds Mad2. (D) HeLa cell lines expressing inducible 3×Flag-Cdc20, wild type, or RA mutant were treated with siRNA against Cdc20, arrested in prometaphase with 0.33 µM nocodazole + 10 µM MG132, and harvested by mitotic shake off. The APC/C or 3×Flag-Cdc20 was immunoprecipitated and analyzed by quantitative immunoblotting with the indicated antibodies. (E) Means of the relative amounts of indicated proteins in APC4 or 3×Flag-Cdc20 immunoprecipitates calculated from three independent experiments with the amount of protein bound to wild-type Cdc20 set to 1. Means are shown on the bottom. (F and G) The ΔKILR mutant cannot substitute for wild-type Cdc20. (F) Cdc20 was depleted by siRNA in cells expressing wild type, R132A, or ΔKILR mutants of Cdc20 from an inducible promoter, and ectopically expressed Cyclin B1–Venus was analyzed by time-lapse DIC and fluorescence microscopy. As controls, noninduced cells were treated with siRNA against Cdc20, and cells expressing wild-type Cdc20 were treated with siRNA against GAPDH (siCTR). The fluorescence of individual cells was measured, the value at NEBD was set to 1, and the means ± SD for all cells were plotted. Cyclin B1 destruction in Cdc20-depleted and noninduced cells, in cells induced for wild-type Cdc20, the RA, the ΔKILR mutants, or in cells expressing wild-type Cdc20 and treated with control siRNA are plotted on the same graph. n = number of cells analyzed in two independent experiments. (G) The rate of Cyclin B1 destruction is plotted as a box and whisker chart. The maximal rate of cyclin degradation was obtained from the data in F by nonlinear regression analysis assuming a sigmoidal dose–response (variable slope). The center lines are the medians, boxes correspond to the range between 25 and 75% of all the data, and the whiskers correspond to the minimum and maximum of all the data. Means and SDs were calculated from three independent experiments. Ave, average; end, endogenous; WT, wild type.
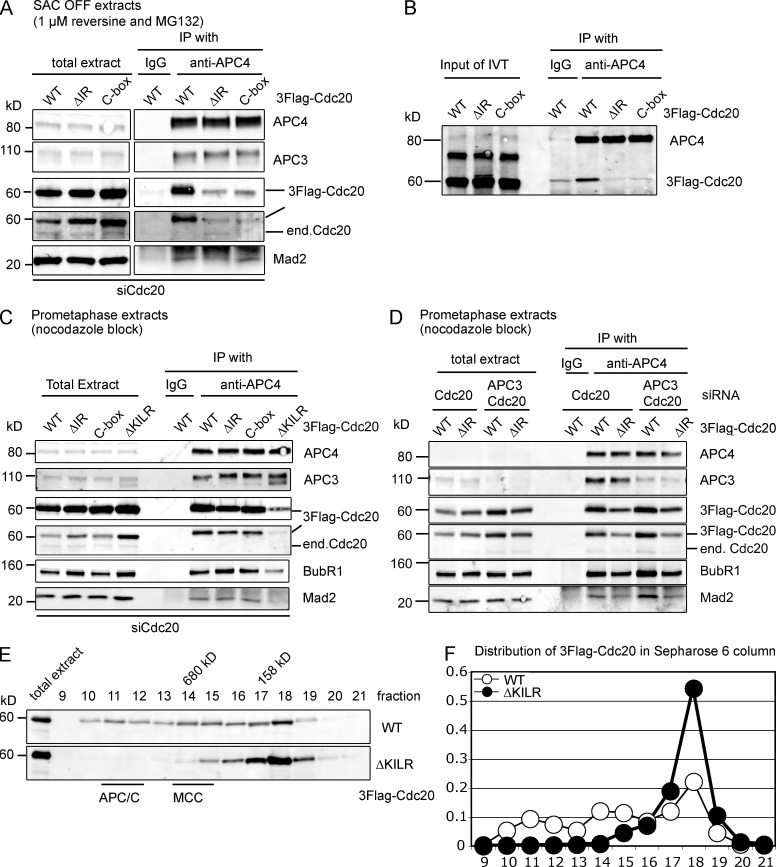
Figure 3.
The ΔKILR motif interacts with the APC/C in a different manner compared with the IR tail and the C box. (A and B) The IR tail and C box motif in Cdc20 are required to interact with the APC/C. (A) HeLa cells expressing 3×Flag–wild-type, ΔIR, or C box mutant Cdc20 from an inducible promoter were treated with siRNA against Cdc20 and synchronized at mitosis as in Fig. 2 B. The APC/C was immunoprecipitated with anti-APC4 antibodies and analyzed by quantitative immunoblotting. Results are representative of three independent experiments. (B) In vitro translated (IVT) full-length 3×Flag–wild type, ΔIR, or C box mutants of Cdc20 were tested for their ability to bind to the APC/C as in Fig. 2 C. Results are representative of four independent experiments. (C and D) The ΔIR and C box mutants of Cdc20 can still interact with the APC/C when part of the MCC. (C) HeLa cell line expressing inducible 3×Flag-tagged wild-type, ΔIR, C box, or ΔKILR mutant Cdc20 were treated with siRNA against Cdc20, arrested at prometaphase with 0.33 µM nocodazole, and harvested by mitotic shake off. The APC/C was immunoprecipitated using anti-APC4 antibodies and analyzed by quantitative immunoblotting. Results are representative of three independent experiments. (D) HeLa cell lines expressing inducible 3×Flag-tagged wild-type or ΔIR mutant Cdc20 were treated with siRNA against Cdc20 or Cdc20 and APC3, arrested at prometaphase, and analyzed as in C. (E and F) The ΔKILR mutant is not able to form the MCC or bind to the APC/C. (E) HeLa cell lines expressing inducible 3×Flag-Cdc20wt or Cdc20ΔKILR were treated with siRNA against Cdc20, arrested in prometaphase with 0.33 µM nocodazole, and harvested by mitotic shake off. Extracts were analyzed by size-exclusion chromatography on a Sepharose 6 column, and fractions were analyzed by quantitative immunoblotting with antibodies against Cdc20. The peaks of APC/C and MCC migration are indicated. Results are representative of two independent experiments. (F) Distributions of wild-type Cdc20 and ΔKILR mutant with the sum of Cdc20 intensities set to 1. Immunoblotting with antibodies against APC3, Cdc20, BubR1, and Mad2 is shown in Fig. S3. end, endogenous; IP, immunoprecipitation; WT, wild type.
Cdc20 binds and activates the APC/C through the KILR motif
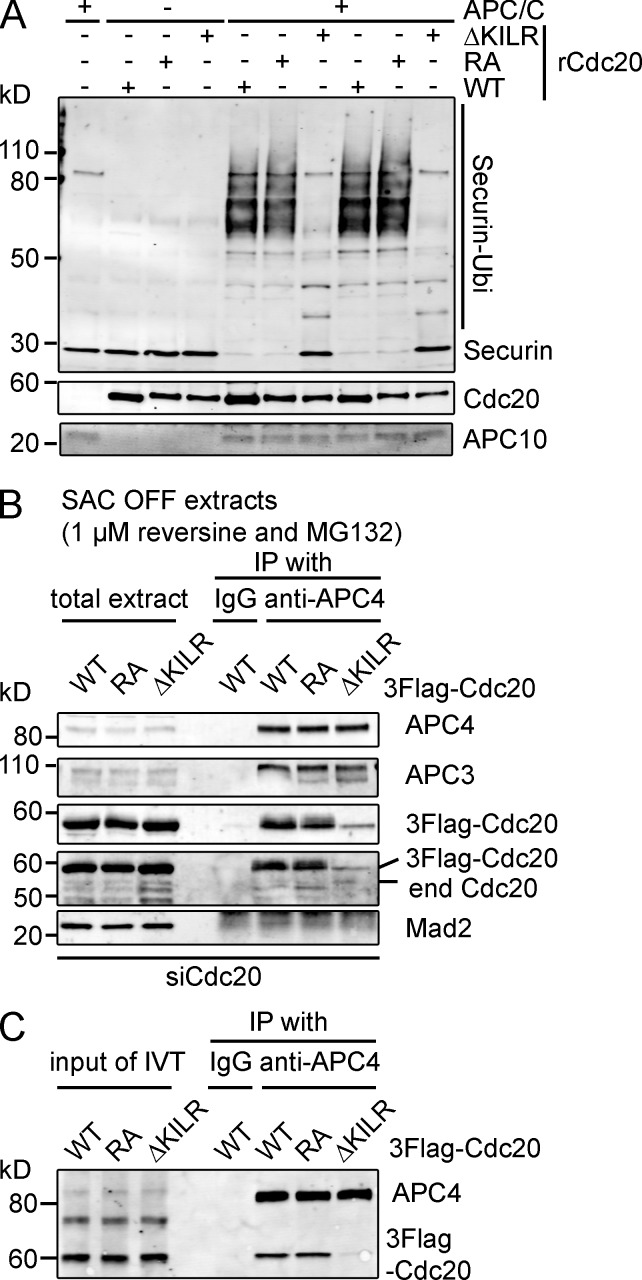
These rescue experiments indicated that Cdc20ΔKILR was defective in its ability to activate the APC/C. To test this, we assayed the ability of the Cdc20ΔKILR and Cdc20R132A mutants to activate the APC/C in vitro (see Materials and methods; Fig. 2 A). These assays revealed that Cdc20ΔKILR had little or no ability to activate the APC/C, whereas both wild-type and Cdc20R132A activated the APC/C to a similar extent (Fig. 2 A).
Figure 2.
The ΔKILR motif is required for Cdc20 to bind and activate the APC/C. (A) The ΔKILR mutant cannot activate the APC/C in vitro. Wild-type, RA, or ΔKILR mutants of Cdc20 were purified from baculovirus-infected insect cells, and their ability to activate the APC/C was assayed using securin as a substrate. The APC/C was prepared from mitotic cells depleted of Cdc20. Results are representative of two experiments. (B and C) The ΔKILR mutant is defective in binding to the APC/C. (B) HeLa cell lines expressing inducible 3×Flag–wild type, RA, or ΔKILR mutant Cdc20 were treated with siRNA against Cdc20 for 48 h. 9 h after release from a thymidine block, cells were treated with 1 µM reversine + 10 µM MG132 for 3 h and harvested by mitotic shake off. The APC/C was immunoprecipitated with anti-APC4 antibodies and analyzed by quantitative immunoblotting. (C) In vitro translated (IVT) 3×Flag–wild-type, RA, or ΔKILR mutant Cdc20 was incubated with mitotic extracts depleted of endogenous Cdc20, and the APC/C was immunoprecipitated with anti-APC4 antibodies before immunoblotting with anti-APC4 and anti-Flag epitope antibodies. Results in B and C are representative of three independent experiments. end, endogenous; IP, immunoprecipitation; Ubi, ubiquitin; WT, wild type.
To determine why the KILR motif was required for activity, we tested whether it was required for Cdc20 to bind to the APC/C. Cdc20 has two previously described motifs, the C box and the isoleucine-arginine (IR) tail, which are needed for Cdc20 to bind to the APC/C as a coactivator (see following paragraph; Schwab et al., 2001; Vodermaier et al., 2003). To remove potential indirect effects, we depleted endogenous Cdc20 by siRNA and prevented the assembly of the MCC with an Mps1 inhibitor (reversine; Santaguida et al., 2010). In reversine-treated cells, Cdc20 binds to the APC/C solely as a coactivator, as demonstrated by its requirement for the APC3 subunit (Fig. S2 A; Izawa and Pines, 2011). In reversine-treated cells, both wild type and Cdc20R132A could bind the APC/C, whereas Cdc20ΔKILR could not (Fig. 2 B). Cdc20ΔKILR was also defective in its ability to bind to the APC/C in an in vitro binding assay (Fig. 2 C) using in vitro translated Cdc20 (wild-type, R132A, and ΔKILR mutants) that binds to the APC/C through its APC3-dependent metaphase binding site (Fig. S2 B; Izawa and Pines, 2011). This evidence supported the conclusion that the KILR motif is essential to a previously undescribed APC/C binding motif.
The KILR motif interacts with the APC/C in a different manner from the IR tail and the C box
We next compared the contribution to binding the APC/C of the KILR motif with those of the known APC/C interaction motifs: the IR tail and the C box. The IR tail and the C box motifs were essential for Cdc20 to bind to the APC/C as a coactivator (Fig. 3 A) and in vitro (Fig. 3 B) but not when the SAC was active in prometaphase (Fig. 3 C). This agreed with our previous results that Cdc20 bound to different sites on the APC/C depending on whether or not it was part of the MCC (Izawa and Pines, 2011). In support of this, depleting APC3 did not interfere with the binding of wild type or Cdc20ΔIR to the APC/C in SAC-arrested cells (Fig. 3 D). In contrast to the IR tail and C box motifs, however, the KILR motif was necessary for Cdc20 to bind to the APC/C when the SAC was active (Fig. 3 C), most likely because it was needed for Cdc20 to be incorporated into the MCC through interaction with Mad2. In support of this, Cdc20ΔKILR did not co-migrate with either the APC/C or the MCC in size-exclusion chromatography (Fig. 3, E and F; and Fig. S3). We conclude that, unlike the C box and the IR tail, the KILR motif is required both for Cdc20 to bind to the APC/C and to form the MCC.
The APC/C and Mad2 compete for Cdc20 through the KILR motif
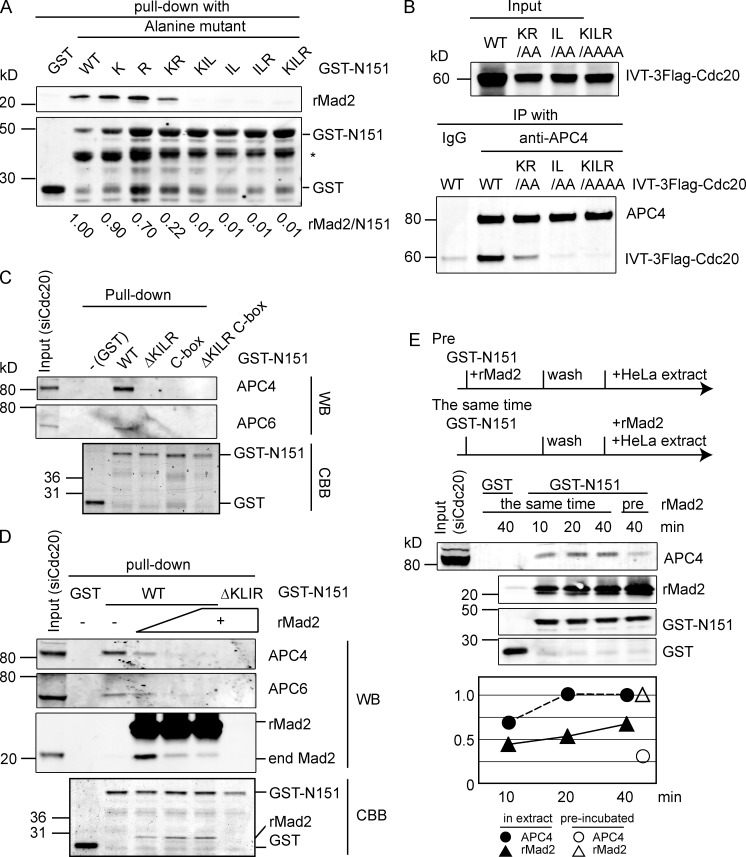
To define the KILR motif further, we made a series of point mutations and tested their ability to bind to Mad2 (Fig. 4 A). Mutating the positively charged residues (K129 or R132) only weakly affected binding to Mad2 (Fig. 4 A). By comparison, mutating the hydrophobic residues dramatically reduced binding to Mad2 (Fig. 4 A) and impaired the ability of Cdc20 to bind to the APC/C (Fig. 4 B). Indeed, the Cdc20IL/AA and the Cdc20ΔKILR mutants were equally defective in binding to the APC/C in vitro. Thus, a very small region on Cdc20 is critical to interact with both Mad2 and the APC/C.
Figure 4.
Mad2 prevents Cdc20 from interacting with the APC/C. (A) The hydrophobic core of the KILR motif is essential to bind Mad2. E. coli extracts expressing GST, GST fused to the N terminus of wild-type Cdc20, or the indicated mutants were incubated with recombinant human Mad2 for 30 min at 4°C and purified with glutathione–Sepharose, and the amount of Mad2 was analyzed by quantitative immunoblotting. Relative amount of Mad2 bound is shown at the bottom. Results are representative of three independent experiments. Asterisk shows a truncated form of Cdc20 that does not bind Mad2. (B) The IL motif is required to bind to the APC/C. In vitro translated (IVT) full-length 3×Flag–wild-type Cdc20 or the indicated mutants were analyzed as in Fig. 2 C. Results are representative of three independent experiments. (C) The KILR and C box motifs are required for the N terminus of Cdc20 to interact with the APC/C. GST fusion proteins of the N terminus of Cdc20 (N151), wild type, and the indicated mutants were incubated for 40 min at 4°C with mitotic extracts depleted of endogenous Cdc20. Proteins retained on the beads were analyzed by quantitative immunoblotting with the indicated antibodies. Results are representative of three independent experiments. (D and E) Mad2 competes with the APC/C for binding to the KILR motif. (D) GST or GST fusion proteins of the N terminus of wild-type Cdc20 or the ΔKILR mutant were incubated with recombinant Mad2 and mitotic HeLa cell extracts as in C. The APC/C bound to the beads was analyzed by quantitative immunoblotting with the indicated antibodies (Western blot [WB]). Recombinant proteins were also detected by Coomassie blue staining (CBB). (E) GST or GST fusion proteins were prebound to gluthatione–Sepharose and incubated with mitotic extract plus recombinant Mad2. “Pre” indicates the fusion protein was incubated with Mad2 before the mitotic extract. Mad2 was added in fourfold excess over the amount of GST-N151. Samples were analyzed by quantitative immunoblotting with the indicated antibodies. The relative amount of the APC/C and Mad2 bound to the beads is shown on the bottom where the amount of APC/C bound at 40 min or the amount of Mad2 bound at 40 min in the “pre” sample is set to 1. Binding assays in D and E are representative of three experiments. end, endogenous; IP, immunoprecipitation; WT, wild type.
A previous study had suggested that Mad2 and the APC/C bind to overlapping but distinct binding sites on Cdc20 (Zhang and Lees, 2001); however, our results indicated that Mad2 and the APC/C compete for exactly the same binding site on Cdc20. To test this, we set up a competition assay using the first 151 amino acids of Cdc20 (N151). When incubated in mitotic extracts, human Cdc20N151 stably bound to the APC/C (Fig. 4 C). Binding required the C box motif (Fig. 4 C), in agreement with observations in Xenopus laevis extracts (Kimata et al., 2008), but also required the KILR motif (Fig. 4 C). Consistent with the idea that Mad2 and the APC/C compete for the KILR motif, preincubating Cdc20N151 with recombinant Mad2 prevented Cdc20N151 from interacting with the APC/C (Fig. 4 D). Next, we incubated Cdc20N151 with recombinant Mad2 and the APC/C at the same time to assay their relative affinities (Fig. 4 E). This revealed that preincubating Cdc20N151 with Mad2 (Fig. 4 E, pre 40 min) was more effective than simultaneous incubation (Fig. 4 E, 40 min) at blocking binding to the APC/C.
Stable interaction of Cdc20 with Mad2 is required to maintain SAC signaling
Our results revealed how Mad2 prevented Cdc20 from binding to the APC/C in a competitive manner in vitro; therefore, we asked whether Mad2 contributed to Cdc20 inhibition in vivo. Despite the very stable Mad2–Cdc20 complex crystallized in vitro (Luo et al., 2002; Sironi et al., 2002), very little Mad2–Cdc20 dimer was detected in cells (Nilsson et al., 2008), and Mad2 was not able to inhibit Cdc20 in vivo when BubR1 was depleted (Meraldi et al., 2004). Recent studies indicated that Mad2 binding to Cdc20 was destabilized by the p31comet protein (Hagan et al., 2011; Jia et al., 2011; Mansfeld et al., 2011; Teichner et al., 2011; Varetti et al., 2011; Westhorpe et al., 2011), and overexpressing a Mad2 mutant that cannot bind to p31comet could delay mitosis (Westhorpe et al., 2011). We obtained similar results and found that although the mitotic delay correlated with the expression level of Mad2 (Fig. S3 A), BubR1 was still required to delay mitosis (Fig. S3 B). Thus, partially stabilizing the Mad2–Cdc20 complex will delay mitosis but primarily through the ability of Mad2 to promote binding to BubR1 (Nilsson et al., 2008).
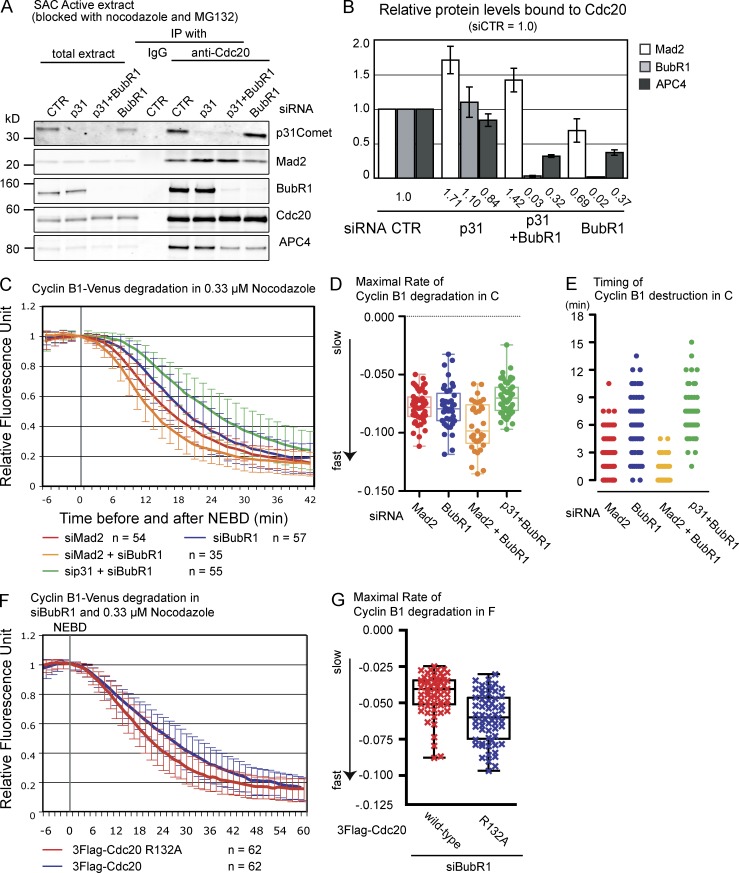
We next analyzed the inhibitory activity of Mad2 in the absence of BubR1 by partially stabilizing the binding between Mad2 and Cdc20 through depleting p31comet. Consistent with previous studies (Hagan et al., 2011; Jia et al., 2011; Mansfeld et al., 2011; Teichner et al., 2011; Varetti et al., 2011; Westhorpe et al., 2011), the amount of Mad2 bound to Cdc20 increased in cells when p31comet was depleted (Fig. 5, A and B, quantification). In addition, a Mad2–Cdc20 complex accumulated in cells when p31comet and BubR1 were codepleted, compared with depleting BubR1 alone (Fig. 5, A and B). When BubR1 was depleted, the amount of APC/C bound to Cdc20 dramatically decreased (Fig. 5, A and B), indicating that the Mad2–Cdc20 complex required BubR1 to bind to the APC/C. The residual Cdc20 that bound to the APC/C was most likely bound as a coactivator; consistent with this, little Mad2 was coimmunoprecipitated with the APC/C in the absence of BubR1 (Fig. S3 C).
Figure 5.
Mad2 inhibits Cdc20 in vivo. (A) The binding between Cdc20 and Mad2 is stabilized by depleting p31comet. HeLa cells were treated with siRNA against GAPDH (control), p31comet, BubR1, or p31comet and BubR1 for 72 h, arrested at prometaphase with nocodazole + MG132, and harvested by mitotic shake off. Cdc20 was immunoprecipitated and analyzed by quantitative immunoblotting. (B) Quantification of the levels of Mad2 and BubR1 bound to Cdc20 calculated from three independent experiments with the amount of protein in control siRNA set to 1. Mean values are shown at the bottom. Means and SDs were calculated from three independent experiments. (C) Cyclin B1 destruction in cells depleted of Mad2, BubR1, Mad2 + BubR1, or BubR1 + p31comet. HeLa cells were treated with siRNA against Mad2, BubR1, or Mad2 + BubR1 for 48 h or against BubR1 for 48 h and against p31comet for 72 h, and the level of ectopically expressed Cyclin B1–Venus was analyzed by time-lapse DIC and fluorescence microscopy at 90-s intervals in the presence of 0.33 µM nocodazole. The means ± SD for all cells from three independent experiments are plotted. n = number of cells analyzed. (D) The rates of Cyclin B1–Venus degradation in C were analyzed as in Fig. 1 G. (E) The timing of Cyclin B1–Venus degradation from NEBD in C was analyzed plotted as a box and whisker chart. (F) Cyclin B1 destruction in the absence of BubR1 is accelerated by reducing Mad2 binding to Cdc20. HeLa cells expressing 3×Flag-Cdc20, wild type, or the R132A mutant were treated with siRNA against BubR1, and Cyclin B1 destruction was assayed as in C. n = number of cells analyzed in three independent experiments. (G) The rate of Cyclin B1–Venus degradation in E was measured and plotted as in D. The center lines are the medians, boxes correspond to the range between 25 and 75% of all the data, and the whiskers correspond to the minimum and maximum of all the data. CTR, control; IP, immunoprecipitation.
To assay the effect of Cdc20 binding to Mad2 on the activity of the APC/C, we analyzed the rate of destruction of Cyclin B1–Venus in cells in which we depleted Mad2, BubR1, Mad2 and BubR1, or p31comet and BubR1 (Fig. 5, C–E, quantified in D and E). Consistent with a role for Mad2 as an inhibitor, the rate of destruction of Cyclin B1 in BubR1-depleted cells increased when we codepleted Mad2 (Fig. 5, C and D) and decreased when we codepleted p31comet, accompanied by an increase in the proportion of Cdc20 bound to Mad2 (P = 0.0205). These results were consistent with the possibility that Mad2 contributed to the inhibition of Cdc20 in parallel with BubR1 in vivo (with the caveat that siRNA treatment might have depleted BubR1 to different extents in the different experiments and that the exact mechanism by which p31comet affects Mad2 has not been definitively determined).
Finally, we tested the converse condition by depleting BubR1 to leave Mad2 as the sole inhibitor in the cell and compared the activity of wild-type Cdc20 with Cdc20R132A, which could not make a stable complex with Mad2 in vivo (Fig. 1 D; although this mutant does bind to recombinant Mad2 in vitro [Fig. 4 A]). This showed that Cyclin B1 was degraded more quickly in cells expressing Cdc20R132A than in cells with wild-type Cdc20 (Fig. 5, F and G, quantification, P < 0.0001), supporting the conclusion that Mad2 inhibited Cdc20 in parallel with BubR1 in vivo.
Our results have implications for understanding of how Cdc20 activates the APC/C. Previous studies identified two APC/C-binding motifs, the C box and the C-terminal dipeptide IR tail (Schwab et al., 2001; Vodermaier et al., 2003), which are conserved in Cdh1. To these, we can add the KILR motif, which appears to be specific to Cdc20, perhaps because of its additional role in the SAC. All three motifs are needed for Cdc20 to bind Cdc20 the APC/C in metaphase and in vitro because mutating any one motif is sufficient to impair the interaction. Current evidence indicates that the IR tail interacts with APC3 (Vodermaier et al., 2003) and the C box binds to APC2 in yeast (Thornton et al., 2006) or to human APC3 (Kraft et al., 2005). It is unclear which APC/C subunit recognizes the KILR motif, but APC8 is a strong candidate (Matyskiela and Morgan, 2009; Izawa and Pines, 2011).
Recent structural data add credence to the idea that Cdc20 binds next to APC10 to form a bipartite degron receptor. A putative D box binding site on Cdc20 was identified in fission yeast MCC as a conserved channel between blades 1 and 7 on the rim of the WD40 domain (Chao et al., 2012). When modeled onto the pseudoatomic structure of the APC/C, this site would be correctly positioned in relation to APC10 to form a degron receptor (Buschhorn et al., 2011; da Fonseca et al., 2011; Schreiber et al., 2011). It is, however, unclear why Cdc20 should require three different motifs to bind in this position, unless binding to multiple subunits on the same complex alters the conformation of the APC/C and thereby induces activity.
Several early studies showed that Mad2 directly binds to Cdc20 and can inhibit APC/CCdc20 in vitro (Fang et al., 1998; Hwang et al., 1998; Kallio et al., 1998; Kim et al., 1998; Yang et al., 2008). Recently, it was demonstrated that Mad2 alone can inhibit Cdc20 in budding yeast but only when the two proteins are tethered together; under normal conditions, the binding between Mad2 and Cdc20 is stabilized by Mad3 (Lau and Murray, 2012). The mechanism by which Mad2 inhibited APC/CCdc20, however, was not identified. We show here that Mad2 and the APC/C compete for the same binding site on Cdc20, and the structure of the MCC shows that this site on Cdc20 is bound tightly by the “safety belt” of Mad2, which would prevent it interacting with the APC/C (Chao et al., 2012). Therefore, our identification of the KILR motif as a site that can be bound by Mad2 or the APC/C indicates a mechanism during MCC assembly by which Mad2 would ensure that Cdc20 cannot bind to the APC/C at the same time as promoting binding to BubR1.
Materials and methods
Cell culture and synchronization
HeLa cells were maintained in Advanced DME with 10% FBS. For synchronization at the beginning of S-phase HeLa cells, 2.5 mM thymidine was added to the culture medium for 24 h (Izawa and Pines, 2011). For prometaphase enrichment, cells were released from a thymidine block and, 6 h later, treated with nocodazole at a final concentration of 0.1 ng/µl for 6–12 h. For SAC inactivated samples, cells were released from a nocodazole block into medium including 1 µM reversine and 10 µM MG132 for a further 1 h.
Transfection of DNA and siRNA
The following ON-TARGETplus (Thermo Fisher Scientific) oligonucleotides (oligos) were used: Cdc20, 5′-CGGAAGACCUGCCGUUACAUU-3′; Mad2, 5′-GGAAGAGUCGGGACCACAGUU-3′; BubR1, 5′-GAUGGUGAAUUGUGGAAUA-3′; APC3, 5′-GGAAAUAGCCGAGAGGUAAUU-3′; p31comet, 5′-UUCUUCGGACUUCUCAUACCACUCC-3′; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; D-001830-01). Cells were transfected with 20–100 nM oligos using Oligofectamine or Lipofectamine RNAiMAX (Invitrogen). For transfection of DNA plasmid and siRNA oligos at the same time, Lipofectamine 2000 was used following the manufacturer’s instructions.
Microscopy
Before imaging, the culture medium was replaced with Leibovitz’s L-15 medium (Gibco) supplemented with 10% FBS and penicillin/streptomycin. HeLa cells were imaged using a 40×, 1.35 NA Plan Apochromat lens on a microscope (DeltaVision Core; GE Healthcare) equipped with an EM charge-coupled device (CCD) camera (Cascade II; Photometrics) an environmental chamber at 37°C (Applied Precision) or in Delta T dishes at 37°C with a 40× Plan Apochromat 1.25 NA lens on a microscope (DMIRB; Leica) equipped with an EM CCD camera (QuantEM 512C; Photometrics) and Lambda LS illumination (Sutter Instrument) as previously described (Izawa and Pines, 2011). For Cyclin B1–Venus destruction assays, images were captured at 1.5- or 3-min intervals using softWoRx (for the DeltaVision; Applied Precision) or SlideBook (Intelligent Imaging Innovations) software, and the fluorescence intensities were measured and analyzed using ImageJ software (National Institutes of Health) as previously described (Izawa and Pines, 2011). In brief, a region of interest was drawn around a cell, and the total fluorescence was measured using ImageJ software. This value was divided by the area, and after background subtraction, the value at NEBD was set to 1. Statistical analysis was performed using Prism 4 or 5 software (GraphPad Software). The rate of Cyclin B1 destruction was determined using Prism software by nonlinear regression analysis assuming a sigmoidal dose–response (variable slope). Significance was determined using a two-tailed Student’s t test.
Inducible cell line
A HeLa–flippase recognition target cell line (gift of S. Taylor, University of Manchester, Manchester, England, UK) was transfected using the Flp-In system (Invitrogen) to generate stable inducible cell lines using the ORF of siRNA-resistant Cdc20 generated by DNA 2.0 (Nilsson et al., 2008) and cloned into a modified version of pcDNA5/flippase recognition target/tetracycline on (Invitrogen). To induce Cdc20 expression, cells were treated with 1 µg/ml tetracycline (EMD Millipore) 36 h before harvesting.
Immunoprecipitation and size-exclusion chromatography
Protein complexes were immunoprecipitated with antibodies covalently coupled to Dynabeads (Invitrogen) using Hepes buffer (150 mM KaCl, 40 mM Hepes, pH 7.8, 10 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM DTT, inhibitor cocktail tablet [Complete; Roche], 0.2 µM microcystin, and 1 mM PMSF) for incubation and washing. Cells for immunoprecipitation were lysed with Hepes buffer for 10 min on ice and clarified by a 20,000 g spin for 10 min. For size-exclusion chromatography analysis, cells were resuspended in buffer A (140 mM NaCl, 30 mM Hepes, pH 7.8, 6 mM MgCl2, 5% glycerol, 1 mM DTT, Complete inhibitor cocktail tablet, 0.2 µM microcystin, and 1 mM PMSF) at a 1:1 ratio of buffer to cells and lysed by nitrogen cavitation (1,000 lbs/in2 for 30 min; Parr Instrument). Lysed cells were centrifuged at 20,000 g for 10 min and 259,000 g for 10 min before loading onto a Superose 6 PC 3.2/30 column (GE Healthcare). The column was run at a flow rate of 25 µl/min−1 in buffer B (140 mM NaCl, 30 mM Hepes, pH 7.8, 5% glycerol, and 1 mM DTT), and 100 µl fractions were collected.
Antibodies
The following antibodies were used at the indicated dilutions: Cdc20 (sc-13162; Santa Cruz Biotechnology, Inc.) 1:500, Cdc20 (A301-180A; Bethyl Laboratories, Inc.) 1:500, BubR1 (A300-386A; Bethyl Laboratories, Inc.), Mad2 (A300-301A; Bethyl Laboratories, Inc.) 1:500, p31comet (clone E29.19.14; a gift of A. Musacchio, Max Plank Institute of Molecular Physiology, Dortmund, Germany) 1:200, Bub3 (611730; BD) 1:500, Cyclin A (mAb AT10.3; Cancer Research UK) 1:1,000, APC3 (610455; BD) 1:500, APC4 (monoclonal antibody raised against a C-terminal peptide) 1:500, APC6 (sc-6395; Santa Cruz Biotechnology, Inc.), GST (sc-138; Santa Cruz Biotechnology, Inc.) 1:500, and anti-Flag epitope (M2; Sigma-Aldrich) 1:5,000. For secondary antibodies, Alexa Flour 680 rabbit anti–goat (A21088; Invitrogen), IRDye 680 donkey anti–mouse (926–322227; LI-COR Biosciences), Alexa Fluor 680 goat anti–rabbit (A21076; Invitrogen), IRDye 800CW donkey anti–mouse (926–32212; LI-COR Biosciences), and IRDye 800CW donkey anti–rabbit (926–32213; LI-COR Biosciences) were all used at 1:10,000.
Quantitative immunoblotting
After blotting with primary antibodies, blots were incubated with fluorescently labeled secondary antibodies, and the fluorescence was measured using a CCD scanner (Odyssey; LI-COR Biosciences) according to the manufacturer’s instructions.
Protein expression and purification
His6-Mad2 was expressed in BL21 (DE3) RIL cells at 37°C and purified by nickel affinity chromatography (QIAGEN) followed by size-exclusion chromatography on a Superdex 75 column. Wild-type Cdc20, Cdc20RA, and Cdc20ΔKILR were cloned into pFAST–bacterial artificial chromosome vector, were expressed in Sf9 cells according to the manufacturer’s instructions (Invitrogen), and purified by nickel affinity chromatography.
In vitro ubiquitylation assays
In vitro ubiquitylation assays were performed as described previously (Garnett et al., 2009) except for the preparation of the APC/C. In brief, HeLa cells were treated with siRNA against Cdc20 for 48 h, arrested at prometaphase by nocodazole treatment, and harvested by mitotic shake. Cell extracts were prepared by nitrogen cavitation (see Immunoprecipitation and size-exclusion chromatography), and the APC/C was purified using an anti-APC3 (AF3.1) antibody and elution with the peptide antigen (CMTDADDTQLHAAESDEF) in buffer A. Ubiquitylation reactions contained E1 ligase, UbcH10, Cdc20, ubiquitin, ATP, ATP regenerating system, and securin as a substrate in QA buffer (100 mM NaCl, 30 mM Hepes, pH 7.8, 2 mM ATP, 2 mM MgCl2, 0.1 µg/µl BSA, and 1 mM DTT). Samples were incubated at 37°C for 30 min and stopped by adding SDS sample buffer.
APC/C binding and competition assays
GST-Cdc20N151 was purified from Escherichia coli extract with glutathione–Sepharose 4B (GE Healthcare) for 40 min at 4°C in PBS with 0.2% NP-40. After washing twice with 0.2% NP-40 PBS, the beads were incubated with mitotic HeLa extract in Hepes buffer for 1 h at 4°C and washed three times with Hepes buffer before analysis. To prebind Mad2 in Fig. 4 D, His6-Mad2 was added into E. coli extract expressing GST-Cdc20N151 before purification on glutathione–Sepharose 4B.
Online supplemental material
Fig. S1 shows that the inducible cell lines express similar amounts of Cdc20 and that the Cdc20ΔKILR mutant binds to Cyclin A. Fig. S2 shows that reversine causes the MCC to dissociate from the APC/C, that APC3 is required to bind in vitro translated Cdc20, and that wild type but not the ΔKILR mutant cannot bind to the APC/C when analyzed by size-exclusion chromatography. Fig. S3 shows that a Mad2 mutant that does not bind to p31comet still requires BubR1 to delay mitosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205170/DC1.
Acknowledgments
We thank Andrea Musacchio and Steven Taylor for reagents, Jörg Mansfeld for extensive discussions, and all members of the laboratory for constructive criticisms.
D. Izawa was supported by projects grants from the American Institute for Cancer Research and subsequently Cancer Research UK, and J. Pines was supported by a program grant from Cancer Research UK. J. Pines acknowledges the core funding provided by the Wellcome Trust (092096) and Cancer Research UK (C6946/A14492).
Footnotes
Abbreviations used in this paper:
APC/C
anaphase-promoting complex/cyclosome
CCD
charge-coupled device
DIC
differential interference contrast
GAPDH
glyceraldehyde 3-phosphate dehydrogenase
MCC
mitotic checkpoint complex
NEBD
nuclear envelope breakdown
SAC
spindle assembly checkpoint
References
- Burton J.L., Solomon M.J. 2007. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 21:655–667 10.1101/gad.1511107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn B.A., Petzold G., Galova M., Dube P., Kraft C., Herzog F., Stark H., Peters J.M. 2011. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat. Struct. Mol. Biol. 18:6–13 10.1038/nsmb.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. 2012. Structure of the mitotic checkpoint complex. Nature. 484:208–213 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- da Fonseca P.C.A., Kong E.H., Zhang Z., Schreiber A., Williams M.A., Morris E.P., Barford D. 2011. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 470:274–278 10.1038/nature09625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J., Harris L.D., Goorha R. 2006. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp. Cell Res. 312:1831–1842 10.1016/j.yexcr.2006.02.018 [DOI] [PubMed] [Google Scholar]
- Di Fiore B., Pines J. 2010. How cyclin A destruction escapes the spindle assembly checkpoint. J. Cell Biol. 190:501–509 10.1083/jcb.201001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M.L., Benezra R., Sorger P.K. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 101:635–645 10.1016/S0092-8674(00)80875-2 [DOI] [PubMed] [Google Scholar]
- Elowe S., Dulla K., Uldschmid A., Li X., Dou Z., Nigg E.A. 2010. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J. Cell Sci. 123:84–94 10.1242/jcs.056507 [DOI] [PubMed] [Google Scholar]
- Fang G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 13:755–766 10.1091/mbc.01-09-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M.W. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871–1883 10.1101/gad.12.12.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M.J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A.R. 2009. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 11:1363–1369 10.1038/ncb1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Skaar J.R., Pagano M. 2009. APC/C- and Mad2-mediated degradation of Cdc20 during spindle checkpoint activation. Cell Cycle. 8:167–171 10.4161/cc.8.1.7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan R.S., Manak M.S., Buch H.K., Meier M.G., Meraldi P., Shah J.V., Sorger P.K. 2011. p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol. Biol. Cell. 22:4236–4246 10.1091/mbc.E11-03-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W. 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 279:1041–1044 10.1126/science.279.5353.1041 [DOI] [PubMed] [Google Scholar]
- Izawa D., Pines J. 2011. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat. Cell Biol. 13:223–233 10.1038/ncb2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Li B., Warrington R.T., Hao X., Wang S., Yu H. 2011. Defining pathways of spindle checkpoint silencing: functional redundancy between Cdc20 ubiquitination and p31(comet). Mol. Biol. Cell. 22:4227–4235 10.1091/mbc.E11-05-0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M., Weinstein J., Daum J.R., Burke D.J., Gorbsky G.J. 1998. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 141:1393–1406 10.1083/jcb.141.6.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Pines J. 2010. Centromere tension: a divisive issue. Nat. Cell Biol. 12:919–923 10.1038/ncb1010-919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T. 1998. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 279:1045–1047 10.1126/science.279.5353.1045 [DOI] [PubMed] [Google Scholar]
- Kimata Y., Baxter J.E., Fry A.M., Yamano H. 2008. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol. Cell. 32:576–583 10.1016/j.molcel.2008.09.023 [DOI] [PubMed] [Google Scholar]
- Kops G.J., van der Voet M., Manak M.S., van Osch M.H., Naini S.M., Brear A., McLeod I.X., Hentschel D.M., Yates J.R., III, van den Heuvel S., Shah J.V. 2010. APC16 is a conserved subunit of the anaphase-promoting complex/cyclosome. J. Cell Sci. 123:1623–1633 (published erratum appears in J. Cell Sci. 2010. 123:1815) 10.1242/jcs.061549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Vodermaier H.C., Maurer-Stroh S., Eisenhaber F., Peters J.M. 2005. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell. 18:543–553 10.1016/j.molcel.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau D.T., Murray A.W. 2012. Mad2 and Mad3 cooperate to arrest budding yeast in mitosis. Curr. Biol. 22:180–190 10.1016/j.cub.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Murray A.W. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531 10.1016/0092-8674(81)90015-5 [DOI] [PubMed] [Google Scholar]
- Luo X., Fang G., Coldiron M., Lin Y., Yu H., Kirschner M.W., Wagner G. 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7:224–229 10.1038/73338 [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 9:59–71 10.1016/S1097-2765(01)00435-X [DOI] [PubMed] [Google Scholar]
- Maciejowski J., George K.A., Terret M.-E., Zhang C., Shokat K.M., Jallepalli P.V. 2010. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 190:89–100 10.1083/jcb.201001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J., Collin P., Collins M.O., Choudhary J.S., Pines J. 2011. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat. Cell Biol. 13:1234–1243 10.1038/ncb2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela M.E., Morgan D.O. 2009. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol. Cell. 34:68–80 10.1016/j.molcel.2009.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Morrow C.J., Tighe A., Johnson V.L., Scott M.I., Ditchfield C., Taylor S.S. 2005. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J. Cell Sci. 118:3639–3652 10.1242/jcs.02487 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Yekezare M., Minshull J., Pines J. 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10:1411–1420 10.1038/ncb1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. 2011. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12:427–438 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- Rahmani Z., Gagou M.E., Lefebvre C., Emre D., Karess R.E. 2009. Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 187:597–605 10.1083/jcb.200905026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A., Stengel F., Zhang Z., Enchev R.I., Kong E.H., Morris E.P., Robinson C.V., da Fonseca P.C., Barford D. 2011. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 470:227–232 10.1038/nature09756 [DOI] [PubMed] [Google Scholar]
- Schwab M., Neutzner M., Möcker D., Seufert W. 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20:5165–5175 10.1093/emboj/20.18.5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczaniecka M., Feoktistova A., May K.M., Chen J.S., Blyth J., Gould K.L., Hardwick K.G. 2008. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C). J. Biol. Chem. 283:23039–23047 10.1074/jbc.M803594200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K.T., Musacchio A. 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21:2496–2506 10.1093/emboj/21.10.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj R., Li B., Yu H. 2001. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1:227–237 10.1016/S1534-5807(01)00019-3 [DOI] [PubMed] [Google Scholar]
- Teichner A., Eytan E., Sitry-Shevah D., Miniowitz-Shemtov S., Dumin E., Gromis J., Hershko A. 2011. p31comet promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc. Natl. Acad. Sci. USA. 108:3187–3192 10.1073/pnas.1100023108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B.R., Ng T.M., Matyskiela M.E., Carroll C.W., Morgan D.O., Toczyski D.P. 2006. An architectural map of the anaphase-promoting complex. Genes Dev. 20:449–460 10.1101/gad.1396906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton A.R., Wang K., Link L., Bellizzi J.J., Huang H., Yen T., Liu S.T. 2011. BUBR1 and closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. J. Biol. Chem. 286:21173–21179 10.1074/jbc.M111.238543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varetti G., Guida C., Santaguida S., Chiroli E., Musacchio A. 2011. Homeostatic control of mitotic arrest. Mol. Cell. 44:710–720 10.1016/j.molcel.2011.11.014 [DOI] [PubMed] [Google Scholar]
- Vodermaier H.C., Gieffers C., Maurer-Stroh S., Eisenhaber F., Peters J.M. 2003. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 13:1459–1468 10.1016/S0960-9822(03)00581-5 [DOI] [PubMed] [Google Scholar]
- Wang Q., Liu T., Fang Y., Xie S., Huang X., Mahmood R., Ramaswamy G., Sakamoto K.M., Darzynkiewicz Z., Xu M., Dai W. 2004. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 103:1278–1285 10.1182/blood-2003-06-2158 [DOI] [PubMed] [Google Scholar]
- Westhorpe F.G., Tighe A., Lara-Gonzalez P., Taylor S.S. 2011. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J. Cell Sci. 124:3905–3916 10.1242/jcs.093286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R., Clay-Farrace L., van Zon W., Yekezare M., Koop L., Ogink J., Medema R., Pines J. 2008. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell. 30:290–302 10.1016/j.molcel.2008.02.027 [DOI] [PubMed] [Google Scholar]
- Yang M., Li B., Liu C.-J., Tomchick D.R., Machius M., Rizo J., Yu H., Luo X. 2008. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 6:e50 10.1371/journal.pbio.0060050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lees E. 2001. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell. Biol. 21:5190–5199 10.1128/MCB.21.15.5190-5199.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]