Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology (original) (raw)
. Author manuscript; available in PMC: 2012 Oct 19.
Abstract
Chloroplast genomes defied the laws of Mendelian inheritance at the dawn of plant genetics, and continue to defy the mainstream approach to biotechnology, leading the field in an environmentally friendly direction. Recent success in engineering the chloroplast genome for resistance to herbicides, insects, disease and drought, and for production of biopharmaceuticals, has opened the door to a new era in biotechnology. The successful engineering of tomato chromoplasts for high-level transgene expression in fruits, coupled to hyper-expression of vaccine antigens, and the use of plant-derived antibiotic-free selectable markers, augur well for oral delivery of edible vaccines and biopharmaceuticals that are currently beyond the reach of those who need them most.
Chloroplast transformation is an environmentally friendly approach to plant genetic engineering that minimizes out-crossing of transgenes to related weeds or crops [1,2] and reduces the potential toxicity of transgenic pollen to non-target insects [3]. Because the plastid genome is highly polyploid, transformation of chloroplasts permits the introduction of thousands of copies of foreign genes per plant cell, and generates extraordinarily high levels of foreign protein [3]. Chloroplast transformation vectors use two targeting sequences that flank the foreign genes and insert them, through homologous recombination, at a precise, predetermined location in the organelle genome (Fig. 1). This results in uniform transgene expression among transgenic lines and eliminates the ‘position effect’ often observed in nuclear transgenic plants. Gene silencing, frequently observed in nuclear transgenic plants, has not been observed in genetically engineered chloroplasts. The ability to express foreign proteins at high levels in chloroplasts and chromoplasts, and to engineer foreign genes without the use of antibiotic resistant genes [4,5],make this compartment ideal for the development of edible vaccines [6]. Moreover, the ability of chloroplasts to form disulfide bonds and to fold human proteins has opened the door to high-level production of biopharmaceuticals in plants [7]. Furthermore, foreign proteins observed to be toxic in the cytosol are non-toxic when accumulated within transgenic chloroplasts [6,8]. Chloroplast and nuclear genetic engineering are compared in Table 1.
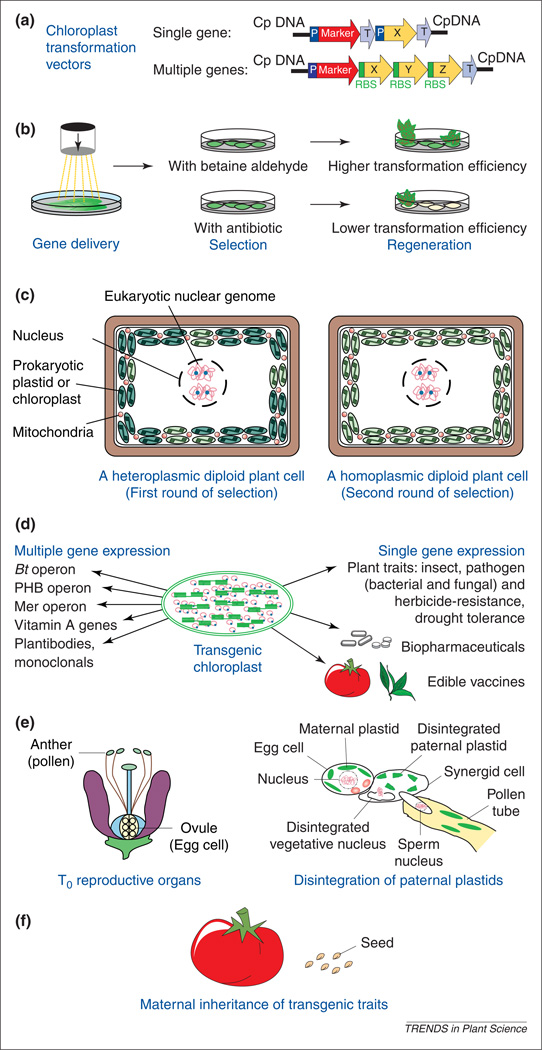
Fig. 1.
Various steps in chloroplast genetic engineering. (a) In single-gene (X) chloroplast transformation vectors, coding sequences are flanked by independent 5′ and 3′ regulatory regions, which encompass promoters (P) and terminators (T), respectively. In multiple gene (X, Y, Z) constructs, a single promoter (P) regulates expression of the operon, and individual ribosome binding sites (RBS) are engineered upstream of each open-reading-frame. (b) Gene delivery is usually performed by particle bombardment of chloroplast vectors. Antibiotic selection is performed with spectinomycin or streptomycin, whereas betaine aldehyde is used for plant-derived antibiotic-free selection. (c) The first round of selection generally results in heteroplasmy (left) whereas the second round achieves homoplasmy (right). (d) High-level expression of all the single and multiple gene traits listed here have been achieved, except for PHB and vitamin A operons. This is because of high ploidy of transgenes (5000–10 000 copies per cell). (e) In T0 reproductive organs, after meiosis, haploid egg and sperm cells are formed (left). The zygote contains only maternal plastids because the paternal plastids disintegrate in the synergid cell (right). Maternal inheritance of transgenes offers biological containment because of lack of gene flow through pollen. Transgenic pollen generally does not contain the foreign gene product. (f) When transgenic seeds are germinated on selectable agents, 100% germination is observed. In contrast with Mendelian segregation of traits, chloroplast transgenes are maternally transmitted without any segregation.
Table 1.
Comparison of chloroplast and nuclear genetic engineering
| Transgenic | Chloroplast genome | Nuclear genome |
|---|---|---|
| Transgene copy number | 10–100 plastid genome (single circular chromosome) copies per plastid, and 10–100 plastids per cell depending upon age and type of tissue, resulting in many transgene copies (up to 10 000) per cell | Chromosome number is species-specific, but two copies of each of many chromosomes present per cell results in a few copies of the transgene per cell |
| Level of gene expression | Polyploidy results in abundant transgene transcripts and a high accumulation of foreign proteins (up to 47% of total soluble protein) | Gene regulation determines the rate of transcription, and accumulation of foreign protein is often a limitation |
| Gene arrangement and transcription | Genes are often arranged in operons and transcribed into polycistronic RNA so that multiple transgenes can be introduced and expressed in a single transformation event | Each transgene is independently inserted into the chromosome and transcribed into a monocistronic mRNA |
| Position effect | Site-specific insertion through two homologous recombination events eliminates position effects on transgene expression | Random insertions result in variable transgene expression levels |
| Gene silencing | Not reported | Gene silencing results in decrease or elimination of transgene expression. Both transcriptional and post-transcriptional gene silencing have been reported |
| Gene containment | Maternal gene inheritance in most crop plants results in natural gene containment | Paternal transgene inheritance results in out-crossing among crops and weeds |
| Folding and disulfide bond formation | Chloroplasts form disulfide bonds and correctly fold human proteins, making them ideal for development of edible vaccines, pharmaceuticals and plantibodies | For disulfide bond formation, proteins are targeted to the endoplasmic reticulum |
| Toxicity of foreign proteins | Adverse effects of toxic proteins might be minimized by chloroplast compartmentalization | Toxic proteins accumulating within the cytosol might result in serious pleiotropic effects |
| Transgenic lines | Uniform gene expression | Highly variable gene expression |
| Homogeneity at ploidy level | Chloroplast transgenic lines are mostly homoplasmic (all genome copies are homogeneous for the transgene). Homoplasmy is mostly achieved by repetitive selection and regeneration | Nuclear transgenic lines are either heterozygous or homozygous. Homozygosity is achieved either by selfing or crossing |
Brief history of chloroplast genetic engineering
When the concept of chloroplast genetic engineering was developed in the 1980s, it was possible to introduce isolated intact chloroplasts into protoplasts and regenerate transgenic plants. Therefore, early investigations of chloroplast transformation in vascular plants focused on the development of chloroplast systems capable of efficient, prolonged protein synthesis and expression of foreign genes [9]. The development of the gene gun as a transformation device by John Sanford (reviewed in Ref. [10]), enabled plant chloroplasts to be transformed without the use of isolated plastids. Chloroplast genetic engineering was accomplished through autonomously replicating chloroplast vectors in dicot plastids [11] and transient expression in monocot plastids [12], and by stable integration of selectable marker genes into the chloroplast genomes of Chlamydomonas reinhardtii and tobacco using the gene gun [13,14]. However, it is only recently that genes conferring agronomically valuable traits have been introduced via chloroplast genetic engineering. For example, integrating the cry genes into the chloroplast genome generated plants that were insecticidal to Bacillus thuringiensis (Bt)-sensitive [15] and highly resistant insects [16]. Chloroplasts have also been engineered recently to generate plants tolerant to bacterial and fungal diseases [17], drought [8] or herbicides [2,18–20]. Transgenes recently engineered via the chloroplast genome are listed in Table 2.
Table 2.
Foreign gene expression in chloroplasts of higher plantsa
| Genes and use | Gene products and use | Refs |
|---|---|---|
| Selectable markers and reporters | ||
| aadA | Aminoglycoside-3′-adenylyltransferase | [14] |
| nptII | Neomycin phosphotransferase | [52] |
| codA | Cytosine deaminase | [53] |
| BADH | Betaine aldehyde dehydrogenase | [4] |
| uidA | β-glucoronidase | [12] |
| cat | Chloramphenicol acetyl transferase | [9,11] |
| gfp | Green fluorescent protein | [24,54] |
| aadA:gfp | Selectable or screenable fusion protein | [47] |
| Plant traits: herbicide resistance | ||
| aroA | Glyphosate resistance | [2,19] |
| bar | Bialaphos resistance | [18,20] |
| Insect resistance | ||
| Cry1Ac | Bacillus thuringiensis (Bt) toxin | [15] |
| Cry2Aa2 | Bacillus thuringiensis (Bt) toxin | [16] |
| Cry2Aa2 operon | Bacillus thuringiensis (Bt) toxin | [3] |
| Pathogen resistance | ||
| msi-99 | Bacterial, fungal resistance | [17] |
| Drought or salt tolerance | ||
| tps1 | Trehalose phosphate synthase | [8] |
| BADH | Betaine aldehyde dehydrogenase | [4] |
| Amino acid biosynthesis | ||
| EPSPS | 5-enol-pyruvyl shikimate-3-phosphate synthase | [2,19] |
| ASA2 | Anthranilate synthase (AS) α-subunit | [55] |
| Phytoremediation | ||
| mer A | Mercuric ion reductase | b |
| mer B | Organomercurial lyase | b |
| Non-plant traits: biopharmaceuticals | ||
| hST | Human somatotropin | [7] |
| HSA | Human serum albumin | c |
| msi 99 | Anticancer, lytic antibiotic | [17] |
| proinsulin | Human insulin α, β chains | d |
| IFN α5 | Human interferon α5 | e |
| Monoclonals | ||
| Guy's 13 | For dental caries against Streptococcus mutans | c |
| Biomedical polymer | ||
| gvgvp-120 | Bioelastic protein-based polymer | [21] |
| Edible vaccines | ||
| ctxB | Cholera toxin β-subunit | [6] |
Exceptionally high accumulation of foreign proteins (up to 46% of total soluble protein) has been reported recently for chloroplast transgenes [3]. This feature should make the compartment ideal for low-cost production of biopharmaceuticals. Stable expression of a pharmaceutical protein in tobacco chloroplasts was first reported for GVGVP, a protein-based polymer with varied medical applications [21], and subsequently for human somatotropin [7]. Additionally, chloroplast genomes have been engineered to express pharmaceutical peptides [17], human serum albumin (HSA), monoclonals (H. Daniell et al., www.publish.csiro.au/books/bookpage.cfm?PID=3051&TXT=DES#DES) and antigens composed of oligomeric proteins with stable disulfide bridges [6]. Several recent advances should make chloroplasts even more attractive as biopharmaceutical reactors. These include engineering multiple foreign genes as operons [3]; transformation without use of antibiotic markers [4,5]; elimination of resistance genes subsequent to transformation [18,22,23]; and the transformation of edible crops such as potato and tomato [24,25]. This review highlights these and other recent achievements.
Engineering the chloroplast genome for herbicide resistance
Glyphosate is a potent, broad-spectrum herbicide that is highly effective against grasses and broad-leaf weeds. Glyphosate works by competitive inhibition of an enzyme in the aromatic amino acid biosynthetic pathway, 5-enol-pyruvyl shikimate-3-phosphate synthase (EPSPS). Unfortunately, like most commonly used herbicides, glyphosate does not distinguish crops from weeds, thereby restricting its use. Engineering crop plants for resistance to the herbicide is a standard strategy to overcome the lack of herbicide selectivity. However, this approach raises the concern that if the engineered resistance gene escapes via pollen dispersal, it might result in resistant weeds or might cause genetic pollution among other crops [26,27]. Engineering foreign genes through chloroplast genomes (which are maternally inherited in most crops) offers a solution to this problem. Although pollen from plants with maternal chloroplast inheritance can contain metabolically active plastids, the plastid DNA is lost during pollen maturation and hence is not transmitted to the next generation [4]. In addition, the target proteins for many herbicides are compartmentalized within the chloroplast. The chloroplast genome can be engineered to confer herbicide resistance by expressing a petunia EPSPS nuclear gene via the chloroplast genome [2]. The resultant transgenic plants are resistant to tenfold higher levels of glyphosate than the lethal dosage, and the transgene is maternally inherited.
Recently, the Agrobacterium EPSPS gene (C4) was expressed in tobacco plastids and resulted in 250-fold higher levels of the glyphosate-resistant C4 protein than were achieved via nuclear transformation [19]. Even though C4 expression in plastids was enhanced more than nuclear expression levels, field tolerance to glyphosate remained the same, showing that higher levels of expression do not always proportionately increase herbicide tolerance. In two different studies, transgenic tobacco plants expressing bar genes via the chloroplast genome exhibited field-level tolerance to phosphinothricin (PPT) [18,20]. In this case, even plants with the lowest levels of bar expression were resistant to the highest levels of PPT tested, and no pollen transmission of the transgene was detected. However, high-level expression of bar genes in the chloroplast was not sufficient to allow direct selection of chloroplast transformants on medium containing PPT [20].
Engineering bacterial operons via chloroplast genomes
Typical plant nuclear mRNAs are monocistronic (i.e. only one polypeptide chain is translated per messenger molecule). This poses a serious drawback when engineering multiple genes [28]. For example, to express the polyhydroxybutyrate polymer or Guy’s 13 antibody, single genes were first introduced into individual plants, which were then backcrossed to reconstitute the entire pathway or the complete protein [29,30]. Similarly, in a seven-year effort, three genes were introduced in rice to generate a biosynthetic pathway for β-carotene expression [31]. By contrast, most chloroplast genes are co-transcribed as polycistronic RNAs (i.e. they encode multiple proteins that are separately translated from the same mRNA molecule), which are subsequently processed to form translatable transcripts [28]. Therefore, introduction of multiple chloroplast transgenes arranged in an operon should allow expression of entire pathways in a single transformation event.
Recently, the _Bt cry_2Aa2 operon was used as a model system to test the feasibility of multigene operon expression in engineered chloroplasts [3]. _Cry_2Aa2 is the distal gene of a three-gene operon. The open-reading-frame immediately upstream of _cry_2Aa2 encodes a putative chaperonin that facilitates the folding of Cry proteins into stable cuboidal crystals [32]. Operon-derived Cry2Aa2 protein accumulates in transgenic chloroplasts as cuboidal crystals, to a level of 45.3% of the total soluble protein and remains stable even in senescing leaves (46.1%). These data suggest that chaperonin-mediated folding might assist large-scale production of foreign proteins within chloroplasts. The crystals enhance protein stability and facilitate single-step purification. This is the highest level of foreign gene expression ever reported in transgenic plants, killing insects that are exceedingly difficult to control. Importantly, this study also showed that there was no insecticidal protein present in pollen, thus eliminating potential harm to non-target insects. This first demonstration of bacterial operon expression in transgenic plants opens the door to engineer novel pathways in a single transformation event. Subsequently, the mer operon has been used to express mercuric ion reductase and organomercurial lyase to achieve phytoremediation of mercury (O. Ruiz, MSthesis, University of Central Florida, 2001).
Engineering the chloroplast genome for pathogen resistance
Because plant diseases have plagued global crop production, it is highly desirable to engineer plants that are resistant to pathogenic bacteria and fungi. Amphipathic peptides such as magainin have known antimicrobial properties [33], but their possible uses in agriculture have not been fully explored yet. Recently, a synthetic antimicrobial peptide (MSI-99) was expressed via the chloroplast genome [17]. MSI-99 is an amphipathic α-helical molecule with affinity for the negatively charged phospholipids found in the outer-membrane of bacteria and fungi. Upon contact with these membranes, individual peptides aggregate to form pores, resulting in microbial lysis. Because of the concentration-dependent action of antimicrobial peptides, MSI-99 was expressed via the chloroplast genome to accomplish high-dose release at the point of infection. In vitro and in planta assays with T0, T1 and T2 plants confirmed that the peptide was expressed at high levels (up to 21.5% total soluble protein) and retained biological activity against Pseudomonas syringae, a major plant pathogen. In addition, leaf extracts from transgenic plants inhibited the growth of pre-germinated spores of three fungal species Aspergillus flavus, Fusarium moniliforme and Verticillium dahliae by >95% compared with untransformed controls – these observations were confirmed by in planta assays. Importantly, growth and development of the transgenic plants were unaffected by hyper-expression of MSI-99 within chloroplasts. Because the outer membrane is an essential and highly conserved part of all microbial cells, microorganisms are unlikely to develop resistance against these peptides. Therefore, these results give a new option in the battle against phytopathogens.
Pharmaceutical companies are exploring the use of lytic peptides as broad-spectrum topical and systemic antibiotics. Previous studies have reported that analogs of magainin are effective against hematopoietic, melanoma, sarcoma and ovarian teratoma lines. Cystic fibrosis makes patients susceptible to respiratory infections, especially multidrug-resistant Pseudomonas aeruginosa. When MSI-99 was tested against P. aeruginosa in vitro, cell-free extracts of T1 chloroplast transgenic plants displayed a 96% level of growth inhibition against P. aeruginosa (H. Daniell et al., www.publish.csiro.au/books/bookpage.cfm?PID=3051&TXT=DES#DES). These studies show that MSI-99 has the potential to become an alternative to current antibiotic treatments for microbial infections.
Engineering the chloroplast genome for drought tolerance
Water stress caused by drought, salinity or freezing is a major limiting factor in plant growth and development. Trehalose is a non-reducing disaccharide of glucose whose synthesis is mediated by the trehalose-6-phosphate (T6P) synthase and trehalose-6-phosphate phosphatase complex in Saccharomyces cerevisiae. Because it accumulates under stress conditions such as freezing, heat, salt or drought, there is general consensus that trehalose protects against damage imposed by these stresses [34]. Therefore, engineering high levels of trehalose in plants might confer drought tolerance.
Gene containment in transgenic plants is a serious concern when plants are genetically engineered for drought tolerance because of the possibility of creating drought-tolerant weeds and passing on undesired pleiotropic traits to related crops. To prevent these consequences, it is desirable to engineer crop plants for drought tolerance via the chloroplast genome instead of the nuclear genome. Recently, the yeast trehalose phosphate synthase (TPS1) gene was introduced into the tobacco chloroplast and nuclear genomes to study the resultant phenotypes [8]. Although the chloroplast transgenic and the nuclear transgenic plants expressed significant TPS1 enzyme activity, chloroplast transgenic plants showed up to 25-fold higher accumulation of trehalose than nuclear transgenic plants. Nuclear transgenic plants (T0) with significant amounts of trehalose accumulation exhibited a stunted phenotype, sterility and other pleiotropic effects, whereas chloroplast transgenic plants (T1, T2 and T3) grew normally and had no visible pleiotropic effects. Chloroplast transgenic plants also showed a high degree of drought tolerance by growing in 6% polyethylene glycol, whereas respective control plants were bleached. Air-dried chloroplast transgenic seedlings and transgenic plants under extreme drought (not watered for 24 days) successfully rehydrated, whereas similarly treated control plants died. Investigations have confirmed that trehalose functions by protecting the integrity of biological membranes rather than regulating water potential. Therefore, this study shows that compartmentalization of trehalose within chloroplasts confers drought tolerance without undesirable phenotypes.
Engineering the chloroplast genome without antibiotic resistance genes
One disadvantage of chloroplast genetic engineering technology is the use of the antibiotic resistance gene, aadA, as a selectable marker. The aadA gene product inactivates its selective agents, spectinomycin and streptomycin, by transferring the adenyl moiety of ATP to the antibiotics. Because these antibiotics are commonly used to control bacterial infection in humans and animals, there is concern that their over-use might lead to the development of resistant bacteria. Therefore, several studies have explored strategies for engineering chloroplasts that are free of antibiotic-resistance markers. The spinach betaine aldehyde dehydrogenase (BADH) gene has been developed as a plant-derived selectable marker to transform chloroplast genomes [4]. The selection process involves conversion of toxic betaine aldehyde (BA) by the chloroplast-localized BADH enzyme to non-toxic glycine betaine, which also serves as an osmoprotectant [35]. Because the BADH enzyme is present only in chloroplasts of a few plant species adapted to dry and saline environments [35,36], it is suitable as a selectable marker in many crop plants. The transformation study showed that BA selection was 25-fold more efficient than spectinomycin, exhibiting rapid regeneration of transgenic shoots within two weeks. This is the first report of chloroplast engineering without the use of antibiotic resistance genes. Use of selectable markers that are naturally present in spinach, in addition to gene containment, should ease public concerns over genetically modified crops.
Another approach to develop marker-free transgenic plants is to eliminate the antibiotic resistance gene after transformation. Strategies have been developed to remove genes using endogenous chloroplast recombinases that delete the marker genes via engineered direct repeats [18]. Early experiments with Chlamydomonas reinhardtii showed that it is possible to exploit these recombination events to eliminate introduced selectable marker genes. Homologous recombination between two direct repeats, engineered to flank a selectable marker, enabled marker removal under non-selective growth conditions [37]. A similar approach applied in tobacco was effective in generating _aadA_-free T0 transplastomic lines while leaving a third unflanked transgene, bar, in the genome to confer herbicide resistance [18]. Recently, another strategy to eliminate selectable marker genes has been developed, using the P1 bacteriophage CRE-lox site-specific recombination system. In two separate studies, a marker gene flanked by lox-sites was introduced into the tobacco chloroplast genome. Its removal was subsequently induced by expressing the CRE protein from the nucleus [22,23]. These reports show that efficient removal of selectable marker(s) from chloroplast genomes is feasible.
Engineering the chloroplast genome to overproduce biopharmaceuticals
Although pharmaceutical proteins have been synthesized from plant nuclear transgenes, expression levels (particularly of human proteins) are generally disappointingly low [38]. Chloroplasts, with their highly polyploid genomes offer an ideal compartment for overproduction of foreign proteins. An additional significant advantage of using chloroplasts is their potential to process eukaryotic proteins, including folding and formation of disulfide bridges. Such folding and assembly can minimize the need for expensive in vitro processing of pharmaceutical proteins after their extraction. For example, 60% of the total production cost of human insulin is associated with in vitro processing [39].
Somatotropin (hST), a human therapeutic protein, is used in the treatment of hypopituitary dwarfism, Turner syndrome, chronic renal failure and HIV wasting syndrome. hST has been expressed in tobacco chloroplasts to levels of between 0.2 and 7.0% of total soluble protein in plants, depending upon the plastid translation signals used [7]. Chloroplast-expressed hST was shown to be correctly disulfide-bonded and biologically active. Recently, we have achieved high-level accumulation of HSA in transgenic tobacco chloroplasts in spite of the protein’s extreme susceptibility to proteolytic degradation (H. Daniell et al., www.publish.csiro.au/books/bookpage.cfm?PID=3051&TXT=DES#DES). HSA accounts for 60% of the total protein in blood serum and is the most widely used intravenous protein in several human therapies. In transgenic chloroplasts, HSA was expressed between 0.02% and 11.10% total soluble protein depending upon the regulatory signals used. HSA accumulation is so high that inclusion bodies increase the size of the transgenic chloroplasts, facilitating purification by centrifugation. This is the highest level of pharmaceutical protein expressed in transgenic chloroplasts, 500-fold higher than previous reports of HSA expression in nuclear transgenic plants.
Engineering the chloroplast genome to develop edible vaccines
Another major cost of biopharmaceutical production lies in purification; for example, in insulin production, chromatography accounts for 30% of the production cost and 70% of the set-up cost [39]. Therefore, oral delivery of properly folded and fully functional biopharmaceuticals should significantly cut down the production cost. Bioencapsulation of pharmaceutical proteins within plant cells offers protection against digestion in the stomach but allows successful delivery to the target tissues [40].
The β subunits of enterotoxigenic E. coli (LTB) and cholera toxin of Vibrio cholerae (CTB) are candidate vaccine antigens. Integration of an unmodified CTB gene into the tobacco chloroplast genome results in accumulation of up to 4.1% of total soluble leaf protein as functional CTB oligomers (410-fold higher than the unmodified LTB gene expressed via the nuclear genome) [6]. In addition, binding assays confirm that chloroplast-synthesized CTB binds to the intestinal membrane GM1-ganglioside receptor, indicating correct folding and disulfide bond formation of the plant-derived CTB pentamers. Increased production of an efficient transmucosal carrier molecule and delivery system, such as CTB, in transgenic chloroplasts makes plant-based oral pharmaceuticals commercially feasible. Because the quaternary structure of many proteins is essential for their function, this investigation shows the potential for other foreign multimeric proteins to be properly expressed and assembled in transgenic chloroplasts.
Engineering the chloroplast genome of edible crop plants
To exploit the chloroplast compartment for the production of orally delivered pharmaceuticals, it is important to extend chloroplast transformation to edible crops. For this reason, the recent development of chloroplast transformation for both potato [24] and tomato [25] is particularly exciting. Western analysis of transgenic potato tissues revealed that the chloroplast transgene (green fluorescent protein, GFP) was expressed to high levels in leaves (5.00% total soluble protein), but accumulated much lower levels in the microtubers (0.05% total soluble protein). Tomato plants with transformed plastids yielded more encouraging results; although the transgene (aadA) expression level was not quantified, western blot analysis showed that the expression levels in tomato fruits was about half that of leaves [25]. This study predicts the feasibility of expressing high-levels of foreign proteins in the plastids of edible plant organs.
Optimizing foreign gene expression in chloroplasts
Regulatory sequences have been analyzed in chloroplasts by fusion to reporter genes. This approach has identified strong chloroplast promoters as well as UTR sequences mediating RNA stability [41]. Because reporter protein levels do not always correlate with steady-state transcript abundance, it is evident that translation is a crucial control step for optimizing protein production. Several studies have revealed a minimal effect of 3′UTR sequences on chloroplast gene translation [42] although, recently, the petD 3′UTR was shown to influence reporter protein expression [43]. By contrast, many 5′UTRs mediate enhanced translation of downstream open-reading-frames [44]. One of the most effective of these, the tobacco psbA 5′UTR, directs strong, light-regulated, translation of chimeric transcripts regardless of which coding sequence, 3′UTR or promoter it is combined with [42].
Additional translational-enhancing elements have been engineered from non-plant sequences. For example, the bacteriophage T7 gene_10_ leader sequence (G_10_L), which enhances translation of foreign genes in bacteria, has the same effect in chloroplasts [7,19,21,45]. Accumulation of a herbicide-tolerant EPSPS protein was increased 200-fold when its chloroplast transgene included the G_10_L sequence [19]. This translational enhancement was attributed to complementarity between the G10L ribosome-binding site (rbs) and the anti-rbs on chloroplast 16S rRNA [7,19]. However, complementarity of a foreign transcript to 16S rRNA is not always beneficial. The first 24 nucleotides of the T7 gene 10 coding sequence are partially complementary to the chloroplast 16S rRNA. In bacteria, genes with this feature are highly translated. By contrast, in chloroplasts, fusion of these 24 nucleotides to a reporter sequence reduced its translation, and mutagenizing these nucleotides to improve their complementarity to the16S rRNA actually reduced reporter gene transcript and protein levels [45]. Therefore, the 5′ end of a transcript’s coding region influences expression of chloroplast transgenes. In support of this conclusion, fusion of the 14 N-terminal amino acids of the chloroplast rbcL and atpB genes to a reporter sequence resulted in different levels of reporter protein that did not reflect differences in transcript abundance [46]. Furthermore, silent mutations in the fused N-terminal coding sequences were found to decrease reporter protein accumulation without influencing RNA level [46]. In a separate study, fusion of sequences encoding the 14 N-terminal amino acids of GFP to a synthetic EPSPS gene increased EPSPS protein accumulation 33-fold without influencing transcript levels [19]. Therefore, when designing foreign genes for maximal expression in chloroplasts, one must optimize not only 5′UTR elements but also sequences downstream of the initiation codon. By changing these sequences it is possible to manipulate levels of foreign protein over an extraordinary 10 000-fold range [19].
Current limitations of chloroplast genetic engineering
One barrier to developing plastid transformation for crop plants, including cereals, has been their regeneration from non-green embryonic cells (containing proplastids) rather than leaf cells (containing chloroplasts). Plastid transformation in rice has been shown using vectors containing a translational fusion of aadA with gfp [47]. Inspection of streptomycin-resistant rice shoots by confocal microscopy revealed a few proplastids with GFP fluorescence. Although this report only showed heteroplasmy, it represents a step in the right direction towards monocot plastid transformation. A major limitation in accomplishing this goal might be the low level of marker gene expression in non-green plastids because of low genome copy number and low rates of protein synthesis. Identification of promoters and UTRs active in nongreen tissues should help to overcome this limitation.
Another limitation is the lack of information on genome sequences for several important crop species to locate intergenic sequences for integration of transgenes. Because of the conservation of the plastid genome sequence across many plant species, the concept of using targeting sequences from one species to transform the plastid genome of another unknown species was developed [2]. Both potato [24] and tomato [26] were transformed using chloroplast vectors with tobacco targeting sequences, and tobacco was transformed with petunia targeting sequences [17].
Yet another limitation is the challenge of delivering foreign DNA through the double plastid membrane. Among the methods used to transform plant chloroplasts, only particle bombardment has proven to be efficient in transforming different plant species. PEG treatment of protoplasts has been used to stably transform tobacco chloroplast genomes [48] and although laborious, it is inexpensive when compared to particle bombardment. Microinjection is effective for foreign gene expression in chloroplasts [49], although application of this technology for stable transformation of chloroplast genomes awaits further investigation. Development of new methods of gene delivery systems into chloroplasts should facilitate further progress in this field.
Epilogue
The accomplishments of chloroplast molecular biologists have spearheaded the plant biotechnology revolution, including the cloning and sequencing of the first plant gene [50], the development of the first useful plant trait (herbicide resistance) [50] and the determination of the first complete sequence of a plant genome [51]. Recent success in engineering the chloroplast genome for resistance to herbicides, insects, disease and drought, and for production of biopharmaceuticals, has opened the door to a new era in biotechnology. These recent developments suggest that commercial products of chloroplast genetic engineering are now on the horizon. In spite of being enslaved by the nucleus, this endosymbiont defiantly marches on, leading the biotechnology field in new directions.
Contributor Information
Henry Daniell, Dept of Molecular Biology and Microbiology, University of Central Florida, Orlando, FL 32826-3227, USA.
Muhammad S. Khan, Dept of Molecular Biology and Microbiology, University of Central Florida, Orlando, FL 32826-3227, USA
Lori Allison, Dept of Biochemistry, N258 Beadle Center, University of Nebraska, Lincoln, NE 68588-0664, USA.
References
- 1.Scott SE, Wilkinson MJ. Low probability of chloroplast movement from oilseed rape (Brassica napus) into wild Brassica rapa. Nat. Biotechnol. 1999;17:390–392. doi: 10.1038/7952. [DOI] [PubMed] [Google Scholar]
- 2.Daniell H, et al. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat. Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Cosa B, et al. Overexpression of the Bt Cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniell H, et al. Marker free transgenic plants: engineering the chloroplast genome without the use of antibiotic selection. Curr. Genet. 2001;39:109–116. doi: 10.1007/s002940100185. [DOI] [PubMed] [Google Scholar]
- 5.Daniell H, et al. Antibiotic free chloroplast genetic engineering – an environmentally friendly approach. Trends Plant Sci. 2001;6:237–239. doi: 10.1016/s1360-1385(01)01949-5. [DOI] [PubMed] [Google Scholar]
- 6.Daniell H, et al. Expression of cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J. Mol. Biol. 2001;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staub JM, et al. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat. Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- 8.Lee SB, et al. Drought tolerance conferred by the yeast trehalose-6 phosphate synthase gene engineered via the chloroplast genome. Transgenic Res. (in press). [Google Scholar]
- 9.Daniell H, McFadden BA. Uptake and expression of bacterial and cyanobacterial genes by isolated cucumber etioplasts. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6349–6353. doi: 10.1073/pnas.84.18.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniell H. Foreign gene expression in chloroplasts of higher plants mediated by tungsten particle bombardment. Methods Enzymol. 1993;217:536–556. doi: 10.1016/0076-6879(93)17088-m. [DOI] [PubMed] [Google Scholar]
- 11.Daniell H, et al. Transient foreign gene expression in chloroplast of cultured tobacco cells after biolistic delivery of chloroplast vectors. Proc. Natl. Acad. Sci. U. S. A. 1990;87:88–92. doi: 10.1073/pnas.87.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniell H, et al. Expression of β-glucuronidase gene in different cellular compartments following biolistic delivery of foreign DNA into wheat leaves and calli. Plant Cell Rep. 1991;9:615–619. doi: 10.1007/BF00231800. [DOI] [PubMed] [Google Scholar]
- 13.Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svab Z, Maliga P. High frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. U. S. A. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBride KE, et al. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Bio/Technology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- 16.Kota M, et al. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeGray G, et al. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- 18.Iamtham S, Day A. Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat. Biotechnol. 2000;18:1172–1176. doi: 10.1038/81161. [DOI] [PubMed] [Google Scholar]
- 19.Ye G-N, et al. Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 2001;25:261–270. doi: 10.1046/j.1365-313x.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- 20.Lutz KA, et al. Expression of bar in the plastid genome confers herbicide resistance. Plant Physiol. 2001;125:1585–1590. doi: 10.1104/pp.125.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guda C, et al. Stable expression of biodegradable protein based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- 22.Hajdukiewicz PT, et al. Multiple pathways for Cre/lox-mediated recombination in plastids. Plant J. 2001;27:161–170. doi: 10.1046/j.1365-313x.2001.01067.x. [DOI] [PubMed] [Google Scholar]
- 23.Corneille S, et al. Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox site-specific recombination system. Plant J. 2001;27:171–178. doi: 10.1046/j.1365-313x.2001.01068.x. [DOI] [PubMed] [Google Scholar]
- 24.Sidorov VA, et al. Technical advance: stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- 25.Ruf S. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat. Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- 26.Daniell H. Environmentally friendly approaches to genetic engineering. In Vitro Cell. Dev. Biol. Plant. 1999;35:361–368. [Google Scholar]
- 27.Daniell H. Genetically modified food crops: current concerns and solutions for next generation crops. Biotechnol. Genet. Eng. Rev. 2000;17:327–352. doi: 10.1080/02648725.2000.10647997. [DOI] [PubMed] [Google Scholar]
- 28.Bogorad L. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 2000;18:257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- 29.Nawrath C, et al. Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12760–12764. doi: 10.1073/pnas.91.26.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma JK, et al. Generation and assembly of secretory antibodies in plants. Science. 1995;268:716–719. doi: 10.1126/science.7732380. [DOI] [PubMed] [Google Scholar]
- 31.Ye X, et al. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 32.Ge B, et al. Differential effects of helper proteins encoded by the cry2A and cry11A operons on the formation of Cry2A inclusions in Bacillus thuringiensis. FEMS Microbiol. Lett. 1998;165:35–41. doi: 10.1111/j.1574-6968.1998.tb13124.x. [DOI] [PubMed] [Google Scholar]
- 33.Jacob L, Zasloff M. Potential therapeutic applications of magainins and other antimicrobial agents of animal origin. Ciba Found. Symp. 1994;186:197–223. doi: 10.1002/9780470514658.ch12. [DOI] [PubMed] [Google Scholar]
- 34.Sharma SC. A possible role of trehalose in osmotolerance and ethanol tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1997;152:11–15. doi: 10.1111/j.1574-6968.1997.tb10402.x. [DOI] [PubMed] [Google Scholar]
- 35.Rathinasabapathy B, et al. Metabolic engineering of glycine betaine synthesis: plant betaine aldehyde dehydrogenases lacking typical transit peptides are targeted to tobacco chloroplasts where they confer aldehyde resistance. Planta. 1994;193:155–162. doi: 10.1007/BF00192524. [DOI] [PubMed] [Google Scholar]
- 36.Nuccio ML, et al. Metabolic engineering of plants for osmotic stress resistance. Curr. Opin. Plant Biol. 1999;2:128–134. doi: 10.1016/s1369-5266(99)80026-0. [DOI] [PubMed] [Google Scholar]
- 37.Fischer N, et al. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 1996;251:373–380. doi: 10.1007/BF02172529. [DOI] [PubMed] [Google Scholar]
- 38.Daniell H, et al. Medical molecular farming: production of antibodies, biopharmaceuticals and edible vaccines in plants. Trends Plant Sci. 2001;6:219–226. doi: 10.1016/S1360-1385(01)01922-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrides D, et al. Computer-aided process analysis and economic evaluation for biosynthetic human insulin production – a case study. Biotechnol. Bioeng. 1995;48:529–541. doi: 10.1002/bit.260480516. [DOI] [PubMed] [Google Scholar]
- 40.Walmsley AM, Arntzen CJ. Plants for delivery of edible vaccines. Curr. Opin. Biotechnol. 2000;11:126–129. doi: 10.1016/s0958-1669(00)00070-7. [DOI] [PubMed] [Google Scholar]
- 41.Monde RA, et al. Processing and degradation of chloroplast RNA. Biochimie. 2000;82:573–582. doi: 10.1016/s0300-9084(00)00606-4. [DOI] [PubMed] [Google Scholar]
- 42.Eibl C, et al. In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J. 1999;19:333–345. doi: 10.1046/j.1365-313x.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- 43.Monde RA, et al. The sequence and secondary structure of the 3′-UTR affect 3′-end maturation, RNA accumulation, and translation in tobacco chloroplasts. Plant Mol. Biol. 2000;44:529–542. doi: 10.1023/a:1026540310934. [DOI] [PubMed] [Google Scholar]
- 44.Zerges W. Translation in chloroplasts. Biochimie. 2000;82:583–601. doi: 10.1016/s0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]
- 45.Kuroda H, Maliga P. Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res. 2001;29:970–975. doi: 10.1093/nar/29.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuroda H, Maliga P. Sequences downstream of the translation initiation codon are important determinants of the translation efficiency in chloroplasts. Plant Physiol. 2001;125:430–436. doi: 10.1104/pp.125.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan MS, Maliga P. Fluorescent antibiotic resistance marker to trace plastid transformation in higher plants. Nat. Biotechnol. 1999;17:910–915. doi: 10.1038/12907. [DOI] [PubMed] [Google Scholar]
- 48.Koop H-U, et al. Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta. 1996;199:193–201. doi: 10.1007/BF00196559. [DOI] [PubMed] [Google Scholar]
- 49.Knoblauch M, et al. A galinstan expansion femtosyringe for microinjection of eukaryotic organelles and prokaryotes. Nat. Biotechnol. 1999;17:906–909. doi: 10.1038/12902. [DOI] [PubMed] [Google Scholar]
- 50.Bogorad L, et al. Biosynthesis of the Photosynthetic Apparatus: Molecular Biology, Development and Regulation. Alan R: Liss Inc.; 1984. The organization of maize plastid chromosome: properties, and expression of its genes; pp. 257–272. [Google Scholar]
- 51.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrer H, et al. Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet. 1993;241:49–56. doi: 10.1007/BF00280200. [DOI] [PubMed] [Google Scholar]
- 53.Serino G, Maliga P. A negative selection scheme based on the expression of cytosine deaminase in plastids. Plant J. 1997;12:697–701. doi: 10.1046/j.1365-313x.1997.00697.x. [DOI] [PubMed] [Google Scholar]
- 54.Hibberd JM, et al. Transient expression of green fluorescent protein in various plastid types following microprojectile bombardment. Plant J. 1998;16:627–632. [Google Scholar]
- 55.Zhang XH, et al. Targeting a nuclear anthranilate synthase α subunit gene to the tobacco plastid genome results in enhanced tryptophan biosynthesis. Return of a gene to its pre-endosymbiotic origin. Plant Physiol. 2001;127:131–141. doi: 10.1104/pp.127.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]