Stable transformation of the cotton plastid genome and maternal inheritance of transgenes (original) (raw)
. Author manuscript; available in PMC: 2012 Oct 26.
Published in final edited form as: Plant Mol Biol. 2004 Sep;56(2):203–216. doi: 10.1007/s11103-004-2907-y
Abstract
Chloroplast genetic engineering overcomes concerns of gene containment, low levels of transgene expression, gene silencing, positional and pleiotropic effects or presence of vector sequences in transformed genomes. Several therapeutic proteins and agronomic traits have been highly expressed via the tobacco chloroplast genome but extending this concept to important crops has been a major challenge; lack of 100% homologous species-specific chloroplast transformation vectors containing suitable selectable markers, ability to regulate transgene expression in developing plastids and inadequate tissue culture systems via somatic embryogenesis are major challenges. We employed a ‘Double Gene/Single Selection (DGSS)’ plastid transformation vector that harbors two selectable marker genes (_aph_A-6 and _npt_II) to detoxify the same antibiotic by two enzymes, irrespective of the type of tissues or plastids; by combining this with an efficient regeneration system via somatic embryogenesis, cotton plastid transformation was achieved for the first time. The DGSS transformation vector is at least 8-fold (1 event/2.4 bombarded plates) more efficient than ‘Single Gene/Single Selection (SGSS)’ vector (_aph_A-6; 1 event per 20 bombarded plates). Chloroplast transgenic lines were fertile, flowered and set seeds similar to untransformed plants. Transgenes stably integrated into the cotton chloroplast genome were maternally inherited and were not transmitted via pollen when out-crossed with untransformed female plants. Cotton is one of the most important genetically modified crops ($ 120 billion US annual economy). Successful transformation of the chloroplast genome should address concerns about transgene escape, insects developing resistance, inadequate insect control and promote public acceptance of genetically modified cotton.
Keywords: chloroplast genetic engineering, genetically modified crops, transgene containment, transgenic cotton
Introduction
Chloroplasts genetic engineering approach offers a number of attractive advantages, including a high-level of transgene expression (Daniell et al., 2002a, b), multi-gene engineering in a single transformation event (DeCosa et al., 2001; Ruiz et al., 2003; Daniell and Dhingra, 2002), transgene containment via maternal inheritance (Daniell et al., 1998; Daniell, 2002; Daniell et al., 2004b), lack of gene silencing (Lee et al., 2003; DeCosa et al., 2001), vector sequences (Daniell et al., 2002b), position effect due to site specific transgene integration (Daniell et al., 2004b) and pleiotropic effects due to sub-cellular compartmentalization of transgene products (Daniell et al., 2001; Lee et al., 2003). However, plastid transformation has been so far highly efficient only in tobacco; 5–15 events per bombarded leaf have been reported (Daniell et al., 2001; Fernandez-San Millan et al., 2003; Dhingra and Daniell, 2004). Plastid transformation in tomato was a rare event and only six transgenic calli were obtained from 520 selection plates (60 bombarded leaves) and it took 2 years to regenerate transgenic plants (Ruf et al., 2001). Transformation of potato chloroplast genome with a vector harboring two genes (_aad_A/gfp) resulted in 3 events out of 104 bombarded plates. Two were chimeric transgenic calli and one was a transgenic line. The calli produced shoots that expressed GFP uniformly. When the _aad_A gene alone was used for transformation, 3 potato chloroplast transgenic lines were obtained from 46 bombarded plates (Sidorov et al., 1999). Arabidopsis chloroplast transgenic plants, though homoplasmic, were sterile (Sikdar et al., 1998). Homoplasmy or maternal inheritance could not be achieved in oilseed rape (Hou et al., 2003). Although foreign gene expression in rice (Khan and Maliga, 1999) and wheat (Daniell et al., 1991) were reported in plastids, stable integration or homoplasmy or transfer of traits to progeny was never achieved. Recently efficient plastid transformation using non-green tissues has been accomplished in carrot; in this case chloroplast transgenic lines were generated via somatic embryogenesis from tissues containing proplastids (Kumar et al., 2004).
Cotton is the single most important textile fiber in the world and is grown in more than 90 countries. The US accounts for 21% of the total world fiber production and is the leading exporter in global trade of raw cotton. Cotton is the most important cash crop in the US and annual business revenue stimulated by cotton in the US economy is about $120 billion each year, making cotton America’s number one value-added crop. In 2002–2003, US farmers planted ~73% transgenic cotton in 5.6 million hectares modified via the nuclear genome, compared to 81% of genetically modified (GM) soybean and 40% GM corn (http://www.ers.usda.gov/Briefing/). Upland cotton, Gossypium hirsutum, has the potential to hybridize with Hawaiian cotton, G. tomentosum, and feral populations of G. hirsutum in the Florida Keys, and of G. hirsutum/G. barbadense on the US Virgin Islands and Puerto Rico. Therefore, restrictions on field plot experimental use permits and commercial planting of Bt-cotton have been instituted in these areas. Similarly, GM cotton is now planted only in the regions of the world where there are no wild relatives in order to avoid potential outcross with related weeds. Dispersal of pollen from transgenic cotton plants to surrounding non-transgenic plants has been reported (Llewellyn and Fitt, 1996). Umbeck et al. (1991) investigated pollen dispersal from transgenic cotton embedded in a field of conventional cotton in the US and observed up to 5.7% outcrossing rates in spite of buffer rows. Transgene escape could be avoided via chloroplast genetic engineering because of maternal inheritance of transgenes in cotton. Although movement of chloroplast integrated transgenes to the nucleus has been recently reported, the frequency is rather low; 1/16,000 pollen derived from chloroplast transgenic line carried the transgene that was integrated into the plastid genome (Huang et al., 2003a, Stegemann et al., 2003). However, experimental details and conclusions drawn from these studies have been questioned and debated (Daniell and Parkinson, 2003; Huang et al., 2003b). Most importantly, transgenes integrated into the nuclear genome were non-functional (Huang et al., 2003a) and therefore had no impact on biotechnology applications (Daniell and Parkinson, 2003). In the rare event where transgene escape occurs, engineering male sterility via chloroplast genome should provide a failsafe mechanism; such a cytoplasmic male sterility system has been developed recently (Daniell et al., 2004b).
Another concern about GM crops expressing Bt toxins is that suboptimal production of toxins might result in an increased risk of pests developing Bt resistance (Daniell, 2000). There are also reports that Bt-cotton failed to control Heliothines armigera in Australia and cotton bollworm on at least 20 000 acres in Texas and herbicide resistant transgenic lines suffered a similar failure (Hilder and Boulter, 1999). The Bt-cotton is not fully protected from the insect-pest attack and it needs repeated pesticide applications on crop fields to minimize the yield loss (http://www.epa.gov/scipoly/sap/2000/october/brad7_cotton_final.pdf). Among the potential environmental concerns, transgene containment, insects developing resistance and impact of non-target insects are of paramount importance. These studies suggest the need for higher levels of cry gene expression. The cry2Aa2 gene engineered via the chloroplast genome was shown to kill insects that developed resistance (up to 40 000-fold) to insecticidal proteins (Kota et al., 1999). In another study expression of the _Cry_IIa5 in transgenic chloroplasts provided a complete protection against the larvae of Helicoverpa armigera irrespective of its development stage (Reddy et al., 2002). Multigene engineering via the chloroplast genome was used, in a single transformation event, to express very high levels of CRY proteins (up to 46.1% tsp) and this resulted in chaperone assisted formation of cuboidal crystals that prevented proteolytic degradation and this led to high levels of insecticidal protein accumulation even in bleached old leaves that killed insects that are highly tolerant or resistant to CRY proteins. In spite of such high levels of expression in leaves, CRY protein was not expressed in pollen as determined by lack of toxicity to insects fed on pollen (DeCosa et al., 2001). This opens the possibility of engineering multiple genes for insect resistance to delay the onset of resistance or engineer multiple agronomic traits (e.g. insect, herbicide, disease resistance, drought tolerance) in a single transformation event via the chloroplast genome.
There is an urgent need to develop the concept of chloroplast transformation in economically important crop species. Some of the major obstacles to extend this technology to major crop species include inadequate tissue culture and regeneration protocols, selectable markers and inability to express transgenes in developing plastids (Daniell et al., 2002b; Bogorad, 2000). Major challenges to achieve cotton plastid transformation include, the use of appropriate regulatory sequences and selectable markers that function in non-green and green plastids, the ability to regenerate chloroplast transgenic plants in recalcitrant crops via somatic embryogenesis and achieve homoplasmy, which lacks the benefit of subsequent rounds of regeneration offered by organogenesis.
Here we present the first report of stable and reproducible plastid transformation of cotton from the embryogenic calli using novel chloroplast vectors. The selectable markers, _aph_A-6 and _npt_II detoxify the same antibiotic i.e. kanamycin and the employment of appropriate regulatory sequences ensure that detoxification of kanamycin continues in the light and dark, irrespective of the tissue type.
Materials and methods
Construction of cotton plastid transformation vectors
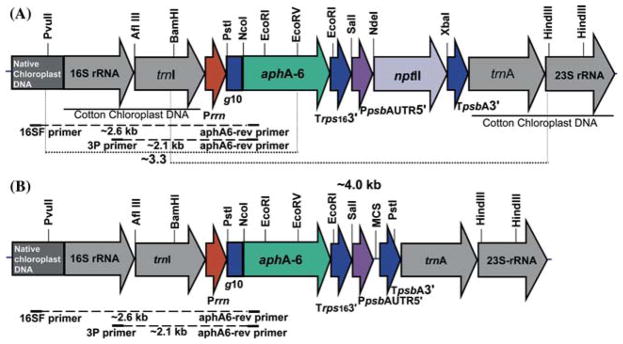
A DNA fragment representing cotton flanking sequence was amplified from cotton genomic DNA that was isolated from the leaves using DNeasy Plant mini kit (Qiagen Inc.) following manufacturer’s protocol. The flanking sequence fragment was amplified with the primers based on tobacco chloroplast genome sequence information using Platinum Pfx DNA polymerase (Invitrogen Inc.). This Pfu based enzyme has been demonstrated to be more efficient than Pfu DNA polymerase and it has the lowest error rate at roughly 1.3 × 10−6 error per base pair (Cline et al., 1996). The forward primer, ADLF and the reverse primer, ADLR amplified a 4.0 kb DNA fragment representing the 16S/_trn_I-trn_A/23S region of the cotton chloroplast genome. The PCR amplified DNA fragment was treated with T4 polynucleotide kinase (Promega) and cloned into PvuII digested pBluescript II KS, dephosphorylated with Shrimp Alkaline phosphatase (Promega). The kinase and dephosphorylation reactions were performed as per the manufacturer’s instructions. The chloroplast promoters and regulatory sequences were amplified using PCR based on the information available for the tobacco chloroplast genome (Accession # NC_001879). The primers used were as follows: ADLF (5′ CACTCTGCTGGGCCGACACTGACAC 3′); ADLR (5′ CACTAGCCGACCTTGACCCCTGTT 3′); P_rrn (Forward: 5′ATCGATGAGCCTGATTATCCTAAG3′; Reverse 5′CAGCAGGTAGACAAAGCGGATTC3′), P_p_-_sb_A (Forward 5′ GATATCGTCGACGTAGAGAAGTCCG 3′; Reverse 5′ CATATGAAAATCTTGGTTTATTTAA3′); T_psb_A (Forward 5′ TCTAGAGCGATCCTGGCCTAG3′; Reverse 5′GAGCTCGCAGCCCAAACAAATAC 3′); T_rps_16 (Forward 5′ACTAGTCCTAATCAACCGAAATTC3′; Reverse 5′ GAGCTCGAACACGGAATT CAATGGAAGC3′); T7 gene 10 (Forward 5′GGTAACCCCGGGAGAC CACAACGGTTTCCCTCTAGAAATAATTTTGTTTA3′; Reverse 5′CATATGTATATCTCCTTCTTAAAGTTA3′); 3P (5′AAAACCCGTCCTCAGTTCGGATTGC 3′); 16S F (5′CAGCAGCCGCGGTAATACA GAGGA3′); _aph_A-6 Rev (5′CGAGAGACACACTGTATGTGGTCTCTG3′). The cotton specific chloroplast transformation vector pDD-_Gh_-_aph_A-6/_npt_II (Figure 1A) was constructed by inserting a blunt ended fragment representing the _aph_A-6/_npt_II expression cassette into PvuII site of cotton chloroplast DNA flanking sequences. This site is present in the intergenic region between the _trn_I-_trn_A genes in the cloned flanking region. The chloroplast vector pKD-_Gh-aph_A-6 (Figure 1B) was constructed by excising the _npt_II-coding region along with the _psb_A 3′ UTR from pDD-Gh-_aph_A-6/_npt_II and cloning a PCR generated fragment representing a multiple cloning site (MCS) and _psb_A 3′ UTR. For this PCR, the following primers were used: forward primer 5′ TTAACAT ATGAGGCCTTAGAGCGATCCTGGC 3′ and reverse primer 5′ CAATTGCAAGAGCGGAGC TCTACCAAC 3′. All general bacterial and DNA manipulations were performed as per standard molecular biology protocols.
Figure 1.
(A and B). Physical maps of the cotton chloroplast transformation vectors. (A) Cotton chloroplast transformation vector pDD-Gh-aph_A_6/nptII with double selection marker genes _aph_A-6 and _npt_II. Primer annealing sites and probe used for Southern analysis are shown. (B) Cotton chloroplast transformation vector pKD-_Gh-aph_A-6 with a single selection marker _aph_A-6 gene.
Transformation and regeneration protocol for cotton
Cotton grayish-green friable callus, produced from hypocotyl explants of 5 day-old cotton seedlings (cultivar Coker310FR; Kumar et al., 1998), was spread uniformly on sterile filter papers placed in petri-dishes containing MST1 medium (MS salts; Murashige and Skoog, 1962, B5 vitamins; Gamborg et al., 1968, 0.1 mg/l 2,4-D, 0.5 mg/l kinetin and 3% glucose) as described (Trolinder and Goodin, 1988) and bombarded with gold particles coated with the chloroplast transformation vector pDD_-Gh-aph_A-_6/npt_II, using the helium-driven biolistic particle delivery system (Bio-Rad) (Daniell, 1997). After particle bombardment, these cultures were incubated in the dark for 48 h and then transferred to selection medium (MST1) supplemented with 50 mg/l kanamycin and incubated in 16/8 h day/night cycle at 50 _μ_E light intensity and 27 ± 1 °C temperature. Putative transgenic cell cultures were further multiplied on MST1 medium supplemented with 100 mg/l kanamycin and maintained on MST1 medium supplemented with 100 mg/l kanamycin for 4–5 months. Transformed calli were converted into somatic embryos and plantlets on MST0 medium (MS salts, B5 vitamins) containing additional (1.9 g/l) potassium nitrate. PCR-confirmed chloroplast transgenic plantlets were transferred to a growth chamber for flowering and seed setting.
Optimization of gene delivery in cotton
For optimization of gene delivery, cotton calli were spread on Whatman # 1 filter paper placed on MST1 medium. Gene delivery was optimized by using the pDD_-Gh-aphA-6/npt_II chloroplast transformation vector coated on 0.6 _μ_m gold particles employing different parameters listed in Table 1. The bombarded calli were incubated in dark for 2 days, after which they were plated on selective MST1 medium supplemented with 50 mg/l kanamycin. Kanamycin resistant transgenic calli were tested for site-specific integration of transgenes into plastid genomes by PCR.
Table 1.
Cotton calli (8–10 weeks old) bombarded with the vector pDD-_Gh-aph_A_-6/npt_II and pKD-_Gh_-_aph_A-6, coated on 0.6 _μ_m gold particles using indicated parameters. The transgenic cell lines selected on MST1 medium containing 50 mg/l kanamycin after bombardments were confirmed by PCR for site-specific transgene integration.
| Plastid vector | Rupture disc (psi) | Distancea (cm) | No. of platesb | Total eventsc | Efficiency (%)d |
|---|---|---|---|---|---|
| _aph_A6/_npt_II | 650 | 6 | 15 | 0 | 0 |
| _aph_A6/_npt_II | 650 | 9 | 31 | 13 | 42 |
| _aph_A6/_npt_II | 900 | 9 | 17 | 5 | 29 |
| _aph_A6/_npt_II | 900 | 12 | 18 | 3 | 17 |
| _aph_A6/_npt_II | 1100 | 9 | 32 | 2 | 6 |
| _aph_A6/_npt_II | 1100 | 12 | 46 | 5 | 11 |
| _aph_A6 | 650 | 9 | 40 | 2 | 5 |
Analysis of maternal inheritance in cotton
Emasculated non-transgenic cotton plants were pollinated with pollen derived from chloroplast transgenic cotton plants. Hybrid F1 (♀ non-transgenic × ♂ transgenic) as well as chloroplast transgenic cotton seeds were germinated on 1/2MS basal medium supplemented with 50 mg/l kanamycin to test for maternal inheritance of chloroplast integrated transgenes.
Results and discussion
Construction of cotton plastid transformation vectors
Cotton chloroplast vectors target the expression cassette to the 16S/_trn_I-_trn_A/23S region of the chloroplast genome for integration via homologous recombination. The site of integration is similar to the universal chloroplast transformation vector (pLD CtV) reported earlier from our laboratory (Daniell et al., 1998; Guda et al., 2000). For construction of the cotton chloroplast transformation vector, the flanking region was amplified from cotton genomic DNA. In the absence of chloroplast genome sequence information for cotton, primers were designed based on the sequence information available for tobacco. PCR amplification of the flanking region from cotton resulted in a 4.0 kb DNA fragment that is approximately twice the size of the flanking region used in pLD CtV vector. It has been shown that frequency of homologous recombination is dependent on length and homology (Shen and Huang, 1986; Fujitani et al., 1995) of flanking sequences. Based on these observations, length of the flanking sequences was increased to 2 kb each on either side of the transgene cassette so that it might enhance the frequency of homologous recombination. Cotton specific chloroplast transformation vector (pDD-_Gh_-_aph_A-6/_npt_II, Figure 1A) harbors the _aph_A-6 gene regulated by the 5′ ribosome binding site (rbs) region of the bacteriophage T7 gene 10 leader (Guda et al., 2000; Staub et al., 2000; Dhingra et al., 2004; Kumar et al., 2004)/_rps_16 3′UTR in order to facilitate expression in green as well as non-green tissues. The _npt_II gene was expressed under the regulation of P_psb_A 5′ and 3′ _psb_A UTR in order to facilitate light regulated expression in green tissues (Dhingra et al., 2004, Devine and Daniell, 2004; Daniell et al., 2004a; Fernandez San Millan et al., 2003; Staub and Maliga, 1995). The second cotton specific chloroplast transformation vector (pKD-_Gh_-_aph_A-6, Figure 1B) harbors only the _aph_A-6 gene driven by the 16S rRNA full length promoter and regulated by T7 gene 10 5′ UTR/_rps_16 3′UTR. A P_psb_A 5′UTR and _psb_A 3′ UTR with a multiple cloning site in between the UTRs was inserted in order to clone genes of interest. Transcription of the expression cassettes, in the cotton chloroplast transformation vectors, is driven by the full-length 16S rRNA promoter (Silhavy and Maliga, 1998). The full-length promoter comprises of binding sites for both the plastid-encoded and nuclear-encoded RNA polymerase thereby facilitating transcription in green or non-green tissues. All the 5′ and 3′ regulatory elements were PCR amplified from the tobacco genomic DNA except for the T7 gene 10 5′UTR which was PCR amplified from pET 11 vector (NEB). Details of primers are provided in Materials and methods.
Transformation of cotton plastids and plant regeneration
Embryogenic cell cultures serve as an ideal target material for biolistic transformation (Christou, 1992). However, cotton is particularly challenging to manipulate in-vitro due to the difficulties encountered in plant regeneration through somatic embryogenesis. Coker cultivars used for in vitro regeneration are highly variable in their embryogenic response due to genotype specificity (Trolinder and Chen, 1989; Kumar et al., 1998), which has severely impacted the efficacy of genetic transformation through somatic embryogenesis. Nuclear transformation of elite cotton depends mainly on Coker cultivars that are used as the explant source for in vitro transformation. Availability of pure embryogenic line (Coker 310FR; fully regenerating) was reported to improve the transformation frequency as well as the regeneration of transgenic cotton plants (Kumar et al., 1998).
Friable grayish callus induced from hypocotyl segments of fully embryogenic cotton (Gossypium hirsutum cv. Coker 310FR) was bombarded with cotton chloroplast vector pDD_-Gh-aph_A-_6/npt_II as described (Daniell, 1997; Kumar and Daniell, 2004; Daniell et al., 2004c). Cotton plastid transformation was extensively tried using the _aad_A gene containing cotton chloroplast vectors. One hundred and five cotyledons, 60 leaves and 72 plates of embryogenic callus (1 mm thick layer × 20 mm in diameter) of Gossypium hirsutum cv. Coker 310 were bombarded with cotton chloroplast vector containing the _aad_A gene. Bombarded cotyledons and leaves were cut into small pieces and callus cultures were plated evenly on selection medium containing 25, 75 and 150 mg/l spectinomycin. However, no transgenic calli or plants were recovered over a period of 6 months using spectinomycin as the selection agent due to its lethal effect on cotton cultures.
Embryogenic cotton calli bombarded with chloroplast transformation vector pDD_-Gh_-_aph_A-_6/npt_II produced several transgenic lines (Table 1), selected on MST1 medium containing 0.1 mg/l 2,4-D and 0.5 mg/l kinetin (callus induction medium) and supplemented with 50 mg/l kanamycin (Figure 2A and B). Further, transformed cultures were multiplied on higher concentrations of kanamycin (100 mg/l) in order to increase the number of transgenic chloroplasts in cotton cultures. Transgenic somatic embryos were induced from calli on basal MST0 medium (containing 1.9 g/l extra KNO3) supplemented with 25 mg/l kanamycin in about 3 months. Transgenic embryos placed on Whatman filter paper (No. 1) in a petri-dish containing MST0 medium were matured and elongated into plantlets in about 1 month (Figure 2 D). Transgenic cell cultures and plantlets were tested for stability of site-specific transgene integration using PCR and southern blotting. Confirmed transgenic plants were transferred to a growth chamber for flowering and seed setting.
Figure 2.
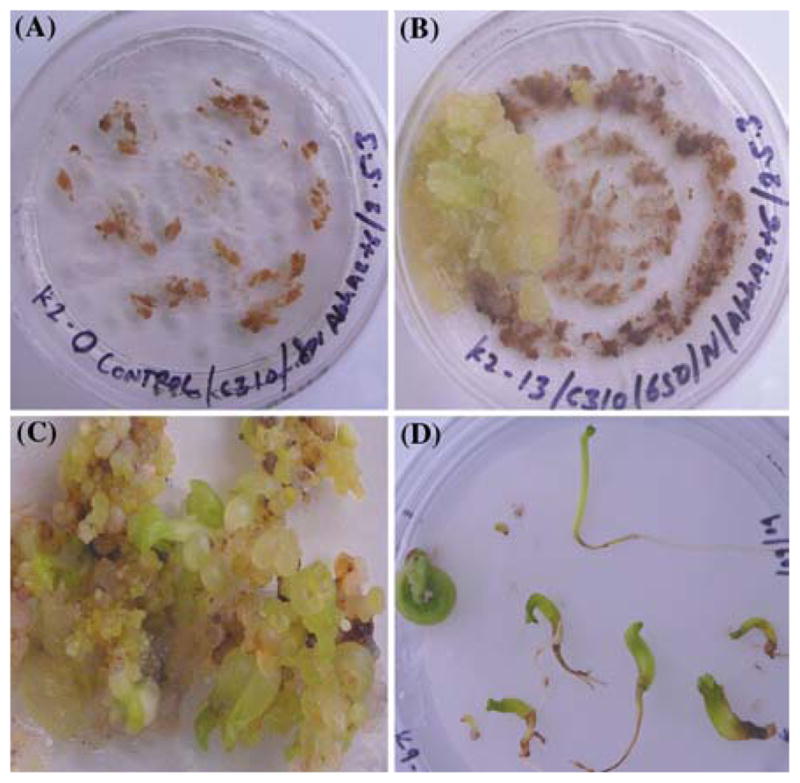
(A–D) Cotton calli transformed with the chloroplast transformation vector pDD_-Gh-aph_A-_6/npt_II. (A) Untransformed control cotton calli and (B) transformed primary cotton calli selected on MST1 medium supplemented with 50 mg/l kanamycin. (C) Transgenic cotton calli converted into somatic embryos. (D) Transgenic elongated somatic embryos that were used to induce homoplasmy by initiating fresh callus from their hypocotyl segments.
Optimization of plastid transformation
Plastid transformation efficiency is very high in tobacco (approximately 5–15 events per bombarded leaf; Daniell et al., 2001; Fernandez San Millan et al., 2003; Dhingra and Daniell, 2004) but has been quite inefficient in other crops, including other solanaceous species (Ruf et al., 2001, Sidorov et al., 1999). The first nuclear transformation of cotton via somatic embryogenesis using particle gun was demonstrated by Finer and McMullen (1990) with 0.7% efficiency. In order to optimize gene delivery in cotton chloroplasts, the chloroplast transformation vector, pDD_-Gh-aph_A-_6/npt_II (Figure 1A) was bombarded using different rupture discs and by varying the distances between rupture discs and target tissues. Maximal transformation efficiency (41.7% or 1 transformation event per 2.4 bombarded plates) was observed when cell cultures (friable; gray in color) were bombarded at 9 cm distance with 650-psi rupture disc (Table 1). These results suggest that higher cell death or production of phenolic compounds at sites of injury may be important determinants in transformation efficiency. The use of double gene single selection (DGSS) plastid vector greatly facilitated optimization of bombardment parameters, transformation conditions and facilitated chloroplast transformation even with a single gene (_aph_A-6) single selection vector (SGSS). Using optimized conditions, the chloroplast vector with a single selectable marker gene (_aph_A-6 gene) yielded only two transformation events out of 40 bombarded plates (Table 1). Therefore, the transformation efficiency with the double selectable marker gene is 8-fold more than single selectable marker gene. For the choice of single selectable marker gene we did not choose _npt_II as it has been used earlier but the efficiency was reported to be very low even in tobacco, where plastid transformation is highly efficient (Carrer et al., 1993). However, the _aph_A-6 gene has been successfully used for chloroplast transformation in Chlamydomonas (Bateman and Purton, 2000) or tobacco protoplasts by PEG-mediated gene delivery (Huang et al., 2002). Even though _aph_A-6 was employed for particle gun mediated chloroplast transformation of tobacco leaves, no chloroplast transgenic lines were obtained and the authors cited sub-optimal selection conditions and the limited number of bombardments as the reason (Huang et al., 2002).
Confirmation of transgene integration into cotton chloroplast genome
The cotton chloroplast transformation vector pDD_-Gh-aph_A-_6/npt_II integrates the aphA6 and _npt_II genes into the 16S-23S-spacer region of the plastid genome by homologous recombination. Integration of _aph_A-6 and _npt_II into the cotton cultures was confirmed by PCR using internal primers, 3P (anneals to the flanking sequence) and _aph_A-6-rev (anneals to the _aph_A-6 coding region). A 2.1 kb size PCR product was amplified and this confirmed integration of the transgenes in different cell cultures of cotton (Figure 3A). In order to distinguish between nuclear and chloroplast transgenic lines, the 16S-F primer was designed to anneal to the native chloroplast genome, 200 bp upstream of integration site and the aphA6-rev primer was designed to anneal to the _aph_A-6 transgene; this yielded a 2.6 kb size PCR product, which confirmed the site-specific integration of the transgenes into cotton chloroplast genome (Figure 3B). Since this PCR product can be obtained in the event of nuclear integration mediated by promiscuous DNA, the possibility of nuclear integration was eliminated by Southern analysis (discussed below). Transgenic calli bombarded with pKD-_Gh-aph_A-6 (Figure 1B; containing single aph_A_-6 marker gene) produced two transgenic lines on MST1 medium supplemented with 50 mg/l kanamycin (Figure 3C, D). Transgene integration was again confirmed using different sets of primers: 3P and _aph_A6-rev (Figure 3C); 16SF and _aph_A6-rev (Figure 3D).
Figure 3.
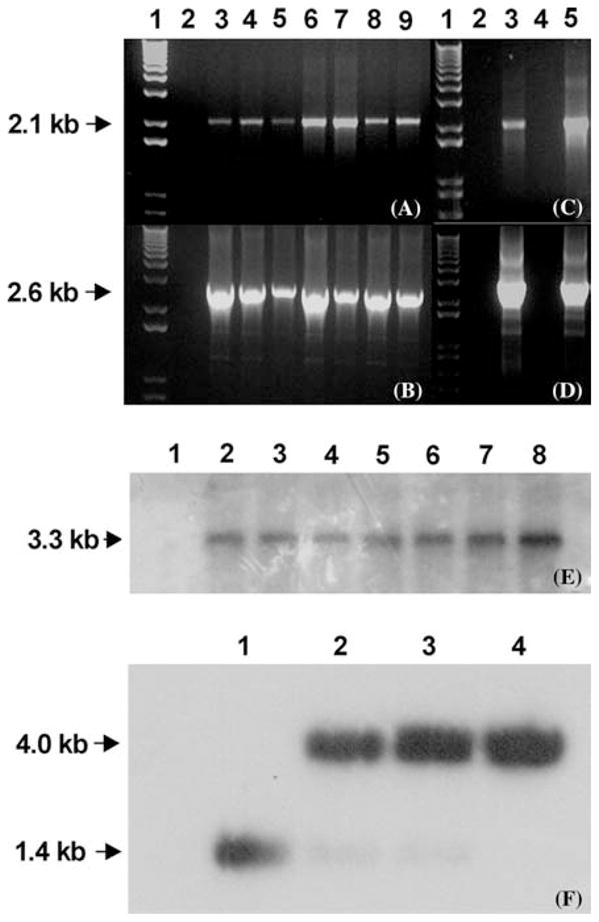
(A–F). Confirmation of transgene integration into the cotton plastid genome by PCR and Southern blot analysis. (A) PCR analysis of cotton calli transformed with chloroplast transformation vector pDD-_Gh-aph_A-_6/npt_II: Primers 3P (anneal to the flanking sequences) and aphA6-rev (anneal to the _aph_A-6 coding region) yield a 2.1 kb PCR product. (B) Primers 16SF (anneal to the native chloroplast genome) and _aph_A-6-rev (anneal to the _aph_A-6 coding region) yield a 2.6 kb PCR product. Lane 1 1kb DNA ladder; Lane 2 Non-transgenic cotton calli; lanes 3–9 Transgenic cotton calli. (C) PCR analysis for cotton calli transformed with vector pKD-_Gh-aph_A-6: Primers 3P and aphA6-rev yield a 2.1 kb PCR product. (D) Primers 16SF and _aph_A-6-rev yield a 2.6 kb PCR product. Lane 1, 1 kb DNA ladder; Lane 2 Non-transgenic cotton calli; lanes 3 and 5 Transgenic cotton calli; lane 4 is blank. (E) Southern blot analysis of cotton (transformed with vector pDD-_Gh-aph_A-_6/npt_II) genomic DNA digested with PvuII-EcoRV and hybridized with probe _aph_A-6 (see figure 1A). Lane 1 Untransformed; Lanes 2–8 In vitro transgenic cell cultures. (F) Southern analysis of genomic DNA digested with BamHI-Hind III and hybridized with flanking probe. Lane 1: untransformed cotton (control); Lanes 2 and 3: Genomic DNA of transgenic plants derived from seeds; Lane 4: Genomic DNA from transgenic cotton culture initiated from transgenic somatic embryos (see figure 2D).
Integration of the transgenes (aph_A_-6 and _npt_II) into cotton plastid genome and homoplasmy were investigated by Southern blot analysis. Genomic DNA from transformed and untransformed cultures was digested with appropriate restriction enzymes, transferred to nitrocellulose membrane and probed with P32-radiolabeled aphA-6 gene fragment (~650 bp, digested with Pst1 and EcoRV from pDD_-Ghaph_A-_6/npt_II). Transformed chloroplast genomic DNA digested with PvuII and EcoRV yielded an expected 3.3 kb size hybridizing fragment, thereby confirming the site-specific stable integration of the transgenes into the chloroplast genome in cell cultures and cotton plants (Figure 3E, Lane 2–8). As expected, no fragment hybridizing with the aph_A_-6 probe was observed in the wild-type untransformed cotton chloroplast genome (Figure 3E, Lane 1). Southern blot analysis was performed using total genomic DNA isolated from untransformed and transgenic cotton lines and calli derived from transgenic somatic embryos.
In order to investigate heteroplasmy or homoplasmy, genomic DNA of cotton plants transformed with pDD_-Gh-aph_A-_6/npt_II was digested with BamHI and HindIII and hybridized with the 1.4 kb radioactive flanking DNA probe (isolated from a vector containing only the flanking regions with BamHI and HindIII). Autoradiogram of the Southern blot showed a 4.0 kb hybridizing fragment representing the flanking and the transgene sequences in the homoplasmic transgenic lines (Figure 3F; lanes 2–4). The wild type untransformed cotton yielded a 1.4 kb fragment representing the flanking sequences (Figure 3F; lane 1). Heteroplasmic transgenic cotton lines should yield both fragments of 4.0 kb and 1.4 kb. Homoplasmic transgenic cotton lines were selected by repetitive subcultures of callus induced from hypocotyls segments of elongated somatic embryos that were regenerated after four cycles of subcultures. All transgenic calli were repeatedly subcultured on MST1 medium supplemented with 100 mg/l kanamycin after initial selection of transgenic calli on MST1 medium supplemented with 50 mg/l kanamycin. This helps to eliminate the untransformed cultures from putatively transgenic cotton cell cultures.
Determination of maternal inheritance in cotton
In vitro produced transgenic cotton lines were grown in the growth chamber along with non-transgenic plants, under similar growth conditions. Growth of chloroplast transgenic lines (Figure 4 A), onset of flowering, floral parts, boll formation and seed setting (Figure 4 C, D, E) were similar to the untransformed cotton plants (Figure 4B, F, G, H). Cotton lines were grown to maturation stage in the growth chamber (Figure 4A) and emasculated flowers of non-transgenic cotton were pollinated with pollen derived from chloroplast transgenic lines. More than 150 seeds obtained from F1 crosses (non-transgenic ♀ × ♂ transgenic) were germinated on ½ MS medium supplemented with 50 mg/l kanamycin. Seedlings from F1 crosses (non-transgenic ♀ × ♂ transgenic chloroplast) germinated on kanamycin selection medium but failed to grow further, whereas the transgenic seeds germinated well, produced copious roots and leaves, thereby confirming resistance to kanamycin (Figure 4I and J). This demonstrates that there is no paternal inheritance in cotton and that the chloroplast transgenic trait is inherited maternally. All seeds derived from self-pollinated chloroplast transgenic plants germinated on kanamycin and therefore, no Mendelian segregation was observed in the tested seeds. Uniparental maternal inheritance of cotton plastid genome has been reported earlier where the mechanism of maternal inheritance was investigated in depth and established (Lax et al., 1987; Hagemann, 2004).
Figure 4.

(A–J). (A) Transgenic and (B) non-transgenic control cotton plants at the stage of flowering and seed setting. (C–E) Different floral parts of transgenic cotton and (F–H) non-transgenic control cotton. I and J: F1 Seedlings germinated on kanamycin (50 mg/l). (I) Cross between chloroplast transgenic ♀ × ♂ non-transgenic cotton (J) Transgenic cotton obtained after self-pollination.
Conclusions
This study reports the first and a reproducible process for generating cotton plastid transgenic lines from transformed callus via somatic embryogenesis. The vectors employed for chloroplast transformation of potato, tomato and Lesquerella contained the flanking sequences from tobacco or Arabidopsis (Sidorov et al., 1999; Ruf et al., 2001; Skarjinskaia et al., 2003). This may be one of the reasons for lower transformation efficiency in these crops. Efficiency of tobacco plastid transformation using homologous native flanking sequences has been quite high (Fernandez San Millan et al., 2003; Dhingra and Daniell, 2004). However, when petunia flanking sequences were used for chloroplast transformation of tobacco, the transformation efficiency decreased drastically (DeGray et al., 2001). We have recently achieved efficient transformation of carrot chloroplast genome (1 event per ~7 bombarded plates) using species-specific carrot chloroplast transformation vector containing homologous flanking sequences (Kumar et al., 2004). Therefore, even though the universal vector concept was proposed several years ago (Daniell et al., 1998), species-specific vectors have been used for demonstration of plastid transformation, especially in recalcitrant crops.
The origin of replication has been mapped in the tobacco chloroplast genome and _ori_A is located within the _trn_I gene that forms the left flank in the chloroplast transformation vector used for cotton transformation. Several methods have been used to study ctDNA replication origins in higher plants including electron microscopy, two-dimensional (2D) agarose gel electrophoresis, primer extension mapping of nascent 5′ ends from total plastid DNA (Kolodner and Tewari, 1975; Meeker et al., 1988; Chiu and Sears, 1992; Hedrick et al., 1993; Nielsen et al., 1993; Kunnimalayaan et al., 1997), and in vitro replication analysis of ctDNA ori subclones (Kunnimalayaan and Nielsen, 1997 a, b). All these studies have identified two replication origins (_ori_A and _ori_B) that flank the 23S rRNA gene in tobacco ctDNA (Kunnimalayaan and Nielsen, 1997a,b). It has been shown that for clones containing a single ctDNA ori, replication occurs by a rolling circle mechanism and for clones containing both replication origins, replication proceeds by the D-loop/theta mechanism (Kunnimalayaan and Nielsen 1997b; Reddy et al., 1994). In vitro replication analysis has revealed that the minimal sequences required for activity of single-ori ctDNA templates in tobacco is 82 bp for _ori_A and 243 bp for _ori_B (Kunnimalayaan and Nielsen, 1997b). There are conflicting reports on the effect of deletions of chloroplast _ori_s (Muhlbauer et al., 2002; Kunnimalayaan and Nielsen, 1997b; Lu et al., 1996; Lugo et al., 2004). It is quite possible that disruption of an ori in vivo is not lethal if another mechanism can take over. It is believed that chloroplast DNA may replicate by a recombinant-dependent mechanism (Bendich, 2004). Therefore, like T4 and lambda phage, it is very likely that ctDNA replicates by more than one mechanism.
When chloroplast vectors with or without _ori_A were bombarded into cultured tobacco cells, only the vector with _ori_A located within the _trn_I gene showed prolonged and higher levels of CAT enzyme activity (Daniell et al., 1990). When chloroplast vectors with or without _ori_A were bombarded with the same transgenes, the vector with _ori_A present within the _trn_I flanking region achieved homoplasmy even in the first round of selection (Guda et al., 2000). Therefore, higher transformation efficiency observed in cotton might be due to the use of long homologous flanking sequences that contain one of the chloroplast origins of replication; one possibility is that this could offer large number of templates within plastids for integration.
The use of non-green explants has often been cited as one of the major obstacles that have limited chloroplast transformation to solanaceous crops (Bogorad, 2000). It has been erroneously claimed earlier that rice plastid transformation was achieved via somatic embryogenesis (Maliga, 2003) but no data was provided to support stable transgene integration into the plastid genome by homoplasmy or maternal inheritance of transgenes (Khan and Maliga, 1999). Understanding and manipulating the somatic embryogenesis system, which lacks the advantage of subsequent rounds of regeneration from heteroplasmic tissues, is a major challenge. Because most of the crop species are regenerated via somatic embryogenesis, methods developed here should help in transforming the plastid genomes of other crop plants. Non-green tissues contain several kinds of plastids namely proplastids, leucoplasts, amyloplasts, etioplasts, chromoplasts, elaioplasts and gerontoplasts in which gene expression and gene regulation systems are quite different from green chloroplasts. During transformation, transformed proplastids should develop into mature chloroplasts and transformed cells should survive the selection process during all stages of development. Therefore, the major challenge is to provide plastids an ability to survive selection in the light and the dark and at different developmental stages. This is absolutely critical because only one or two chloroplasts are transformed in a plant cell after bombardment and these plastids should have the ability to survive the selection pressure, multiply and establish themselves while all other untransformed plastids are eliminated in the selection process. The DGSS plastid vector accomplishes this by using genes coding for two different enzymes capable of detoxifying the same selection agent (or spectrum of selection agents), driven by regulatory signals that are functional in proplastids as well as in mature chloroplasts.
Both aph_A_-6 and aph_A_-2 (_npt_II) genes code for enzymes that belong to the aminoglycoside phosphotransferase family but they originate from different prokaryotic organisms. Both enzymes have similar catalytic activity but the aph_A_-6 gene product has an extended ability to detoxify kanamycin and provides a wider spectrum of aminoglycoside detoxification, including amikacin (Bateman and Purton, 2000; Huang et al., 2002). In nuclear genetic engineering, majority of the crop species have been transformed using aminoglycoside detoxification (kanamycin for dicots and geneticin for monocots). Both transgenes (aph_A_2 or _npt_II, _aph_A-6) are transcribed by the full-length plastid Prrn promoter containing binding site for nuclear-encoded and plastid-encoded RNA polymerase and is expected to function both in proplastids and mature chloroplasts (Silhavy and Maliga, 1998). The _aph_A-6 gene is further regulated by the T7 gene 10 5′UTR capable of efficient translation in the dark, in proplastids present in non-green tissues (i.e. grayish friable culture of cotton initially bombarded with cotton specific chloroplast transformation vector). The _rps_16 3′UTR was used to stabilize _aph_A-6 gene transcripts (Stern and Gruissem, 1987). The T7 gene 10 5′ UTR and _rps_16 3′ UTR facilitated 74.8% transgene expression in non-green edible parts (carrots) containing chromoplasts (grown under the ground in the dark) and 48% in proplastids, compared to chloroplasts in leaves (100%, Kumar et al., 2004). Therefore, it is reasonable to assume that the _aph_A-6 gene is expressed in non-green and green plastids in the light or dark. The _npt_II gene in the cotton plastid transformation vector is driven by the _psb_A 5′ and _psb_A 3′ UTRs, which have been repeatedly shown to be responsible for light regulated expression of transgenes integrated into the plastid genome (Dhingra et al., 2004; Devine and Daniell, 2004; Daniell et al., 2004a, b; Fernandez San Millan et al., 2003; Staub & Maliga, 1995). Thus, it is logical to expect a breakdown of kanamycin in both dark and light conditions Therefore, a combination of both aph_A_-6 and aph_A_-2 genes driven by regulatory signals in the light and in the dark, in both proplastids and chloroplasts, provides continuous protection for transformed plastids/chloroplasts from the selectable agent. These approaches helped in the optimization of transformation procedure of cotton cell culture and to achieve a high frequency of transformation. Such optimized conditions further helped to obtain transformation even with a single selectable marker _aph_A-6. Presence of two antibiotic resistance genes should not pose any problem because several methods are currently available to eliminate marker genes from transformed chloroplast genomes (Fischer et al., 1996; Iamtham and Day, 2000; Corneille et al., 2001; Hajdukiewicz et al., 2001; Klaus et al., 2004).
The number of independent resistant calli obtained with pDD-_Gh_-_aph_A-6/_np_tII vector is shown in the Table 1. All of these independent resistant calli tested positive by PCR without any escape or mutation. Therefore, all tested lines were plastid transformants. Out of seven lines confirmed by Southern blots for site-specific integration (Figure 3E, Line 2–8), three cell lines showed best morphogenic response. These cultures were chosen to produce elongated somatic embryos (see Figure 2D). Hypocotyl of elongated embryos were dissected into small pieces to induce callus and somatic embryos. Plants produced from first two lines were grown in the growth chamber. T1 seedlings from these lines were tested by Southern blots and showed homoplasmy (Figure 3F, Line 2 and 3). Third line maintained in vitro in the callus form, derived from hypocotyl of elongated somatic embryos was also observed to be homoplasmic (Figure 3E, Line 4). About 5–20 somatic embryos were derived from transgenic calli after each subculture. Therefore, generating several transgenic plants from a single culture is quite feasible.
In summary, transgenic calli were selected (on MST1 medium containing 50 mg/l kanamycin) within 8 weeks after bombardment. Selected cell cultures were repeatedly subcultured every month on higher selection pressure (100 mg/l kanamycin), up to 4 months, in order to increase the number of transgenic chloroplasts in cell cultures (visually green in color). To induce somatic embryogenesis, cell cultures were plated for 3 months on medium (MST0 + 25 mg/l kanamycin) and well-differentiated somatic embryos were elongated on a Whatman #1 filter paper placed on medium (MST0 + 25 mg/l kanamycin) for a month. Further, hypocotyls of elongated somatic embryos were dissected into small pieces and placed on selection medium (MST1 + 100 mg/l kanamycin) for 2 months to induce the callus. Induced callus was again plated for induction of embryos and subsequently for elongation of somatic embryos into plantlets. Overall, it took about 18 months to obtain a homoplasmic transgenic plant from the bombarded pro-embryogenic cotton calli. Thus, plastid transformation in cotton is a slow process but it is quite reproducible
Acknowledgments
The authors are thankful to Professor Saul Purton for providing the _aph_A-6 coding sequence and Professor Deepak Pental for providing cotton germplasm Coker310FR. Investigations reported in this article were supported in part by funding from NIH R 01 GM63879 and USDA 3611-21000-017-00D grants to HD.
References
- Bateman JM, Purton S. Tools for chloroplast transformation in Chlamydomonas: expression vectors and a new dominant selectable marker. Mol Gen Genet. 2000;263:404–410. doi: 10.1007/s004380051184. [DOI] [PubMed] [Google Scholar]
- Bendich AJ. Circular chloroplast chromosomes: the grand illusion. Plant Cell. 2004;16:1661–1666. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad L. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 2000;18:257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- Carrer H, Hockenberry TN, Svab Z, Maliga P. Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol Gen Genet. 1993;241:49–56. doi: 10.1007/BF00280200. [DOI] [PubMed] [Google Scholar]
- Chiu WL, Sears BB. Electron microscopic localization of replication origins in Oenothera chloroplast DNA. Mol Gen Genet. 1992;232:33–39. doi: 10.1007/BF00299134. [DOI] [PubMed] [Google Scholar]
- Christou P. Genetic transformation of crop plants using microprojectile bombardment. Plant J. 1992;2:275–281. [Google Scholar]
- Cline J, Braman JC, Hogrefe HH. PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneille S, Lutz K, Svab Z, Maliga P. Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox site-specific recombination system. Plant J. 2001;27:171–178. doi: 10.1046/j.1365-313x.2001.01068.x. [DOI] [PubMed] [Google Scholar]
- Daniell H. Transformation and foreign gene expression in plants mediated by micoprojectile bombardment. Methods Mol Biol. 1997;62:463–489. doi: 10.1385/0-89603-480-1:463. [DOI] [PubMed] [Google Scholar]
- Daniell H. Genetically modified food crops: current concerns and solutions for next generation crops. In: Harding SE, editor. Biotechnology and Genetic Engineering Reviews, Intercept, Andover. Vol. 17. 2000. pp. 327–352. [DOI] [PubMed] [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A. Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol. 2002;13:136–141. doi: 10.1016/s0958-1669(02)00297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Parkinson CL. Jumping genes and containment. Nat Biotechnol. 2003;21:374–375. doi: 10.1038/nbt0403-374. [DOI] [PubMed] [Google Scholar]
- Daniell H, Carmona-Sanchez O, Burns B. Chloroplast derived antibodies, biopharmaceuticals and edible vaccines. In: Schillberg S, editor. Molecular Farming. Wiley-VCH Verlag publishers; Germany: 2004a. pp. 113–133. [Google Scholar]
- Daniell H, Cohill P, Kumar S, Dufourmantel N. Chloroplast genetic engineering. In: Daniell H, Chase C, editors. Molecular Biology and Biotechnology of Plant Organelles. Kluwer Academic Publishers; Dordrecht: 2004b. pp. 423–468. [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A, Allison L. Chloroplast transformation: from basic molecular biology to biotechnology. Rev Plant Physiol Biochem. 2002a;1:1–20. [Google Scholar]
- Daniell H, Khan MS, Allison L. Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci. 2002b;7:84–91. doi: 10.1016/s1360-1385(01)02193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Krishnan M, McFadden BF. Transient expression of beta-glucuronidase in different cellular compartments following biolistic delivery of foreign DNA into wheat leaves and calli. Plant Cell Rep. 1991;9:615–619. doi: 10.1007/BF00231800. [DOI] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO. Expression of cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol. 2001;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Ruiz ON, Dhingra A. Chloroplast genetic engineering to improve agronomic traits in transgenic Plants. Methods Mol Biol. 2004c;286:111–137. doi: 10.1385/1-59259-827-7:111. [DOI] [PubMed] [Google Scholar]
- Daniell H, Vivekananda J, Nielsen BL, Ye GN, Tewari KK, Sanford JC. Transient foreign gene expression in chloroplasts of cultured tobacco cells after biolistic delivery of chloroplast vectors. Proc Natl Acad Sci USA. 1990;87:88–92. doi: 10.1073/pnas.87.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt Cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- Devine A, Daniell H. Chloroplast genetic engineering for enhanced agronomic traits and expression of proteins for medical/industrial applications. In: Møller SG, editor. Plastids. Blackwell publishing; Oxford: 2004. pp. 283–320. [Google Scholar]
- Dhingra A, Daniell H. Chloroplast genetic engineering via organogenesis or somatic embryogenesis. Arabidopsis protocols. 2004;2 doi: 10.1385/1-59745-003-0:245. in press. [DOI] [PubMed] [Google Scholar]
- Dhingra A, Portis AR, Jr, Daniell H. Enhanced translation of a chloroplast expressed RbcS gene restores SSU levels and photosynthesis in nuclear antisense RbcS plants. Proc Natl Acad Sci USA. 2004;101:6315–6320. doi: 10.1073/pnas.0400981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-San Millan A, Mingo-Castel A, Daniell H. Chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J. 2003;1:71–79. doi: 10.1046/j.1467-7652.2003.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer JJ, McMullen MD. Transformation of cotton Gossypium hirsutum L. via particle bombardment. Plant Cell Rep. 1990;8:586–589. doi: 10.1007/BF00270059. [DOI] [PubMed] [Google Scholar]
- Fischer N, Stampacchia O, Redding K, Rochaix JD. Selectable marker recycling in the chloroplast. Mol Gen Genet. 1996;251:373–380. doi: 10.1007/BF02172529. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Yamamoto K, Kobayashi I. Dependence of frequency of homologous recombination on the homology length. Genetics. 1995;140:797–809. doi: 10.1093/genetics/140.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Guda C, Lee SB, Daniell H. Stable expression of biodegradable protein based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- Hagemann R. The sexual inheritance of plant organelles. In: Daniell H, Chase C, editors. Molecular Biology and Biotechnology of Plant Organelles. Kluwer Academic Publishers; Dordrecht: 2004. pp. 87–108. [Google Scholar]
- Hajdukiewicz PTJ, Gilbertson L, Staub JM. Multiple pathways for Cre/lox-mediated recombination in plastids. Plant J. 2001;27:161–170. doi: 10.1046/j.1365-313x.2001.01067.x. [DOI] [PubMed] [Google Scholar]
- Hedrick LA, Heinhorst S, White MA, Cannon GC. Analysis of soybean chloroplast DNA replication by two-dimensional gel electrophoresis. Plant Mol Biol. 1993;23:779–792. doi: 10.1007/BF00021533. [DOI] [PubMed] [Google Scholar]
- Hilder VA, Boulter D. Genetic engineering of crop plants for insect resistance – a critical review. Crop Protection. 1999;18:177–191. [Google Scholar]
- Hou BK, Zhou YH, Wan LH, Zhang ZL, Shen GF, Chen ZH, Hu ZM. Chloroplast transformation in oilseed rape. Transgenic Res. 2003;12:111–114. doi: 10.1023/a:1022180315462. [DOI] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003a;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. Organelle evolution meets biotechnology. Nat Biotechnol. 2003b;21:489–490. doi: 10.1038/nbt0503-489. [DOI] [PubMed] [Google Scholar]
- Huang FC, Klaus S, Herz S, Zou Z, Koop HU, Golds T. Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Mol Gen Genet. 2002;268:19–27. doi: 10.1007/s00438-002-0738-6. [DOI] [PubMed] [Google Scholar]
- Iamtham S, Day A. Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol. 2000;18:1172–1176. doi: 10.1038/81161. [DOI] [PubMed] [Google Scholar]
- Khan MS, Maliga P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol. 1999;17:910–915. doi: 10.1038/12907. [DOI] [PubMed] [Google Scholar]
- Klaus SM, Huang FC, Golds TJ, Koop HU. Generation of marker-free plastid transformants using a transiently cointegrated selection gene. Nat Biotechnol. 2004;22:225–229. doi: 10.1038/nbt933. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Tewari KK. Chloroplast DNA from higher plants replicates by both the Cairns and rolling circle mechanism. Nature. 1975;256:708–711. doi: 10.1038/256708a0. [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski SF, Gould F, William MJ. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Daniell H. Engineering the chloroplast genome for hyper-expression of human therapeutic proteins and vaccine antigens in Recombinant Protein Protocols. Methods Mol Biol. 2004;267:365–383. doi: 10.1385/1-59259-774-2:365. [DOI] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Plastid expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots and leaves confers enhanced salt tolerance. Plant Physiol. 2004;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Sharma P, Pental D. A genetic approach to in vitro regeneration of non regenerating cotton (Gossypium hirsutum L.) cultivars. Plant Cell Rep. 1998;18:59–63. [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL. Chloroplast DNA replication: mechanism, enzymes and replication origins. J Plant Biochem Biotechnol. 1997a;6:1–7. [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL. Fine mapping of replication origins (oriA and oriB) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res. 1997b;25:3681–3686. doi: 10.1093/nar/25.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Shi F, Nielsen BL. Analysis of the tobacco chloroplast DNA replication origin (oriB) downstream of the 23S rRNA gene. J Mol Biol. 1997;268:273–283. doi: 10.1006/jmbi.1997.0972. [DOI] [PubMed] [Google Scholar]
- Lax AR, Vaughn KC, Duke SO, Endrizzi JE. Structural and physiological studies of a plastome cotton mutant with slow sorting out. J Hered. 1987;78:147–152. [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, MO, Daniell H. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breeding. 2003;11:1–13. [Google Scholar]
- Llewellyn D, Fitt G. Pollen dispersal from two field trials of transgenic cotton in the Namoi valley, Australia. Mol Breeding. 1996;2:157–166. [Google Scholar]
- Lu Z, Kunnimalaiyaan M, Nielsen BL. Characterization of replication origins flanking the 23S rRNA gene in tobacco chloroplast DNA. Plant Mol Biol. 1996;32:693–706. doi: 10.1007/BF00020210. [DOI] [PubMed] [Google Scholar]
- Lugo SK, Kunnimalaiyaan M, Singh NK, Nielsen BL. Required sequence elements for chloroplast DNA replication activity in vitro and in electroporated chloroplasts. Plant Science. 2004;166:151–161. [Google Scholar]
- Maliga P. Progress towards commercialization of plastid transformation technology. Trends Biotechnol. 2003;21:20–28. doi: 10.1016/s0167-7799(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Meeker R, Nielsen B, Tewari KK. Localization of replication origins in pea chloroplast DNA. Mol Cell Biol. 1988;8:1216–1223. doi: 10.1128/mcb.8.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlbauer SK, Lossl A, Tzekova L, Zou Z, Koop HU. Functional analysis of plastid DNA replication origins in tobacco by targeted inactivation. Plant J. 2002;32:175–184. doi: 10.1046/j.1365-313x.2002.01408.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nielsen BL, Lu Z, Tewari KK. Characterization of the pea chloroplast DNA oriA region. Plasmid. 1993;30:197–211. doi: 10.1006/plas.1993.1052. [DOI] [PubMed] [Google Scholar]
- Reddy MK, Choudhury NR, Kumar D, Mukherjee SK, Tewari KK. Characterisation and mode of in vitro replication of pea chloroplast oriA sequences. Eur J Biochem. 1994;220:933–941. doi: 10.1111/j.1432-1033.1994.tb18697.x. [DOI] [PubMed] [Google Scholar]
- Reddy VS, Leelavathi S, Selvapandian A, Raman R, Ferraiolo G, Shukla V, Bhatnagar RK. Analysis of chloroplast transformed tobacco plants with cry1Ia5 under rice psbA transcriptional elements reveal high level expression of Bt toxin without imposing yield penalty and stable inheritance of transplastome. Mol Breeding. 2002;9:259–69. [Google Scholar]
- Ruf S, Hermann M, Berger I, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- Ruiz ON, Hussein H, Terry N, Daniell H. Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol. 2003;132:1344–1352. doi: 10.1104/pp.103.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Huang HV. Homologous recombination in Escherichia coli - Dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PTJ, Staub JM, Nehra NS. Technical advance: stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Sikdar SR, Serino G, Chaudhuri S, Maliga P. Plastid transformation in Arabidopsis thaliana. Plant Cell Rep. 1998;18:20–24. [Google Scholar]
- Silhavy D, Maliga P. Mapping of promoters for the nucleus encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr Genet. 1998;33:340–344. doi: 10.1007/s002940050345. [DOI] [PubMed] [Google Scholar]
- Skarjinskaia M, Svab Z, Maliga P. Plastid transformation in Lesquerella fendleri, an oilseed Brassicacea. Transgenic Res. 2003;12:115–122. doi: 10.1023/a:1022110402302. [DOI] [PubMed] [Google Scholar]
- Staub JM, Maliga P. Expression of a chimeric uidA gene indicates that polycistronic messenger-RNAs are efficiently translated in tobacco plastids. Plant J. 1995;7:845–848. doi: 10.1046/j.1365-313x.1995.07050845.x. [DOI] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, Ward D, Ye GN, Russell DA. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- Stegemann S, Hartmann S, Ruf S, Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci USA. 2003;100:8828–8833. doi: 10.1073/pnas.1430924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Gruissem W. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987;51:1145–57. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Trolinder NL, Goodin JR. Somatic embryogenesis in cotton (Gossypium hirsutum) 1. Effects of source of explant and hormone regime. Plant Cell Tiss Org Cul. 1988;12:178–181. [Google Scholar]
- Trolinder NL, Chen XX. Genotype specificity of the somatic embryogenesis in cotton. Plant Cell Rep. 1989;8:133–136. doi: 10.1007/BF00716824. [DOI] [PubMed] [Google Scholar]
- Umbeck PF, Barton KA, Nordheim EV, McCarty JC, Parrot WL, Jenkins JN. Degree of pollen dispersal by insects from a field test of genetically engineered cotton. J Econ Entomol. 1991;84:1943–1950. [Google Scholar]