Rsx, a metatherian RNA with Xist-like properties (original) (raw)
. Author manuscript; available in PMC: 2012 Oct 31.
Published in final edited form as: Nature. 2012 Jul 12;487(7406):254–258. doi: 10.1038/nature11171
Abstract
In female (XX) mammals one of the two X chromosomes is inactivated to ensure an equal dose of X-linked genes with males (XY)1. X-inactivation in eutherian mammals is mediated by the non-coding RNA Xist2. Xist is not found in metatherians3 and how X-inactivation is initiated in these mammals has been the subject of speculation for decades4. Using the marsupial Monodelphis domestica we identify Rsx (_R_NA-on-the-_s_ilent X), an RNA that exhibits properties consistent with a role in X-inactivation. Rsx is a large, repeat-rich RNA that is expressed only in females and is transcribed from, and coats, the inactive X chromosome. In female germ cells, where both X chromosomes are active, Rsx is silenced, linking Rsx expression to X-inactivation and reactivation. Integration of an Rsx transgene on an autosome in mouse embryonic stem cells leads to gene silencing in cis. Our findings permit comparative studies of X-inactivation in mammals and pose questions about the mechanisms by which X-inactivation is achieved in eutherians.
X-dosage compensation mechanisms vary between metazoans5. In metatherians, X chromosome inactivation (XCI) is imprinted, affecting the paternal X chromosome6, but the factors that drive XCI in these mammals are unknown4. The metatherian and eutherian female inactive Xs share common epigenetic features7, 8, 9, suggesting that XCI in these mammals proceeds via a similar mechanism. Notably, the metatherian inactive X is enriched for H3 lysine-27 trimethylation (H3K27me3)7, 8, 9, 10. In eutherians, this H3K27me3 enrichment is _Xist_-dependent11, 12. We therefore proposed that an unidentified X-linked RNA initiates XCI in metatherians8. Xist RNA is expressed in female but not male somatic tissues, coats the inactive X chromosome, and is expressed from the inactive X13, 14, 15, 16. We posited that a regulator of XCI in metatherians would also exhibit these unusual properties.
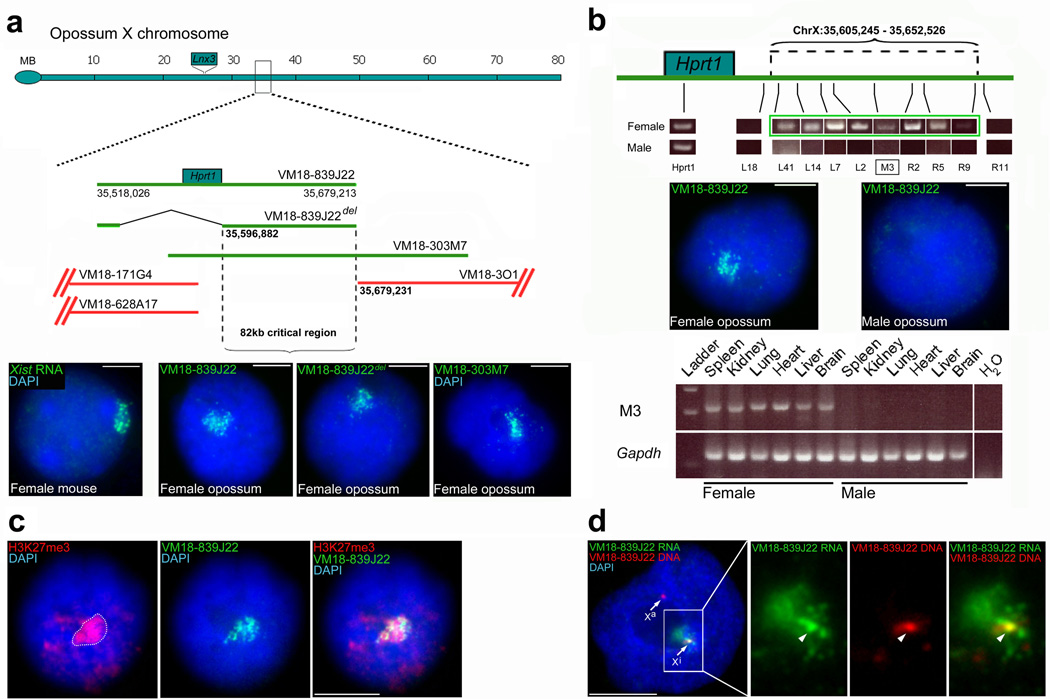
While analyzing XCI in the female brain of Monodelphis domestica, the short-tailed opossum, we used RNA FISH to study the expression of the X-gene Hprt1 with a BAC, VM18-839J22, containing Hprt1 plus 49kb of upstream and 82kb of downstream sequence, and in which no other known genes mapped (Fig 1a). RNA FISH signals usually appear as pinpoint dots. However the RNA signal detected resembled a cloud (Fig. 1a, Supplementary Figure 1) that was reminiscent of the Xist RNA cloud seen in female mouse (Fig. 1a) and human cells15. We observed the same RNA cloud using a modified form of the BAC carrying an Hprt1 deletion (Fig. 1a). The RNA therefore originated from another, uncharacterised gene located within the genomic region defined by VM18-839J22. RNA FISH using additional BACs narrowed down this region to 82kb downstream of Hprt1 (Fig. 1a). We identified the RNA using RTPCR on female brain cDNA with primers located along this critical region (Fig. 1b; Supplementary Table 1), revealing a transcription unit spanning 47kb (Fig. 1b).
Figure 1. Discovery of a candidate X-inactivating RNA in the opossum.
a, RNA FISH mapping of the novel gene using BACs; green BACs give RNA FISH cloud signals, red BACs do not. BAC VM18-839J22 gives a cloud signal (green; second panel) identical to Xist RNA (first panel) as seen in mouse brain cells (additional RNA cloud images Supplementary Figure 1). The cloud is still observed with VM18-839J22_del_ (third panel), deleted for Hprt1 (Supplementary Table 1 for recombineering sequences) and VM18-303M7 (fourth panel). The 82kb critical interval is defined by VM18-839J22_del_ and VM18-3O1. The Lnx3 gene which gave rise to Xist3, maps to a locus distinct from Rsx. MB = megabases b, top: RTPCR identification of a female-specific 47kb transcript (green rectangle) within the 82kb critical interval (primers Supplementary Table 1). Transcript limits shown above the X. middle: RNA FISH images showing RNA clouds in female but not male brain cells. bottom: RTPCR using primer pair M3 (black rectangle in first RTPCR image) shows female-specific expression in all tissues. Gapdh is an autosomal control. c, combined VM18-839J22 RNA FISH / H3K27me3 immunostaining shows the inactive X (marked by dotted line in first panel) coated with the novel RNA. d, combined VM18-839J22 RNA and DNA FISH for the novel RNA. No RNA signal is observed from the active X locus (Xa), but an RNA signal colocalizes with the DNA locus on the inactive X (Xi). . Scale bars 5µm.
We then investigated whether the RNA exhibited other _Xist_-like features. First, we looked for evidence of sexually dimorphic expression. No RNA clouds were detected in male opossum brain by VM18-839J22 BAC RNA FISH (Fig. 1b), demonstrating that in this tissue expression of the RNA was female-specific. Consistent with this, RTPCR on male brain cDNA revealed no expression of the 47kb transcript previously identified in females (Fig. 1b). RTPCR on a broad array of tissues, representing derivatives of endoderm, mesoderm and ectoderm, from both males and females revealed expression of the RNA in all female but not male tissues examined (Fig. 1b).
Next, we established whether the RNA coats the inactive X. We combined VM18-839J22 RNA FISH on female brain cells with immunostaining for the inactive X marker H3K27me3. We observed colocalization of RNA clouds and H3K27me3 signals (Fig. 1c), demonstrating inactive X coating.
To determine if the RNA was expressed from the inactive X, we performed dual RNA/DNA FISH using BAC VM18-839J22. No RNA signal was seen colocalizing with the DNA signal on the active X (Fig. 1d). In contrast, an RNA signal was observed colocalizing with the DNA signal on the inactive X (Fig. 1d). This RNA signal was brighter than others in the surrounding cloud, a feature characteristic of a site of nascent RNA synthesis. Thus, the RNA is expressed only from the inactive X. This must be the paternal X, as this chromosome is always chosen for inactivation6. In summary, like Xist, the RNA that we identified is female-specific, coats the inactive X and is transcribed only from the inactive X. We call the RNA Rsx (_R_NA-on-the-_s_ilent X).
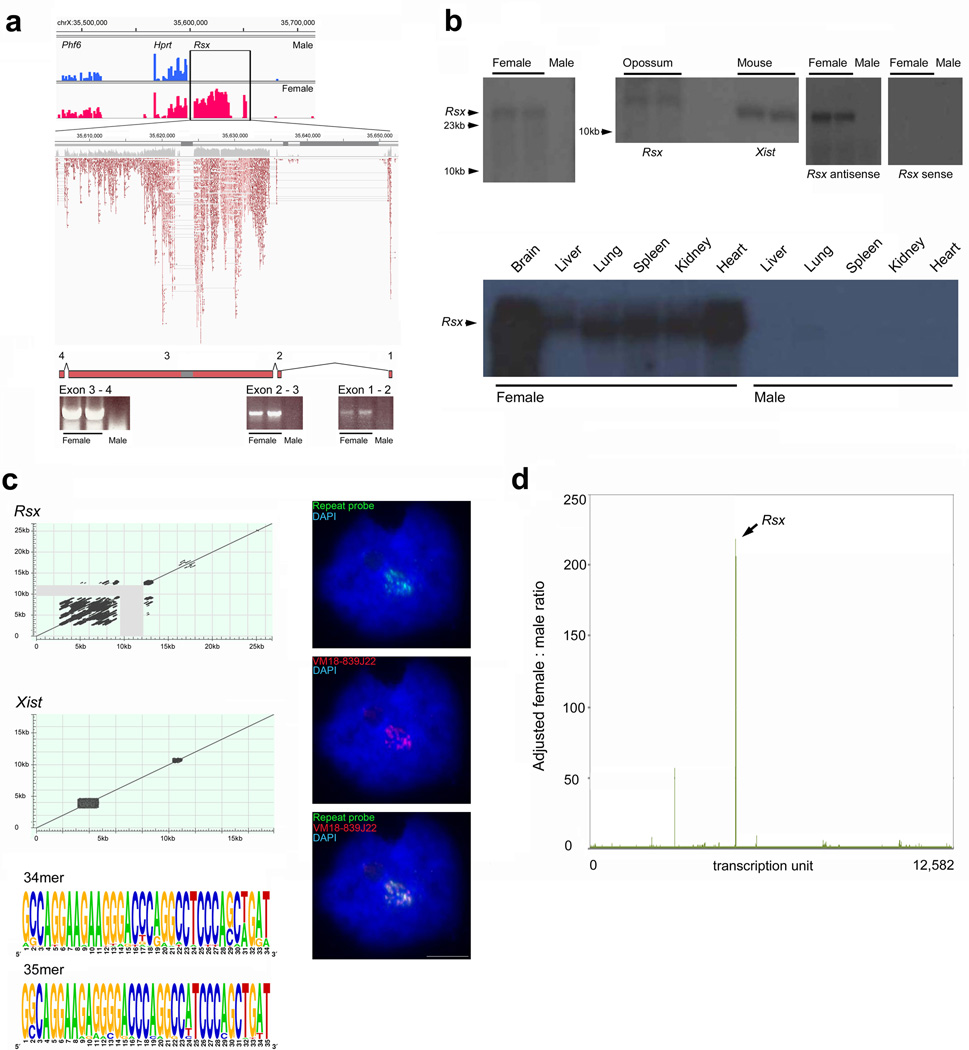
To further characterize Rsx, we performed RNA-sequencing (RNA-Seq) on female opossum brain (Fig. 2a). This confirmed that the Rsx gene generates a precursor RNA of 47kb (UCSC MonDom5 co-ordinates: chrX 35,605,415-35,651,609) transcribed antisense relative to Hprt1. Split RNA reads implied that Rsx encodes a spliced RNA comprised of four exons: this was confirmed by RTPCR (Fig. 2a, Supplementary Table 1). The RNA-Seq data predicted that the mature Rsx RNA is large, approximating 27kb, with 25kb of sequence deriving from a single exon. Northern blots confirmed that Rsx RNA was large, exceeding the 17kb mouse Xist RNA in size, and validated the strandedness, female-specificity and broadness of Rsx expression (Fig. 2b). The level of Rsx expression varied between female tissues, an observation also noted for Xist (Supplementary Figure 2). 3’-RACE demonstrated that Rsx transcripts are polyadenylated.
Figure 2. Characterisation of the Rsx RNA.
a, RNA-Seq shows female specific expression of Rsx,while reads mapping to the Phf6 and Hprt1 genes are found in both sexes. Boxed area shows magnification of Rsx locus. Pale red reads are ambiguous, i.e. they hit multiple repeats within the Rsx RNA. Inferred exons and their verification by RTPCR are shown. Dark grey bars along the chromosome co-ordinate axis represent DNA sequence gaps (Online Methods). b, Northern analysis of Rsx. (controls: Supplementary Figure 2, primers for probes: Supplementary Table 1). Top left panel: female-specific expression of an RNA greater than 23kb. Middle panel: size verification of Rsx RNA by comparison with the 17kb Xist RNA. Right two panels: verification of the strandedness of Rsx transcription (sequences in Supplementary Table 1). Bottom panel: multi-tissue blot showing female-specific Rsx expression in all tissues. c, Comparison of repeat organisation with that of Xist by sequence-similarity plots, window size = 28 nucleotides (grey area represents the unsequenced 2.8kb). The 5-prime 12kb stretch of repeats includes two highly conserved 34 and 35mer motifs (Supplementary Figure 3 for predicted 34mer stem-loops). RNA FISH using a repeat probe (green) co-localises with BAC VM18-839J22 (red; antisense probe, Supplementary Table 1). A sense probe generates no signals (data not shown), confirming the transcription orientation of Rsx. d, Female:male adjusted ratios inferred from brain RNA-Seq data identifying Rsx as a candidate XCI RNA (Online Methods). The second highest ranking RNA, MAPK4K, was found by RTPCR not to be female-specific in other tissues, so can be excluded as an XCI candidate. Scale bars 5µm.
Sequence comparisons between Rsx and Xist revealed no significant homology. Nevertheless Rsx exhibited features reminiscent of Xist. Notably, it was highly enriched in tandem repeats biased towards the 5-prime end of the RNA (Fig. 2c) and exhibiting high GC content. The Rsx repeats included two highly conserved and similar motifs with potential to form stem loop structures (Fig. 2c; Supplementary Figure 3). RNA FISH using an oligonucleotide probe recognising one of these repeats gave a cloud signal indistinguishable from that seen using VM18-839J22 BAC, confirming that the repeats are included in the RNA that coats the inactive X (Fig. 2c). The longest ORF found for Rsx constituted less than five percent of the total RNA length, and was located in the repeat region, suggesting that Rsx functions as a non-coding RNA. We conclude that the Rsx and Xist RNAs display similar features.
RNA-Seq has been used previously to identify novel transcripts17 We speculated that analysis of RNA-Seq data alone would identify Rsx as a candidate XCI RNA. An RNA with a role in XCI would be X-linked and expressed only in females.and so should be evident in a comparison of female and male transcriptomes. To identify X-linked genes with sexually dimorphic expression levels, we compared the numbers of reads mapping to each region of the X in the female with that in the male brain and expressed this as a female:male ratio (Supplementary Table 2; and Methods). When all transcribed regions on the X were examined, Rsx was an outlier, with a female:male ratio exceeding the second-ranked RNA by three-fold (Fig. 2d). We repeated this RNA-Seq approach on liver, in which the level of Rsx expression is low (Fig. 2b). In this analysis, Rsx appeared second (Supplementary Table 2). Thus, RNA-Seq can be used as a preliminary discovery tool to identify RNAs involved in dosage compensation.
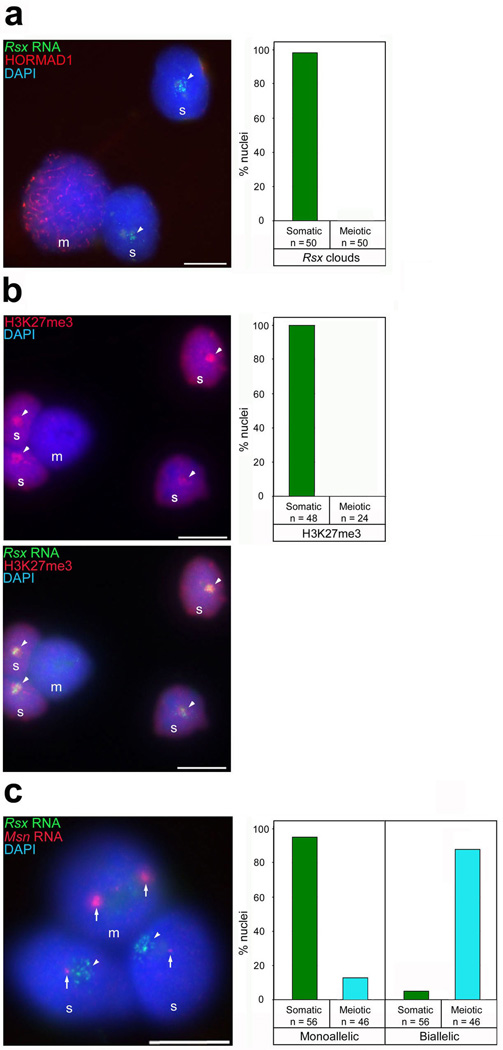
To investigate a link between Rsx RNA and XCI, we examined Rsx expression in the female germ line. In mice, Xist is expressed in somatic tissues but is silenced during oocyte development. This is accompanied by loss of H3K27me3 from the inactive X and X-reactivation18, 19, 20. Similar to other somatic cells, supporting cells in the ovary displayed Rsx clouds (Fig. 3a) and XCI, as shown by X chromosome H3K27me3 enrichment (Fig. 3b) and monoallelic expression of the X-gene Msn (Fig. 3c). However, in germ cells, identified by HORMAD1 immunostaining21, Rsx clouds were absent (Fig. 3a). Consistent with a relationship between Rsx expression and XCI, most meiotic cells had two active Xs, with no X chromosome H3K27me3 accumulation (Fig. 3b), and biallelic Msn expression (Fig. 3c). Rsx expression is therefore linked to X-inactivation and reactivation.
Figure 3. Links between Rsx RNA expression and X-inactivation and reactivation.
a, Rsx clouds (arrowheads) are present in supporting cells (‘s’) but not in meiotic cells (‘m’, labelled with HORMAD1). b, Rsx clouds (arrowheads) colocalise with the XCI marker H3K27me3 which is not observed in meiotic cells. c, RNA FISH for the X-gene Msn shows that while supporting cells undergo XCI (i.e. display a single RNA spot, arrow), meiotic cells have two active Xs. Msn RNA signals are very bright in meiotic cells due to increase in global transcription during this point in germ cell development35. Scale bars 5µm.
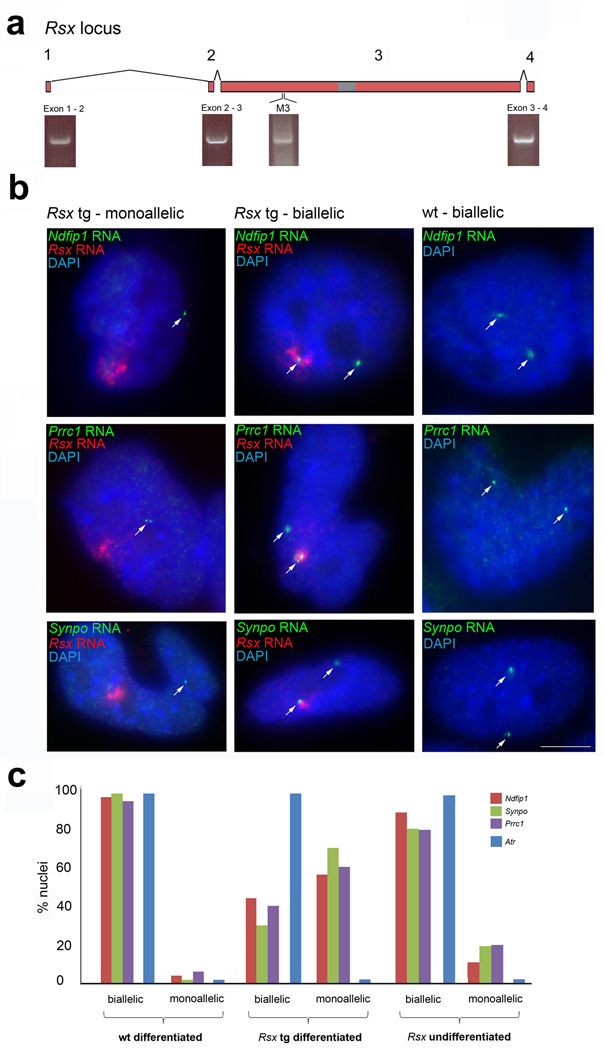
We next carried out experiments to address whether Rsx induces gene silencing. Xist transgenes function as ectopic X-inactivation centres in mouse embryonic stem (ES) cells, with Xist RNA coating the transgenic chromosome and inducing gene silencing in cis22, 23, 24. We generated an XX ES cell line, 303.2, carrying a single-copy chromosome 18-integrated transgene expressing full length Rsx RNA (Fig. 4a).
Figure 4. Autosomal gene silencing in mouse ES cells by an Rsx transgene.
a, in Rsx transgenic female ES cell clone 303.2, the full length Rsx transgene is expressed, as shown by RTPCR spanning all exon-exon boundary and ‘M3’ (see Figure 1) RTPCR. b, Rsx RNA appears as a cloud in 303.2 cells, indicating autosomal coating. RNA FISH for three chromosome 18 genes; Ndfip1, Prrc1 and Synpo, shows that this coating induces gene silencing in differentiated cells (first column), while in others silencing does not occur (middle column). Wild type ES cells show biallelic expression for each chromosome 18 gene (last column). Scale bars 5µm. c, quantitation of gene silencing (n>100 cells / gene) in differentiated 303.2 cells versus controls (PCR primers for chromosome 18 RNA FISH probes, Supplementary Table 1).
We performed RNA FISH for Rsx and three chromosome 18 genes, Ndfip1, Prrc1 and Synpo, mapping near the transgene integration site, in differentiated 303.2 ES cells. We observed coating of the transgenic chromosome by Rsx RNA (Fig. 4b). While Ndfip1, Prrc1 and Synpo were biallelically expressed in control ES cells (Fig. 4b,c), all three genes were silenced in more than one half of 303.2 ES cells (Fig. 4b,c). Silencing also occurred in undifferentiated 303.2 cells, albeit in a lower proportion than seen post-differentiation (Fig. 4c). This finding is reminiscent of that obtained with human XIST transgenes in mouse ES cells25. We conclude that Rsx expression can induce gene silencing in cis.
Finally, we looked for evidence of Rsx conservation among metatherians. Metatherians are divided into the South American and Australasian groups, which diverged 75–80 Mya26. We identified ESTs with homology to opossum Rsx in two Australasian marsupials, the brushtail possum and tammar wallaby (Supplementary Table 3). In support of a role in XCI, RTPCR demonstrated that in both organisms these ESTs were expressed only in females (Supplementary Figure 4, Supplementary Table 1). Rsx therefore originated prior to the major American / Australasian metatherian evolutionary split, implying a common mechanism of XCI in all metatherians.
Here we identify Rsx, an RNA with properties suggestive of a role in metatherian XCI. Our findings indicate that RNA-mediated dosage compensation mechanisms are widespread in the mammals. In eutherians, Xist is one of many non-coding RNAs expressed at the onset of XCI and colocated in the X-inactivation centre (XIC)27. Our work identifies a candidate XIC on the metatherian X and provides a point of focus for the identification of additional RNAs that regulate XCI and ensure that it is imprinted. These studies will deepen our understanding not only of XCI, but also of the evolution and mechanisms controlling genomic imprinting in mammals.
Our results raise questions about the epigenetics of eutherian XCI. Xist RNA has been proposed to spread along the X chromosome via LINE elements, which are abundant on the X chromosome 28. Genomic analyses have concluded that the opossum X is not enriched in LINEs29, however Rsx RNA can nevertheless coat the inactive X. Thus, other factors, such as nuclear matrix and scaffold proteins30, 31, may be more important primary determining factors for Rsx and potentially Xist spreading than LINEs. In addition, a study has found that in mice, imprinted inactivation of some X-linked genes proceeds in the absence of _Xist_32 (but see also33). It is therefore essential to determine whether an Rsx orthologue is present in eutherians and, if so, whether it is expressed and contributes to imprinted XCI in these mammals (Supplementary Discussion).
Previous work has shown that epigenetic features of XCI are conserved between metatherians and eutherians8, 9, 10. Although Rsx and Xist share no significant homology, arising independently during evolution, they exhibit similarities in secondary sequence features. Thus, many aspects of the XCI pathway appear to be have evolved convergently in metatherians and eutherians. With this in mind, it will be interesting to establish whether Rsx and Xist can replace one another in the XCI process. These experiments, as well as those that directly test the role of Rsx in metatherian XCI, would benefit from a genetically manipulable in vitro method for studying XCI in metatherians, akin to the ES cell system used in eutherians. The application of somatic cell reprogramming techniques34 to marsupials should now make this achievable.
Methods
Animals
Material for this study was acquired from opossums maintained at Southwest Foundation for Biomedical Research in San Antonio, Texas, USA (germ cell studies) and from opossums maintained at the MRC NIMR (all other studies) according to UK Home Office regulations. Tammar wallabies of Kangaroo Island, South Australia origin were maintained in a breeding colony in open grassy enclosures. Husbandry, handling and experiments were in accordance with the National Health and Medical Research Council of Australia/Commonwealth Scientific and Industrial Research Organization / Australian Research Council (2004) guidelines and all sampling techniques and collection of tissues were approved by the University of Melbourne Animal Experimentation Ethics Committees. Material from the brushtail possums was obtained from adult male and female possums housed at the Landcare Research Captive Animal Facility, Lincoln, New Zealand following the Landcare Research code of ethical conduct and in accordance with Part 6 of the New Zealand Animal Welfare Act 1999.
RNA FISH / DNA FISH / immunostaining
Combined RNA FISH / DNA FISH / immunostaining was carried out exactly as previously described36. All RNA FISH was carried out on primary cells harvested immediately post-mortem. Germ cells were harvested at both 14 d_pp_ and 17d_pp_.
Recombineering
BAC VM18-839J22 was electroporated into modified DH10B strain SW102 cells followed by selection using chloramphenicol. Competent BAC-containing SW102 cells were grown to competency and then electroporated with a recombineering construct that had been generated from a kanamycin resistance template using primers listed in Supplementary Table 1. Kanamycin resistant colonies were subsequently picked and the Hprt1 deletion verified using primers flanking the deletion site.
RNA-Seq
RNA was sequenced using a strand-specific protocol37 with the exception that total RNA was used instead of polyA+ fraction. Briefly, 3.5 µg of total RNA was reverse transcribed using Superscript II (Invitrogen) in the presence of 10 pmol of T18VN primer, 250 ng random hexamers (Promega), 120 ng of Actinomycin D (Sigma), 40 u RNAzin (Promega), 0.5 mM dNTP in total volume of 20 µl 1x reverse transcription buffer (Invitrogen). Reaction mixture was purified using QIAquick PCR purification kit (Qiagen) and second strand synthesis was performed using SuperScript double-stranded cDNA synthesis kit (Invitrogen) as recommended by the manufacturer with the exception that 200 µM dTTP was replaced with 400 µM dUTP. Double-stranded cDNA was fragmented using Bioruptor (Diagenode) (15 min, Low power, 30 sec ON, 30 sec OFF) in 50 µl volume. Sequencing libraries were prepared from fragmented cDNA using NEBNext DNA sample preparation kit (New England Biolabs) and Illumina PE adapters (PE-102–1003). Before final PCR amplification samples were treated with Uracil-N-Glycosylase (Applied Biosystems). Sequencing was performed on Illumina HiSeq 2000 sequencer in NIDDK Genomics core in 2×50 bp paired-end mode. After initial processing with Illumina pipeline, quality-filtered sequencing reads were aligned to monDom5 version of opossum genome assembly using BWA38. We generated in total 96 M reads for female brain sample and 56 M reads for male brain sample. Data were further processed using Samtools39 and visualized with IGV40.
Sequencing of Rsx RNA
RNA seq gives an overall predicted size of the mature Rsx RNA as 26,800 bp. Note that the predicted transcription start site differs when using RNA-Seq or 5’ RACE (co-ordinates in Supplementary Table 1). Split reads spanning the two sequence gaps on the right, implying that these gaps contain only intronic sequence (Figure 2a). We sequenced 8kb of the gaps located within this intron, and no RNA-Seq reads mapped to this sequence, suggesting that additional exons have not been missed. A third gap resides in the middle of the third exon of Rsx. PCR shows that this gap is 5kb, rather than 8kb according to the MonDom5 version of genome assembly. We sequenced 2.2kb of this gap, and encountered a short and highly repetitive unit that precludes sequencing of the remaining 2.8kb.
Female and male transcriptome analysis
RNA-Seq data was converted into a total of 12582 transcription units (top five hundred listed in Supplementary Table 2). Closely mapping and overlapping reads were amalgamated into 'blocks', where adjacent reads were amalgamated if there was no more than a 'coalescence distance' between them, measured between their nearest inside edges. Initially this was done with a 250 nt coalescence distance, and the male and female sample were analysed separately. A count was kept of the number of reads amalgamated into each block. In order to compare identical loci the sex-specific blocks were then amalgamated with each other using a longer coalescence distance of 2kb. These second level amalgamated blocks were then analysed for the ratio of female:male reads in each block. To prevent divide-by-zero errors, and suppress probable false positives where only small numbers of reads are involved, we added an arbitrary amount of noise (in this case 10 reads) to both female and male read counts for each block, before calculating the ratio. Blocks are then ranked by this 'noise adjusted' ratio. Note that the ratio of 150 for Rsx cited in the text is the average of three different regions of the RNA; individual ratios: 218, 205 and 28 (see also Supplementary Table 2).
Repeat predictions
For repeat prediction, the 5' region of the Rsx transcript was searched for over-represented motifs using CisFinder41. Motifs were then mapped back to the primary transcript using pairwise BLAST.
RNA extraction, RTPCR, cloning, northern blotting ,RACE and qPCR
RNA was extracted using Trizol (GIBCO BRL) according to manufacturer’s instructions. For RTPCR, 2µg of total RNA was treated with DNAse (Promega) for one hour at 37°C, before random hexameric reverse transcription using Superscript II (Gibco BRL) for 1.5 hours at 42°C. PCR primers (Supplementary Table 1) were designed using Primer3 and all PCRs were carried out using the parameters: 1 cycle: 94°C 3 mins, 35 cycles: 96°C 10 sec, 56°C 30 sec, 72°C 30sec, 1 cycle 72°C 10 mins. PCR products spanning exon-exon boundaries (Supplementary Table 1) were cloned into TOPO TA cloning vector (Clontech) for subsequent sequencing.
For northern blot analysis 10µg of total RNA, extracted as described above, was electrophoresed, together with RNA size markers, on a 0.8% agarose gel containing 1.9 M formaldehyde. RNA was transferred to Hybond-N membranes (Amersham) using 20X SSC and the membrane was hybridized overnight at 60°C for α32P-labelled probes and 42°C for γ32P-labelled oligonucleotide probes. For α32P-labelled probe experiments, filters were then washed at 60°C for 20 minutes in 2X SSC, 0.1% SDS then for 30 minutes in 0.5X SSC, 0.1% SDS and finally for 10 minutes in 0.2X SSC, 0.1% SDS. For γ32P-labelled oligonucleotide probe experiments, filters were washed at 42°C for 10 minutes in 6X SSPE, 0.1% SDS, then for 5 minutes in 2X SSC, 0.1% SDS. Note: The reason why Rsx is expressed at varying levels in different tissues is not clear, but may reflect differing requirements for this RNA in X-gene silencing in different cell types. The same phenomenon of variable expression is also observed for Xist (Supplementary Figure 2) so is not peculiar to this RNA.
5’ RACE was performed using the 5’ RACE system (Invitrogen) and 3’ RACE with the SMARTer RACE cDNA Amplification Kit (Clontech), in both cases using 1µg of total RNA from female opossum brain.
qPCR for copy number was carried out as previously described47.
ES cell derivation and differentiation
ES cell lines were established from a mouse transgenic for BAC VM18-303M7, according to published protocols42. Briefly, blastocysts were flushed from the uterus at 3.5d_pc_ and cultured in 2i/LIF for 10 days without feeders, during which time inner cell mass outgrowth occurred. This was dissociated in tryspin and passaged to generate ES cells. ES cells were differentiated using retinoic acid for three days exactly as described43.
Microscopy
Image acquisition was performed with an Olympus IX70 inverted microscope with a 100 W mercury arc lamp and a 1003/1.35 UPLAN APO oil immersion objective. Each fluorochrome image was captured separately as a 12-bit source image with a computer-assisted (SoftWoRx) liquid cooled charge-coupled device (Photometrics CH350L; Kodak KAF1400 sensor, 1317 3 1035 pixels).
Supplementary Material
merged SI data
supp table 1
supp table 2
supp table 3
Acknowledgments
We thank Donald Bell and Robin Lovell-Badge for advice on the characterisation of Rsx, Jeffrey Cloutier, and Grzegorz Polikiewicz for help with germ cell preparations and real-time PCR, NIDDK Genomics core (NIH) for RNA sequencing, Attila Toth (University Dresden) for the HORMAD1 antibody, the Biological Services unit at NIMR for animal husbandry and Julie Cocquet, Louise Reynard, Helen Byers and members of the Turner and Paul Burgoyne labs for critical reading of the manuscript. This work was supported by the Medical Research Council (U117588498, U117597141, U117581331, U117597137), the NIH (HD60858), the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, the New Zealand Foundation for Research, Science and Technology, Possum Biocontrol (C10X0501), the Australian National Health and Medical Research Council (1010453) and the NIDDK (NIH) Intramural Research Program.
Footnotes
Contributions
J.G and J.M.A.T conceived and designed the experiments, performed RNA FISH and RTPCR and wrote the manuscript. P.K. R.D.C-O and M.J.G generated and analysed RNA-Seq data. J.G, G.E and W.T performed repeat analysis. J.M.A.T and S.K.M performed northerns. M.N.S generated the transgenic ES cell line, and H.R determined ES cell transgene copy number. J.D., J.R.McC., J.L.V., and M.B.R provided animals and tissues.
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse, (Mus musculus L.) Nature. 1961;190:372. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Penny GD, et al. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 3.Duret L, et al. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312(5780):1653. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 4.Deakin JE, Chaumeil J, Hore TA, Marshall Graves JA. Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Res. 2009;17(5):671. doi: 10.1007/s10577-009-9058-6. [DOI] [PubMed] [Google Scholar]
- 5.Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- 6.Sharman GB. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature. 1971;230(5291):231. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- 7.Mahadevaiah SK, et al. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr. Biol. 2009;19(17):1478. doi: 10.1016/j.cub.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 8.Rens W, et al. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci U S A. 107(41):17657. doi: 10.1073/pnas.0910322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaumeil J, et al. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals. PLoS One. 6(4):e19040. doi: 10.1371/journal.pone.0019040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 11.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2(7):E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva J, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell. 2003;4(4):481. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 13.Brockdorff N, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 14.Borsani G, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 15.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 16.Brown CJ, et al. The human XIST gene: Analysis of a 17kb inactive X-specific RNA that contains conserved repeats and is highly localised within the nucleus. Cell. 1992;71:527. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10(1):57. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Napoles M, Nesterova T, Brockdorff N. Early loss of Xist RNA expression and inactive X chromosome associated chromatin modification in developing primordial germ cells. PLoS ONE. 2007;2(9):e860. doi: 10.1371/journal.pone.0000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimoto M, Abe K, et al. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3(7):e116. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuva de Sousa Lopes SM, et al. X chromosome activity in mouse XX primordial germ cells. PLoS Genet. 2008;4(2):e30. doi: 10.1371/journal.pgen.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojtasz L, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5(10):e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JT, Strauss WM, Dausman JA, Jaenisch R. A 450kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 23.Lee JT, Jaenisch R, et al. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 2007;386:275. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 24.Herzing LBK, Romer JT, Horn JM, Ashworth A, et al. Xist has properties of the X-inactivation centre. Nature. 1997;386:272. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 25.Heard E, et al. Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells. PNAS. 1999;96(12):6841. doi: 10.1073/pnas.96.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson MA, et al. Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biol. 8(7):e1000436. doi: 10.1371/journal.pbio.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Augui S, Nora EP, Heard E, et al. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12(6):429. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 28.Lyon MF. Do LINEs Have a Role in X-Chromosome Inactivation? J Biomed Biotechnol. 2006;2006(1):59746. doi: 10.1155/JBB/2006/59746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447(7141):167. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa Y, et al. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell. 19(3):469. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
merged SI data
supp table 1
supp table 2
supp table 3