Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences (original) (raw)
Main
Metatherians (‘marsupials’) comprise one of the three major groups of modern mammals and represent the closest outgroup to the eutherian (‘placental’) mammals (Supplementary Fig. 1). Metatherians and eutherians diverged ∼180 million years (Myr) ago, long before the radiation of the extant eutherian clades ∼100 Myr ago1,2. Although the metatherian lineage originally radiated from North America, only one extant species can be found there (the Virginia opossum), whereas all other species are found in South America (including more than 65 species of opossums and shrew opossums) and Australasia (∼200 species, including possums, kangaroos, koalas and many small insectivores and carnivores)3.
All sequenced mammalian genomes until now have come from eutherian species. Although metatherians and eutherians (together, ‘therians’) share many ancient mammalian characteristics, they have each evolved distinctive morphological and physiological traits. Metatherians are particularly noted for the birth of young at a very early stage of development, followed by a lengthy and complex lactational period. Genomic analysis will help reveal the genetic innovations that underlie the distinctive traits of each lineage4,5,6.
Equally important, metatherian genomes can shed light on the human genome. Comparative analysis of eutherians has greatly improved our understanding of the architecture and functional organization of mammalian genomes7,8,9,10. Identification of sequence elements thought to be under purifying selection, on the basis of cross-species sequence conservation, has led to increasingly refined inventories of protein-coding genes11,12, proximal and distal regulatory elements13,14 and putative RNA genes15. Yet, we still know relatively little about the evolutionary dynamics of these and other functional elements: how stable is the complement of protein-coding genes? How rapidly do regulatory sequences appear and disappear? From what substrate do they evolve?
Comparison of the human genome with genomes from distant outgroups such as birds (divergence ∼310 Myr ago) or fish (∼450 Myr ago) has provided valuable information. When similarity between sequences from such distantly related genomes can be detected, it surely signals functional importance; but the high specificity of these signals16 is offset by dramatically reduced sensitivity10,17,18. Simulations have shown that the feasibility of aligning orthologous genomic sequences declines rapidly once their mean genetic distance exceeds 1 substitution per site19. The genome of chicken, the most closely related non-mammalian amniote genome available, is separated from the human genome by approximately 1.7 substitutions per site in orthologous, neutrally evolving sequences20. Even moderately constrained functional elements may therefore be difficult to detect. In contrast, metatherian mammals are well positioned to address this issue: because unconstrained regions of their genomes are separated from that of human by only ∼1 substitution per site (see below), most orthologous, constrained sequence should be readily aligned.
Here we report the first high-quality draft of a metatherian genome sequence, which was derived from a female, grey, short-tailed opossum—Monodelphis domestica. The species was chosen chiefly on the availability and utility of the organism for research purposes. M. domestica is a small rapidly breeding South American species that has been raised in pedigreed colonies for more than 25 years and developed as one of only two laboratory bred metatherians21,22. M. domestica is being actively used as a model system for investigations in mechanisms of imprinting23,24,25, immunogenetics26,27,28, neurobiology, neoplasia and developmental biology (reviewed in ref. 6). For example, newborn opossums are remarkable in that they can heal complete transections of the spinal cord29. Elucidation of the molecular mechanisms underlying this ability promise important insights relevant to regenerative medicine concerning spinal cord or peripheral nerve injuries. Other than human, M. domestica is also the only mammal known in which ultraviolet radiation is a complete carcinogen for malignant melanoma30, and this has led to its establishment as a unique neoplasia model. All of these investigations will directly benefit from the development of genomic resources for this species.
Below we describe the generation of the draft sequence of the opossum genome, analyse its large-scale characteristics, and compare it to previously sequenced amniote genomes. Our key findings include:
• The distinctive features of the opossum genome provide an informative test of current models of genome evolution and support the hypothesis that biased gene conversion has a key role in determining overall nucleotide composition.
• The evolution of random inactivation of the X chromosome in eutherians correlates with acquisition of X-inactive-specific transcript (XIST), elevation in long interspersed element (LINE)/L1 density and suppression of large-scale rearrangements.
• The opossum genome seems to contain 18,000–20,000 protein-coding genes, the vast majority of which have eutherian orthologues. Lineage-specific genes largely originate from expansion and rapid turnover in gene families involved in immunity, sensory perception and detoxification.
• Identification of orthologues of highly divergent immune genes and a novel T-cell receptor isotype challenge previous claims that metatherians possess a ‘primitive’ immune system.
• Of the non-coding sequences conserved among eutherians, ∼20% seem to have evolved after the divergence from metatherians. Of protein-coding sequences conserved among eutherians, only ∼1% seems to be absent in opossum.
• At least 16% of eutherian-specific conserved non-coding elements are clearly derived from transposons, implicating these elements as an important creative force in mammalian evolution.
Extensions to these findings, as well as additional topics, are reported in a series of companion papers31,32,33,34,35,36,37,38,39,40,41.
Genome assembly and single nucleotide polymorphism discovery
We sequenced the genome of a partially inbred female opossum using the whole-genome shotgun (WGS) method7,42. The resulting WGS assembly has a total length of 3,475 megabases (Mb), consistent with size estimates based on flow cytometry (∼3.5–3.6 Gb; Supplementary Notes 1–2 and Supplementary Fig. 2). Approximately 97% of the assembled sequence has been anchored to eight large autosomes and one sex chromosome on the basis of genetic markers mapped by linkage analysis38 or fluorescence in situ hybridization43 (FISH; Supplementary Note 3). The draft genome sequence has high continuity, coverage and accuracy (Table 1; Supplementary Note 4 and Supplementary Tables 1–7).
Table 1 Genome assembly characteristics
To enable genetic mapping studies of opossum, we also created a large catalogue of candidate single nucleotide polymorphisms (SNPs). We identified ∼775,000 SNPs within the sequenced individual by analysing assembled sequence reads. We identified an additional ∼510,000 SNPs by generating and comparing ∼300,000 sequence reads from three individuals from distinct, partially outbred laboratory stocks maintained at the Southwest Foundation for Biomedical Research (San Antonio, Texas)22,44 (Supplementary Note 5). The SNP rates between the different stocks range from 1 per 360 to 1 per 140 bases and correlate with the distance between their geographical origins (Supplementary Table 8–10 and Supplementary Fig. 3).
The data from this study, including the draft genome assembly and SNPs, are freely available on our website (http://www.broad.mit.edu/mammals/opossum/) and have been deposited in appropriate public databases.
Genome landscape
The opossum genome has certain unusual properties that provide an opportunity to test recent models of genome evolution. The opossum autosomes are extremely large: they range from 257 Mb to 748 Mb, with the smallest being larger than the largest chromosome previously sequenced in any amniote (human chromosome 1). In contrast, the X chromosome is only ∼76 Mb long; this is substantially less than the size of the X chromosome in any sequenced eutherian. Studies of G-banding and chromosome painting have also shown that karyotypes and basic chromosomal organization are extraordinarily conserved throughout Metatheria, even between the distantly related American and Australasian lineages (∼55–80 Myr ago)5,45.
Sequence composition
Recent analyses have uncovered two major trends in the evolution of sequence composition in amniote genomes: first, most modern lineages seem to be experiencing a gradual decline in total G+C content relative to their common ancestors46; second, the local rate of recombination is positively correlated with local G+C content and, even more strongly, with the local density of CpG dinucleotides20,47. These observations have led to a proposed model48 whereby sequence composition reflects the balance between a genome-wide, (A+T)-biased mutation process and a localized recombination-mediated (G+C)-biased gene conversion process. This model predicts that the sequence composition of a genomic region is a function of its historical rate of recombination, with the frequency of hypermutable CpG dinucleotides being a particularly sensitive indicator.
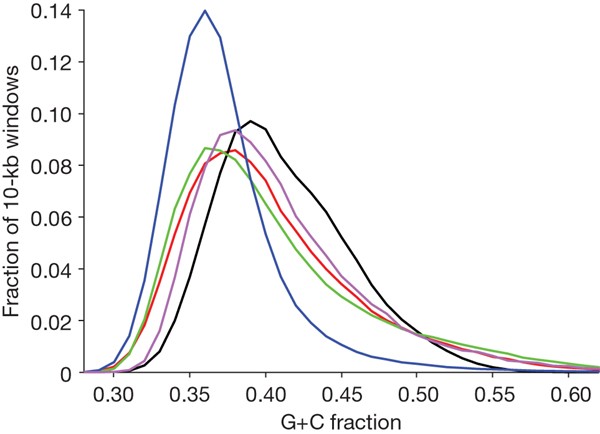
The opossum genome fits the predictions of this model well (see also refs 34, 35). Current linkage data38 show that the average recombination rate for the autosomes (∼0.2–0.3 cM Mb-1) is lower than in other sequenced amniotes (0.5–>3 cM Mb-1). Consistent with the proposed model, the mean autosomal G+C content (37.7%) is also lower than in other sequenced amniotes (40.9–41.8%) and, in particular, the mean autosomal density of CpGs (0.9%) is twofold lower than in other amniotes (1.7–2.2%). Because large-scale patterns of recombination seem to be relatively stable in the absence of chromosomal rearrangements49,50, the stability of the opossum karyotype suggests that the majority of the genome has experienced low recombination rates over an extended period. Indeed, the sequence composition is also more homogeneous than seen in other amniotes (Fig. 1).
Figure 1: Sequence composition in the opossum genome.
Distribution of G+C content in 10-kb windows across the genome in opossum (blue), human (red), mouse (black), dog (green) and chicken (purple).
The subtelomeric regions of autosomes are notable outliers with respect to sequence composition in the opossum genome, providing additional support for the biased gene conversion hypothesis. Cytological studies in opossum51,52 suggest that the rate of chiasmata formation (and hence meiotic recombination) is relatively uniform across each autosome in males, whereas it is strongly biased to subtelomeric regions in females. Consistent with a higher sex-averaged rate of recombination, mean G+C-content (41.6%) and CpG density (1.9%) are significantly elevated within ∼10 Mb of the chromosome ends (Supplementary Fig. 4).
Similarly, the very short X chromosome also supports the biased gene conversion hypothesis. Although few linkage data are currently available for opossum X chromosome, the average effective recombination rate must be at least 0.44 cM Mb-1, and thus larger than for the autosomes. (This estimate follows from the requirement of at least one meiotic crossover per bivalent in the female germ-line53,54.) The mean G+C content (40.9%) and CpG density (1.4%) of the X chromosome are substantially higher than for any of the autosomes (Supplementary Table 11). The opossum pattern is thus the opposite of that seen in eutherians, in which the X chromosome has low recombination and low G+C content and CpG density (Table 2).
Table 2 Comparative analysis of genome landscape in opossum and other amniotes
Segmental duplication
In human and other eutherians, segmental duplications (defined as pairs of regions with ≥90% sequence similarity over ≥1 kb) are associated with chromosomal fragility and syntenic breakpoints55,56. The relative karyotypic stability of metatherians therefore indicated that they might have a low proportion of segmental duplications.
The overall proportion of segmental duplication in opossum (1.7%) is indeed substantially lower than in other sequenced amniotes (2.5–5.3%). The segmental duplications are also relatively short: only 22 exceed 100 kb in opossum as compared with 483 in human (Supplementary Table 12). Additionally, the segmental duplications are more locally distributed: 76% are intrachromosomal (versus 46% for human) and the median distance between related duplications is 175 kb (versus 2.2 Mb for human). We find no indication that correction for over-collapsed duplications in the assembly would significantly alter these estimates (Supplementary Note 6 and Supplementary Table 13).
Transposable elements
Metatherian transposable elements largely belong to families also found in eutherians, but can be divided into more than 500 subfamilies, many of which are lineage specific (catalogued in Repbase57). At least 52% of the opossum genome can be recognized as transposable elements and other interspersed repeats (Table 2)33,35, which is more than in any of the other sequenced amniotes (34–43%). Notably, the opossum genome is significantly enriched in non-long terminal repeat (LTR) retrotransposons (LINEs, 29%), comprising copies of various LINE subfamilies. Given the low abundance of segmental duplications, accumulation of transposable elements seems to be the primary reason for the relatively large opossum genome size. The total euchromatic sequence that is not recognized as transposable elements is rather similar in opossum and human (1638 Mb versus 1568 Mb, respectively). The enrichment of LINEs may be related to the overall low recombination rate in opossum, inasmuch as studies of eutherian genomes have shown that LINEs occur at elevated densities in regions with low local recombination rates47.
Conserved synteny
Identification of syntenic segments between related genomes can facilitate reconstruction of chromosomal evolution and identification of orthologous functional elements. Starting from nucleotide-level, reciprocal-best alignments (‘synteny anchors’), we found that the opossum and human genomes can be subdivided (at a resolution of 500 kb) into 510 collinear segments with an N50 length (size x such that 50% of the assembly is in units of length at least x) of 19.7 Mb, which cover 93% of the opossum genome (Supplementary Fig. 5). If local rearrangements are disregarded, these segments can be further grouped into 372 blocks of large-scale, conserved synteny.
Extending this analysis to additional eutherians (mouse, rat and dog), with chicken as an additional outgroup, we created a high-resolution synteny map that reveals 616 blocks of conserved synteny across the five fully sequenced mammals (Supplementary Note 7, Supplementary Figs 6–7 and Supplementary Table 14). Because the majority of synteny breakpoints between human, mouse, rat and dog are clearly lineage specific (see also ref. 10), genomic regions that were probably contiguous in the last common boreoeutherian ancestor can be inferred by parsimony (Supplementary Note 8). We found that the mammalian synteny blocks can be used to infer 43 connected groups in the ancestral boreoeutherian genome (Supplementary Fig. 8). In fact, the largest 30 groups cover 95% of the human genome (see also ref. 58).
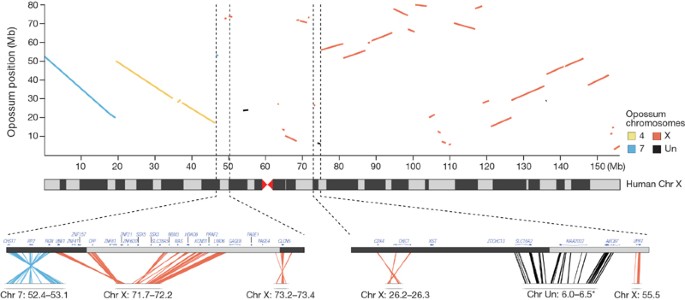
The resulting synteny map can be used to clarify chromosomal rearrangements during early mammalian evolution. For example, limited comparative mapping previously revealed that the eutherian X chromosome contains an ‘X-conserved region’ (XCR) that corresponds to the ancestral therian X chromosome, and an ‘X-added region’ (XAR), which was translocated from an autosome after the split from Metatheria59,60. The exact extent of the XCR has been unclear, however, owing to unclear synteny with non-mammalian out-groups at its boundary61. Using our high-resolution synteny map we can now confidently map the XAR–XCR fusion point to 46.85 Mb on human chromosome band Xp11.3 (Fig. 2).
Figure 2: Opossum–human synteny for the X chromsome.
The dot plot shows correspondence between the human chromosome (Chr) X and opossum chromosomes at a resolution of 300 kb. Expanded views, at a resolution of 50 kb, of the XAR–XCR fusion and the XIC are shown on the bottom left and right, respectively. In the XIC region, the closest contig on the distal flank (*) was not anchored in the monDom5 assembly (see Methods), but has been subsequently mapped near UPRT (opossum X chromosome ∼55 Mb) by FISH40.
X chromosome inactivation
In opossum and other metatherian mammals, dosage compensation for X-linked genes is achieved through inactivity of the paternally derived X chromosome in females62. In contrast, eutherian dosage compensation involves inactivation of the paternal X chromosome at spermatogenesis, reactivation in the early embryo, followed by random and clonally stable inactivation of one of the two X chromosomes in each cell of female embryos63. The random inactivation step is controlled by a complex locus known as the X inactivation centre (XIC). In the early female embryo, the non-coding XIST gene is transcribed from the XIC and coats one chromosome, in cis, to initiate silencing of the majority of its genes. It has been proposed that paternal X chromosome inactivation represents the ancestral therian dosage compensation system, and that random X chromosome inactivation is a recent innovation in the eutherian lineage64,65. The opossum genome sequence provides the first opportunity to test major hypotheses about the evolution of this system.
No XIST homologue in opossum
We searched all assembled and unassembled opossum WGS sequence for homology to the human and mouse XIC non-coding genes but, in agreement with a recent report66, did not find any significant alignments. (In particular, we found no match to the highly conserved 150-bp region overlapping the critical exon 4 of XIST; this region is so strongly conserved in the Eutheria that it should be readily detectable if present40.) Analysis of synteny in the regions surrounding the eutherian XIC also revealed that it has been disrupted by large-scale rearrangements (Fig. 2)40,41. In eutherians, the XIC is flanked by the ancient protein-coding genes CDX4_–_CHIC1 on one side and SLC16A2_–_RNF12 on the other side. In both chicken and frog these four genes are clustered in autosomal XIC homologous regions (which do not contain homologues of the XIC non-coding genes66). On the opossum X chromosome, however, these two pairs of genes are separated by ∼29 Mb (compared with ∼750 Kb in human). Taken together, the evidence strongly suggests that XIST is specific to eutherians40,41,66.
The Lyon repeat hypothesis
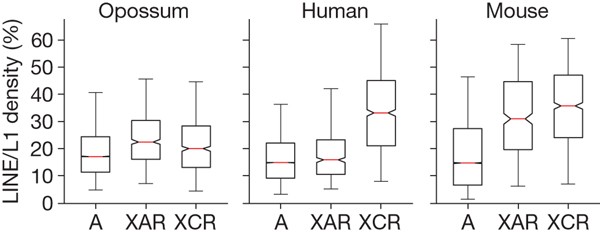
LINE/L1 elements are of particular interest to the study of X chromosome inactivation. These transposable elements have been proposed to act as ‘boosters’ for the spread of X chromosome inactivation in cis from the XIC (reviewed in ref. 67). This hypothesis is supported in part by the observation that in human, LINE/L1 density is significantly elevated in the XCR (33%), where nearly all genes are inactivated, but approximates the autosomal density in the XAR (19%), where many genes escape inactivation (Fig. 3)61,68. In mouse, we found that the LINE/L1 density is elevated in both the XCR (35%) and the XAR (32%), which is consistent with the observation that genes that escape inactivation on the human XAR are often inactivated in mouse69. As previously observed in human68, the LINE/L1 elevation in mouse is particularly dramatic among recent, lineage-specific subfamilies (Supplementary Fig. 9).
Figure 3: Enrichment of LINE/L1 correlates with random X chromosome inactivation.
Box plot of LINE/L1 density in 500-kb intervals across the autosomes (A), the X-added region (XAR) and its homologous regions in opossum, and the X conserved region (XCR). Red bar, median; box edges, 25th and 75th percentiles; whiskers, range.
In contrast to human and mouse, the LINE/L1 density on the opossum X chromosome (22%) is significantly lower than in the eutherian XCR, and is in fact slightly less than in the autosomal regions homologous to the eutherian XAR (23%). This difference between metatherian and eutherian X chromosomes is not readily explained by any simple correlation between LINE/L1 density, recombination or mutation rates. We therefore conclude that LINE/L1 density is unlikely to be a critical factor for X chromosome inactivation in the metatherian lineage, and that the approximately twofold increase on the eutherian X chromosome may be directly related to the acquisition of XIST and random X chromosome inactivation.
Suppression of large-scale rearrangements
Comparative analyses have revealed that the structure of the human X chromosome has remained essentially unchanged since the eutherian radiation10,20,61. A possible reason is that the requirement for XIST transcripts to spread across the chromosome from a central location has led to selection against structural rearrangements. For example, translocation of LINE/L1-poor XAR segments into the XCR could potentially disrupt inactivation at more distal loci. Consistent with this hypothesis, our synteny map reveals that the XAR and XCR homologous regions have experienced several major rearrangements both in the opossum lineage (∼15 lineage-specific synteny breakpoints) and in the eutherian lineage before the eutherian radiation (∼9 lineage-specific breakpoints; Supplementary Table 15). The low rate of rearrangements in the human lineage is therefore unlikely to be due to functions or sequences that were present on the ancestral therian X chromosome, or in early eutherian evolution.
We note that unlike in human, the mouse X chromosome has experienced several rearrangements (with 15 lineage-specific synteny breakpoints), such that the XAR and XCR are no longer two separate segments. This would be consistent with the more comprehensive inactivation in the mouse imposing weaker constraints on rearrangement. Although little is known about the extent of X chromosome inactivation in dog or rat, their X chromosomes are also consistent with this hypothesis. The dog X chromosome is collinear with human and is enriched for LINE/L1 only in the XCR (33.4% versus 16.8% for the XAR). The rat X chromosome has accumulated ∼4 lineage-specific synteny breakpoints after the divergence from mouse61, and is similarly enriched for LINE/L1 in both the XCR (36.7%) and the XAR (34.5%).
Genes
The gene content of metatherian and eutherian genomes provides key information about biological functions. We analysed the gene content of the opossum genome and compared it with that of the human genome. We focused on instances of rapid divergence and duplication of protein-coding genes, which have led to lineage-specific gene complements70.
Gene catalogue
We generated an initial catalogue of 18,648 predicted protein-coding genes and 946 non-coding genes (primarily small nuclear RNA, small nucleolar RNA, microRNA and ribosomal RNA) in opossum34 (Supplementary Note 9 and Supplementary Data). Regularly updated annotations can be obtained from public databases (http://www.ensembl.org and http://genome.ucsc.edu).
We next characterized orthology and paralogy relationships between predicted protein-coding genes in opossum and human11 (Table 3). We could identify unambiguous human orthologues for 15,320 (82%) of the opossum predicted genes, with 12,898 cases having a single copy in each species (1:1 orthologues). Notably, we identified orthologues of key T-cell lineage markers such as CD4 and CD8, which had not been successfully identified by cloning in metatherian species39. Most (2,704) of the remaining genes are homologous to human genes, but could not be assigned to orthologous groups with certainty.
Table 3 Opossum and human gene predictions and projected gene counts
A small number (624) of predicted opossum genes have no clear homologue among the human gene predictions. Inspection revealed that most of these are short (median length of 120 amino acids, compared with 445 for 1:1 orthologues) and probably originate from pseudogenes or spurious open reading frames. Only eight currently have strong evidence of representing functional genes without homologues in humans (Supplementary Table 16). These include CPD-photolyase, which is part of an ancestral photorepair system still active in opossum71, malate synthase72 and inosine/uridine hydrolase. The latter two are ancient genes not previously identified in a mammalian species.
Conversely, approximately ∼1,100 current gene predictions from human have no clear homologue in the initial opossum catalogue (Supplementary Data). Of these, ∼620 can be at least partially aligned to the opossum genome and may not have been annotated as genes owing to imperfections in the draft assembly or high sequence divergence. In particular, manual re-annotation identified orthologues of several rapidly evolving cytokines39. The remaining predictions are dominated by gene families known to have undergone expansion and rapid evolution in the human lineage, such as β-defensins and cancer-testis antigens. On the basis of our comparison, we conclude that the opossum genome probably contains ∼18,000–20,000 protein-coding genes, with the vast majority having eutherian orthologues.
Divergence rates among orthologues
We calculated the synonymous substitution rate (_K_S; substitutions that do not result in amino acid change because of codon redundancy) of 1:1 opossum–human orthologues to approximate the unconstrained divergence rate between the species7,10. The median value of _K_S is 1.02. Consistent with expectation, this value is substantially smaller than the chicken–human _K_S value (1.7), with the ratio being very close to the ratio of prior estimates of the divergence times for the two lineages (∼180 Myr ago for opossum and ∼310 Myr ago for chicken).
Notably, the median _K_S for orthologues located on the XCR is significantly elevated relative to orthologues located on autosomes in both species (1.2 versus 1.0; P < 10-3; see also refs 34, 35). This is the opposite to what is observed within Eutheria10, but is consistent with the expectation that the higher G+C-content and recombination rate on the opossum X chromosome relative to its autosomes implies a higher rate of mutation47. A similar elevation can also be detected in subtelomeric regions34.
Innovation and turnover in gene families
We next studied the evolution of gene family expansions in the metatherian lineage. The opossum gene catalogue contains 2,743 (15%) genes that have probably been involved in one or more duplication or gene conversion event since the last common ancestor with eutherian mammals, as inferred from low _K_S between the copies (median = 0.41). The number of duplications is one-third fewer than the number of human lineage-specific duplications (4,037; 20%), which may reflect the lower rate of segmental duplication in the opossum genome.
We found a large number of lineage-specific copies of genes involved in sensory perception, such as the γ-crystallin family of eye lens proteins73, and taste, odorant74 and pheromone receptors. Other major lineage-specific duplications were found in the rapidly evolving KRAB zinc-finger family, and in genes related to toxin degradation and dietary adaptations, including cytochrome P450 and various gastric enzymes (see also ref. 34).
Innovation in the innate and adaptive immune systems is visible through substantial duplication or gene conversion involving the leukocyte receptor and natural killer complexes, immunoglobulins, type I interferons and defensins32,39. The opossum genome also contains a new T-cell receptor isotype that is expressed early in ontogeny, before conventional T-cell receptors, and may provide early immune function in the altricial young37.
The opossum also shows some surprising gene family expansions that are without precedent in other vertebrates. Notable among these are multiple duplications of the nonsense-mediated decay factors SMG5 and SMG6, and the pre-mRNA splicing factors, KIAA1604 and PRP18. The opossum genome also harbours two adjacent paralogous copies of DNA (cytosine-5)-methyltransferase 1 (DNMT1), which catalyses methylation of CpG dinucleotides. It will be interesting to discover if specialized functions have been adopted by these paralogous genes.
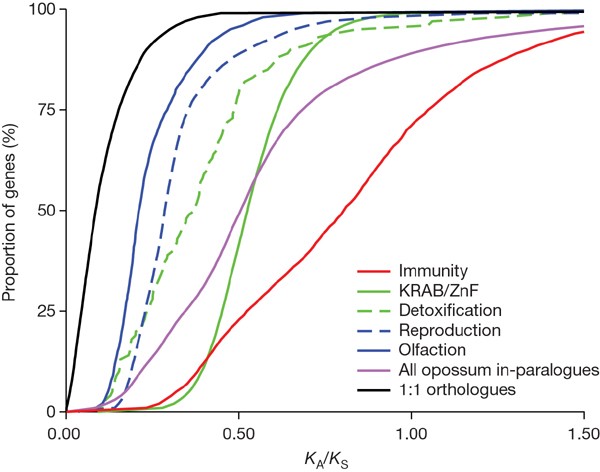
The patterns of evolution among duplicated genes largely mirror those observed in eutherians34,70. The set of opossum paralogues is strongly biased towards recent duplications (_K_S < 0.1) and in general have accumulated a disproportionately high number of nonsynonymous mutations (Fig. 4). The median intraspecies ratio of nonsynonymous to synonymous substitution rates (_K_A/_K_S) between paralogues is 0.51, which is sixfold higher than the interspecies ratio seen for 1:1 orthologues (0.086). This is consistent with the rapid gene birth and death model75, which predicts that duplicated genes either undergo functional divergence in response to positive selection or rapidly degenerate owing to lack of evolutionary benefit.
Figure 4: Cumulative distribution of K A /K S values for duplicated genes.
Estimates are shown for pairs of genes duplicated in opossum (in-paralogues) in the most common functional categories: immunity, KRAB zinc finger (ZnF) transcription factors, detoxification (including cytochrome P450, sulphotransferases), reproduction (including vomeronasal receptors, lipocalins and β-seminoproteins) and olfaction. The total distributions for opossum in-paralogues and opossum–human 1:1 orthologues are shown for comparison.
Conserved sequence elements
The most surprising discovery to emerge from comparative analyses of eutherian genomes is the finding that the majority of evolutionarily conserved sequence does not represent protein-coding genes, but rather are conserved non-coding elements (CNEs)7,10. The opossum genome provides a well-positioned outgroup to study the origin and evolution of these elements.
For simplicity, we will refer to sequence elements as ‘amniote conserved elements’ if they are conserved between chicken and at least one of opossum or human; ‘eutherian conserved elements’ if they are conserved between human and at least one of mouse, rat or dog; and ‘eutherian-specific elements’ if they are eutherian conserved sequence absent from both opossum and chicken. (‘Metatherian-specific elements’ surely also exist, but cannot be identified without additional metatherian genomes.)
Loss of amniote conserved elements in mammals
We first studied the extent to which amniote conserved elements have been lost in the human lineage. We focused on ∼133,000 conserved intervals between opossum and chicken (68 Mb), ∼50% of which overlaps protein-coding regions (Supplementary Data).
Nearly all (97.5%) of these amniote conserved elements can be aligned to the human genome (Fig. 5a). We reasoned that some of the remainder might be orthologous to sequence that lies within gaps in the current human assembly, or which had been missed by the initial genome-wide alignment. We therefore repeated the analysis, focusing only on amniote elements present in opossum and occurring in ‘ungapped intervals’ (that is, syntenic intervals between human and opossum that have no sequence gaps); the ungapped intervals contain 63% of all conserved elements.
Figure 5: Lineage-specific conserved sequence elements.
a, Phylogenetic distribution of amniote conserved elements. b, Distribution for alignment scores of amniote elements, represented by opossum (human), to ungapped syntenic intervals in the human (opossum) genome, for shared (purple) and lineage-specific (orange) elements, and randomly permuted sequences of the same length and base composition (green). Ungapped syntenic intervals are flanked by two synteny anchors (white) and contain no assembly gaps (inset). Md, Monodelphis domestica; Hs, Homo sapiens; Gg, Gallus gallus. c, Phylogenetic distribution of eutherian conserved elements. d, Distribution of alignment scores for eutherian CNEs (log2-odds score ≥ 60), represented by human, to ungapped syntenic intervals in the opossum genome, for shared (blue) and eutherian-specific (red) elements, and randomly permuted sequences of the same length and base composition (green). The bimodal distribution of scores confirms that highly conserved eutherian-specific elements have no significant homology in the syntenic opossum sequence. Mm, Mus musculus; Cf, Canis familiaris. e, The miRNA hsa-mir-194-1 corresponds to an amniote CNE lost in opossum (orange). It is flanked by an unrelated amniote miRNA that is present in opossum (purple). f, A eutherian-specific CNE in the intron of the BCL2 gene (red) overlaps a DNase hypersensitive site in human lymphocytes (black).
We found that 99.0% of amniote elements in ungapped intervals could be unambiguously aligned to the human genome. The remaining 1.0% of amniote elements could not be found even by a more sensitive alignment algorithm (Fig. 5b), and thus seem to have been lost in the human lineage.
We also performed the converse analysis, by aligning the human and chicken genomes to identify amniote conserved elements potentially lost in opossum. The results were similar, with 99.4% of elements in ungapped intervals being readily aligned to opossum.
We conclude that the vast majority of amniote conserved elements encode such fundamental functions that they cannot be lost in either eutherians or metatherians. Nonetheless, the small fractions that have been lost correspond to more than 1,400 elements in total; it will be interesting to investigate their function and the consequence of their loss. Notably, although protein-coding sequence comprises 50% of all amniote conserved elements, they comprise only 4% of the elements lost in one of the lineages.
Eutherian-specific conserved elements
We next explored the appearance of novel conserved elements in the lineage leading from the common therian ancestor to the boreoeutherian ancestor, which could shed light on the origin of such elements in general. We identified a collection of eutherian conserved elements that cover 104 Mb (3.7%) of the human genome, using the phylo-HMM approach14; ∼29% of them overlap protein-coding sequence (Supplementary Data).
Only a small proportion of human conserved protein-coding sequences could not be aligned to the opossum genome (1.1% in ungapped regions; Fig. 5c). In contrast, a much larger proportion of human non-coding elements seem to be eutherian specific (20.5% in ungapped regions). Taking the results from ungapped syntenic intervals as a conservative estimate for the proportion of total innovation, we conclude that approximately 14.8 Mb (1.1% of 30 Mb of coding sequence and 20.5% of 74 Mb of CNEs) of the eutherian conserved elements are eutherian specific.
The amount of apparent innovation is highest among short and moderately conserved elements (median length of 37 bp; median log2-odds score = 22), probably reflecting, in part, that shorter elements may more readily diverge beyond recognition (see also refs 36, 76). Nonetheless, substantial innovation is apparent even among elements that are relatively long and unambiguously conserved within Eutheria. For example, the proportion of eutherian-specific elements is 8.1% among CNEs with log2-odds score ≥ 60, which have a median length of 197 bp (Fig. 5d).
Lineage-specific CNEs correspond to functional elements
To establish the biological relevance of lineage-specific CNEs, we examined the overlap of eutherian and amniote CNEs with two disparate sets of experimentally identified functional elements. If the eutherian-specific CNEs were enriched for false-positive predictions, we would expect them to be substantially under-represented among these functional elements.
We first considered a set of known human microRNAs (miRNAs)77. Of the 51 miRNAs that overlap amniote CNEs, only one (hsa-mir-194-1; ref. 78) seems to have been lost in opossum (Fig. 5e). (The mature form of this miRNA is identical to a second conserved miRNA, hsa-mir-194-2, which does have an opossum orthologue; this apparent redundancy may have made it more susceptible to lineage-specific loss.) Of the 183 miRNAs that overlap eutherian CNEs in ungapped syntenic regions, 27 (15%) correspond to eutherian-specific elements (Supplementary Data). An example is an 87-bp eutherian-specific CNE corresponding to hsa-mir-28; it has previously been detected by northern blot analysis in human and mouse, but not in any non-mammalian species79.
We next considered a genome-wide set of DNase hypersensitive sites from human lymphocytes, which represent a variety of putative regulatory elements80. Of the 290 sites that overlap amniote CNEs present in human, none overlaps instances that are lost in opossum. Of the 2,041 sites that overlap eutherian CNEs in ungapped syntenic regions, 407 (20%) exclusively overlap eutherian-specific elements (Supplementary Data). An example is a 269-bp eutherian-specific CNE in intron 2 of the apoptosis regulator BCL2, which overlaps a DNase hypersensitive site, suggesting it has a _cis_-regulatory function (Fig. 5f).
The fraction of eutherian CNEs overlapping DNase hypersensitive sites that are eutherian specific is strikingly similar to the fraction of all conserved non-coding sequence that is eutherian specific (20.5%). The fraction of miRNAs that correspond to eutherian-specific CNEs is slightly lower (15%), which is consistent with their higher average conservation scores. In particular, the results provide strong evidence that the majority of eutherian-specific CNEs are likely to be genuine functional elements.
Lineage-specific CNEs associated with key developmental genes
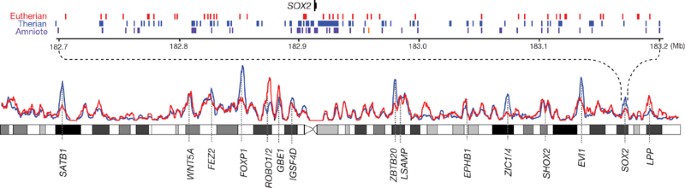
We next explored the distribution of lineage-specific CNEs across the human genome. Overall, there is a strong regional correlation between the density of eutherian CNEs shared with opossum and the density of eutherian-specific CNEs (Spearman’s ρ = 0.82 for 1-Mb windows; Fig. 6). The densities of amniote CNEs present or lost in opossum are also positively correlated (Spearman’s ρ = 0.30).
Figure 6: Lineage-specific CNEs near key developmental genes.
The densities of eutherian CNEs present (blue) or absent (red) in opossum are plotted in 1-Mb sliding windows across human chromosome 3. Peaks in the distributions often correspond to key developmental genes. The expanded view shows positions of amniote CNEs (purple), eutherian CNEs not overlapping amniote CNEs (blue) and eutherian-specific CNEs (red) across a 500-kb gene desert surrounding the SOX2 transcription factor gene. One amniote CNE present in human has been lost in opossum (orange).
Previous studies have shown that both eutherian and amniote CNEs are enriched in certain large, gene-poor regions surrounding genes that have key roles in development, primarily encoding transcription factors, morphogens and axon guidance receptors10,81,82. For example, 35% of all eutherian CNEs and 49% of all amniote CNEs (in ungapped syntenic regions) lie within the 204 largest clusters of CNEs in the human genome (described in ref. 10). The ∼240 key developmental genes in these regions have relatively low rates of amino acid divergence (median K_A/K_S = 0.03) and show little evidence of lineage-specific loss or duplications. In contrast, we found that the rate of gain and loss of CNEs in the same regions is only moderately (∼30%) lower than elsewhere in the genome. Indeed, we identified more than 37,000 lineage-specific CNEs in these developmentally important regions.
Because experimental studies of CNEs in these regions have frequently uncovered _cis_-regulatory functions affecting the nearby developmental genes16,82,83,84,85, the substantial innovations in these regions are candidates for genetic changes underlying differential morphological and neurological evolution in mammalian lineages. This pattern would be consistent with the notion that modification of regulatory networks has been a major force in the evolution of animal diversity86,87,88.
Eutherian-specific CNEs derived from transposable elements
In general, each eutherian-specific element must have arisen by one of three mechanisms: (1) divergence of an ancestral functional element to such an extent that its similarity is no longer detectable; (2) duplication of an ancestral functional element giving rise to an element without a 1:1 orthologue in other clades; or (3) evolution of a novel functional element from sequence that was absent or non-functional in the ancestral genome.
The first mechanism is not likely to account for most of the eutherian-specific CNE sequence, at least among those with high conservation scores—if an ancient functional element underwent such rapid divergence at some point in the eutherian lineage that it is no longer detectable, then there should be concomitant ‘loss’ of an amniote conserved element. But, lineage-specific loss seems to be relatively rare for both amniote elements, as shown above, and for eutherian elements10.The majority of eutherian-specific conserved elements therefore probably arose after the metatherian divergence, either by adaptive evolution of new or previously non-functional sequence, or by duplication of ancestral elements.
One intriguing source for eutherian-specific CNEs is transposable elements. A number of researchers have argued that transposable elements offer an obvious and ideal substrate for the evolution of lineage-specific functions89,90,91,92,93. Transposable elements contain a variety of functional subunits that can be exapted and modified by the host genome89,91, and they can mediate duplication of existing CNEs to distant genomic locations through transduction or chimaerism92. Individual instances of CNEs derived from transposable elements have been described previously14,94,95. However, these cases together comprise only a trivial fraction of the CNEs in the human genome. It has thus been unclear whether the evolution of CNEs from transposable elements represents a general mechanism or a rare exception.
When we examined the set of eutherian-specific CNEs, we found a striking overlap with transposable elements. In ungapped syntenic intervals, at least 16% of eutherian-specific CNEs overlap currently recognized transposable elements in human. The fraction is similar (14%) if we focus only on the most highly conserved elements (phylo-HMM log2-odds score) ≥ 60, see above). The overlapping transposable elements originate from most major transposon families found in eutherians (Table 4), and are not clearly differentiated from other CNEs in terms of distribution across the genome. This implies that transposable-element-mediated evolution has been a significant creative force in the emergence of recent CNEs. The fact that sequences from transposable elements themselves can be identified within these CNEs also implies that exaptation of at least a portion of the transposable element, rather than simply incidental transduction of adjacent sequence, has been a frequent occurrence.
Table 4 Eutherian-specific conserved non-coding elements derived from transposons
In contrast, the eutherian CNEs that are present in opossum (and thus are more ancient) only rarely show overlap with recognizable transposable elements (∼0.7%). We speculate that many of these CNEs also arose from transposable elements, but that they are difficult to recognize as such owing to substantial divergence. In fact, three large families of ancient paralogous CNEs have recently been discovered that were clearly distributed around the genome as parts of transposable elements96,97,98. In each case, only a minority of the family members still retain evidence of transposon-like features. We also previously described ∼100 smaller CNE families that pre-date the eutherian radiation, but which had no members associated with known transposable elements98. For all but two of these families, we can find orthologues in the opossum genome for the majority of their members (Supplementary Note 10 and Supplementary Fig. 10). Moreover, closer inspection reveals previously unrecognized transposon-like features in several of these and other ancient CNE families33.
Strikingly, the proportion of eutherian-specific CNEs recognizable as transposable-element-derived (16%) is very similar to the proportion of the total aligned sequence between the human, mouse and dog genomes recognizable as ancestral transposable elements (∼17% of ∼812 Mb; the vast majority of which is inactive)10. It is widely suspected that the latter proportion is a significant underestimate owing to the difficulty of recognizing transposable elements that inserted more than ∼100–200 Myr ago7,33. In cases where the transposable-element-related sequence hallmarks are not essential to the subsequent CNE, or where evolution of a new function did not follow immediately after the transposable element insertion, exapted sequences would be expected to have diverged to the point that they can no longer be readily recognized at a rate similar to inactive insertions. Because this seems to have occurred for most of the families of ancient CNEs described above, it is likely that the proportion of all eutherian (not just eutherian-specific) CNEs derived from transposable elements is substantially higher than the observed proportion of 16%.
Conclusions
The generation of the first complete genome sequence for a marsupial, Monodelphis domestica, provides an important resource for genetic analysis in this unique model organism, as well as the first reference sequence for metatherian mammals. Our initial results demonstrate the usefulness of this sequence for comparative analyses of the architecture and functional organization of mammalian genomes.
The relationship of sequence composition, segmental duplications and transposable element density with the large and stable karyotype of the opossum genome has provided new support for an emerging, general model of chromosome evolution in mammals. In addition, comparison of the opossum and eutherian X chromosomes revealed that the evolution of random X chromosome inactivation correlates with acquisition of XIST, elevation in LINE/L1 density and suppression of large-scale rearrangements.
Comparative analysis of protein-coding genes showed that the eutherian complement is largely conserved in opossum. Lineage-specific genes seem to be largely limited to gene families that are rapidly turning over in all mammals, although improved annotations that do not rely on homology to distant species will be required to complete the opossum gene catalogue. Identification of a wide array of both conserved and lineage-specific immune genes is particularly notable because limited success in isolating these genes by cloning has led to claims that the metatherian immune system is relatively ‘primitive’. Availability of the genome sequence now facilitates more systematic study of the metatherian immune response39.
At timescales longer than the characteristic time of loss for gene duplications, it is clear that innovation in non-coding elements has been substantially more common relative to protein-coding sequences, at least during eutherian evolution. The opossum genome sequence has provided the first estimate of the genome-wide rate of CNE innovation in eutherian evolution, as well as identification of tens of thousands of lineage-specific elements. It has also provided evidence that exaptation of transposable elements has a much greater role in the evolution of novel CNEs than has been previously realized.
Sequencing of additional metatherian genomes would be helpful for extending our results by allowing detection of metatherian-specific coding and non-coding elements. In addition, sampling of both the American and Australasian lineages would allow the reconstruction of the genome of their common ancestor, which would complement ongoing efforts for the boreoeutherian ancestral genome58. The shorter genetic distance between the ancestral metatherian and boreoeutherian genomes (∼0.6–0.7 substitutions per site) would facilitate a more comprehensive analysis of short and weakly conserved functional elements, for which the phylogenetic distribution and evolutionary origins are still difficult to ascertain.
Methods summary
WGS sequencing and assembly
Approximately 38.8 million high-quality sequence reads were assembled using an interim version of ARACHNE2+ (http://www.broad.mit.edu/wga/).
SNP discovery
The SNP discovery was performed using ARACHNE and SSAHA-SNP99. Linkage disequilibrium was assessed using Haploview100.
Genome alignment and comparisons
Synteny maps were generated using standard methods7,10.
Gene prediction and phylogeny
Opossum protein-coding and non-coding RNA genes were predicted using a modified version of the Ensembl genebuild pipeline101, followed by several rounds of refinement using Exonerate102 and manual curation. Orthology and paralogy were inferred using the PhyOP pipeline11,34.
Conserved element prediction
Amniote conserved elements were inferred from pairwise BLASTZ alignment blocks with more than 75% identity for ≥100 bp. Eutherian conserved elements were inferred using phastCons14. Eutherian elements that did not fall within a 10-kilobase or longer synteny ‘net’103 were ignored.
Phylogeny of conserved elements
For amniote conserved elements, pairwise best-in-genome BLASTZ alignments of opossum to human and vice versa were used to infer their phylogenetic distributions. For eutherian conserved elements, concomitant BLASTZ/MULTIZ alignments to opossum and chicken were used. A conserved element was called absent from a species if it was not covered by a single aligned nucleotide in the relevant alignment.
Correction for assembly gaps and initial alignment artefacts
A conserved element was considered to be in an ungapped syntenic interval if it was flanked by two synteny anchors within 200 kb on the same contigs in both the human and opossum assemblies. All conserved elements in ungapped syntenic intervals were realigned using water (http://emboss.sourceforge.net). Putatively eutherian-specific elements, including XIST, were also searched against all opossum sequencing reads using MegaBLAST.
Online Methods
WGS sequencing and assembly
Approximately 38.8 million high-quality sequence reads were derived from paired-end reads of 4- and 10-kb plasmids, fosmid and BAC clones, prepared from primary tissue DNA from a single female opossum. The reads were assembled using an interim version of ARACHNE2+ (http://www.broad.mit.edu/wga/). No comparative data were used in the assembly process. An intermediate assembly (monDom4) was used for the majority of the analyses reported here. The most recent version (monDom5) has identical sequence content and scaffold structure, but includes additional FISH data as described in Supplementary Note 2.
SNP discovery
The SNP discovery was performed using ARACHNE by comparison of the two haplotypes derived from the opossum assembly using only high-quality discrepancies supported by two or more reads each. Sequence reads from three additional individuals were also aligned to the reference assembly, and SNPs were discovered using SSAHA-SNP99. Linkage disequilibrium was assessed using Haploview100.
Genome alignment and comparisons
The assembly versions used in all comparative analyses were hg17 or hg18 (human), mm8 (mouse), rn4 (rat), canFam2 (dog), monDom4 or monDom5 (opossum) and galGal3 (chicken). The number of aligned nucleotides was counted directly from unfiltered, pairwise BLASTZ alignments (obtained from http://genome.ucsc.edu). Synteny maps were generated using standard methods7,10, starting from 320,000 reciprocal-best syntenic anchors identified by PatternHunter104 (see Supplementary Note 7). Reconstruction of the boreoeutherian ancestral karyotype is described in Supplementary Note 8.
Gene prediction and phylogeny
Opossum protein-coding and non-coding RNA genes were predicted using a modified version of the Ensembl genebuild pipeline101, followed by several rounds of refinement using Exonerate102 and manual curation. Orthology and paralogy were inferred using the PhyOP pipeline with all predicted opossum and human (Ensembl v40) gene transcripts as input and _K_S as the distance metric11,34. Coding regions were aligned according to their amino acid sequences using BLASTP. _K_A and _K_S were estimated using the codeml program105, with default settings and the F3X4 codon frequency model. Functional categories were identified using the Gene Ontology106.
Conserved element prediction
Amniote conserved elements were inferred directly from pairwise BLASTZ alignments of chicken to opossum or human. Every alignment block with more than 75% identity for ≥100 bp was classified as an amniote conserved element. Eutherian conserved elements were inferred using phastCons14 on BLASTZ/MULTIZ107,108 alignments of human to mouse, rat and dog. The nonconserved model was fitted to fourfold degenerate sites from 15,900 human RefSeqs projected onto the same alignments, using phyloFit and REV. A separate model was fitted for the X chromosome. The scaling parameter for the conserved model was estimated by phastCons. Target coverage and expected element length were set to 12.5% and 12 bp, respectively. Predicted eutherian conserved elements that did not fall within a 10-kb or longer synteny ‘net’103 between human, mouse and dog were ignored. The coding status of each element was inferred from ≥1 nucleotide overlap with entries in the UCSC human ‘known genes’ track109. Proportions are reported out of the total length of the elements considered. Eutherian CNEs were classified as transposable-element-derived if they showed more than 20% nucleotide overlap (median = 100% for all elements, 54% for elements with log2-odds score ≥ 60) with human RepeatMasker annotations.
Phylogeny of conserved elements
For amniote conserved elements, pairwise best-in-genome BLASTZ alignments of opossum to human and vice versa were used to infer their phylogenetic distributions. For eutherian conserved elements, concomitant BLASTZ/MULTIZ alignments to opossum and chicken were used. A conserved element was called absent from a species if it was not covered by a single aligned nucleotide in the relevant BLASTZ alignment.
Correction for assembly gaps and initial alignment artefacts
A conserved element was considered to be in an ungapped syntenic interval if it was flanked by two PatternHunter synteny anchors within 200-kb of each other on the same contigs in both the human and opossum assemblies. All conserved elements (represented by human or opossum, as appropriate) in ungapped syntenic intervals were realigned to the unmasked genome sequence (in opossum or human) using the water program (http://emboss.sourceforge.net) with default parameters and a gap extension penalty of 4. A randomly permuted version of each element was also realigned. For amniote conserved elements, only the longest interval with ≥75% identity from within the originating alignment block (see above) was realigned. Amniote elements were called lost, and eutherian elements were called eutherian-specific if their Smith–Waterman realignment score, divided by the length of the element, did not exceed the corresponding score for the permuted element plus one. (Conservatively calling an element found if its score simply exceeded the score of the permuted element resulted in 15% of eutherian CNEs in ungapped regions and 8% of those with log2-odds score ≥ 60 being called eutherian-specific.) Putatively eutherian-specific elements, including XIST, were also searched against all opossum sequencing reads using discontiguous MegaBLAST.
References
- Kumar, S. & Hedges, S. B. A molecular timescale for vertebrate evolution. Nature 392, 917–920 (1998)
ADS CAS PubMed Google Scholar - Woodburne, M. O., Rich, T. H. & Springer, M. S. The evolution of tribospheny and the antiquity of mammalian clades. Mol. Phylogenet. Evol. 28, 360–385 (2003)
CAS PubMed Google Scholar - Tyndale-Biscoe, C. H. Life of Marsupials (CSIRO Publishing, Collingwood, Victoria, 2005)
Google Scholar - Wakefield, M. J. & Graves, J. A. M. Marsupials and monotremes sort genome treasures from junk. Genome Biol. 6, 218 (2005)
PubMed PubMed Central Google Scholar - Graves, J. A. M. & Westerman, M. Marsupial genetics and genomics. Trends Genet. 18, 517–521 (2002)
PubMed Google Scholar - Samollow, P. B. Status and applications of genomic resources for the gray, short-tailed opossum, Monodelphis domestica, an American marsupial model for comparative biology. Aust. J. Zool. 54, 173–196 (2006)
Google Scholar - Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002)
Google Scholar - Rat Genome Sequencing Project Consortium Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521 (2004)
Google Scholar - Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005)
- Lindblad-Toh, K. et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005)
ADS CAS PubMed Google Scholar - Goodstadt, L. & Ponting, C. P. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput. Biol. 2, e133 (2006)
ADS PubMed PubMed Central Google Scholar - Clamp, M. et al. Gene content of the human genome. Nature (submitted).
- Xie, X. et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434, 338–345 (2005)
ADS CAS PubMed PubMed Central Google Scholar - Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005)
CAS PubMed PubMed Central Google Scholar - Pedersen, J. S. et al. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2, e33 (2006)
ADS CAS PubMed PubMed Central Google Scholar - Nobrega, M. A., Ovcharenko, I., Afzal, V. & Rubin, E. M. Scanning human gene deserts for long-range enhancers. Science 302, 413 (2003)
CAS PubMed Google Scholar - Ovcharenko, I., Stubbs, L. & Loots, G. G. Interpreting mammalian evolution using Fugu genome comparisons. Genomics 84, 890–895 (2004)
CAS PubMed Google Scholar - Prabhakar, S. et al. Close sequence comparisons are sufficient to identify human _cis_-regulatory elements. Genome Res. 16, 855–863 (2006)
CAS PubMed PubMed Central Google Scholar - Margulies, E. H. et al. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc. Natl Acad. Sci. USA 102, 4795–4800 (2005)
ADS CAS PubMed PubMed Central Google Scholar - Hillier, L. W. et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 (2004)
ADS CAS Google Scholar - VandeBerg, J. L. The gray short-tailed opossum. (Monodelphis domestica) as a model didelphid species for genetic research. Aust. J. Zool. 37, 235–247 (1990)
Google Scholar - VandeBerg, J. L. in UFAW Handbook on the Management of Laboratory Animals. Vol. 1 Terrestrial Vertebrates (eds Poole, T. & English, P.) 193–209 (Blackwell Science, Oxford, 1999)
Google Scholar - Murphy, S. K. & Jirtle, R. L. Imprinting evolution and the price of silence. Bioessays 25, 577–588 (2003)
CAS PubMed Google Scholar - Rapkins, R. W. et al. Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet. 2, e182 (2006)
PubMed PubMed Central Google Scholar - Weidman, J. R. et al. Phylogenetic footprint analysis of IGF2 in extant mammals. Genome Res. 14, 1726–1732 (2004)
CAS PubMed PubMed Central Google Scholar - Deakin, J. E. et al. Evolution and comparative analysis of the MHC Class III inflammatory region. BMC Genomics 7, 281 (2006)
PubMed PubMed Central Google Scholar - Deakin, J. E., Olp, J. J., Graves, J. A. & Miller, R. D. Physical mapping of immunoglobulin loci IGH@, IGK@, and IGL@ in the opossum (Monodelphis domestica). Cytogenet. Genome Res. 114, 94H (2006)
CAS PubMed Google Scholar - Belov, K. et al. Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol. 4, e46 (2006)
PubMed PubMed Central Google Scholar - Wintzer, M. et al. Strategies for identifying genes that play a role in spinal cord regeneration. J. Anat. 204, 3–11 (2004)
CAS PubMed PubMed Central Google Scholar - VandeBerg, J. L. et al. Genetic analysis of ultraviolet radiation-induced skin hyperplasia and neoplasia in a laboratory marsupial model (Monodelphis domestica). Arch. Dermatol. Res. 286, 12–17 (1994)
CAS PubMed Google Scholar - Baker, M. L. et al. Analysis of a set of Australian northern brown bandicoot expressed sequence tags with comparison to the genome sequence of the south American grey short-tailed opossum. BMC Genom. 8, 50 (2007)
Google Scholar - Belov, K. et al. Characterization of the opossum immune genome provides insights into the evolution of the mammalian immune system. Genome Res. doi: 10.1101/gr.6121807 (2007)
- Gentles, A. J. et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. doi: 10.1101/gr.6070707 (2007)
- Goodstadt, L., Heger, A., Webber, C. & Ponting, C. P. An analysis of the gene complement of a marsupial Monodelphis domestica: Evolution of lineage-specific genes and giant chromosomes. Genome Res. doi: 10.1101/gr.6093907 (2007)
- Gu, W. et al. Phylogenetic detection, population genetics, and distribution of active SINEs in the genome of Monodelphis domestica. Gene doi: 10.1016/j.gene.2007.02.028 (2007).
- Mahony, S., Corcoran, D. L., Feingold, E. & Benos, P. V. Regulatory conservation of protein coding and miRNA genes in vertebrates: lessons from the opossum genome. Genome Biol. (in the press).
- Parra, Z. E. et al. A new T-cell receptor discovered in marsupials. Proc. Natl Acad. Sci. USA (submitted).
- Samollow, P. B. et al. A microsatellite-based, physically anchored linkage map for the gray, short-tailed opossum (Monodelphis domestica). Chromosome Res. advance online publication, doi:10.1007/s10577-007-1123-4 (25 February 2007)
- Wong, E. S., Young, L. J., Papenfuss, A. T. & Belov, K. In silico identification of opossum cytokine genes suggests the complexity of the marsupial immune system rivals that of eutherian mammals. Immunome Res. 2, 4 (2006)
PubMed PubMed Central Google Scholar - Hore, T., Koina, E., Wakefield, M. J. & Graves, J. A. M. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 15, 147–161 (2007)
CAS PubMed Google Scholar - Davidow, L. S. et al. The search for a marsupial XIC reveals a break with vertebrate synteny. Chromosome Res. 15, 137–146 (2007)
CAS PubMed Google Scholar - Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001)
ADS CAS PubMed Google Scholar - Duke, S. E. et al. Integrated cytogenetic BAC map of the genome of the gray short-tailed opossum, Monodelphis domestica. Chromosome Res. advance online publication, doi:10.1007/s10577-007-1131-4 (6 April 2007)
Google Scholar - VandeBerg, J. L. The laboratory opossum (Monodelphis domestica) in laboratory research. ILAR J. 38, 4–12 (1997)
PubMed Google Scholar - Rens, W. et al. Karyotype relationships between distantly related marsupials from South America and Australia. Chromosome Res. 9, 301–308 (2001)
ADS CAS PubMed Google Scholar - Belle, E. M., Duret, L., Galtier, N. & Eyre-Walker, A. The decline of isochores in mammals: an assessment of the GC content variation along the mammalian phylogeny. J. Mol. Evol. 58, 653–660 (2004)
ADS CAS PubMed Google Scholar - Jensen-Seaman, M. I. et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14, 528–538 (2004)
CAS PubMed PubMed Central Google Scholar - Duret, L., Eyre-Walker, A. & Galtier, N. A new perspective on isochore evolution. Gene 385, 71–74 (2006)
CAS PubMed Google Scholar - Dumas, D. & Britton-Davidian, J. Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and robertsonian populations of the house mouse. Genetics 162, 1355–1366 (2002)
PubMed PubMed Central Google Scholar - Myers, S. et al. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005)
ADS CAS PubMed Google Scholar - Hope, R. M. Selected features of marsupial genetics. Genetica 90, 165–180 (1993)
CAS PubMed Google Scholar - Sharp, P. J. & Hayman, D. L. An examination of the role of chiasma frequency in the genetic system of marsupials. Heredity 60, 77–85 (1988)
PubMed Google Scholar - Holm, P. B. Ultrastructural analysis of meiotic recombination and chiasma formation. Tokai J. Exp. Clin. Med. 11, 415–436 (1986)
CAS PubMed Google Scholar - Samollow, P. B. et al. First-generation linkage map of the gray, short-tailed opossum, Monodelphis domestica, reveals genome-wide reduction in female recombination rates. Genetics 166, 307–329 (2004)
CAS PubMed PubMed Central Google Scholar - Bailey, J. A. et al. Hotspots of mammalian chromosomal evolution. Genome Biol. 5, R23 (2004)
PubMed PubMed Central Google Scholar - Webber, C. & Ponting, C. P. Hotspots of mutation and breakage in dog and human chromosomes. Genome Res. 15, 1787–1797 (2005)
CAS PubMed PubMed Central Google Scholar - Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005)
CAS PubMed Google Scholar - Ma, J. et al. Reconstructing contiguous regions of an ancestral genome. Genome Res. 16, 1557–1565 (2006)
CAS PubMed PubMed Central Google Scholar - Kohn, M. et al. Wide genome comparisons reveal the origins of the human X chromosome. Trends Genet. 20, 598–603 (2004)
CAS PubMed Google Scholar - Graves, J. A. Sex chromosome specialization and degeneration in mammals. Cell 124, 901–914 (2006)
PubMed Google Scholar - Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005)
ADS CAS PubMed PubMed Central Google Scholar - Cooper, D. W., Johnston, P. G., Graves, J. A. & Watson, J. M. X-inactivation in marsupials and monotremes. Sem. Dev. Biol. 4, 117–128 (1993)
Google Scholar - Heard, E. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16, 247–255 (2004)
CAS PubMed Google Scholar - Wakefield, M. J., Keohane, A. M., Turner, B. M. & Graves, J. A. Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc. Natl Acad. Sci. USA 94, 9665–9668 (1997)
ADS CAS PubMed PubMed Central Google Scholar - Reik, W. & Lewis, A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nature Rev. Genet. 6, 403–410 (2005)
CAS PubMed Google Scholar - Duret, L. et al. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312, 1653–1655 (2006)
ADS CAS PubMed Google Scholar - Lyon, M. F. Do LINEs have a role in X–chromosome inactivation? J. Biomed. Biotechnol. 2006, 59746 (2006)
PubMed PubMed Central Google Scholar - Bailey, J. A., Carrel, L., Chakravarti, A. & Eichler, E. E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl Acad. Sci. USA 97, 6634–6639 (2000)
ADS CAS PubMed PubMed Central Google Scholar - Disteche, C. M., Filippova, G. N. & Tsuchiya, K. D. Escape from X inactivation. Cytogenet. Genome Res. 99, 36–43 (2002)
CAS PubMed Google Scholar - Emes, R. D., Goodstadt, L., Winter, E. E. & Ponting, C. P. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 12, 701–709 (2003)
CAS PubMed Google Scholar - Kato, T. et al. Cloning of a marsupial DNA photolyase gene and the lack of related nucleotide sequences in placental mammals. Nucleic Acids Res. 22, 4119–4124 (1994)
CAS PubMed PubMed Central Google Scholar - Kondrashov, F. A. et al. Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct 1, 31 (2006)
PubMed PubMed Central Google Scholar - Wistow, G. et al. γN-crystallin and the evolution of the βγ-crystallin superfamily in vertebrates. FEBS J. 272, 2276–2291 (2005)
CAS PubMed Google Scholar - Grus, W. E., Shi, P., Zhang, Y. P. & Zhang, J. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc. Natl Acad. Sci. USA 102, 5767–5772 (2005)
ADS CAS PubMed PubMed Central Google Scholar - International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004)
ADS Google Scholar - Dermitzakis, E. T. & Clark, A. G. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 19, 1114–1121 (2002)
CAS PubMed Google Scholar - Griffiths-Jones, S. et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34 (database issue). D140–D144 (2006)
CAS PubMed Google Scholar - Michael, M. Z. et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1, 882–891 (2003)
CAS PubMed Google Scholar - Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 (2001)
ADS CAS PubMed Google Scholar - Crawford, G. E. et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16, 123–131 (2006)
CAS PubMed PubMed Central Google Scholar - Sandelin, A. et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genom. 5, 99 (2004)
Google Scholar - Woolfe, A. et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7 (2005)
PubMed Google Scholar - Bailey, P. J. et al. A global genomic transcriptional code associated with CNS-expressed genes. Exp. Cell Res. 312, 3108–3119 (2006)
CAS PubMed Google Scholar - de la Calle-Mustienes, E. et al. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 15, 1061–1072 (2005)
CAS PubMed PubMed Central Google Scholar - Pennacchio, L. A. et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 (2006)
ADS CAS PubMed Google Scholar - Carroll, S. B. Evolution at two levels: on genes and form. PLoS Biol. 3, e245 (2005)
PubMed PubMed Central Google Scholar - Davidson, E. H. & Erwin, D. H. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 (2006)
ADS CAS PubMed Google Scholar - Stathopoulos, A. & Levine, M. Genomic regulatory networks and animal development. Dev. Cell 9, 449–462 (2005)
CAS PubMed Google Scholar - Britten, R. J. Mobile elements inserted in the distant past have taken on important functions. Gene 205, 177–182 (1997)
CAS PubMed Google Scholar - Britten, R. J. & Davidson, E. H. Gene regulation for higher cells: a theory. Science 165, 349–357 (1969)
ADS CAS PubMed Google Scholar - Brosius, J. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica 107, 209–238 (1999)
CAS PubMed Google Scholar - Kazazian, H. H. Mobile elements: drivers of genome evolution. Science 303, 1626–1632 (2004)
ADS CAS PubMed Google Scholar - Marino-Ramirez, L., Lewis, K. C., Landsman, D. & Jordan, I. K. Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet. Genome Res. 110, 333–341 (2005)
CAS PubMed Google Scholar - Cooper, G. M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005)
CAS PubMed PubMed Central Google Scholar - Silva, J. C. et al. Conserved fragments of transposable elements in intergenic regions: evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet. Res. 82, 1–18 (2003)
ADS CAS PubMed Google Scholar - Bejerano, G. et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441, 87–90 (2006)
ADS CAS PubMed Google Scholar - Nishihara, H., Smit, A. F. & Okada, N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 16, 864–874 (2006)
CAS PubMed PubMed Central Google Scholar - Xie, X., Kamal, M. & Lander, E. S. A family of conserved noncoding elements derived from an ancient transposable element. Proc. Natl Acad. Sci. USA 103, 11659–11664 (2006)
ADS CAS PubMed PubMed Central Google Scholar - Ning, Z., Cox, A. J. & Mullikin, J. C. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001)
CAS PubMed PubMed Central Google Scholar - Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005)
CAS PubMed Google Scholar - Birney, E. et al. Ensembl 2006. Nucleic Acids Res. 34 (database issue). D556–D561 (2006)
CAS PubMed Google Scholar - Slater, G. S. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005)
PubMed PubMed Central Google Scholar - Kent, W. J. et al. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA 100, 11484–11489 (2003)
ADS CAS PubMed PubMed Central Google Scholar - Ma, B., Tromp, J. & Li, M. PatternHunter: faster and more sensitive homology search. Bioinformatics 18, 440–445 (2002)
CAS PubMed Google Scholar - Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997)
CAS PubMed Google Scholar - Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nature Genet. 25, 25–29 (2000)
CAS PubMed Google Scholar - Blanchette, M. et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004)
CAS PubMed PubMed Central Google Scholar - Schwartz, S. et al. Human-mouse alignments with BLASTZ. Genome Res. 13, 103–107 (2003)
CAS PubMed PubMed Central Google Scholar - Hsu, F. et al. The UCSC known genes. Bioinformatics 22, 1036–1046 (2006)
CAS PubMed Google Scholar
Acknowledgements
Generation of the Monodelphis domestica sequence at the Broad Institute of MIT and Harvard was supported by grants from the National Human Genome Research Institute (NHGRI). For work from other members of the Opossum Genome Sequencing Consortium, we acknowledge the support of the National Institutes of Health (NHGRI, NIAID, NLM), the National Science Foundation, the Robert J. Kleberg Jr and Helen C. Kleberg Foundation, the State of Louisiana Board of Regents Support Fund, State of Colorado support funds, the Pittsburgh Foundation, TATRC/DoD, the UK Medical Research Council and the Australian Research Council. We thank colleagues at the UCSC genome browser for providing data (BLASTZ/MULTIZ alignments, synteny nets, and annotations). We thank L. Gaffney for assistance in preparing the manuscript and figures, and J. Danke for flow cytometry data.
All analysed data sets can be obtained from http://www.broad.mit.edu/mammals/opossum/. This Monodelphis domestica whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under NCBI accession code AAFR00000000. SNPs have been deposited in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Author information
Authors and Affiliations
- Broad Institute of MIT and Harvard, 7 Cambridge Center, Cambridge, Massachusetts 02142, USA,
Tarjei S. Mikkelsen, Jean L. Chang, Manuel Garber, Michael Kamal, Evan Mauceli, Ted Sharpe, Michele Clamp, April Cook, James Cuff, Manfred Grabherr, Michael Kleber, Claire M. Wade, Xiaohui Xie, Michael C. Zody, Eric S. Lander & Kerstin Lindblad-Toh - Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA,
Tarjei S. Mikkelsen - Bioinformatics Division, The Walter & Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville Victoria 3050, Australia,
Matthew J. Wakefield, Anthony T. Papenfuss & Terence P. Speed - The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK,
Bronwen Aken & Stephen M. J. Searle - Molecular Genetics Program, Benaroya Research Institute at Virginia Mason, 1201 Ninth Avenue, Seattle, Washington 98101, USA,
Chris T. Amemiya - Department of Molecular Biomedical Sciences, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, North Carolina 27606, USA,
Shannon Duke & Matthew Breen - Stanford University School of Medicine, P060 Lucas Center, Stanford, California 94305, USA,
Andrew J. Gentles - Genetic Information Research Institute, 1925 Landings Drive, Mountain View, California 94043, USA,
Andrew J. Gentles & Jerzy Jurka - Department of Physiology, MRC Functional Genetics Unit, Anatomy and Genetics, University of Oxford, South Parks Road, Oxford OX1 3QX, UK,
Leo Goodstadt, Andreas Heger, Caleb Webber & Chris P. Ponting - Department of Biology, Center for Evolutionary and Theoretical Immunology, University of New Mexico, Albuquerque, New Mexico 87131, USA,
Michelle L. Baker, Robert D. Miller & Zuly E. Parra - Department of Biological Sciences, Biological Computation and Visualization Center, Center for Bio-Modular Multi-Scale Systems, Louisiana State University, 202 Life Sciences Building, Baton Rouge, Louisiana 70803, USA,
Mark A. Batzer & Jerilyn A. Walker - Department of Computational Biology, University of Pittsburgh, 3501 Fifth Avenue, Suite 3064, BST3, Pittsburgh, Pennsylvania 15260, USA,
Panayiotis V. Benos & Shaun Mahony - Faculty of Veterinary Science, University of Sydney, New South Wales 2006, Australia,
Katherine Belov - Department of Radiation Oncology, Duke University Medical Center, Box 3433, Durham, North Carolina 27710, USA,
Radhika Das, Randy L. Jirtle & Jennifer R. Weidman - Department of Molecular Biology, and Department of Genetics, Hughes Medical Institute, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts 02114, USA,
Lance Davidow & Jeannie T. Lee - ARC Centre for Kangaroo Genomics, Research School of Biological Sciences, The Australian National University, Canberra, ACT 2601, Australia,
Janine E. Deakin, Timothy A. Hore, Edda Koina, Katherine Thompson, Paul D. Waters & Jennifer A. Marshall Graves - Department of Medicine (Hematology) and Molecular Genetics, Albert Einstein College of Medicine, Ullmann 911, 1300 Morris Park Avenue, Bronx, New York 10461, USA,
Melissa J. Fazzari, Jacob L. Glass, John M. Greally & Mayumi Oda - Department of Biochemistry and Molecular Genetics, University of Colorado Health Sciences Center, MS 8101, 12801 17th Avenue, Aurora, Colorado 80045, USA,
Wanjun Gu & David D. Pollock - John Curtin School of Medical Research, The Australian National University, Canberra, ACT 0200, Australia,
Gavin A. Huttley - Genome Sciences Centre, British Columbia Cancer Agency, 570 West 7th Avenue, Vancouver, British Columbia V5Z 4S6, Canada,
Marco A. Marra & Jacqueline E. Schein - Department of Pediatrics, Research Center Children’s Hospital of Pittsburgh, 3460 Fifth Avenue, Room 2109, Rangos, Pittsburgh, Pennsylvania 15213, USA,
Robert D. Nicholls - Department of Biology, West Virginia University, Morgantown, West Virginia 26505, USA,
David A. Ray - Department of Genetics and Southwest National Primate Research Center, Southwest Foundation for Biomedical Research, San Antonio, Texas 78245, USA,
John L. VandeBerg - Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge Street, Boston, Massachusetts 02114, USA,
Claire M. Wade - Center for Comparative Medicine and Translational Research, North Carolina State University, 4700 Hillsborough Street, Raleigh, North Carolina 27606, USA,
Matthew Breen - Department of Veterinary Integrative Biosciences, Texas A&M University, 4458 TAMU, College Station, Texas 77843, USA,
Paul B. Samollow - Whitehead Institute for Biomedical Research, 9 Cambridge Center, Cambridge, Massachusetts 02142, USA,
Eric S. Lander - Broad Institute of MIT and Harvard, 7 Cambridge Center, Cambridge, Massachusetts 02142, USA.,
Jennifer Baldwin, Amr Abdouelleil, Jamal Abdulkadir, Adal Abebe, Brikti Abera, Justin Abreu, St Christophe Acer, Lynne Aftuck, Allen Alexander, Peter An, Erica Anderson, Scott Anderson, Harindra Arachi, Marc Azer, Pasang Bachantsang, Andrew Barry, Tashi Bayul, Aaron Berlin, Daniel Bessette, Toby Bloom, Jason Blye, Leonid Boguslavskiy, Claude Bonnet, Boris Boukhgalter, Imane Bourzgui, Adam Brown, Patrick Cahill, Sheridon Channer, Yama Cheshatsang, Lisa Chuda, Mieke Citroen, Alville Collymore, Patrick Cooke, Maura Costello, Katie D'Aco, Riza Daza, Georgius De Haan, Stuart DeGray, Christina DeMaso, Norbu Dhargay, Kimberly Dooley, Erin Dooley, Missole Doricent, Passang Dorje, Kunsang Dorjee, Alan Dupes, Richard Elong, Jill Falk, Abderrahim Farina, Susan Faro, Diallo Ferguson, Sheila Fisher, Chelsea D. Foley, Alicia Franke, Dennis Friedrich, Loryn Gadbois, Gary Gearin, Christina R. Gearin, Georgia Giannoukos, Tina Goode, Joseph Graham, Edward Grandbois, Sharleen Grewal, Kunsang Gyaltsen, Nabil Hafez, Birhane Hagos, Jennifer Hall, Charlotte Henson, Andrew Hollinger, Tracey Honan, Monika D. Huard, Leanne Hughes, Brian Hurhula, M. Erii Husby, Asha Kamat, Ben Kanga, Seva Kashin, Dmitry Khazanovich, Peter Kisner, Krista Lance, Marcia Lara, William Lee, Niall Lennon, Frances Letendre, Rosie LeVine, Alex Lipovsky, Xiaohong Liu, Shangtao Liu, Tashi Lokyitsang, Yeshi Lokyitsang, Rakela Lubonja, Annie Lui, Pen MacDonald, Vasilia Magnisalis, Kebede Maru, Charles Matthews, William McCusker, Susan McDonough, Teena Mehta, James Meldrim, Louis Meneus, Oana Mihai, Atanas Mihalev, Tanya Mihova, Rachel Mittelman, Valentine Mlenga, Anna Montmayeur, Leonidas Mulrain, Adam Navidi, Jerome Naylor, Tamrat Negash, Thu Nguyen, Nga Nguyen, Robert Nicol, Choe Norbu, Nyima Norbu, Nathaniel Novod, Barry O'Neill, Sahal Osman, Eva Markiewicz, Otero L. Oyono, Christopher Patti, Pema Phunkhang, Fritz Pierre, Margaret Priest, Sujaa Raghuraman, Filip Rege, Rebecca Reyes, Cecil Rise, Peter Rogov, Keenan Ross, Elizabeth Ryan, Sampath Settipalli, Terry Shea, Ngawang Sherpa, Lu Shi, Diana Shih, Todd Sparrow, Jessica Spaulding, John Stalker, Nicole Stange-Thomann, Sharon Stavropoulos, Catherine Stone, Christopher Strader, Senait Tesfaye, Talene Thomson, Yama Thoulutsang, Dawa Thoulutsang, Kerri Topham, Ira Topping, Tsamla Tsamla, Helen Vassiliev, Andy Vo, Tsering Wangchuk, Tsering Wangdi, Michael Weiand, Jane Wilkinson, Adam Wilson, Shailendra Yadav, Geneva Young, Qing Yu, Lisa Zembek, Danni Zhong, Andrew Zimmer, Zac Zwirko, David B. Jaffe, Pablo Alvarez, Will Brockman, Jonathan Butler, CheeWhye Chin & Sante Gnerre
Authors
- Tarjei S. Mikkelsen
You can also search for this author inPubMed Google Scholar - Matthew J. Wakefield
You can also search for this author inPubMed Google Scholar - Bronwen Aken
You can also search for this author inPubMed Google Scholar - Chris T. Amemiya
You can also search for this author inPubMed Google Scholar - Jean L. Chang
You can also search for this author inPubMed Google Scholar - Shannon Duke
You can also search for this author inPubMed Google Scholar - Manuel Garber
You can also search for this author inPubMed Google Scholar - Andrew J. Gentles
You can also search for this author inPubMed Google Scholar - Leo Goodstadt
You can also search for this author inPubMed Google Scholar - Andreas Heger
You can also search for this author inPubMed Google Scholar - Jerzy Jurka
You can also search for this author inPubMed Google Scholar - Michael Kamal
You can also search for this author inPubMed Google Scholar - Evan Mauceli
You can also search for this author inPubMed Google Scholar - Stephen M. J. Searle
You can also search for this author inPubMed Google Scholar - Ted Sharpe
You can also search for this author inPubMed Google Scholar - Michelle L. Baker
You can also search for this author inPubMed Google Scholar - Mark A. Batzer
You can also search for this author inPubMed Google Scholar - Panayiotis V. Benos
You can also search for this author inPubMed Google Scholar - Katherine Belov
You can also search for this author inPubMed Google Scholar - Michele Clamp
You can also search for this author inPubMed Google Scholar - April Cook
You can also search for this author inPubMed Google Scholar - James Cuff
You can also search for this author inPubMed Google Scholar - Radhika Das
You can also search for this author inPubMed Google Scholar - Lance Davidow
You can also search for this author inPubMed Google Scholar - Janine E. Deakin
You can also search for this author inPubMed Google Scholar - Melissa J. Fazzari
You can also search for this author inPubMed Google Scholar - Jacob L. Glass
You can also search for this author inPubMed Google Scholar - Manfred Grabherr
You can also search for this author inPubMed Google Scholar - John M. Greally
You can also search for this author inPubMed Google Scholar - Wanjun Gu
You can also search for this author inPubMed Google Scholar - Timothy A. Hore
You can also search for this author inPubMed Google Scholar - Gavin A. Huttley
You can also search for this author inPubMed Google Scholar - Michael Kleber
You can also search for this author inPubMed Google Scholar - Randy L. Jirtle
You can also search for this author inPubMed Google Scholar - Edda Koina
You can also search for this author inPubMed Google Scholar - Jeannie T. Lee
You can also search for this author inPubMed Google Scholar - Shaun Mahony
You can also search for this author inPubMed Google Scholar - Marco A. Marra
You can also search for this author inPubMed Google Scholar - Robert D. Miller
You can also search for this author inPubMed Google Scholar - Robert D. Nicholls
You can also search for this author inPubMed Google Scholar - Mayumi Oda
You can also search for this author inPubMed Google Scholar - Anthony T. Papenfuss
You can also search for this author inPubMed Google Scholar - Zuly E. Parra
You can also search for this author inPubMed Google Scholar - David D. Pollock
You can also search for this author inPubMed Google Scholar - David A. Ray
You can also search for this author inPubMed Google Scholar - Jacqueline E. Schein
You can also search for this author inPubMed Google Scholar - Terence P. Speed
You can also search for this author inPubMed Google Scholar - Katherine Thompson
You can also search for this author inPubMed Google Scholar - John L. VandeBerg
You can also search for this author inPubMed Google Scholar - Claire M. Wade
You can also search for this author inPubMed Google Scholar - Jerilyn A. Walker
You can also search for this author inPubMed Google Scholar - Paul D. Waters
You can also search for this author inPubMed Google Scholar - Caleb Webber
You can also search for this author inPubMed Google Scholar - Jennifer R. Weidman
You can also search for this author inPubMed Google Scholar - Xiaohui Xie
You can also search for this author inPubMed Google Scholar - Michael C. Zody
You can also search for this author inPubMed Google Scholar - Jennifer A. Marshall Graves
You can also search for this author inPubMed Google Scholar - Chris P. Ponting
You can also search for this author inPubMed Google Scholar - Matthew Breen
You can also search for this author inPubMed Google Scholar - Paul B. Samollow
You can also search for this author inPubMed Google Scholar - Eric S. Lander
You can also search for this author inPubMed Google Scholar - Kerstin Lindblad-Toh
You can also search for this author inPubMed Google Scholar
Consortia
Broad Institute Genome Sequencing Platform
- Jennifer Baldwin
- , Amr Abdouelleil
- , Jamal Abdulkadir
- , Adal Abebe
- , Brikti Abera
- , Justin Abreu
- , St Christophe Acer
- , Lynne Aftuck
- , Allen Alexander
- , Peter An
- , Erica Anderson
- , Scott Anderson
- , Harindra Arachi
- , Marc Azer
- , Pasang Bachantsang
- , Andrew Barry
- , Tashi Bayul
- , Aaron Berlin
- , Daniel Bessette
- , Toby Bloom
- , Jason Blye
- , Leonid Boguslavskiy
- , Claude Bonnet
- , Boris Boukhgalter
- , Imane Bourzgui
- , Adam Brown
- , Patrick Cahill
- , Sheridon Channer
- , Yama Cheshatsang
- , Lisa Chuda
- , Mieke Citroen
- , Alville Collymore
- , Patrick Cooke
- , Maura Costello
- , Katie D'Aco
- , Riza Daza
- , Georgius De Haan
- , Stuart DeGray
- , Christina DeMaso
- , Norbu Dhargay
- , Kimberly Dooley
- , Erin Dooley
- , Missole Doricent
- , Passang Dorje
- , Kunsang Dorjee
- , Alan Dupes
- , Richard Elong
- , Jill Falk
- , Abderrahim Farina
- , Susan Faro
- , Diallo Ferguson
- , Sheila Fisher
- , Chelsea D. Foley
- , Alicia Franke
- , Dennis Friedrich
- , Loryn Gadbois
- , Gary Gearin
- , Christina R. Gearin
- , Georgia Giannoukos
- , Tina Goode
- , Joseph Graham
- , Edward Grandbois
- , Sharleen Grewal
- , Kunsang Gyaltsen
- , Nabil Hafez
- , Birhane Hagos
- , Jennifer Hall
- , Charlotte Henson
- , Andrew Hollinger
- , Tracey Honan
- , Monika D. Huard
- , Leanne Hughes
- , Brian Hurhula
- , M. Erii Husby
- , Asha Kamat
- , Ben Kanga
- , Seva Kashin
- , Dmitry Khazanovich
- , Peter Kisner
- , Krista Lance
- , Marcia Lara
- , William Lee
- , Niall Lennon
- , Frances Letendre
- , Rosie LeVine
- , Alex Lipovsky
- , Xiaohong Liu
- , Jinlei Liu
- , Shangtao Liu
- , Tashi Lokyitsang
- , Yeshi Lokyitsang
- , Rakela Lubonja
- , Annie Lui
- , Pen MacDonald
- , Vasilia Magnisalis
- , Kebede Maru
- , Charles Matthews
- , William McCusker
- , Susan McDonough
- , Teena Mehta
- , James Meldrim
- , Louis Meneus
- , Oana Mihai
- , Atanas Mihalev
- , Tanya Mihova
- , Rachel Mittelman
- , Valentine Mlenga
- , Anna Montmayeur
- , Leonidas Mulrain
- , Adam Navidi
- , Jerome Naylor
- , Tamrat Negash
- , Thu Nguyen
- , Nga Nguyen
- , Robert Nicol
- , Choe Norbu
- , Nyima Norbu
- , Nathaniel Novod
- , Barry O'Neill
- , Sahal Osman
- , Eva Markiewicz
- , Otero L. Oyono
- , Christopher Patti
- , Pema Phunkhang
- , Fritz Pierre
- , Margaret Priest
- , Sujaa Raghuraman
- , Filip Rege
- , Rebecca Reyes
- , Cecil Rise
- , Peter Rogov
- , Keenan Ross
- , Elizabeth Ryan
- , Sampath Settipalli
- , Terry Shea
- , Ngawang Sherpa
- , Lu Shi
- , Diana Shih
- , Todd Sparrow
- , Jessica Spaulding
- , John Stalker
- , Nicole Stange-Thomann
- , Sharon Stavropoulos
- , Catherine Stone
- , Christopher Strader
- , Senait Tesfaye
- , Talene Thomson
- , Yama Thoulutsang
- , Dawa Thoulutsang
- , Kerri Topham
- , Ira Topping
- , Tsamla Tsamla
- , Helen Vassiliev
- , Andy Vo
- , Tsering Wangchuk
- , Tsering Wangdi
- , Michael Weiand
- , Jane Wilkinson
- , Adam Wilson
- , Shailendra Yadav
- , Geneva Young
- , Qing Yu
- , Lisa Zembek
- , Danni Zhong
- , Andrew Zimmer
- & Zac Zwirko
Broad Institute Whole Genome Assembly Team
- David B. Jaffe
- , Pablo Alvarez
- , Will Brockman
- , Jonathan Butler
- , CheeWhye Chin
- , Sante Gnerre
- & Iain MacCallum
Corresponding authors
Correspondence toTarjei S. Mikkelsen, Eric S. Lander or Kerstin Lindblad-Toh.
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Tables S1-S17, Supplementary Figures S1- S10 with Legends, Supplementary Data and additional references. (PDF 2761 kb)
Rights and permissions
About this article
Cite this article
Mikkelsen, T., Wakefield, M., Aken, B. et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences.Nature 447, 167–177 (2007). https://doi.org/10.1038/nature05805
- Received: 05 December 2006
- Accepted: 03 April 2007
- Issue Date: 10 May 2007
- DOI: https://doi.org/10.1038/nature05805