Structural Analyses of Human Toll-like Receptor 4 Polymorphisms D299G and T399I (original) (raw)
Background: TLR4 polymorphism replacing Asp-299 with Gly and Thr-399 with Ile (D299G/T399I) causes LPS hyporesponsiveness.
Results: TLR4SNPs·MD-2·LPS exhibits an agonistic 2:2:2 architecture. Local structural differences were observed around D299G, but not around T399I, SNP site.
Conclusion: These local differences cause the modulation of surface properties of TLR4, which may affect ligand binding.
Significance: This study provides structural evidence of the functionality of the mutant TLR4 carrying the SNPs.
Keywords: Crystal Structure, Innate Immunity, Lipopolysaccharide (LPS), Toll-like Receptors (TLR), X-ray Crystallography, MD-2, SNPs
Abstract
Toll-like receptor 4 (TLR4) and its coreceptor MD-2 recognize bacterial lipopolysaccharide (LPS) and signal the innate immune response. Two single nucleotide polymorphisms (SNPs) of human TLR4, D299G and T399I, have been identified and suggested to be associated with LPS hyporesponsiveness. Moreover, the SNPs have been proposed to be associated with a variety of infectious and noninfectious diseases. However, how the SNPs affect the function of TLR4 remains largely unknown. Here, we report the crystal structure of the human TLR4 (D299G/T399I)·MD-2·LPS complex at 2.4 Å resolution. The ternary complex exhibited an agonistic “m”-shaped 2:2:2 architecture that was similar to that of the human wild type TLR4·MD-2·LPS complex. Local structural differences that might affect the binding of the ligands were observed around D299G, but not around T399I, SNP site.
Introduction
The innate immune system is the first line of defense against microbial infection. Microbial components such as lipoprotein, lipopolysaccharide (LPS), and nucleic acids are recognized by the germ line-encoded, cell surface Toll-like receptors (TLRs)2 and trigger the innate immune response, including the production of proinflammatory cytokines and type I interferon (1). TLR4 is one of the most extensively investigated TLRs and recognizes endotoxic LPS in the outer membrane of Gram-negative bacteria (2). Excessive signaling in response to LPS often causes uncontrolled amplification of inflammatory responses, leading to fatal septic shock (3, 4).
TLR4 is a type I transmembrane protein with an extracellular domain composed of 22 leucine-rich repeats (LRRs), which mediate LPS recognition and receptor dimerization; a transmembrane domain; and the Toll/IL-1 receptor domain (TIR domain) that is essential for downstream signal transduction (5). TLR4 alone does not directly bind LPS and requires the coreceptor MD-2. MD-2 is associated with the extracellular domain of TLR4 and directly binds LPS, conferring LPS responsiveness on TLR4 (6, 7).
The lipid A moiety of LPS, which anchors LPS to the outer membrane of Gram-negative bacteria, is responsible for the immunostimulatory activity of LPS (8, 9). Lipid A consists of a 1,4′-bisphosphorylated diglucosamine backbone to which variable lengths and numbers of acyl chains are covalently linked (9). The acyl chains, as well as the two phosphate groups, determine the agonistic activity of lipid A. For example, Escherichia coli lipid A is usually hexa-acylated and acts as a potent agonist for all mammalian cells, whereas tetra-acylated lipid IVa, the precursor of E. coli LPS, acts as an agonist only for some mammalian species (10, 11). In addition, deletion of either phosphate group of lipid A reduces its endotoxic activity (12, 13).
The crystal structures of human TLR4·MD-2·LPS (14) and mouse TLR4·MD-2·LPS (15) revealed the detailed molecular mechanism of LPS recognition by the TLR4·MD-2 complex, in which five of the six acyl chains of LPS were buried in the MD-2 cavity and the sixth acyl chain was partially exposed to the solvent. LPS binding to the TLR4·MD-2 complex induced the “m”-shaped 2:2:2 TLR4·MD-2·LPS complex, where two TLR4 molecules are in close proximity at their C-terminal convex faces. This arrangement suggested subsequent dimerization of intracellular TIR domains for the initiation of downstream signal transduction (16, 17).
Two single nucleotide polymorphisms (SNPs), A896G and C1196T, have been identified in the human TLR4 gene; these result in the amino acid changes, D299G and T399I, respectively, and have been shown to cause LPS hyporesponsiveness (18). The frequency of these SNPs is reported to be up to 18% (19). The TLR4 SNPs have been reported to be associated with a susceptibility to infections with Gram-negative bacteria (18) or respiratory syncytial virus (20). The TLR4 SNPs have also been linked to reduced risk of noninfectious diseases, such as atherosclerosis (21) and rheumatoid arthritis (22). However, the molecular mechanisms by which the TLR4 SNPs affect receptor function are largely controversial. D299G/T399I mutations in TLR4 markedly reduce extracellular accumulation of functional TLR4 in human embryonic kidney (HEK293T) cells (24). Compromised LPS responses elicited by the D299G variant were originally associated with decreased expression of the mutant TLR4 species in human airway epithelial cells (18). In contrast, another study failed to find impaired cell surface expression of polymorphic TLR4 (23). It was also reported that D299G or T399I TLR4 mediates diminished NF-κB activation and cytokine gene expression in response to LPS, respiratory syncytial virus F protein, or chlamydial Hsp60 under conditions of equal total and cell surface expression of transfected TLR4 species in HEK293T cells (23, 34).
Prohinar et al. (24) reported that the mutant TLR4 (D299G/T399I) has high affinity for LPS, comparable with that of wild-type TLR4, but is more dependent on coexpression of MD-2 in the proper cell surface expression. The D299G mutation has been shown to alter TLR4 signaling by interfering with recruitment of MyD88 and TIR domain-containing adaptor inducing interferon-β (TRIF) (25).
Here, we present the crystal structure of human TLR4 (D299G/T399I)·MD-2·Re-LPS (incorporating the Re chemotype of E. coli LPS) complex at 2.4 Å resolution. The ternary complex exhibited an agonistic m-shaped 2:2:2 architecture that was similar to that of the human wild-type TLR4·MD-2·LPS complex. This study provides structural evidence of the functionality of the mutant TLR4 carrying the SNPs and describes the conformational change in TLR4 that is induced by the SNPs.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
The extracellular domain of hTLR4 (residues 23–629) was inserted into the expression vector pMT/BiP/V5-His of the Drosophila Expression System (Invitrogen). Human MD-2 (residues 19–160), which was fused to a C-terminal thrombin cleavage site located upstream of protein A tags, was also inserted into the vector. The SNP mutations A896G and C1196T were introduced into the hTLR4 vector by PCR-based mutagenesis. Drosophila S2 cells were co-transfected with the hTLR4SNPs, hMD-2, and pCoHygro vectors. Stably transfected cells were selected in Sf-900II-SFM medium containing 300 μg/ml hygromycin. Protein expression was induced by the addition of 0.5 mm CuSO4, and the culture supernatant was harvested 100–120 h after induction.
To prepare the Re-LPS complex, purified hTLR4SNPs·MD-2 was concentrated to 1–2 mg/ml. The purified complex was added to Re-LPS, at a protein:ligand molar ratio of 1:3, in 10 mm Tris, pH 8.0, containing 0.15 m NaCl and 0.5% Triton X-100 and incubated for 1 h at 37 °C. Subsequently, the Re-LPS complex of hTLR4SNPs·MD-2 was purified by HiTrap Q anion exchange chromatography followed by Superdex 200 gel filtration chromatography. The hTLR4SNPs·MD-2·Re-LPS complex was concentrated to 8.1 mg/ml in a crystallization buffer of 10 mm Tris, pH 8.0, and 0.15 m NaCl.
Crystallization experiments were performed using sitting-drop vapor-diffusion at 20 °C. Crystals were obtained with reservoir solution containing 26–30% (w/v) PEG 1000 and 0.1 m Tris-HCl (pH 8.0).
Data Collection and Structure Determination
Diffraction datasets were collected on beamlines BL32XU and BL41XU at SPring-8 (Hyogo, Japan) under cryogenic conditions at 100 K. Crystals of the hTLR4SNPs·MD-2·Re-LPS complex were cryoprotected in reservoir solution supplemented with 25% (v/v) glycerol. The datasets were processed with the HKL2000 package (26). The initial phases were determined with the molecular replacement method using the program Molrep (27) and the coordinates of the hTLR4wt·MD-2·Ra-LPS complex (Protein Data Bank (PDB) ID: 3FXI). Atomic models of Re-LPS were built into strong and continuous electron densities found in and above the MD-2 cavity. The structural models of hTLR4SNPs·MD-2·Re-LPS were refined at 2.4 Å resolution, with stepwise cycles of manual model building, using the program COOT (28), and restrained refinement, with REFMAC (29). The quality of the final structure was evaluated with PROCHECK (30). The data collection and refinement statistics are summarized in Table 1. Figures representing structures were prepared using PyMOL (31).
TABLE 1.
Data collection and refinement statistics
| hTLR4SNPs·MD-2·Re-LPS | ||
|---|---|---|
| Data collection | ||
| Beamline | SPring-8 BL32XU | |
| Wavelength (Å) | 1.0000 | |
| Resolution range (Å) | 39.9–2.4 | |
| Space group | C2 | |
| Lattice parameters a, b, c (Å) | 158.0, 124.7, 109.1 | |
| Lattice parameter β (°) | 115.7 | |
| No. of measured reflections | 467,456 | |
| No. of unique reflections | 74,485 | |
| Multiplicity | 6.3 (6.0)a | |
| Average I | σ | (I) | 9.8 (2.8)a |
| Completeness (%) | 100.0 (99.9)a | |
| _R_merge(I)b | 0.129 (0.605)a | |
| Refinement | ||
| Resolution range (Å) | 39.2–2.40 | |
| No. of reflections used | 70,730 | |
| Model | 2:2:2 TLR4SNPs·MD-2·Re-LPS | |
| Average _B_-factor (Å2) | 48.4 | |
| R (%)c | 19.9 | |
| _R_free (%)d | 24.9 | |
| r.m.s. deviations | ||
| Bond length (Å) | 0.008 | |
| Bond angles (°) | 1.46 |
Native Page
Purified TLR4·MD-2 proteins (1.5 μg) were mixed with various amounts of Re-LPS (0.0125, 0.05, and 0.2 μg) in 10 mm Tris-HCl (pH 7.5), 0.15 m NaCl, and 0.1% Triton X-100 in the absence or presence of hCD14 (0.75 μg) and incubated for 1 h at 37 °C. After incubation, loading buffer (30% glycerol/Tris-HCl (pH 8.0), bromphenol blue) was added to the samples. Samples were subjected to 12.5% native PAGE and stained with Coomassie Brilliant Blue.
RESULTS
Oligomerization State of the TLR4SNPs·MD-2 Complex
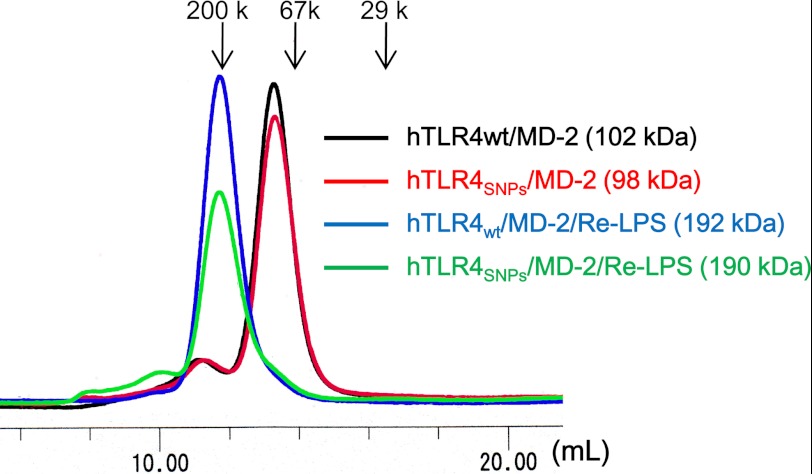
To analyze whether TLR4 SNPs (D299G/T399I) affect LPS binding and LPS-induced oligomerization of TLR4·MD-2, purified wild-type TLR4·MD-2 and TLR4 (D299G/T399I)·MD-2, hereafter referred to as TLR4WT and TLR4SNPs, respectively, were prepared. As we recently reported, TLR4SNPs·MD-2 and TLR4WT·MD-2 bind to LPS with similar efficiency, as judged by native PAGE analysis (32). The TLR4SNPs·MD-2· complex formed a dimer in solution, as determined by gel filtration chromatography (Fig. 1), consistent with the previous observation that LPS binding induces the dimerization of hTLR4WT·MD-2 (33).
FIGURE 1.
Oligomerization state of the TLR4SNPs·MD-2 complex. Gel filtration chromatography of hTLR4SNPs·MD-2 is shown. The hTLR4WT·MD-2 (black), hTLR4SNPs·MD-2 (red), hTLR4WT·MD-2·Re-LPS (blue), and hTLR4SNPs·MD-2·Re-LPS (green) complexes were analyzed by Superdex 200 gel filtration chromatography. Columns were calibrated with the molecular mass standards β-amylase (200 kDa (200 k)), bovine serum albumin (67 kDa (67 k)), and carbonic anhydrase (29 kDa (29 k)), as indicated by arrows. The apparent molecular mass of the proteins was calculated from the elution volumes.
Overall Structure of the TLR4SNPs·MD-2· Complex
To address whether TLR4SNPs induces local or overall conformational change in the TLR4·MD-2 complex, we determined the crystal structure of the TLR4SNPs·MD-2·Re-LPS complex at a resolution of 2.4 Å (Table 1). Structures were determined by the molecular replacement method with the coordinates of the TLR4WT·MD-2·LPS complex (PDB ID: 3FXI). Lipid A and one 3-deoxy-d-_manno_-oct-2-ulosonic acid (Kdo) moiety of Re-LPS were unambiguously modeled into the continuous electron densities observed in and above the hydrophobic cavity of MD-2. The outer Kdo residue of Re-LPS was not modeled because of poor electron density.
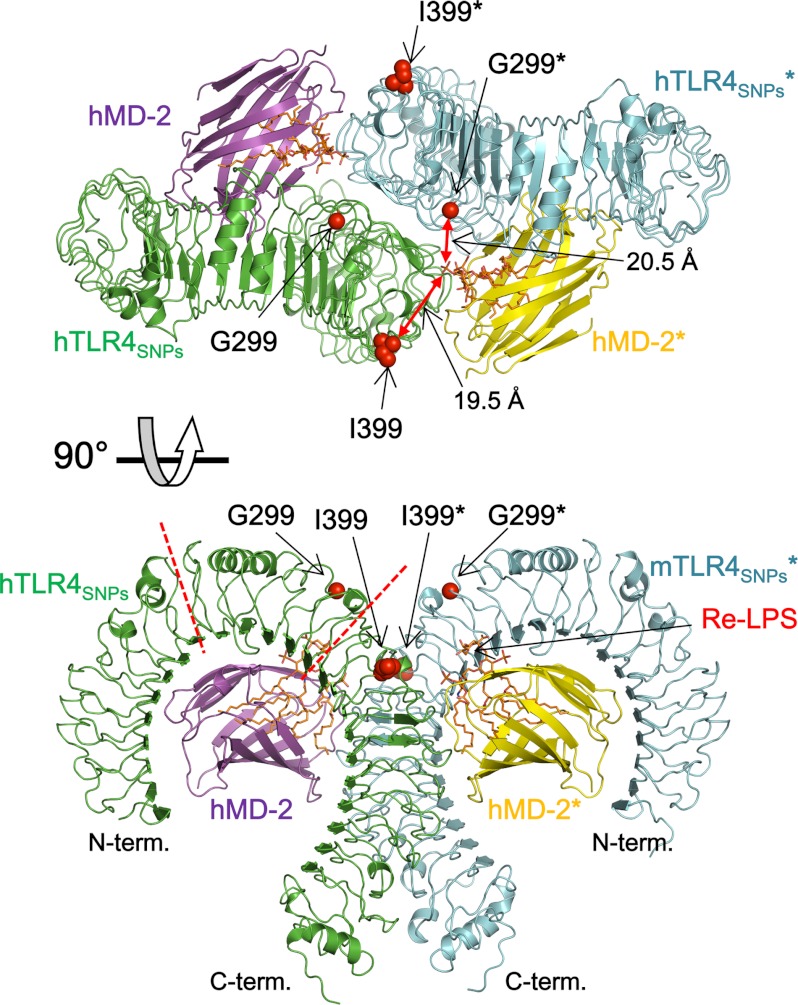
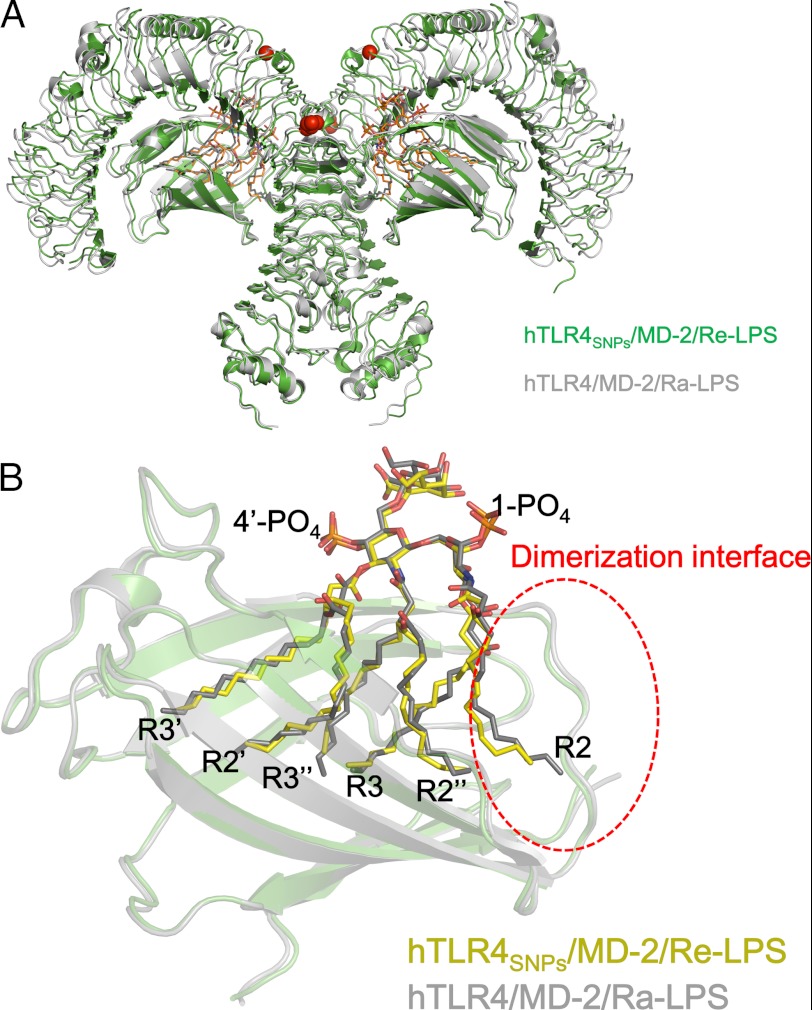
The TLR4SNPs·MD-2·Re-LPS complex crystallized in space group C2, and the crystallographic asymmetric unit contained two copies of the 1:1:1 TLR4SNPs·MD-2·Re-LPS complex related by noncrystallographic 2-fold symmetry, generating a 2:2:2 TLR4SNPs·MD-2· complex (Fig. 2). As expected, because TLR4SNPs·MD-2 and TLR4WT·MD-2 bound to lipid A with comparable efficiency and dimerization was induced by binding to lipid A (32), the overall structure of the 2:2:2 TLR4SNPs·MD-2·Re-LPS complex was very similar to that of the previously reported agonistic form of the 2:2:2 hTLR4wt·MD-2·Ra-LPS complex (14), with root mean square deviation (r.m.s.d.) values of 1.6 Å for all 1,620 Cα atoms of the 2:2:2 TLR4·MD-2·LPS complexes (Fig. 3A). The Re-LPS molecules bound to the hydrophobic cavity of MD-2 in the hTLR4SNPs·MD-2·Re-LPS complex exhibited conformations nearly identical to those of Ra-LPS molecules in the hTLR4wt·MD-2·Ra-LPS complex, with the acyl chain R2 partially exposed to the solvent, and mediated dimerization by interacting with another TLR4SNPs molecule (Fig. 3B). The 2:2:2 TLR4SNPs·MD-2·Re-LPS complex showed m-shaped tail-to-tail organization, in which the N termini of two TLR4SNPs faced in opposite directions and the C termini approached at the center. The undistinguished overall organization of the LPS-bound forms of TLR4WTMD-2·Ra-LPS and TLR4SNPs·MD-2·Re-LPS suggests that TLR4SNPs, like TLR4WT, could recognize LPS and transduce a signal to the intracellular compartment, although the efficiency may be impaired. Throughout this study, the second TLR4, MD-2, LPS, and their residues in the 2:2:2 TLR4·MD-2·LPS complex are indicated by asterisks to distinguish them from those of the 1:1 TLR4·MD-2 complex.
FIGURE 2.
Overall structure of the hTLR4SNPs·MD-2·Re-LPS complex. hTLR4 and hMD-2 are shown in green and magenta, respectively, and their dimerization partners are shown in blue and yellow, respectively. Re-LPS molecules bound to the hydrophobic cavity of MD-2 are shown in orange as stick representations. The SNP residues Gly-299 and Ile-399 are shown in space-filling representations in red. The dimerization partners of the 1:1:1 hTLR4SNPs·MD-2·Re-LPS complex are indicated by asterisks.
FIGURE 3.
Comparison of hTLR4SNPs·MD-2·Re-LPS with hTLR4wt·MD-2·Ra-LPS. A, superposition of hTLR4SNPs·MD-2·Re-LPS (green) and hTLR4WT·MD-2·Re-LPS (gray, PDB ID 3FXI) in the same orientation as in the lower panel of Fig. 2. B, structural comparison of the LPS ligand in the MD-2 cavity. Superposition of MD-2 and LPS ligands of hTLR4SNPs·MD-2·Re-LPS (carbon atoms of LPS in yellow) and hTLR4·MD-2·Ra-LPS (carbon atoms of LPS in gray) is shown. LPS ligands are shown in stick representation with oxygen, nitrogen, and phosphorus atoms in red, blue, and orange, respectively.
SNP Sites Are Far from the LPS Binding Site
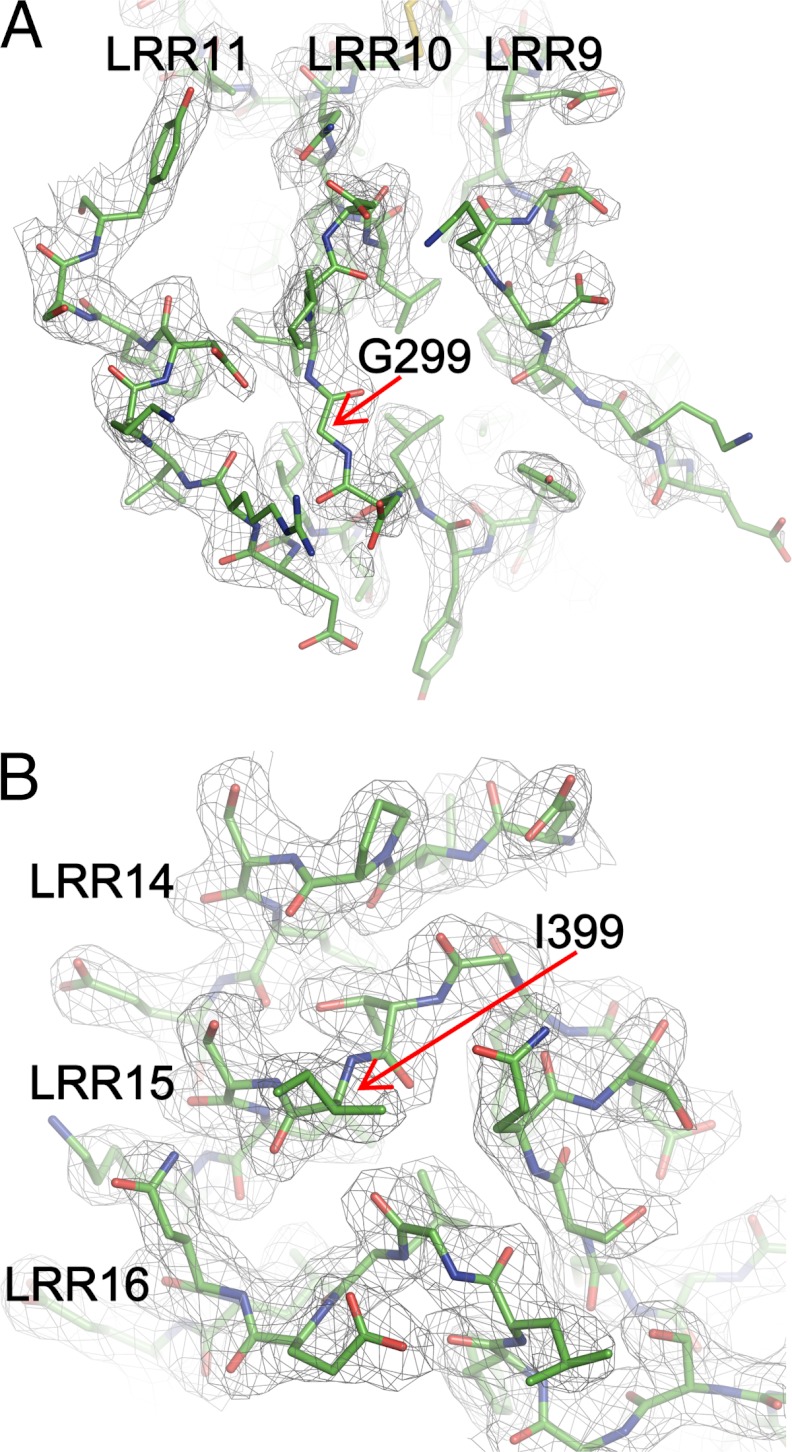
The quality of the electron density map is sufficiently high to allow modeling of the two SNP residues, Gly-299 and Ile-399, and the residues surrounding them with confidence (Fig. 4). The D299G SNP is located in the loop region of LRR10, on the convex face of TLR4 (Fig. 2). The T399I SNP at position 3 of the consensus LRR motif in LRR15 is located on the lateral face of TLR4 before the β-sheet of the concave face (Fig. 2). These two residues are not involved in the direct recognition of LPS and MD-2; the distances between the 1-phosphate atom of Re-LPS* and the Cα atoms of Gly-299* and Ile-399 are 20.5 and 19.2 Å, respectively. These observations suggest that the SNPs do not directly affect binding to either LPS (at least in the case of Re-LPS) or MD-2.
FIGURE 4.
SNP structures. Electron densities are contoured at the 1.0-σ level in the 2_Fo_ − Fc map around the D299G (A) and T399I (B) sites. LRRs containing the SNP residues and nearby LRRs are shown.
Structural Comparison of TLR4WT and TLR4SNPs
The structures of the TLR4 monomers in TLR4SNPs and TLR4WT within the TLR4WT·MD-2·Ra-LPS complex could be well superposed on top of each other, with an r.m.s.d. value of 0.78 Å for all Cα atoms. Specifically, the r.m.s.d. values were 0.45, 0.85, and 0.65 Å for the N-terminal domain (LRRNT and LRR1–6, residues 27–202), the central domain (LRR7–12, residues 203–348), and the C-terminal domain (LRR13–22 and LRRCT, residues 349–631), respectively. D299G and T399I SNPs are located in the central and C-terminal domains, respectively (Fig. 2).
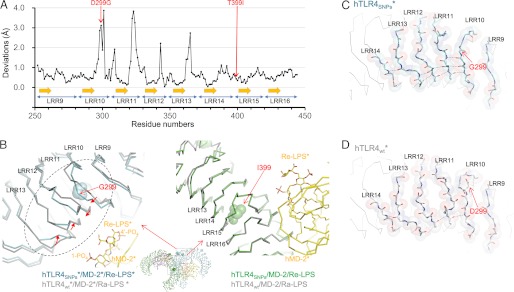
Although the overall arrangements of the 2:2:2 TLR4SNPs·MD-2·Re-LPS complex and the 2:2:2 TLR4WT·MD-2·Ra-LPS complex are very similar, local structural differences were observed around the D299G SNP site. Noticeable positional deviations for the loop regions of LRR10 to LRR13 were observed when the two structures were superposed on each other at TLR4 residues 250–450, whereas the consensus β strands of LRRs for these regions were well superposed (Fig. 5A). The loop structures of LRR10–13 of TLR4SNPs are apparently different from those of TLR4WT in the hTLR4wt·MD-2·Ra-LPS (Fig. 5B, left). The largest deviations for each LRR were 3.9 (LRR10), 3.8 (LRR11), 2.2 (LRR12), and 2.7 Å (LRR13). In the TLR4SNPs structure, the loop regions of LRR10 (residues 298–299) and LRR11 (residues 322–324) were shifted toward LRR12, and LRR12 (residue 343) and LRR13 (residues 362–365) were shifted toward LRR11 with respect to the TLR4WT structure. As a result, LRR11 makes contact with LRR12 to form new van der Waals interactions between the side chain of Val-323 (LRR11) and the side chains of Phe-342 and Pro-346 (LRR12). In the TLR4SNPs structure, eight hydrogen bonds between the neighboring LRRs were observed in the region shown in Fig. 5B (left, dashed circle), whereas in the TLR4WT structure, only three hydrogen bonds were observed (Fig. 5, C and D). In contrast to the structural difference observed around the D299G SNP site, the structure around the T399I SNP site is almost identical in TLR4SNPs and TLR4WT, with an r.m.s.d. value between TLR4SNPs and TLR4wt in the hTLR4wt·MD-2·Ra-LPS complex of 0.45 Å for 120 Cα atoms in LRR13–17 (residues 349–468) (Fig. 5B, right).
FIGURE 5.
Structural differences induced by the SNPs. A, Cα positional deviations after superposition of TLR4SNPs with TLR4WT in TLR4 residues 250–450. B, superposition of hTLR4SNPs·MD-2·Re-LPS (cyan in left panel and green in right panel) and hTLR4wt·MD-2·Ra-LPS (gray) showing the D299G (left) and T399I (right) SNP sites. TLR4 and MD-2 are shown in Cα representations. C and D, close-up view of the dashed circle region in B showing the main-chain interactions around the D299G SNP site of hTLR4SNPs (in C) and hTLR4wt (in D). The main-chain atoms and the side chain of Asp-299 in TLR4wt are shown in stick representations with semitransparent space-filling representations. Hydrogen bonds are shown in dashed lines.
Effect of TLR4 Polymorphism on LPS Responses
To investigate the effect of TLR4 polymorphism on LPS responses, we carried out the experiment of ligand-dependent dimerization in the presence and absence of CD14 by using native PAGE. The addition of 0.05 μg of lipid A induces the dimerization of TLR4WT·MD-2 both in the presence and in the absence of CD14. The dimerization of TLR4SNPs·MD-2 was also induced to the same degree as TLR4WT·MD-2 (supplemental Fig. S1). Thus, we concluded that TLR4 polymorphism does not change the efficiency of the transfer of LPS from CD14 to TLR4. This is consistent with the result reported by Prohinar et al. (24). They reported that D299G/T399I mutations in TLR4 have little or no effect on interaction of TLR4·MD-2 with endotoxin·CD14 complex. The apparent Kd values of this interaction were estimated to be 196 and 186 pm for TLR4WT and TLR4SNPs, respectively. Moreover, both TLR4WT and TLR4SNPs demonstrated similar affinity interaction with endotoxin MD-2 (24). It is possible that the conformational change of TLR4·MD-2 by the D299G/T399I polymorphism influences LPS binding to the complex, but we could not detect any difference between TLR4WT·MD-2 and TLR4SNPs·MD-2 in LPS binding (32).
DISCUSSION
The present structural study clearly shows that the D299G and T399I polymorphisms of TLR4 do not affect its dimerization; however, significant local conformational changes are observed, particularly in the D299G SNP site. In support of the structural difference between TLR4WT and TLR4SNPs, we recently reported that the reactivity of the anti-TLR4 antibody HTA125 with TLR4·MD-2 on Ba/F3 cells was significantly impaired by introduction of D299G/T399I SNPs to TLR4WT, although the surface expression level was not changed, which suggests a conformational difference between TLR4WT·MD-2 and TLR4SNPs·MD-2 in the HTA125 epitope region (32). We also reported that, based on native PAGE analysis, the efficiency of TLR4SNPs·MD-2 dimerization by a weak agonist, monophosphoryl lipid A, is apparently lower than that of TLR4WT·MD-2 (32). The loop regions of LRR10–13 in which conformational changes occurred between TLR4SNPs and TLR4WT were located near (about 10 Å from) both phosphate groups of LPS stabilized by several basic residues. Hence, although not directly involved in LPS recognition, the structural difference may affect the binding efficiency or proper positioning of the LPS ligands for dimerization. The weak agonist monophosphoryllipid A, composed of a monophosphate, would be stabilized to a lesser extent, resulting in reduced dimerization capacity.
The loop conformation of TLR4SNPs is likely not induced by a molecular packing artifact in the crystal because the two TLR4SNPs molecules in the asymmetric unit, despite being in different environments, have essentially the same conformation, with an r.m.s.d. value of 0.26 Å for 124 Cα atoms in LRR8–12 (residues 225–348). To examine the effect of crystal packing, we carried out the further analysis about crystal contacts. We mapped the residues on the protein surface that make contact with the symmetrical molecule in the crystal in accordance with the amount of buried surface area (supplemental Fig. S2_A_, supplemental Table S1). In the TLRWT structure (PDB ID: 3FXI), the two TLR4WT molecules exhibit identical conformation, and the convex region of LRR10–11 of both molecules made little contact with the symmetrical molecule. On the other hand, the molecule B in the TLRSNPs structure interacted with the symmetrical molecule, whereas the molecule A made little contact with it. Most of the LRRs show a small buried surface area (below ∼100 Å2) (supplemental Table S1). LRR12 in the molecule B of the TLRSNPs structure exhibits the largest buried surface area (242 Å2), but this contact is unlikely to induce the significant conformational change. In fact, the two TLR4SNPs molecules in the asymmetric unit, despite being in different environments, have essentially the same conformation.
We also calculated buried surface area around the convex region of LRR13–16 near Thr-399 SNP site (supplemental Fig. S2_B_). This region of all molecules made no contacts with the symmetrical molecules. Therefore, the crystal packing effect would be negligible. On the other hand, the convex region of TLR4 LRR10–11 displays the structural difference between human and mouse TLR4. We solved mouse TLR4 in complex with MD-2 in two forms (PDB IDs: 3VQ1 and 3VQ2). The crystals contain two molecules in the asymmetric unit and belong to the different space group (3VQ1, P212121; 3VQ2, P21). Each molecule existed in the different environment in the crystals, but no obvious conformational changes were observed around LRR10–11. Therefore, the conformation in this region would be inherent for mouse TLR4. Taken together, the observed conformational difference was thought to be the result of the introduction of the D299G SNP although the convex region of LRR10–11 is highly flexible.
In the TLR4wt structure, the side chain of Asp-299 formed a hydrogen bond with the backbone amide nitrogen of Asp-302, stabilizing the helical conformation of this region. Introduction of the glycine residues at this position would cause rearrangement of the loop structure of LRR10, with the effect of this rearrangement then propagating to the nearby LRRs. However, the possibility that the difference is induced by the different chemotype of the LPS ligand used in the structural studies cannot be excluded. The lipid A moiety of the Re-LPS used here has two additional Kdo residues, whereas Park et al. (14) used an Ra-LPS that has 10 additional saccharide residues. We consider this possibility unlikely because the additional saccharides in Ra-LPS are not directly involved in the interaction with TLR4.
In summary, we have determined the crystal structure of TLR4SNPs·MD-2·Re-LPS and clearly demonstrated that the overall organization of the LPS-bound agonistic form of TLR4SNPs·MD-2 is almost identical to that of TLR4WT·MD-2. The substitution of more flexible and neutral residues by the Asp-299 to Gly-299 SNP induced a structural change in the loop region of LRR10–12, causing the modulation of surface properties of TLR4, which may affect ligand binding, as well as folding efficiency, cell surface expression, and protein stability. The effect of this change may be more apparent when TLR4SNPs is stimulated by ligands with weak agonistic activity, such as monophosphoryl lipid A. The impact of the T399I SNP is relatively minor, with almost no structural difference observed. The introduction of more hydrophobic residues by the T399I SNP may influence the folding efficiency, cell surface expression, and protein stability of TLR4.
Supplementary Material
Supplemental Data
Acknowledgments
We thank the beamline staff at SPring-8 for assistance with data collection.
*
This work was supported by Grant-in-Aid for Scientific Research (B) and Grant-in-Aid for Scientific Research on Innovative Areas (3106) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The atomic coordinates and structure factors (code 4G8A) have been deposited in the Protein Data Bank (http://wwpdb.org/).
2
The abbreviations used are:
TLR
Toll-like receptor
hTLR4
human TLR4
TIR
Toll/IL-1 receptor domain
LRR
leucine-rich repeat
r.m.s.d.
root mean square deviation
Kdo
3-deoxy-d-_manno_-oct-2-ulosonic acid.
REFERENCES
- 1.Janeway C. A., Jr., Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 2.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 3.Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., Pinsky M. R. (2001) Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 4.Rangel-Frausto M. S. (2005) Sepsis: still going strong. Arch. Med. Res. 36, 672–681 [DOI] [PubMed] [Google Scholar]
- 5.Gay N. J., Gangloff M. (2007) Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 76, 141–165 [DOI] [PubMed] [Google Scholar]
- 6.Nagai Y., Akashi S., Nagafuku M., Ogata M., Iwakura Y., Akira S., Kitamura T., Kosugi A., Kimoto M., Miyake K. (2002) Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat. Immunol. 3, 667–672 [DOI] [PubMed] [Google Scholar]
- 7.Shimazu R., Akashi S., Ogata H., Nagai Y., Fukudome K., Miyake K., Kimoto M. (1999) MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 189, 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schletter J., Heine H., Ulmer A. J., Rietschel E. T. (1995) Molecular mechanisms of endotoxin activity. Arch. Microbiol. 164, 383–389 [DOI] [PubMed] [Google Scholar]
- 9.Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golenbock D. T., Hampton R. Y., Qureshi N., Takayama K., Raetz C. R. (1991) Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J. Biol. Chem. 266, 19490–19498 [PubMed] [Google Scholar]
- 12.Rietschel E. T., Kirikae T., Schade F. U., Ulmer A. J., Holst O., Brade H., Schmidt G., Mamat U., Grimmecke H. D., Kusumoto S., et al. (1993) The chemical structure of bacterial endotoxin in relation to bioactivity. Immunobiology 187, 169–190 [DOI] [PubMed] [Google Scholar]
- 13.Rietschel E. T., Kirikae T., Schade F. U., Mamat U., Schmidt G., Loppnow H., Ulmer A. J., Zähringer U., Seydel U., Di Padova F., et al. (1994) Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8, 217–225 [DOI] [PubMed] [Google Scholar]
- 14.Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 15.Ohto U., Fukase K., Miyake K., Shimizu T. (2012) Structural basis of species-specific endotoxin sensing by TLR4/MD-2. Proc. Natl. Acad. Sci. U.S.A. 109, 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin M. S., Lee J. O. (2008) Structures of the Toll-like receptor family and its ligand complexes. Immunity 29, 182–191 [DOI] [PubMed] [Google Scholar]
- 17.Kumar H., Kawai T., Akira S. (2009) Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 388, 621–625 [DOI] [PubMed] [Google Scholar]
- 18.Arbour N. C., Lorenz E., Schutte B. C., Zabner J., Kline J. N., Jones M., Frees K., Watt J. L., Schwartz D. A. (2000) TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 25, 187–191 [DOI] [PubMed] [Google Scholar]
- 19.Ferwerda B., McCall M. B., Alonso S., Giamarellos-Bourboulis E. J., Mouktaroudi M., Izagirre N., Syafruddin D., Kibiki G., Cristea T., Hijmans A., Hamann L., Israel S., ElGhazali G., Troye-Blomberg M., Kumpf O., Maiga B., Dolo A., Doumbo O., Hermsen C. C., Stalenhoef A. F., van Crevel R., Brunner H. G., Oh D. Y., Schumann R. R., de la Rúa C., Sauerwein R., Kullberg B. J., van der Ven A. J., van der Meer J. W., Netea M. G. (2007) TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc. Natl. Acad. Sci. U.S.A. 104, 16645–16650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tulic M. K., Hurrelbrink R. J., Prêle C. M., Laing I. A., Upham J. W., Le Souef P., Sly P. D., Holt P. G. (2007) TLR4 polymorphisms mediate impaired responses to respiratory syncytial virus and lipopolysaccharide. J. Immunol. 179, 132–140 [DOI] [PubMed] [Google Scholar]
- 21.Kiechl S., Lorenz E., Reindl M., Wiedermann C. J., Oberhollenzer F., Bonora E., Willeit J., Schwartz D. A. (2002) Toll-like receptor 4 polymorphisms and atherogenesis. N. Engl. J. Med. 347, 185–192 [DOI] [PubMed] [Google Scholar]
- 22.Radstake T. R., Franke B., Hanssen S., Netea M. G., Welsing P., Barrera P., Joosten L. A., van Riel P. L., van den Berg W. B. (2004). Arthritis Rheum. 50, 999–1001 [DOI] [PubMed] [Google Scholar]
- 23.Rallabhandi P., Bell J., Boukhvalova M. S., Medvedev A., Lorenz E., Arditi M., Hemming V. G., Blanco J. C., Segal D. M., Vogel S. N. (2006) Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J. Immunol. 177, 322–332 [DOI] [PubMed] [Google Scholar]
- 24.Prohinar P., Rallabhandi P., Weiss J. P., Gioannini T. L. (2010) Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on coexpression of MD-2 than does wild-type TLR4. J. Immunol. 184, 4362–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueroa L., Xiong Y., Song C., Piao W., Vogel S. N., Medvedev A. E. (2012) The Asp299Gly polymorphism alters TLR4 signaling by interfering with recruitment of MyD88 and TRIF. J. Immunol. 188, 4506–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27.Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 28.Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31.DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 32.Yamakawa N., Ohto U., Akashi-Takamura S., Takahashi K., Saitoh S., Tanimura N., Suganami T., Ogawa Y., Shibata T., Shimizu T., Miyake K. (2012) Human TLR4 polymorphism D299G/T399I alters TLR4/MD-2 conformation and response to a weak ligand monophosphoryl lipid A. Int. Immunol., in press [DOI] [PubMed] [Google Scholar]
- 33.Prohinar P., Re F., Widstrom R., Zhang D., Teghanemt A., Weiss J. P., Gioannini T. L. (2007) Specific high affinity interactions of monomeric endotoxin·protein complexes with Toll-like receptor 4 ectodomain. J. Biol. Chem. 282, 1010–1017 [DOI] [PubMed] [Google Scholar]
- 34.Awomoyi A. A., Rallabhandi P., Pollin T. I., Lorenz E., Sztein M. B., Boukhvalova M. S., Hemming V. G., Blanco J. C., Vogel S. N. (2007) Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J. Immunol. 179, 3171–3177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data