Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders (original) (raw)
Abstract
Mutations in the CHD7 gene cause human developmental disorders including CHARGE syndrome. Genetic studies in model organisms have further established CHD7 as a central regulator of vertebrate development. Functional analysis of the CHD7 protein has been hampered by its large size. We used a dual-tag system to purify intact recombinant CHD7 protein and found that it is an ATP-dependent nucleosome remodeling factor. Biochemical analyses indicate that CHD7 has characteristics distinct from SWI/SNF- and ISWI-type remodelers. Further investigations show that CHD7 patient mutations have consequences that range from subtle to complete inactivation of remodeling activity, and that mutations leading to protein truncations upstream of amino acid 1899 of CHD7 are likely to cause a hypomorphic phenotype for remodeling. We propose that nucleosome remodeling is a key function for CHD7 during developmental processes and provide a molecular basis for predicting the impact of disease mutations on that function.
Keywords: SNF2H, idiopathic hypogonadotropic hypogonadism, chromodomains
Mutation of the Chromodomain Helicase DNA-binding (CHD) 7 gene was identified as the major cause of CHARGE syndrome (1). The acronym CHARGE (formerly Hall-Hittner syndrome) was coined to describe a nonrandom occurrence of typical clinical signs including Coloboma of the eye, Heart defects, Atresia of the choanae, Retardation of growth, Genital and Ear abnormalities including hearing loss (2). This syndrome affects about 1 child of 10,000 born and leads to death before the age of 5 in ∼30% of the cases (3). CHD7 has features that relate it to the trithorax-group (TrxG) of genes. TrxG genes were initially identified in a screen for dominant suppressors of the phenotype caused by a Polycomb gene (Pc) mutation in Drosophila (4). _Pc_-group (PcG) genes prevent the inappropriate expression of homeotic genes (which are developmental master regulators). Consequently many mutations in PcG genes cause homeotic transformations. One of the dominant suppressors of this Pc phenotype was called kismet (kis). This gene was then shown to be essential and the loss of maternal kis causes dramatic embryonic segmentation defects (5).
These studies in Drosophila presaged further studies implicating the kis homolog, CHD7, in vertebrate development and in human disorders beyond CHARGE. Whereas Chd7 −/− mice die at embryonic day E10.5, heterozygous mice develop CHARGE-like symptoms (6, 7). Other organisms such as frog and zebrafish also present similar developmental defects when a dominant negative form of CHD7 mRNA is injected in tadpoles or when CHD7 levels are depleted using morpholinos, respectively (8, 9). The impact of altering CHD7 activity extends beyond the typical CHARGE features (For reviews (3, 10) and refs. therein). In some cases, patients present limb anomalies as severe as monodactily or the absence of a tibia (11). In addition, CHD7 knockdown experiments in zebrafish and CHD7 polymorphisms in humans have been linked to scoliosis (8, 12). Other CHD7 mutations cause Idiopathic Hypogonadotropic Hypogonadism (IHH) or Kallmann Syndrome (KS) (10, 13–15) and analyses of Chd7 heterozygous mice showed that CHD7 plays important roles in the regulation of puberty and reproductive organ formation (16, 17). A recent study has also extended the potential roles for CHD7 to small-cell lung cancer (18).
The fact that CHD7 mutations lead to such a vast spectrum of phenotypes may be explained, at least in part, by the ubiquitous expression of CHD7 in human fetal tissues by 22 d (3), the involvement of CHD7 in the modulation of embryonic stem cell-specific gene expression (19, 20) and the transcription of ribosomal DNA genes (21). Furthermore, CHD7 is important for osteoblastogenesis (22) and for neural crest cell specification and migration (9). The Sox2 transcription factor was identified as a Chd7 binding partner and both proteins were shown to regulate a set of common target genes that are mutated in Alagille, Pallister-Hall, and Feingold syndromes. These findings may account for the fact that all these syndromes, together with SOX2 anophthalmia, share (to various degrees) common developmental features with CHARGE (ref. 23 and references therein). In addition, although mutations in CHD7 are dominant, the phenotypic penetrance of these mutations is variable.
Altogether, the above-mentioned studies show that CHD7 and KISMET (KIS) are key regulators of development. To understand how these proteins achieve their function and how their mutation leads to developmental disorders, it is critical to characterize these proteins at the molecular level. To date, neither KIS nor CHD7 has ever been characterized for activity, as both proteins are extremely large and have resisted purification. We have developed a method to express and purify recombinant CHD7 protein and we have examined its function. We show that CHD7 is, as anticipated from its protein domain composition, a nucleosome remodeling factor. Although KIS/CHD7 mutants share phenotypic similarities with members of the SWI/SNF family remodelers in flies and in humans, we find that CHD7 displays biochemical properties that are distinct from hSWI/SNF remodeling. We performed a structure-function analysis of CHD7 and examined point mutants reported in human patients. Mutations involved in human conditions show a range of impacts on remodeling activity, raising the possibility that even partial impairment of remodeling function has a significant impact on human biology.
Results
CHD7 Is an ATP-Dependent Nucleosome Remodeling Factor.
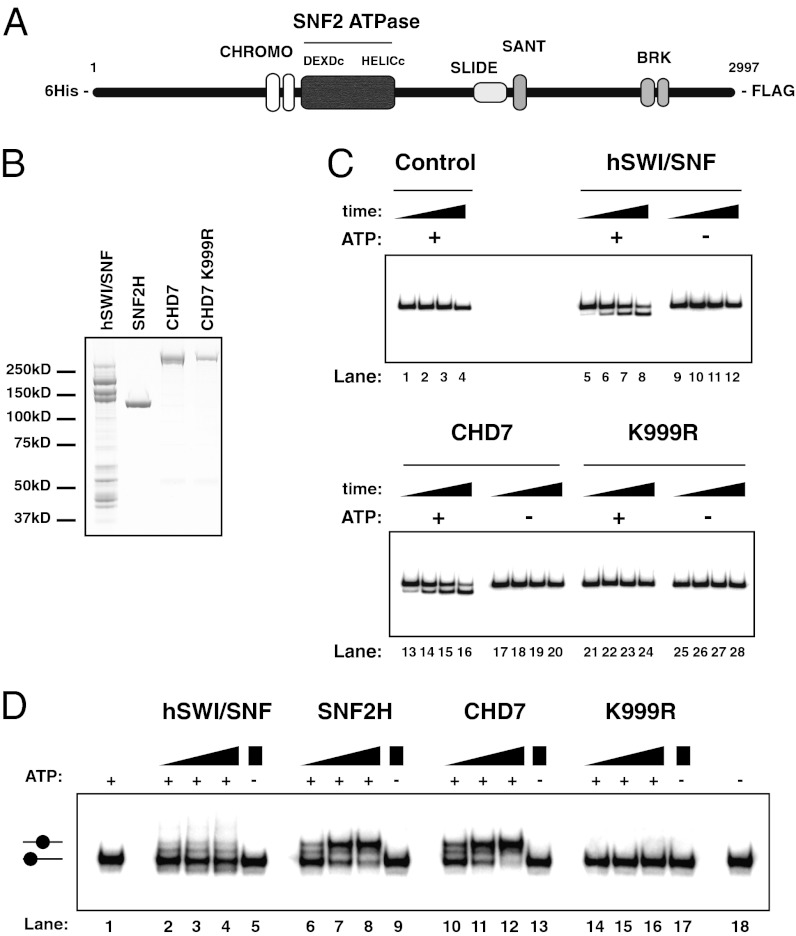
Analysis of the CHD7 protein primary sequence indicates that it is a member of the SNF2-protein superfamily and that it belongs to the CHD family (Fig. 1_A_). Many of these proteins have been shown to be nucleosome/chromatin remodeling factors (24–27). Therefore, we sought to test the ability of CHD7 to remodel nucleosomes. To this end, we cloned the CHD7 full-length cDNA and expressed recombinant CHD7 protein in insect cells using the baculovirus system. Expression of large proteins such as CHD7 (2997 aa, ∼336 kDa) can be challenging due to the increased likelihood of degradation. Indeed, single-tag purifications of CHD7 yielded high levels of degradation products (Fig. S1_A_). To recover full-length protein, we engineered an N-terminal 6-Histidine-tag and a C-terminal FLAG-tag and purified CHD7 by serial use of these epitope tags (Fig. 1_B_).
Fig. 1.
CHD7 is an ATP-dependent nucleosome remodeling factor. (A) Schematic representation of CHD7. For domain terminology see text. (B) Proteins used in assays were separated by SDS-PAGE and stained with Coomassie blue. (C) Analysis of nucleosome remodeling activities by restriction enzyme accessibility (REA) assay. The assay measured the ability of the remodeling factors to expose an MfeI restriction site in the nucleosome at +28-bp (see methods for details). Control (buffer only), remodeling factors (hSWI/SNF ∼75 nM, CHD7 10 nM, CHD7 K999R 10 nM) were incubated with radiolabeled nucleosomes (20 nM) with a 50-bp DNA overhang, in the presence or absence of ATP (2 mM) as indicated. (D) Nucleosome mobilization assay. Increasing concentrations of enzymes (hSWI/SNF: ∼6 nM, ∼12 nM, ∼25 nM, SNF2H: 42 nM, 85 nM, 170 nM; CHD7 and CHD7 K999R: 0.25 nM, 0.5 nM, 1 nM) were incubated with a radiolabeled end-positioned +50-bp nucleosome (20 nM) in the absence or presence of ATP, as indicated. Migration of the end- or centrally positioned nucleosome (filled circle) is indicated on the left.
To determine whether CHD7 is a nucleosome remodeling factor, we tested the recombinant protein using the Restriction Enzyme Accessibility (REA) protocol. Wrapping of DNA around histones to form nucleosomes renders the DNA inaccessible to many proteins, including restriction enzymes. The REA assay measures the ability of a factor to expose nucleosomal DNA to cleavage (28–30). As a control, the hSWI/SNF complex caused a significant increase in DNA accessibility over time, in an ATP-dependent manner (Fig. 1_C_). Notably, addition of CHD7 to the mononucleosome substrate efficiently increased DNA accessibility. Again, this increase was ATP-dependent as no activity was detected in the absence of ATP (Fig. 1_C_).
To exclude that the remodeling activity was due to a contaminating polypeptide in CHD7 protein preparations and to further confirm that ATP hydrolysis is required for nucleosome remodeling by CHD7, we mutated the lysine residue at position 999 to arginine (K999R). This lysine residue is found in a highly conserved putative ATP-binding motif (31–33). Mutation of the equivalent lysine residue in other SNF2-superfamily proteins has been shown to abolish ATPase and consequently nucleosome remodeling activities (27, 34). As shown in Fig. 1_C_, the CHD7 K999R mutant protein did not exhibit any detectable activity in the REA assay (lanes 21–28). We conclude that CHD7’s intrinsic ATP-dependent activity is responsible for the observed changes in nucleosomal DNA accessibility.
To examine the characteristics of remodeling by CHD7, we used a protocol that detects nucleosome “sliding,” also referred to as nucleosome “mobilization.” This assay reveals alterations to histone-DNA contacts (such as nucleosome repositioning) that are significant enough to result in changes in nucleosome electrophoretic mobility. Addition of hSWI/SNF to a mononucleosome positioned at the end of a DNA fragment resulted in subtle changes in electrophoretic mobility. This was expected because hSWI/SNF primarily relocates histone octamers to DNA ends when mononucleosomes are used as a substrate (Fig. 1_D_, lanes 2–4; ref. 35). In contrast, SNF2H caused evident changes in electrophoretic mobility due to “sliding” of the histone octamer from one end to the center of the DNA fragment, as seen previously for this distinct class of remodeling factor (Fig. 1_D_, lanes 6–8). Interestingly, CHD7 caused changes in nucleosome mobility similar to SNF2H, suggesting that CHD7 repositions the histone octamer to the center of the DNA fragment. Again, the CHD7 K999R ATPase mutant protein did not exhibit any detectable activity (Fig. 1_D_, lanes 14–16). We conclude that CHD7 is an efficient ATP-dependent nucleosome remodeling factor.
CHD7 Displays Characteristics That Are Distinct from SWI/SNF-Family Enzymes.
The observation that CHD7 and hSWI/SNF differed in their behavior in the nucleosome “sliding” protocol (Fig. 1_D_) indicated that these proteins might differ more generally in their functional characteristics. This possibility is interesting because mutation of the Drosophila homologs of these remodelers causes similar developmental phenotypes (5, 36), suggesting that these distinct types of remodeling function might both be needed for appropriate development. We were therefore interested in further comparing the functions of these two remodeling activities.
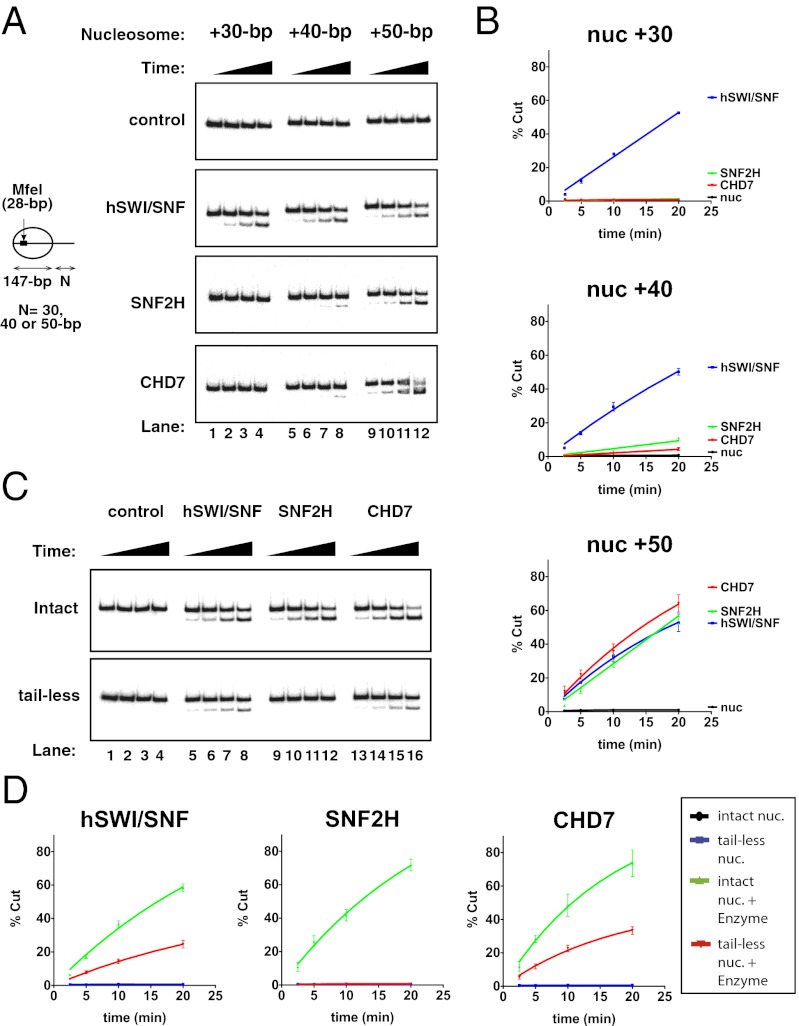
Typically, SWI/SNF-type enzymes can remodel nucleosome core substrates, whereas ISWI-type enzymes and Chd1p require extranucleosomal DNA protruding from the nucleosome core to bind efficiently to nucleosomes and perform the remodeling reaction (37–41). Hence, we investigated whether CHD7 requires a minimal nucleosomal DNA length for remodeling. To this end, we assembled nucleosomes using DNA fragments of increasing length and used these as substrates in REA assays (Fig. 2_A_, control panel). This approach revealed that, in contrast to hSWI/SNF, CHD7 is unable to efficiently increase DNA accessibility on nucleosome core particles or nucleosomes harboring up to 30 bp extending from the nucleosome core (Fig. 2 A and B and Fig. S1_B_). However, CHD7 efficiently remodeled nucleosomes bearing a DNA linker longer than 40-bp similarly to SNF2H, in these conditions (Fig. 2 A and B). Consistent with the sliding assay (Fig. 1_D_), these data suggest that, like ISWI-type enzymes and Chd1p, CHD7 requires interacting with DNA protruding from the nucleosome core to perform the remodeling reaction. Altogether, these results show that CHD7 shares common biochemical properties with ISWI-type enzymes and Chd1p and that it differs biochemically from SWI/SNF-type enzymes.
Fig. 2.
CHD7 presents biochemical properties that are distinct from SWI/SNF- and ISWI-type enzymes. (A) REA assay. The assay measured the ability of the remodeling factors (control: buffer only, hSWI/SNF: 8 nM, SNF2H: 32 nM, CHD7: 1 nM, as indicated) to expose an MfeI restriction site in radiolabeled nucleosomes (4 nM) with a 30-bp, 40-bp, or 50-bp DNA overhang, as indicated. Aliquots of the reactions were quenched at 2.5 min, 5 min, 10 min, and 20 min, and cut/uncut DNAs were separated by PAGE. (B) Quantification of gels in (A). Error bars reflect SEM and were derived from 3 independent experiments. (C) REA assay. These experiments were performed as in A using radiolabeled +50-bp nucleosomes only and nucleosomes were assembled using either untreated (intact) or partially trypsinized (tail-less) histones from HeLa cells, as indicated. (D) Quantification of gels in C. Error bars reflect SEM and were derived from three independent experiments.
We also tested CHD7 for its requirement for intact histone tails, an area where remodeling enzymes also differ. In a nucleosome, the histone N and C termini (referred to as “histone tails”) are believed to form disordered structures that extend away from the nucleosome core structure. Previous analyses have shown that, unlike hSWI/SNF and Drosophila Mi-2, both ISWI enzymes and to some extent yeast Chd1p require the presence of the histone H4 N-terminal tail to accomplish the nucleosome remodeling reaction (37, 42–45). To examine whether nucleosome remodeling by CHD7 depends on any of the histone tails, we purified histones from partially trypsinized chromatin; this treatment leads to digestion of the protruding histone tails (ref. 46; Fig. S1_C_). We then used these “tail-less” histones for reconstitution of nucleosome substrates and compared them to “intact” nucleosomes. As shown in Fig. 2 C and D, hSWI/SNF, SNF2H, and CHD7 all readily increased DNA accessibility over time when we used intact nucleosomes as a substrate. Consistent with previous studies, hSWI/SNF also remodeled tail-less nucleosomes (although less efficiently) and SNF2H did not display any remodeling activity. Interestingly, removal of histone tails did not prevent CHD7 from remodeling the nucleosomes, although like hSWI/SNF, CHD7 was about 3 times slower (Fig. 2 C and D). These results separate CHD7 from SNF2H and suggest that proteins from all three CHD subfamilies can, at least to some extent, function independently of the histone H4 tail.
Mutations Leading to C-Terminal Protein Truncations Upstream of the Amino Acid Residue 1899 Produce Proteins with Very Low or No Remodeling Abilities.
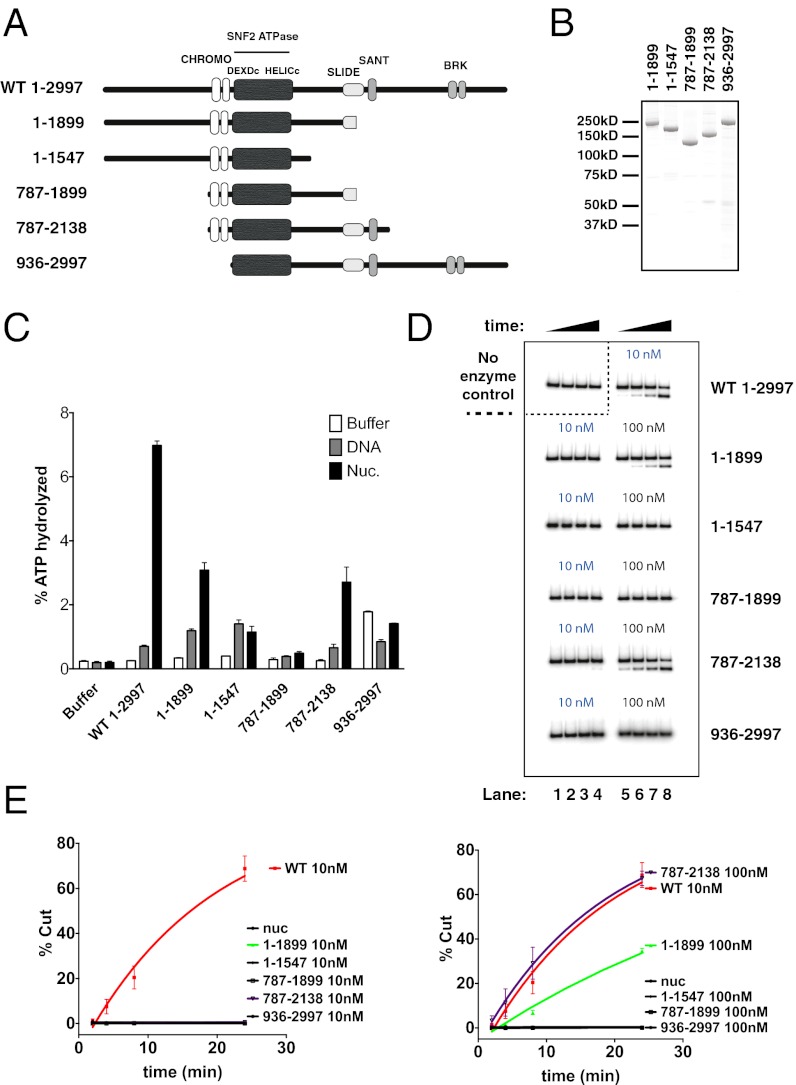
CHARGE syndrome appears to arise from gene expression defects (3) which are expected to be caused, at least in part, by lower levels of CHD7 enzymatic activity at CHD7 target loci. A large collection of CHD7 gene mutations has been described in CHARGE syndrome patients (refs. 3 and 47 and www.chd7.org). Nearly 80% of these patients have either a nonsense or a frameshift mutation (44% and 34%, respectively; ref. 47), which, if the mutated transcript escapes nonsense-mediated mRNA decay (NMD), will cause truncation of the CHD7 protein. To help delineate the consequence(s) that CHARGE mutations might have on the CHD7 protein, we investigated the function of CHD7 protein domains. We generated a set of protein deletion mutants (Fig. 3_A_), expressed them using the baculovirus system (Fig. 3_B_) and analyzed them in ATPase and nucleosome remodeling assays (Fig. 3 C and D).
Fig. 3.
Mutations leading to C-terminal protein truncations upstream of the amino acid residue 1899 produce proteins with very low or no remodeling abilities. (A) Schematic representation of WT CHD7 and deletion mutants. Numbers refer to amino acid residues. (B) Deletion mutant protein preparations were separated by SDS-PAGE and stained with Coomassie blue. (C) ATPase assay. Ten nM of WT CHD7 or deletion mutants were incubated in the presence of buffer only (white bars), “+50-bp” DNA (60 nM; gray bars) or +50-bp nucleosomes (60 nM; black bars) and 20 μM ATP/ 60 μM Mg2+ supplemented with γ-[32P]ATP. Reactions were quantified using TLC plates at various time points; the 24’ time point is displayed as a bar graph. Error bars reflect SEM derived from at least two independent experiments. (D) Analysis of WT CHD7 and deletion mutants in REA assays. REA reactions: Control (buffer only) or remodeling factors: WT CHD7 (10 nM), deletion mutants (10 and 100 nM as indicated on top of the gel lanes) were incubated with radiolabeled +50-bp nucleosomes (20 nM). (E) Quantification of gels shown in D. Error bars reflect SEM and were derived from at least two independent experiments.
SMART (Simple Modular Architecture Research Tool; ref. 48) analysis of the CHD7 protein sequence identifies a pair of chromo (chromatin organization modifier) domains (49, 50) followed by an ATPase domain (comprising DEXDc and HELICc domains; ref. 31), a SANT (SWI3, ADA2, N-CoR and TFIIIB; ref. 51) and a pair of BRK (BRM and KIS; ref. 5) domains. In addition, the Owen–Hughes laboratory recently described the presence of a SLIDE domain (SANT-like ISWI domain; ref. 52) attached to the SANT domain in CHD family proteins (53).
C-terminal deletion of about a third of the CHD7 protein (construct 1–1899, deleting the last putative α-helix of the SLIDE, and the SANT and BRK domains) reduced nucleosome-stimulated ATPase by about half (Fig. 3_C_). However, this deletion impaired nucleosome remodeling activity more significantly as no remodeling was detected when using the same concentration as wild-type (WT) protein (Fig. 3 D and E). Using much higher concentrations of this deletion mutant reveals that, although important, the CHD7 C terminus is not essential to catalyzing nucleosome remodeling (Fig. 3 D and E). These data suggest that the CHD7 C terminus up to the predicted last α-helix of the SLIDE domain (53) is important to efficiently translate ATP hydrolysis into nucleosome remodeling. Further deletion of the C terminus which removes the rest of the SLIDE domain-containing region (construct 1–1547), or additional deletion of the N terminus (construct CHD7 787–1899), abolished the ability of the proteins to catalyze nucleosome remodeling (Fig. 3 D and E). These observations indicate that the regions flanking the central ‘chromodomains-ATPase’ region might play redundant roles in nucleosome remodeling by CHD7. Consistent with this notion, adding back the complete “SLIDE-SANT” module, created a polypeptide (787–2138) that behaved similarly to 1–1899 in both ATPase and REA assays (Fig. 3 C_–_E). These results support published studies indicating that the SANT and SLIDE domains play an important role in remodeling by the CHD family of proteins.
Chromodomains also play a critical role in remodeling by CHD-family proteins (45, 54). We therefore tested the impact of removing the CHD7 N terminus including the chromodomains (construct 936–2997). This completely prohibited CHD7 from remodeling nucleosomes, suggesting that the entire CHD7 C terminus cannot compensate for the absence of the chromodomains (Fig. 3 D and E). Therefore, we conclude that the central “chromodomains-ATPase” module requires both an N-terminal region and the SLIDE-SANT region to efficiently couple ATP hydrolysis to nucleosome remodeling.
Collectively, our data suggest that, combined with the degradation of mutant mRNAs via the NMD pathway, the vast majority of CHARGE patients have a high probability of being haploinsufficient for CHD7.
CHD7 Point Mutations Reported in CHARGE Syndrome and Idiopathic Hypogonadotropic Hypogonadism Patients Variably Affect the Nucleosome Remodeling Activity of CHD7.
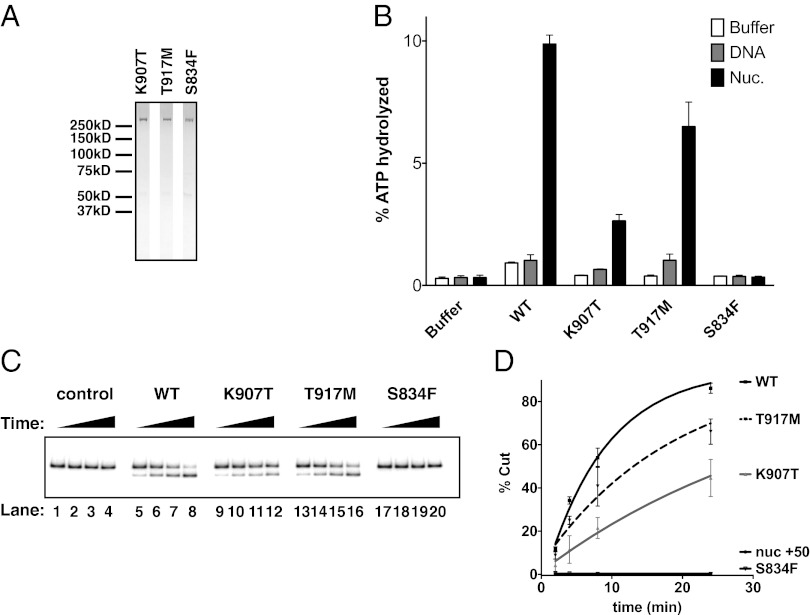
Mutations in the CHD7 gene are found in about 60% of all CHARGE patients (55) and in up to 90% of patients with typical CHARGE symptoms (56). About 8% of these patients have missense mutations (47). To date, the impact of any individual mutation on CHD7 function has not been investigated. We used site-directed mutagenesis to engineer CHD7 point mutations that had been identified in patients (Fig. 4_A_) and measured the impact of single amino acid substitutions on ATPase and nucleosome remodeling by CHD7. Two patient mutations fall in the second chromodomain of CHD7 at residues that are highly conserved across CHD proteins. One of these is a lysine to threonine substitution at the residue 907 (K907T) and the other mutation at position 917 mutates a threonine to a methionine (T917M; ref. 55). Interestingly, the K907T mutant protein showed a major reduction (about 3.5-fold, Fig. 4_B_) in ATPase activity compared with WT, whereas the T917M protein showed a limited decrease (about 1.5-fold). Consistent with these findings, these mutants also have reduced nucleosome-remodeling capabilities (Fig. 4 C and D).
Fig. 4.
CHD7 point mutations reported in CHARGE syndrome and IHH patients variably affect the nucleosome remodeling activity of CHD7. (A) CHD7 point mutant protein preparations were separated by SDS-PAGE and stained with Coomassie blue. (B) ATPase assay. Ten nM of WT or mutant CHD7 proteins were incubated and processed as in Fig. 3_C_. (C) Analysis of WT CHD7 and point mutants in REA assays. Control (buffer only) or WT CHD7 and point mutants (10 nM) were incubated at with radiolabeled +50-bp nucleosomes (1 nM), in the presence of ATP (2 mM). Samples and data were processed as in Fig. 3_D_. (D) Quantification of gels shown in C. Error bars reflect SEM and were derived from at least two independent experiments.
Two independent studies described a serine to phenylalanine substitution at the CHD7 residue 834 (S834F) in two CHARGE patients (57) and in a patient with IHH (14). This missense mutation occurs in a highly conserved sequence motif within the first chromodomain (56, 58). Interestingly, this single amino acid substitution completely abolished both ATPase and nucleosome remodeling activities by CHD7 (Fig. 4 B and C, lanes 17–20, and D). These results indicate that, in patients carrying this mutation, CHARGE syndrome and IHH arise from having one allele that is nonfunctional for remodeling.
Discussion
In this study, we describe the ability of recombinant CHD7 protein to function as an ATP-dependent nucleosome remodeler. We show that many mutations involved in human developmental syndromes impact the chromatin remodeling function of CHD7, thereby tying a function known to be important to gene regulation to these human pathologies.
ATP-dependent nucleosome remodeling proteins are found in several classes that show important functional distinctions in how they render DNA accessible to regulatory proteins. We find that CHD7 differs from hSWI/SNF as it requires mononucleosomes with a DNA overhang longer than 40-bp for efficient remodeling (Fig. 2 A and B). It is possible that in vivo CHD7 might not be able to efficiently remodel closely packed nucleosomes whereas hSWI/SNF might. Alternatively, binding of a factor to DNA near a nucleosome might interfere with CHD7 but not hSWI/SNF remodeling. A second important difference is that CHD7 slides nucleosomes differently than hSWI/SNF (Fig. 1_D_) and this might also impact biological function. The behavior of CHD7 with regards to DNA linker length and sliding is similar to remodelers that can space nucleosomes, suggesting that this might be a role that CHD7 plays during regulation of gene expression. These functional distinctions might partly explain why, although the kis gene and several Drosophila SWI/SNF complex subunits were functionally classified in the same TrxG of genes, embryos require both remodeling activities to execute developmental programs properly. Additionally, our data provide a potential biochemical justification for why these classes of remodelers colocalize extensively in Drosophila (59) and in mouse embryonic stem cells (9).
CHD7 contains a SANT-SLIDE module, which is found in several remodeling proteins including ISWI and CHD proteins. This module has been proposed to bind extranucleosomal DNA (53) and “pump” it toward the histone octamer (41). Although the putative SLIDE domain is found upstream of the SANT domain in the CHD7 primary sequence, it is predicted to fold similarly (53). Our analysis indicates that CHD7 uses the SANT–SLIDE to efficiently remodel nucleosomes because its deletion causes a significant reduction in nucleosome remodeling activity (Fig. 3). This indicates that patients with mutations in CHD7 that cause the loss of the C-terminal region containing the SLIDE domain will likely have levels of CHD7 remodeling similar to heterozygous null patients.
Most mutations (∼80%) identified in CHARGE patients create frameshifts or introduce a premature stop codon in the CHD7 sequence. Of 528 mutations listed by Jansen et al., at least 250 would remove regions upstream of amino acid 1899 if the mutant mRNAs were translated, and this would be predicted to severely impair remodeling by CHD7 (47). Missense point mutations are a smaller class (∼8%) in CHARGE patients. We analyzed three of these mutations and found a range of impact on the activity of CHD7, despite the occurrence of these mutations at highly conserved residues. It is possible that the in vitro conditions used did not assess the full impact on remodeling that these mutations would display in vivo. In addition, it is anticipated that some mutations may not affect the activity of CHD7 but instead disrupt important protein-protein interactions. It is noteworthy that two CHD7 splice variants encoding polypeptides that lack the ATPase domain (and other regions) have been identified (60, 61). These variants might play key biological roles that are ATP-independent; the impact of mutations on these protein isoforms has yet to be determined.
In conclusion, we have shown that CHD7 is a nucleosome remodeling protein with a distinct set of characteristics. Analysis of CHD7 mutants suggests that haploinsufficiency is a likely explanation for the pathogenicity of about 250 CHARGE patient mutations. These analyses allow a basis for predicting the impact of new human patient mutations on CHD7’s remodeling activity, and for studies that assess the impact of these mutations in cell lines derived from these patients.
Experimental Procedures
Cloning.
The CHD7 cDNA (NCBI NM_017780) was cloned from MegaMan Human Transcriptome Library (Agilent Technologies) by RT-PCR (primer sequences available upon request). The N-terminal 6× His-tag, a 5× Gly spacer and a TEV protease cleavage site were introduced upstream of the first ATG and the C-terminal FLAG-tag was introduced upstream the STOP codon by PCR. In CHD7 deletion mutants, only a C-terminal FLAG-tag was introduced. CHD7 point mutation changes were made according to changes found in patients (14, 55, 57) using the QuikChange Site-Directed Mutagenesis kit according to the manufacturer’s instructions.
Protein Expression and Purification.
The full-length double-tag CHD7 cDNA and CHD7 mutant baculoviruses were produced according to the Bac-to-Bac Baculovirus Expression Systems manual (Life Technologies). Proteins only bearing a FLAG-tag were essentially purified as in Bouazoune et al. (62) with minor modifications. Double-tagged full-length CHD7 proteins were purified either from 4L Sf9 cell cultures or from cells used to produce P3 viruses. Cells were resuspended in BC 250 buffer + 5 mM imidazole before adding Ni-NTA resin (Qiagen) for 1 h at 4 °C. Extracts were then poured into Econo columns (Bio-Rad). The flow-through was reapplied to resin before washing 2× with >10-column volume (CV) of BC 250 + 10 mM imidazole. Bound proteins were eluted with BC 250 + 200mM imidazole. Imidazole concentration was diluted down to 50 mM before adding M2-affinity gel for 3h and samples were subsequently processed as FLAG-tagged proteins above. The hSWI/SNF complex was essentially purified as described previously using a cell line expressing FLAG-tagged Ini1 (63).
Nucleosome Substrate Preparations.
DNA fragments used for nucleosome reconstitutions were produced using the ‘601’ strong nucleosome-positioning sequence (64). All DNAs were amplified by PCR (primer sequences available upon request) and radiolabeled with α-32P-dCTP. Constructs were purified using the QIAEX II resin purification kit (Qiagen) according to the manufacturer’s instruction. DNAs were assembled into mononucleosomes by standard salt dialysis using histones purified from HeLa cells. In Fig. 2, tail-less nucleosomes were assembled using trypsinized HeLa histones. Trypsinization was performed as described in ref. 46. All nucleosome assemblies were purified over 10–30% (vol/vol) glycerol gradients.
Restriction Enzyme-Accessibility Assays.
REAs were performed essentially as described in ref. 29, with minor modifications. Here, we measured the ability of remodeling factors to expose an MfeI restriction site at +28-bp inside the nucleosomes and used 1.5 U/μL of MfeI-HF (New England Biolabs). hSWI/SNF complex concentrations are given assuming a Molecular Mass of ∼1.2 MDa). Data were quantified using the ImageQuantTL (GE Healthcare Life Sciences) and cut DNA intensity was normalized to cytosine content before calculating cut/uncut ratios and plotting the data using GraphPad Prism.
Nucleosome Mobilization Assay.
The nucleosome mobilization assay was carried out using 20 nM radiolabeled end-positioned nucleosome +50-bp, in the same conditions as the REA assays except that the restriction enzyme mix was replaced by BC 100 buffer. Increasing concentrations of enzymes were incubated in the absence or presence of ATP/Mg2+ (2 mM/6 mM). An aliquot was taken out after 20 min at 30 °C, and the reaction was stopped by addition of unlabeled competitor DNA (1.5 μg/μL)/EDTA (125 mM), incubated on ice for 10 min before 5% native PAGE in 0.5× TBE. Gel was dried, exposed to “storage phosphor screen” and visualized using a Typhoon trio + imager (Amersham Biosciences).
ATPase Assays.
Ten nM of enzyme were incubated at 30 °C in the presence of 10% glycerol gradient buffer only (Figs. 3 and 4, white bars), 601 +50-bp DNA (60 nM; Figs. 3 and 4, gray bars) or saturating concentration of 601 +50-bp nucleosomes (60 nM; Figs. 3 and 4, black bars) and 20 μM ATP/60 μM Mg2+ and traces of gel-purified radiolabeled γ-[32P]ATP. Reactions were spotted on TLC plates (Baker-flex' Cellulose PEI-F) at 2, 4, 8, 16, 24, and 48 min, and hydrolyzed phosphates were separated from non- hydrolyzed ATP by developing plates in 0.5 M LiCl/1 M formic acid buffer. Plates were dried, exposed to storage phosphor screen, visualized using a Typhoon trio + imager (Amersham Biosciences), and quantified using ImageQuant. Data were plotted using GraphPad Prism.
Supplementary Material
Supporting Information
Acknowledgments
We thank Joe Garlick for technical assistance; Dr. Daniel Grau for sharing reagents; and Dr. Karim-Jean Armache for sharing helpful expertise about protein purification. This work was supported by National Institutes of Health Grant 5R37GM048405-20.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Vissers LE, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36(9):955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 2.Pagon RA, Graham JM, Jr, Zonana J, Yong SL. Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J Pediatr. 1981;99(2):223–227. doi: 10.1016/s0022-3476(81)80454-4. [DOI] [PubMed] [Google Scholar]
- 3.Zentner GE, Layman WS, Martin DM, Scacheri PC. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A. 2010;152A(3):674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci USA. 1988;85(21):8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daubresse G, et al. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999;126(6):1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- 6.Hurd EA, et al. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mammalian Genome. 2007;18(2):94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 7.Bosman EA, et al. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005;14(22):3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 8.Patten SA, et al. Role of Chd7 in zebrafish: A model for CHARGE syndrome. PLoS ONE. 2012;7(2):e31650. doi: 10.1371/journal.pone.0031650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajpai R, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463(7283):958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergman JE, et al. The results of CHD7 analysis in clinically well-characterized patients with Kallmann syndrome. J Clin Endocrinol Metab. 2012;97(5):E858–E862. doi: 10.1210/jc.2011-2652. [DOI] [PubMed] [Google Scholar]
- 11.Van de Laar I, et al. Limb anomalies in patients with CHARGE syndrome: An expansion of the phenotype. Am J Med Genet A. 2007;143A(22):2712–2715. doi: 10.1002/ajmg.a.32008. [DOI] [PubMed] [Google Scholar]
- 12.Gao X, et al. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet. 2007;80(5):957–965. doi: 10.1086/513571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jongmans MC, et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome—the clinical overlap with CHARGE syndrome. Clin Genet. 2009;75(1):65–71. doi: 10.1111/j.1399-0004.2008.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HG, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83(4):511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pallais JC, Au M, Pitteloud N, Seminara S, Crowley WF. Kallmann Syndrome. In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, editors. GeneReviews. Seattle: Univ of Washington; 1993. [Google Scholar]
- 16.Bergman JE, Bosman EA, van Ravenswaaij-Arts CM, Steel KP. Study of smell and reproductive organs in a mouse model for CHARGE syndrome. Eur J Hum Genet. 2010;18(2):171–177. doi: 10.1038/ejhg.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Layman WS, Hurd EA, Martin DM. Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet. 2011;20(16):3138–3150. doi: 10.1093/hmg/ddr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463(7278):184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnetz MP, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19(4):590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnetz MP, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6(7):e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zentner GE, et al. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet. 2010;19(18):3491–3501. doi: 10.1093/hmg/ddq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takada I, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9(11):1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 23.Engelen E, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43(6):607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 24.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 25.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11(2):148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 26.Hota SK, Bartholomew B. Diversity of operation in ATP-dependent chromatin remodelers. Biochim Biophys Acta. 2011;1809(9):476–487. doi: 10.1016/j.bbagrm.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol. 2008;28(12):3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logie C, Peterson CL. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16(22):6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol Cell. 2001;8(6):1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 30.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254(2):130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 31.Bork P, Koonin EV. An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res. 1993;21(3):751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23(14):2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7(4):583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 35.Jaskelioff M, Gavin IM, Peterson CL, Logie C. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol Cell Biol. 2000;20(9):3058–3068. doi: 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brizuela BJ, Elfring L, Ballard J, Tamkun JW, Kennison JA. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137(3):803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brehm A, et al. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 2000;19(16):4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gangaraju VK, Bartholomew B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol. 2007;27(8):3217–3225. doi: 10.1128/MCB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23(6):1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13(12):1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 41.McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol. 2011;31(23):4746–4759. doi: 10.1128/MCB.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21(3):875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Mol Cell Biol. 2007;27(11):4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guyon JR, Narlikar GJ, Sif S, Kingston RE. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol Cell Biol. 1999;19(3):2088–2097. doi: 10.1128/mcb.19.3.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39(5):711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ausio J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206(3):451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 47.Janssen N, et al. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat. 2012;33(8):1149–1160. doi: 10.1002/humu.22086. [DOI] [PubMed] [Google Scholar]
- 48.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eissenberg JC. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene. 2001;275(1):19–29. doi: 10.1016/s0378-1119(01)00628-x. [DOI] [PubMed] [Google Scholar]
- 50.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88(1):263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aasland R, Stewart AF, Gibson T. The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21(3):87–88. [PubMed] [Google Scholar]
- 52.Grüne T, et al. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12(2):449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 53.Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J. 2011;30(13):2596–2609. doi: 10.1038/emboj.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouazoune K, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21(10):2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartels CF, Scacheri C, White L, Scacheri PC, Bale S. Mutations in the CHD7 gene: The experience of a commercial laboratory. Genetic Testing Molecular Biomarkers. 2010;14(6):881–891. doi: 10.1089/gtmb.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergman JE, et al. A novel classification system to predict the pathogenic effects of CHD7 missense variants in CHARGE syndrome. Hum Mutat. 2012;33(8):1251–1260. doi: 10.1002/humu.22106. [DOI] [PubMed] [Google Scholar]
- 57.Delahaye A, et al. Familial CHARGE syndrome because of CHD7 mutation: clinical intra- and interfamilial variability. Clin Genet. 2007;72(2):112–121. doi: 10.1111/j.1399-0004.2007.00821.x. [DOI] [PubMed] [Google Scholar]
- 58.Flanagan JF, et al. Molecular implications of evolutionary differences in CHD double chromodomains. J Mol Biol. 2007;369(2):334–342. doi: 10.1016/j.jmb.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4(10):e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colin C, Tobaruella FS, Correa RG, Sogayar MC, Demasi MA. Cloning and characterization of a novel alternatively spliced transcript of the human CHD7 putative helicase. BMC Res Notes. 2010;3:252. doi: 10.1186/1756-0500-3-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kita Y, Nishiyama M, Nakayama KI. Identification of CHD7(S) as a novel splicing variant of CHD7 with functions similar and antagonistic to those of the full-length CHD7(L) Genes Cells. 2012;17(7):536–47. doi: 10.1111/j.1365-2443.2012.01606.x. [DOI] [PubMed] [Google Scholar]
- 62.Bouazoune K, Miranda TB, Jones PA, Kingston RE. Analysis of individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res. 2009;37(16):5279–5294. doi: 10.1093/nar/gkp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12(18):2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276(1):19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information