ENCODE Data in the UCSC Genome Browser: year 5 update (original) (raw)
Abstract
The Encyclopedia of DNA Elements (ENCODE), http://encodeproject.org, has completed its fifth year of scientific collaboration to create a comprehensive catalog of functional elements in the human genome, and its third year of investigations in the mouse genome. Since the last report in this journal, the ENCODE human data repertoire has grown by 898 new experiments (totaling 2886), accompanied by a major integrative analysis. In the mouse genome, results from 404 new experiments became available this year, increasing the total to 583, collected during the course of the project. The University of California, Santa Cruz, makes this data available on the public Genome Browser http://genome.ucsc.edu for visual browsing and data mining. Download of raw and processed data files are all supported. The ENCODE portal provides specialized tools and information about the ENCODE data sets.
INTRODUCTION
The mission and scope of the Encyclopedia of DNA Elements (ENCODE) Project is well described in previous publications by the ENCODE Consortium (1–2), and results from coordinated analysis of ENCODE results (3) are also available. Previous manuscripts in this publication (4–6) have described the project’s progress since 2007 and detailed how the ENCODE Data Coordination Center at the University of California, Santa Cruz, (UCSC) has worked with ENCODE laboratories worldwide to import its production data, supporting documentation and metadata, and has made the data accessible to the broader biomedical community. A companion article in this issue, ‘The UCSC Genome Browser database: Extensions and updates 2013’, provides background information about the UCSC Genome Browser database and infrastructure (7–8) that underlies ENCODE support at UCSC. This article focuses on ENCODE data and access tools introduced during 2012, the fifth and final year of the initial whole-genome production phase of the project.
DATA AVAILABILITY
All ENCODE production data for the 5-year initial production phase of the project have now been submitted to the ENCODE Data Coordination Center at UCSC. UCSC has performed quality review and publicly released all conforming ENCODE data sets along with metadata, as both tracks for browsing and downloadable files for data mining. In the human genome, 288 cell and tissue types are now represented, covering 32 assays. Chromatin features and sites of DNA binding are mapped for >300 factors and marks. In mouse, 81 cell and tissue types were surveyed in five experimental assays.
Human genome
The results of five new experiment types were released during the fifth year: chromatin interactions based on chromosome conformation capture carbon copy (5C) and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) methods, proteogenomics and DNA replication timing by both sequencing and microarray methods.
Although DNA is a linear molecule, it is packed and organized inside the nucleus in a 3D milieu, and gene regulation can be affected by interactions from elements located hundreds of kilobases distant in the genome. Long-range chromatin looping interactions can be detected using various techniques, including chromosome conformation capture (9) and chromatin interaction analysis with paired-end tag (10). The ENCODE chromatin interactions data sets comprise experiments in 14 cell types.
Proteogenomic methods differ from conventional mass spectrometry proteomic methods that identify peptides by comparing them with peptides produced from known proteins. In contrast, proteogenomic methods compare peptides with all peptides that might be produced by the six translation frames of the genome to identify the genomic region from which the peptides were produced. Study of proteogenomic data offers insights into regulatory mechanisms, including translation, pre-messenger RNA (mRNA) splicing and transcript diversity, nonsense-mediated decay and transcription of novel protein-coding genes. The ENCODE protegenomics data are available in four cell types. Figure 1 presents a Genome Browser session that includes proteogenomics data in conjunction with ENCODE gene, transcriptome and regulatory data sets.
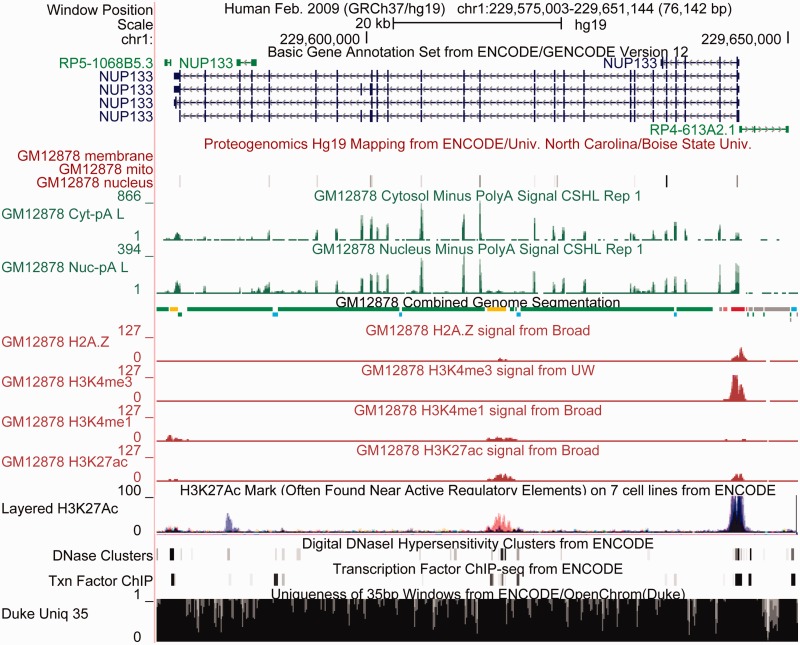
Figure 1.
ENCODE data displayed in the UCSC Genome Browser together with annotations from the ENCODE Analysis Hub in the region of the nucleoporin gene NUP133 demonstrate the power this diversity of data provides for visual interpretation. The GENCODE Basic gene set shows this gene having four protein-coding splice variants and three smaller non-coding transcripts nearby. The proteogenomics track shows support for many of the coding exons, with protein localized in the nucleus, but not in plasma membrane or mitochondria. The long polyA RNA signal shows strong peaks over the exons and low intron signal in the cytosol, with greater signal in the nucleus. This is expected because nuclear mRNAs are not all completely spliced. The Combined Genome Segmentation integrates signal from many histones and classifies regions into those with characteristics of promoters (red), enhancers (yellow), insulators (blue), transcribed regions (green) and repressed (gray). Below are signal tracks from four of the eight histone modifications used as input to the segmentation. The promoter and transcribed regions agree with the RNA evidence, and like the RNA evidence show no evidence of transcription of the non-coding gene to the right of NUP133. Underneath the GM12878 histone signals is a track that overlays one of the histone signals, H3K27Ac, in seven different cell lines (with GM12878 shown in red). A peak in H3K27Ac appears at the enhancer, but as is often the case with enhancers, this appears to be relatively cell specific in contrast to the larger peak near the promoter, where the black coloration indicates the peak is shared by many cell types. The DNAse hypersensitivity and transcription factor tracks also provide evidence for both promoter and enhancer. Finally the mappability track indicates regions where short reads are not uniquely mappable, indicating the data are incomplete and therefore harder to interpret. Although most of this region is mappable, there are many small regions throughout and one larger region on the right where mapping is problematic. Overall, the ENCODE data in this region show strong evidence that this is a nuclear-localized protein-coding gene with a promoter that is used in a wide variety of cell types, and is likely to be regulated by tissue-specific enhancers as well.
The order in which DNA is duplicated during the synthesis phase of the cell cycle is correlated with the expression of genes and the structure of chromosomes; replication timing is known to be an important feature for epigenetic control of gene expression. ENCODE ‘Repli-chip’ (microarray) experiments are available in 9 cell types, and ‘Repli-seq’ (sequencing) experiments in 15.
The encyclopædia of genes and gene variants (GENCODE) gene set (11) is a fundamental resource produced by ENCODE, providing high-quality manual annotation from the Human and Vertebrate Analysis and Annotation (HAVANA) group merged with evidence-based automated annotation from Ensembl (12) across the human genome. For the final release (V12), the data organization and display were improved to make the data more accessible and intuitive. Annotations are now categorized according to their function and level of support. Color coding reflects non-coding, coding, pseudogene or problem status. To complement the ‘Comprehensive’ gene set, a new ‘Basic’ subset provides a simplified view intended for the majority of users, by filtering incomplete and problem annotations while still ensuring that at least one annotation is displayed at every locus. For researchers who require more detail regarding the degree of evidence supporting individual coding transcripts, a five-level scoring metric is provided, based on assessment of alignments of mRNAs and expressed sequence tags (ESTs) across the full length of the annotation. Filtering options allow tuning of the display based on the basic biological function of the transcript (coding, non-coding, etc.), annotation method (manual versus automated) or specific biotype characterization (http://www.gencodegenes.org/gencode_biotypes.html). Finally, two additional annotation subtracks are provided: the ‘2-Way Pseudogene’ subtrack shows consensus pseudogenes predicted by two pipelines [Yale Pseudopipe (13) and UCSC Retrofinder (14)], and the ‘PolyA’ subtrack presents polyA signals and sites manually annotated on the genome based on transcribed evidence.
The majority of ENCODE primary data submitted and released in the past year for the human genome expanded the existing tracks with additional experiments; cell types, subcellular fractions, transcription factors or histone marks were also mapped. The total complement of data sets available is summarized in Tables 1 and 2. All links mentioned in this publication are collected in Table 3.
Table 1.
The full complement of ENCODE data sets summarized by cell type [types annotated as cancer are marked with asterisk (*)]
| Cell type | Tissue | Description | Data sets | |||
|---|---|---|---|---|---|---|
| TF/His | RNA | Other | Total | |||
| Tier 1 initial | ||||||
| GM12878 | Blood | Lymphoblastoid | 137 | 27 | 49 | 213 |
| K562* | Blood | Leukemia | 247 | 45 | 80 | 372 |
| Tier 1 added in 2011 | ||||||
| H1-hESC | Embryonic stem | Embryonic stem | 96 | 14 | 23 | 133 |
| Tier 2 initial | ||||||
| HeLa-S3* | Uterine cervix | Cervical carcinoma | 93 | 14 | 30 | 137 |
| HepG2* | Liver | Liver carcinoma | 118 | 19 | 26 | 163 |
| HUVEC | Umbilical endothelium | Umbilical vein endothelial | 37 | 13 | 16 | 66 |
| Tier 2 added in 2011 | ||||||
| A549* | Lung | Lung carcinoma | 89 | 22 | 12 | 123 |
| CD14+ | Blood | Monocyte | 17 | 4 | 4 | 25 |
| IMR90 | Lung | Lung fibroblast | 11 | 16 | 10 | 37 |
| MCF-7* | Breast | Breast carcinoma | 50 | 15 | 32 | 97 |
| SK-N-SH* | Brain | Neuroblastoma | 36 | 16 | 7 | 59 |
| Tier 2 added in 2012 | ||||||
| CD20+ | Blood | B cell | 11 | 5 | 4 | 20 |
| H1-neuron | Neuron | H1ES-derived neuron | 5 | 3 | 1 | 9 |
| LHCN-M2 | Muscle | Myoblast | 7 | 2 | 4 | 13 |
| Human: totals | ||||||
| Tier1 + Tier2 (14) | 954 | 215 | 298 | 1467 | ||
| Tier 3 (274) | 591 | 94 | 734 | 1419 | ||
| All (288) | 1545 | 309 | 1032 | 2886 | ||
| Mouse | ||||||
| All (81) | 381 | 102 | 100 | 583 |
Table 2.
The full complement of ENCODE data sets summarized by assay
| Data type/assay | Experiments |
|---|---|
| Chromatin interactions | |
| 5C | 14 |
| ChIA-PET | 8 |
| DNA methylation | |
| Methyl array | 125 |
| Methyl RRBS | 95 |
| Methyl-seq | 20 |
| Histone modifications | |
| ChIP-seq | 315 |
| ChIP-seq (MOUSE) | 179 |
| Open chromatin | |
| DNase-DGF | 56 |
| DNase-DGF (MOUSE) | 22 |
| DNase-seq | 221 |
| Dnase-seq (MOUSE) | 55 |
| FAIRE-seq | 39 |
| RNA profiling | |
| CAGE | 78 |
| Exon array | 158 |
| RNA-chip | 26 |
| RNA-PET | 31 |
| RNA-seq | 245 |
| RNA-seq (MOUSE) | 102 |
| Transcription factor binding sites | |
| ChIP-seq | 1229 |
| ChIP-seq (MOUSE) | 202 |
| Other | |
| DNA cleavage | 1 |
| DNA-PET | 6 |
| GENCODE genes | 7 |
| Genotype | 64 |
| Negative regulatory elements | 2 |
| Nucleosome positioning | 2 |
| Proteogenomics | 14 |
| Replication timing | 24 |
| Replication timing (MOUSE) | 18 |
| RNA binding proteins | 47 |
| Short read mapability | 13 |
| Short read mapability (MOUSE) | 5 |
Table 3.
All links mentioned in this publication are collected in this table
Much of the primary human data (January 2011 data freeze) has been processed uniformly and used as the basis of the September 2012 published integrated analyses performed by the ENCODE analysis group. Results from this processing and analysis are accessible in the UCSC browser via a UCSC public Track Data Hub (ENCODE Analysis Hub) accessed at http://genome.ucsc.edu/cgi-bin/hgHubConnect. All data have been reprocessed using the ENCODE uniform processing pipeline, including signal tracks corrected for read length and mappability (http://code.google.com/p/align2rawsignal), peak calls from the SPP (15) and PeakSeq (16) peak callers filtered using the irreproducible discovery rate (17), computed RNA contigs from seven cellular localizations (18) and genome segmentations for the six Tier 1 and 2 cell lines. A total of 2876 data sets are included in the hub, with a reduced set displayed by default. The track organization for the analysis hub is illustrated in Figure 2. Additional background and resources related to the ENCODE analysis effort are provided on the portal’s Integrative Analysis page, described later.
Figure 2.
The ENCODE Analysis Hub at the EBI hosts over 2800 ENCODE data sets, organized in six tracks controlled via the track menu shown here.
Mouse genome
Although the ENCODE project aims to discover all DNA sequences in the human genome with biochemical function under the expectation that these will likely be functional, extending the analysis to use comparative genomics approaches was identified as a fruitful direction for the project. Thus, in the third year, a Mouse ENCODE Project was inaugurated (19). Assays identical to those being used in the ENCODE project are performed in cell types in mouse that are similar or homologous to those studied in the human project. The comparison will be used to discover which epigenetic features are conserved between mice and humans.
The past year marked the expansion of the UCSC Mouse browser (mm9/NCBI37) from a few preliminary ENCODE tracks to a full representation of Mouse ENCODE data production. A total of 20 tracks of ChIP-seq, RNA-seq and DNase-seq were released, reflecting 583 experiments. Figure 3 provides a graphical view of the Mouse ENCODE data availability.
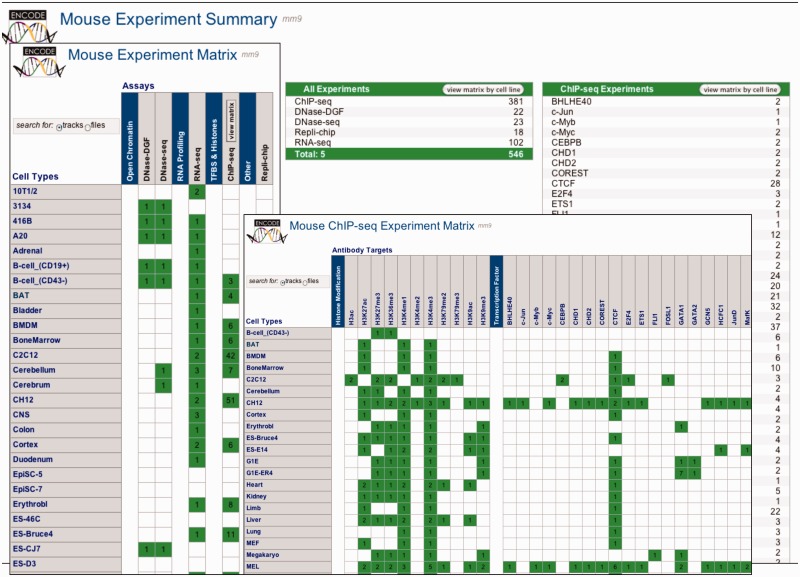
Figure 3.
All three screens of the Experiment Matrix for mouse are shown overlaid. The Data Summary screen lists experiments by data type, and provides launching to the two matrix screens that organize the data by assay and cell type. Clicking the appropriate table row or matrix cell launches a Track or File search tool (based on the Track/File selector control) that allows further refinement of the selection for browsing or download.
Data distribution
The ENCODE Data Coordination Center at UCSC (DCC) has accessioned all relevant ENCODE data at the Gene Expression Omnibus (GEO) (20) and the Short Read Archive. Since September 2011, the DCC has archived a total of 3346 GEO Samples across 53 GEO Series for humans (GRCh37/hg19) and mice (NCBI37/mm9). ENCODE data now encompasses a total of 4835 GEO Samples and 98 GEO Series. ENCODE GEO submissions are listed on the GEO ENCODE summary page, http://www.ncbi.nlm.nih.gov/geo/info/ENCODE.html. ENCODE has been assigned National Center for Biotechnology Information (NCBI) BioProject identifiers to further organize the data: PRJNA30707 for Human ENCODE (with the subproject PRJNA63443 for production phase data) and PRJNA50617 for Mouse ENCODE. Data in each project are further categorized as ‘epigenomic’, ‘functional genomics’ or ‘transcriptome’. Both UCSC and GEO are archival sites for 2007–12 ENCODE data, and user choice of repository is largely a matter of preferred interface.
All released data from ENCODE are tagged with permanent DCC accessions. Each accession represents a logical experiment, and therefore groups-related files representing different levels of results (e.g. sequence files, Binary Alignment/Map (BAM) alignments, signal graphs and peaks) for multiple replicates. ENCODE data sets at UCSC include GEO accessions in the accessible metadata.
ACCESS TOOLS AND FEATURES
A key focus of the past year has been to make the breadth of ENCODE data more readily visible and accessible. Specifically, UCSC has provided new web tools for locating and selecting data of interest, expanded the web portal to include new resources that aid in the interpretation of ENCODE data and developed new tutorial and outreach materials.
Experiment Matrix web tools
The ‘Experiment Matrix’ tool on the ENCODE portal is a set of three web pages that provide an up-to-date view of the breadth of ENCODE data available, along with an interface for selecting experiments for viewing in the browser or downloading as files for analysis. The main Experiment Matrix page shows the number of experiments for each cell type/assay pairing. The ChIP-seq Experiment Matrix page provides a view of the transcription factor and histone modification data sets, showing experiments by cell type and antibody target. The Experiment Summary page lists the number of experiments by assay type alone and includes annotations that are cell type independent (annotations on the reference genome). Each page has a file/track selector. Clicking an experiment item produces a search window listing the resulting files or tracks listed. Details about a file, including GEO accessions, are viewed from the search results listing by clicking the down arrow glyph next to the file of interest. An ‘Overview’ link provides help. Experiment Matrix tools are available for both human and mouse (Figure 3) data. The experiment collections presented in matrix form are also available in list form, with downloads provided in Comma Separated Value (CSV) and Excel (XLS) formats via the ‘Experiment List’ pages on the ENCODE portal.
Portal resources
Integrative analysis
The completion of a comprehensive and coordinated integrative analysis of human ENCODE data by the consortium analysis group resulted in numerous resources published in September 2012, described at http://encodeproject.org/ENCODE/analysis.html. Key resources include a publications package of 30 articles with coordinated publication in three journals, a Nature ‘microsite’ and a virtual machine analysis platform with access to code bundles allowing user replication of the ENCODE analysis results.
Quality metrics
The ENCODE Consortium analysis working group has devoted considerable resources to analyzing the quality of the data produced using a variety of metrics. The resulting quality metrics are available from the ENCODE portal (http://encodeproject.org/ENCODE/qualityMetrics.html). The Quality Metrics page displays downloadable spreadsheets showing quality metrics for human ENCODE data sets, along with descriptions of the metrics and what they appear to measure. Data set metrics are included for DNase I hypersensitive sites sequencing (DNase-seq), formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq), chromatin immunoprecipitation sequencing of transcription factor binding sites (TFBS ChIP-seq), Histone ChIP-seq and ChIP Controls. The software used to generate the quality metrics is described on the Software Tools page of the ENCODE portal.
Software tools
A significant product of the ENCODE project has been the evaluation and development of software to process and analyze the data sets (largely high-throughput sequencing). This section of the portal http://encodeproject.org/ENCODE/softwareTools.html provides descriptions and references for 22 software packages used to create the primary data sets generated by the ENCODE production laboratories and the downstream analyses of the ENCODE analysis group. These include peak callers [e.g. model-based analysis of ChIP-Seq (MACS) (21), SPP (15) and PeakSeq (16)], signal generators (e.g. Wiggler), multi-data type integration tools [e.g. Segway (22) and ChromHMM (23)] and quality assessment tools [e.g. irreproducible discovery rate (17)]. A companion page http://encodeproject.org/ENCODE/analysisTools.html describes and references seven additional tools of interest to users who are analyzing and using ENCODE data in their own research [e.g. Factorbook (24)].
Data standards: experiment guidelines
A notable product of the ENCODE project has been the definition of guidelines for conducting and reporting functional genomics experiments performed using high-throughput sequencing technologies. Guidelines and ‘Best Practices’ developed by the ENCODE project for ChIP-seq (25), RNA-seq, DNase-seq, FAIRE-seq and DNA methylation, along with cell culture guidelines, are available from this page.
Data standards: platform characterization
This new portal page (http://encodeproject.org/ENCODE/platform_characterization.html) provides summary descriptions and references to studies that illuminate the research technologies underpinning the ENCODE resource. Factors affecting ChIP-seq and RNA-seq experimentation are highlighted, and provide valuable aid to interpretation of the data.
Publications
The Publications page, http://encodeproject.org/ENCODE/pubs.html was extensively expanded this year to comprise a comprehensive list of 150 publications covering methods, resources and biological findings produced by the ENCODE Consortium and from ENCODE-funded projects. A subsidiary page has references to 111 publications based on ENCODE data that were written by authors outside the consortium.
Tutorial and outreach materials
A second more detailed tutorial on accessing ENCODE data was developed this year and is available from http://openhelix.com/ENCODE2. The original introductory tutorial remains available at http://openhelix.com/ENCODE. OpenHelix also offers a Quick Reference Card. The Quick Reference Card gives an overview of the site and the numerous methods to access and view ENCODE data within the UCSC Genome Browser using screen shots and callouts to highlight the various features and functions. Cards may be ordered at http://www.openhelix.com/cgi/qrcOrder.cgi.
FUTURE WORK
Although the main production coordination of data for the next phase of ENCODE will be coordinated and managed by Stanford, UCSC will continue to provide data hosting and access tools for current and future ENCODE data. UCSC will incorporate integrated and ‘best-of-breed’ ENCODE data sets from this phase into the UCSC Genome Browser database. An update of the ‘ENCODE Regulation’ track set to incorporate newer data is in progress, and a companion track set is planned for the Mouse genome. Although the bulk of the primary data from the next phase of ENCODE will not be imported into the UCSC browser database and thus will not be available as native tracks, UCSC will provide download access to the data, and visualization via the Browser’s Track Data Hub feature.
CONTACT INFORMATION
General questions and feedback about ENCODE data at UCSC should be directed to the UCSC browser mailing list: genome@soe.ucsc.edu. Specific questions about details of laboratory methods or data interpretation should be directed to the ENCODE laboratory contact listed on the description page for that data set. We announce releases of new ENCODE data via the ENCODE announcement list. To subscribe, visit https://groups.google.com/a/soe.ucsc.edu/forum/#!forum/encode-announce.
FUNDING
National Human Genome Research Institute [5P41HG002371-12, 3P41HG002371-12S1 to the UCSC Center for Genomic Science; 5U41HG004568-04, 3U41HG004568-03S1 to the UCSC ENCODE Data Coordination Center]; Howard Hughes Medical Institute (to D.H.). Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement: The authors receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
ACKNOWLEDGEMENTS
The authors thank the systems administration staff at the Center for Biomolecular Science and Engineering: Jorge Garcia, Erich Weiler and Gary Moro, for their on-going support maintaining our site reliability, capacity, and performance. They also thank members of the ENCODE Consortium for providing these valuable data sets. Special appreciation to Michael Pazin of NHGRI for extensive contributions to the ENCODE portal content.
REFERENCES
- 1.ENCODE Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 2.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2011;40:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2011;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nature Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 10.Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Carriero N, Zheng D, Karro J, Harrison PM, Gerstein M. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics. 2006;22:1437–1439. doi: 10.1093/bioinformatics/btl116. [DOI] [PubMed] [Google Scholar]
- 14.Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J. Retrocopy contributions to the evolution of the human genome. BMC Genomics. 2008;9:466. doi: 10.1186/1471-2164-9-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotech. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotech. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li QH, Brown JB, Huang HY, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 2011;5:1752–1779. [Google Scholar]
- 18.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mouse ENCODE Consortium. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All ENCODE production data for the 5-year initial production phase of the project have now been submitted to the ENCODE Data Coordination Center at UCSC. UCSC has performed quality review and publicly released all conforming ENCODE data sets along with metadata, as both tracks for browsing and downloadable files for data mining. In the human genome, 288 cell and tissue types are now represented, covering 32 assays. Chromatin features and sites of DNA binding are mapped for >300 factors and marks. In mouse, 81 cell and tissue types were surveyed in five experimental assays.
Human genome
The results of five new experiment types were released during the fifth year: chromatin interactions based on chromosome conformation capture carbon copy (5C) and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) methods, proteogenomics and DNA replication timing by both sequencing and microarray methods.
Although DNA is a linear molecule, it is packed and organized inside the nucleus in a 3D milieu, and gene regulation can be affected by interactions from elements located hundreds of kilobases distant in the genome. Long-range chromatin looping interactions can be detected using various techniques, including chromosome conformation capture (9) and chromatin interaction analysis with paired-end tag (10). The ENCODE chromatin interactions data sets comprise experiments in 14 cell types.
Proteogenomic methods differ from conventional mass spectrometry proteomic methods that identify peptides by comparing them with peptides produced from known proteins. In contrast, proteogenomic methods compare peptides with all peptides that might be produced by the six translation frames of the genome to identify the genomic region from which the peptides were produced. Study of proteogenomic data offers insights into regulatory mechanisms, including translation, pre-messenger RNA (mRNA) splicing and transcript diversity, nonsense-mediated decay and transcription of novel protein-coding genes. The ENCODE protegenomics data are available in four cell types. Figure 1 presents a Genome Browser session that includes proteogenomics data in conjunction with ENCODE gene, transcriptome and regulatory data sets.
Figure 1.
ENCODE data displayed in the UCSC Genome Browser together with annotations from the ENCODE Analysis Hub in the region of the nucleoporin gene NUP133 demonstrate the power this diversity of data provides for visual interpretation. The GENCODE Basic gene set shows this gene having four protein-coding splice variants and three smaller non-coding transcripts nearby. The proteogenomics track shows support for many of the coding exons, with protein localized in the nucleus, but not in plasma membrane or mitochondria. The long polyA RNA signal shows strong peaks over the exons and low intron signal in the cytosol, with greater signal in the nucleus. This is expected because nuclear mRNAs are not all completely spliced. The Combined Genome Segmentation integrates signal from many histones and classifies regions into those with characteristics of promoters (red), enhancers (yellow), insulators (blue), transcribed regions (green) and repressed (gray). Below are signal tracks from four of the eight histone modifications used as input to the segmentation. The promoter and transcribed regions agree with the RNA evidence, and like the RNA evidence show no evidence of transcription of the non-coding gene to the right of NUP133. Underneath the GM12878 histone signals is a track that overlays one of the histone signals, H3K27Ac, in seven different cell lines (with GM12878 shown in red). A peak in H3K27Ac appears at the enhancer, but as is often the case with enhancers, this appears to be relatively cell specific in contrast to the larger peak near the promoter, where the black coloration indicates the peak is shared by many cell types. The DNAse hypersensitivity and transcription factor tracks also provide evidence for both promoter and enhancer. Finally the mappability track indicates regions where short reads are not uniquely mappable, indicating the data are incomplete and therefore harder to interpret. Although most of this region is mappable, there are many small regions throughout and one larger region on the right where mapping is problematic. Overall, the ENCODE data in this region show strong evidence that this is a nuclear-localized protein-coding gene with a promoter that is used in a wide variety of cell types, and is likely to be regulated by tissue-specific enhancers as well.
The order in which DNA is duplicated during the synthesis phase of the cell cycle is correlated with the expression of genes and the structure of chromosomes; replication timing is known to be an important feature for epigenetic control of gene expression. ENCODE ‘Repli-chip’ (microarray) experiments are available in 9 cell types, and ‘Repli-seq’ (sequencing) experiments in 15.
The encyclopædia of genes and gene variants (GENCODE) gene set (11) is a fundamental resource produced by ENCODE, providing high-quality manual annotation from the Human and Vertebrate Analysis and Annotation (HAVANA) group merged with evidence-based automated annotation from Ensembl (12) across the human genome. For the final release (V12), the data organization and display were improved to make the data more accessible and intuitive. Annotations are now categorized according to their function and level of support. Color coding reflects non-coding, coding, pseudogene or problem status. To complement the ‘Comprehensive’ gene set, a new ‘Basic’ subset provides a simplified view intended for the majority of users, by filtering incomplete and problem annotations while still ensuring that at least one annotation is displayed at every locus. For researchers who require more detail regarding the degree of evidence supporting individual coding transcripts, a five-level scoring metric is provided, based on assessment of alignments of mRNAs and expressed sequence tags (ESTs) across the full length of the annotation. Filtering options allow tuning of the display based on the basic biological function of the transcript (coding, non-coding, etc.), annotation method (manual versus automated) or specific biotype characterization (http://www.gencodegenes.org/gencode_biotypes.html). Finally, two additional annotation subtracks are provided: the ‘2-Way Pseudogene’ subtrack shows consensus pseudogenes predicted by two pipelines [Yale Pseudopipe (13) and UCSC Retrofinder (14)], and the ‘PolyA’ subtrack presents polyA signals and sites manually annotated on the genome based on transcribed evidence.
The majority of ENCODE primary data submitted and released in the past year for the human genome expanded the existing tracks with additional experiments; cell types, subcellular fractions, transcription factors or histone marks were also mapped. The total complement of data sets available is summarized in Tables 1 and 2. All links mentioned in this publication are collected in Table 3.
Table 1.
The full complement of ENCODE data sets summarized by cell type [types annotated as cancer are marked with asterisk (*)]
| Cell type | Tissue | Description | Data sets | |||
|---|---|---|---|---|---|---|
| TF/His | RNA | Other | Total | |||
| Tier 1 initial | ||||||
| GM12878 | Blood | Lymphoblastoid | 137 | 27 | 49 | 213 |
| K562* | Blood | Leukemia | 247 | 45 | 80 | 372 |
| Tier 1 added in 2011 | ||||||
| H1-hESC | Embryonic stem | Embryonic stem | 96 | 14 | 23 | 133 |
| Tier 2 initial | ||||||
| HeLa-S3* | Uterine cervix | Cervical carcinoma | 93 | 14 | 30 | 137 |
| HepG2* | Liver | Liver carcinoma | 118 | 19 | 26 | 163 |
| HUVEC | Umbilical endothelium | Umbilical vein endothelial | 37 | 13 | 16 | 66 |
| Tier 2 added in 2011 | ||||||
| A549* | Lung | Lung carcinoma | 89 | 22 | 12 | 123 |
| CD14+ | Blood | Monocyte | 17 | 4 | 4 | 25 |
| IMR90 | Lung | Lung fibroblast | 11 | 16 | 10 | 37 |
| MCF-7* | Breast | Breast carcinoma | 50 | 15 | 32 | 97 |
| SK-N-SH* | Brain | Neuroblastoma | 36 | 16 | 7 | 59 |
| Tier 2 added in 2012 | ||||||
| CD20+ | Blood | B cell | 11 | 5 | 4 | 20 |
| H1-neuron | Neuron | H1ES-derived neuron | 5 | 3 | 1 | 9 |
| LHCN-M2 | Muscle | Myoblast | 7 | 2 | 4 | 13 |
| Human: totals | ||||||
| Tier1 + Tier2 (14) | 954 | 215 | 298 | 1467 | ||
| Tier 3 (274) | 591 | 94 | 734 | 1419 | ||
| All (288) | 1545 | 309 | 1032 | 2886 | ||
| Mouse | ||||||
| All (81) | 381 | 102 | 100 | 583 |
Table 2.
The full complement of ENCODE data sets summarized by assay
| Data type/assay | Experiments |
|---|---|
| Chromatin interactions | |
| 5C | 14 |
| ChIA-PET | 8 |
| DNA methylation | |
| Methyl array | 125 |
| Methyl RRBS | 95 |
| Methyl-seq | 20 |
| Histone modifications | |
| ChIP-seq | 315 |
| ChIP-seq (MOUSE) | 179 |
| Open chromatin | |
| DNase-DGF | 56 |
| DNase-DGF (MOUSE) | 22 |
| DNase-seq | 221 |
| Dnase-seq (MOUSE) | 55 |
| FAIRE-seq | 39 |
| RNA profiling | |
| CAGE | 78 |
| Exon array | 158 |
| RNA-chip | 26 |
| RNA-PET | 31 |
| RNA-seq | 245 |
| RNA-seq (MOUSE) | 102 |
| Transcription factor binding sites | |
| ChIP-seq | 1229 |
| ChIP-seq (MOUSE) | 202 |
| Other | |
| DNA cleavage | 1 |
| DNA-PET | 6 |
| GENCODE genes | 7 |
| Genotype | 64 |
| Negative regulatory elements | 2 |
| Nucleosome positioning | 2 |
| Proteogenomics | 14 |
| Replication timing | 24 |
| Replication timing (MOUSE) | 18 |
| RNA binding proteins | 47 |
| Short read mapability | 13 |
| Short read mapability (MOUSE) | 5 |
Table 3.
All links mentioned in this publication are collected in this table
Much of the primary human data (January 2011 data freeze) has been processed uniformly and used as the basis of the September 2012 published integrated analyses performed by the ENCODE analysis group. Results from this processing and analysis are accessible in the UCSC browser via a UCSC public Track Data Hub (ENCODE Analysis Hub) accessed at http://genome.ucsc.edu/cgi-bin/hgHubConnect. All data have been reprocessed using the ENCODE uniform processing pipeline, including signal tracks corrected for read length and mappability (http://code.google.com/p/align2rawsignal), peak calls from the SPP (15) and PeakSeq (16) peak callers filtered using the irreproducible discovery rate (17), computed RNA contigs from seven cellular localizations (18) and genome segmentations for the six Tier 1 and 2 cell lines. A total of 2876 data sets are included in the hub, with a reduced set displayed by default. The track organization for the analysis hub is illustrated in Figure 2. Additional background and resources related to the ENCODE analysis effort are provided on the portal’s Integrative Analysis page, described later.
Figure 2.
The ENCODE Analysis Hub at the EBI hosts over 2800 ENCODE data sets, organized in six tracks controlled via the track menu shown here.
Mouse genome
Although the ENCODE project aims to discover all DNA sequences in the human genome with biochemical function under the expectation that these will likely be functional, extending the analysis to use comparative genomics approaches was identified as a fruitful direction for the project. Thus, in the third year, a Mouse ENCODE Project was inaugurated (19). Assays identical to those being used in the ENCODE project are performed in cell types in mouse that are similar or homologous to those studied in the human project. The comparison will be used to discover which epigenetic features are conserved between mice and humans.
The past year marked the expansion of the UCSC Mouse browser (mm9/NCBI37) from a few preliminary ENCODE tracks to a full representation of Mouse ENCODE data production. A total of 20 tracks of ChIP-seq, RNA-seq and DNase-seq were released, reflecting 583 experiments. Figure 3 provides a graphical view of the Mouse ENCODE data availability.
Figure 3.
All three screens of the Experiment Matrix for mouse are shown overlaid. The Data Summary screen lists experiments by data type, and provides launching to the two matrix screens that organize the data by assay and cell type. Clicking the appropriate table row or matrix cell launches a Track or File search tool (based on the Track/File selector control) that allows further refinement of the selection for browsing or download.
Data distribution
The ENCODE Data Coordination Center at UCSC (DCC) has accessioned all relevant ENCODE data at the Gene Expression Omnibus (GEO) (20) and the Short Read Archive. Since September 2011, the DCC has archived a total of 3346 GEO Samples across 53 GEO Series for humans (GRCh37/hg19) and mice (NCBI37/mm9). ENCODE data now encompasses a total of 4835 GEO Samples and 98 GEO Series. ENCODE GEO submissions are listed on the GEO ENCODE summary page, http://www.ncbi.nlm.nih.gov/geo/info/ENCODE.html. ENCODE has been assigned National Center for Biotechnology Information (NCBI) BioProject identifiers to further organize the data: PRJNA30707 for Human ENCODE (with the subproject PRJNA63443 for production phase data) and PRJNA50617 for Mouse ENCODE. Data in each project are further categorized as ‘epigenomic’, ‘functional genomics’ or ‘transcriptome’. Both UCSC and GEO are archival sites for 2007–12 ENCODE data, and user choice of repository is largely a matter of preferred interface.
All released data from ENCODE are tagged with permanent DCC accessions. Each accession represents a logical experiment, and therefore groups-related files representing different levels of results (e.g. sequence files, Binary Alignment/Map (BAM) alignments, signal graphs and peaks) for multiple replicates. ENCODE data sets at UCSC include GEO accessions in the accessible metadata.