The oscillating miRNA 959-964 cluster impacts Drosophila feeding time and other circadian outputs (original) (raw)
. Author manuscript; available in PMC: 2013 May 7.
Abstract
We sequenced Drosophila head RNA to identify a small set of miRNAs that undergo robust circadian cycling. We concentrated on a cluster of six miRNAs, mir-959-964, all of which peak at about ZT12 or lights-off. The cluster pri-miRNA is transcribed under bona fide circadian transcriptional control, and all 6 mature miRNAs have short half-lives, a requirement for cycling. A viable Gal4 knock-in strain localizes prominent cluster miRNA expression to the adult head fat body. Analysis of cluster knock-out and over-expression strains indicates that innate immunity, metabolism, and feeding behavior are under cluster miRNA regulation. Manipulation of food intake also affects the levels and timing of cluster miRNA transcription with no more than minor effects on the core circadian oscillator. These observations indicate a feedback circuit between feeding time and cluster miRNA expression-function as well as a surprising role of post-transcriptional regulation in the circadian control of these phenotypes.
Keywords: miRNAs, circadian, feeding, Drosophila, immunity
INTRODUCTION
Many organisms have circadian pacemakers that control their physiology and behavior (Golombek and Rosenstein, 2010). These clocks are entrained by external cues like light and temperature (Zeitgebers) and maintain a rhythm of about 24 hr or precisely 24 hr under entrainment conditions. Transcriptional feedback loops play an important role in timekeeping. Positive factors like CLOCK (CLK) and CYCLE (CYC) in D.melanogaster or CLK and BMAL1 in mammals lead to the transcription of negative regulators like PERIOD (PER) and TIMELESS (TIM), or PER and CRYPTOCHROME (CRY), respectively. The negative regulators gain entry to the nucleus, collaborate with kinases and chromatin factors, and then repress CLK-CYC- and CLK-BMAL1-mediated transcription. The negative regulators decay, and circadian transcription begins anew (Allada and Chung, 2010).
In contrast to the abundant information on the transcriptional regulation of circadian rhythms, less is known about post-transcriptional regulation, for example, the circadian regulation of mRNA turnover (So and Rosbash, 1997; Woo et al., 2010; Woo et al., 2009). However, recent work from several labs has addressed the contribution of miRNAs to circadian rhythmicity (see below).
miRNAs are endogenous, ~ 22 nucleotide small non-coding RNAs. They function predominantly by binding either in the 3’ UTR or open reading frame (ORF) of a target mRNA and affect translational regulation and/or lead to decreases in target mRNA levels (Guo et al., 2010; Karginov et al., 2010). miRNAs are generated by cleavage reactions (Ghildiyal and Zamore, 2009; Miyoshi et al., 2010). Drosha processes the pri-miRNA primary transcript within the nucleus to a single hairpin-containing pre-miRNA transcript. It is exported to the cytoplasm where it is processed by Dicer and loaded into an effector RNP complex (RISC = RNA-induced silencing complex). The miRNA-containing RISC complex then interacts with target mRNAs.
In mice, Cheng et al., (2007) highlighted the role of two brain specific miRNAs, miR-219 and miR-132 and their contribution to circadian clock modulation. miR-132 has also been shown to target a number of genes involved in chromatin remodeling and translational control, which then modulate Period gene activity (Alvarez-Saavedra et al., 2010). The liver specific miR-122 has been shown to play a role in the rhythmic expression of the circadian deadenylase nocturnin in mice (Kojima et al., 2010). Rhythmic expression of chicken mir-26a has been shown to modulate the protein expression of photoreceptor L-type voltage-gated calcium channel alpha1C subunit (Shi et al., 2009).
Two Drosophila miRNAs dme-miR-263a and dme-miR-263b, have been reported to exhibit circadian oscillations and are predicted to target clk and or cwo (Yang et al., 2008). A more recent study from our laboratory demonstrated that clk translation is modulated by the developmental miRNA bantam, thus affecting the core circadian clock (Kadener et al., 2009).
To further investigate the role of miRNAs in the Drosophila circadian system, we used the Illumina platform to sequence 18–29 nt RNA and compared six circadian time points. Although most miRNAs showed little or no significant oscillations, there were a few exceptions. We focused on a cluster of six miRNAs, all of which showed high amplitude cycling. The miRNA oscillations are under circadian regulation, as they disappear in three arrhythmic strains: clkAR, clkJRK and per01. The cluster pri-miRNA also shows comparable circadian cycling and becomes arrhythmic in the per01 strain, suggesting that circadian transcriptional regulation makes a major contribution to the miRNA oscillations. Starvation as well as restricted feeding indicates that pri-miRNA circadian transcription is under nutritional/feeding control and disassociates it from the core clock. Identification of target mRNAs using knock-out as well as over-expression strains shows significant overlap and implicates genes involved in various physiological functions including metabolism, oxidative stress, reproductive behavior, peptidase/proteases and immune function. Consistent with some of these functions, the normal regulation of feeding behavior, immune function and possibly stress responses is compromised in the knock-out and/or over-expression strains. Our findings suggest that the cluster miRNAs are synthesized in response to nutritional signals acquired by feeding and then serve to regulate a number of physiological and behavioral responses. These include feeding itself, which suggests a post-transcriptional feedback loop involved in the timing of feeding.
RESULTS
The miRNA 959-964 cluster is under circadian control
To search for miRNAs under circadian regulation, we sequenced small RNAs around the clock. To this end, small RNA libraries (RNA size 19 to 29 nucleotides long) were prepared from Canton S fly heads collected at six different circadian time points during a light-dark cycle: ZT0 (Zeitgeber time 0), ZT4, ZT8, ZT12, ZT16, and ZT20. Sequencing was via the Illumina deep sequencing platform, and an average of 3.1 million reads per time point mapped to unique locations on the genome. Individual miRNA reads were normalized to the total number of miRNA reads from each time point. To search for cycling miRNAs, the number of individual reads was plotted across the six time points, and the graphs visually inspected.
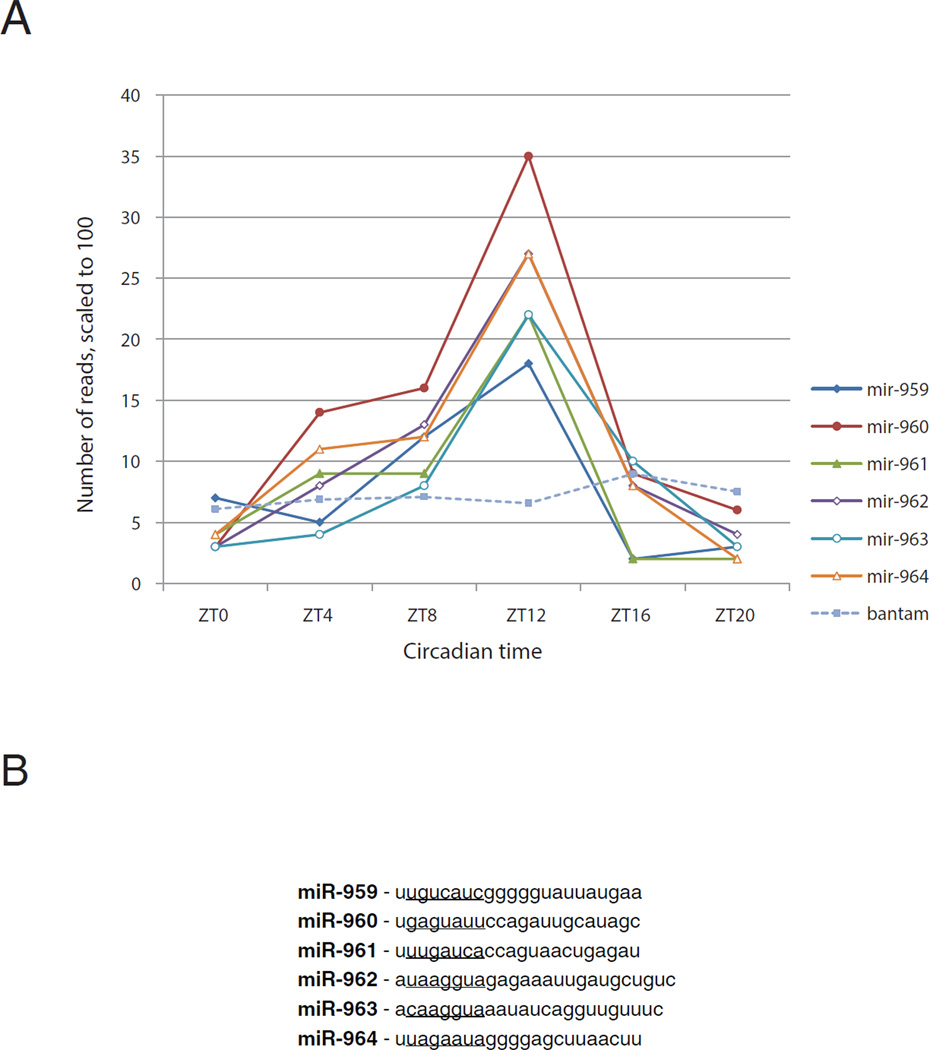
Although most of the 196 known miRNAs did not show any obvious circadian oscillations across the 24 hr cycle, the 6 miRNAs 959–964 correspond to a previously characterized miRNA cluster and showed a similar phase and robust amplitude; they are probably encoded on a single transcription unit (Figure 1A; see below). The six miRNAs are encoded within two adjacent introns of CG31646 (miR-959-962 within one, and miR-963-964 within the other; Supplementary Figure 1). Transcription is anti-sense to CG31646, which has no known regulatory influence on the cluster. Note that the miRNAs 959–964 have different seed sequences (Figure 1B). We focused the rest of this work on these miRNAs and refer to them subsequently as the “cluster miRNAs.”
Figure 1. Identification of cycling miRNAs via Illumina deep sequencing.
Small RNA libraries for Illumina deep sequencing platform were prepared from fly head RNA across six different circadian time points. The number of reads for each miRNA were normalized to total number of reads from each time point and plotted across six circadian time points. A) The abundance of a cluster of six miRNAs (miR-959, miR-960, miR-961, miR-962, miR-963 and miR-964) and the developmental regulator miRNA bantam, is plotted across circadian time. Except bantam, all six candidates show high amplitude of cycling. The actual number of reads for bantam was 10,000 fold more across all six time points than the plotted values. B) The sequences of a cluster of six cycling miRNAs (miR-959 to miR-964). The underlined sequence is the seed region which is different for each member.
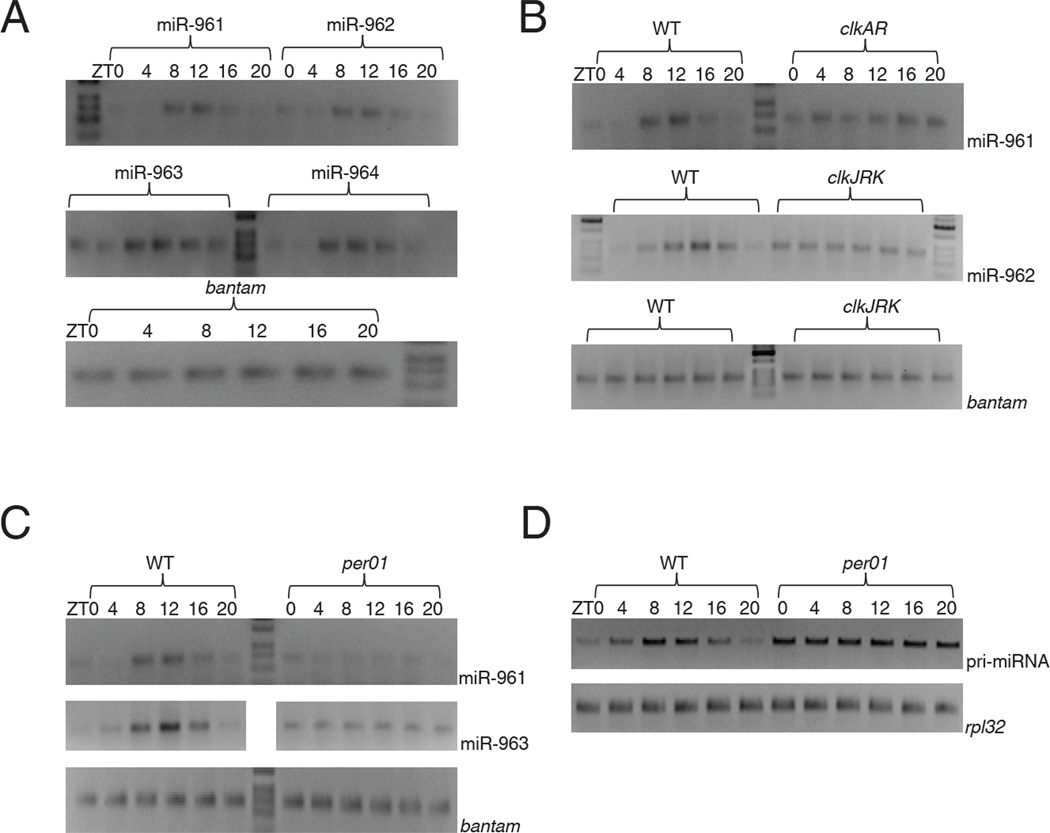
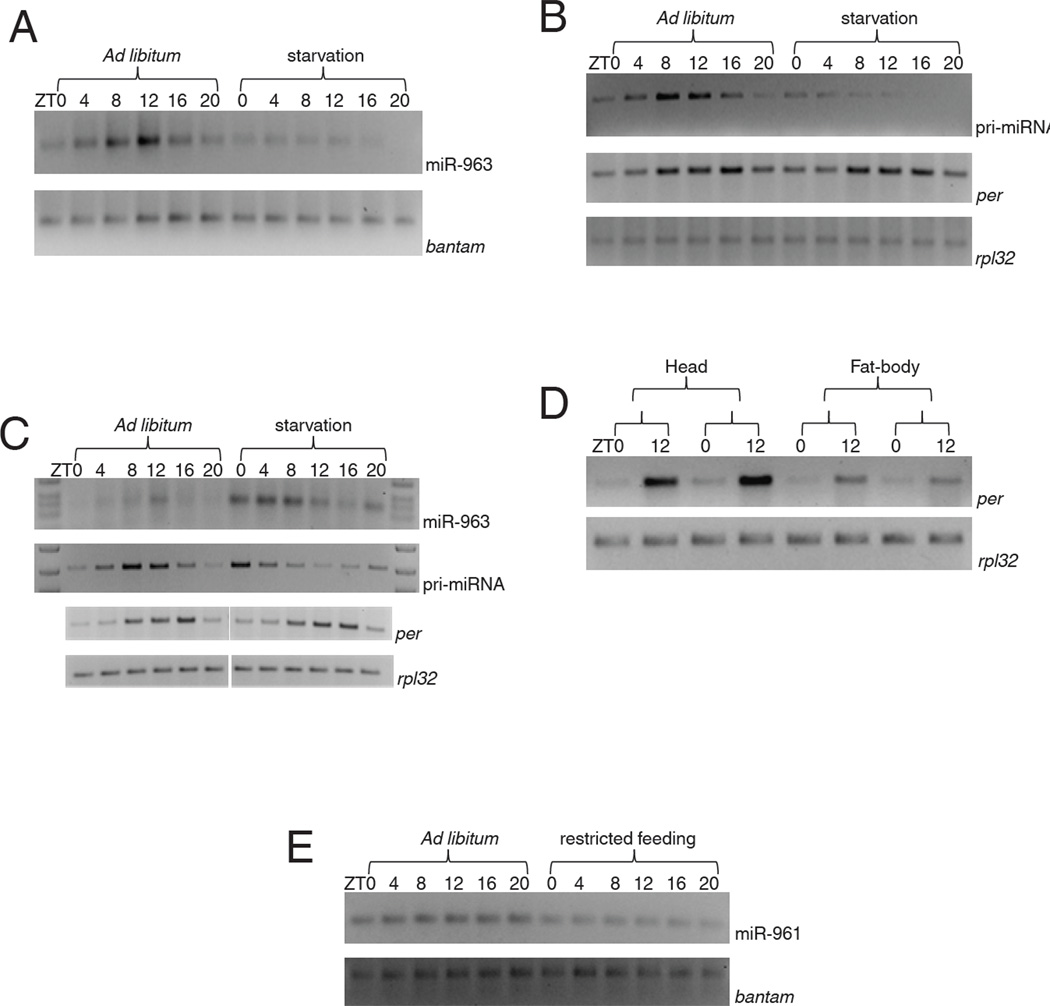
miRNA RT-PCR (Chen et al., 2005) confirmed the sequencing data, namely, the levels of the cluster miRNAs undergo robust cycling with similar amplitude (Figure 2A). Moreover, all miRNA levels peak at about ZT12 (Figure 2A), consistent with the peak time determined by Illumina sequencing (Figure1A for 960). Bantam miRNA was assayed in parallel and was non-cycling or much less obviously cycling across all six time points (Figure 2A), consistent with the sequencing data (Figure 1A). Importantly, there is no cluster miRNA cycling in two classical clk arrhythmic mutant strains, clkAR and clkJRK. This indicates that miRNA cycling requires a functional clock, i.e., an external light: dark cycle is insufficient to drive cluster miRNA cycling (Figure 2B). There was also no apparent cycling in a third arrhythmic mutant strain, (Fig. 2C).
Figure 2. A cluster of six miRNAs show cycling and are under circadian clock control.
Total RNA was extracted from fly heads across six circadian time points and used for miRNA RT-PCR using miRNA specific primers (see materials and methods). A discrete band around 70 base pairs is the PCR product for a mature miRNA. A) Shown here are four representatives from a cluster of six miRNAs – miR-961, miR-962, miR-963 and miR-964 all of which show robust oscillation with a peak around ZT12. The developmental regulator miRNA bantam was used as a control for RT-PCR which did not show any oscillation. B) To investigate the role of circadian clock we assayed cycling and levels in two clk mutants clkAR and clkJRK by RT-PCR. The levels of the mature miRNA in both clkAR and clkJRK were comparable to wild type (WT) and did not show any oscillation in the two mutants. As controls we assayed the levels of bantam in WT and clkJRK flies and did not observe any significant changes. C) Wild type and per01 flies were entrained in LD conditions and fly head RNA was extracted from six circadian time points. The levels of the mature miRNA were assayed using miRNA RT-PCR and as shown earlier the mature miRNA levels showed robust oscillation in WT flies but were arrhythmic in per01 mutant flies. The overall levels of the mature miRNA also showed ~ 2 to 3 fold decrease in the per01 mutant flies. As a control we assayed the levels of bantam miRNA which showed similar levels in WT and per01 flies. D) The levels of the pri-miRNA were analyzed across circadian time in WT and per01. The levels of the pri-miRNA showed robust oscillation in the WT flies but did not oscillate in per01 mutant flies. In fact, pri-miRNA levels were high across all six time points.
To assay pri-miRNA expression, we used primers between miRNA 962 and 963, between the two introns of CG31646 (Fig. 2D). In WT strains, cluster pri-miRNA undergoes a robust circadian oscillation with a phase and amplitude very similar to those of the miRNAs. This suggests that circadian transcriptional regulation is a critical feature of miRNA cycling dynamics. Cycling was notably absent in the arrhythmic per01 strain, suggesting that the clock regulates cluster circadian transcription (Fig. 2D). Because the pri-miRNA level is quite high whereas miRNA levels are quite low in per01 flies, the clock may also affect post-transcriptional processing of the cluster miRNAs (see Discussion).
Generation and Characterization of Cluster Knock-in and Knock-out Mutants
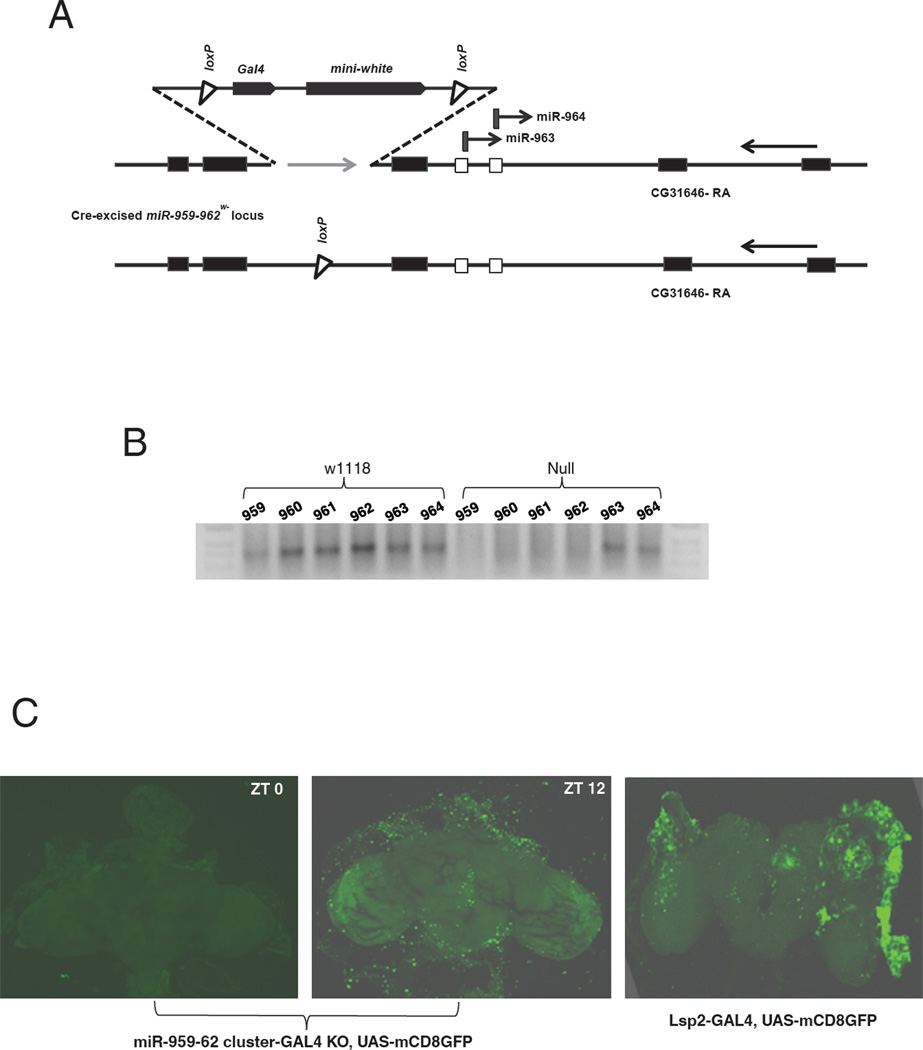
A cluster miRNA mutant missing miR-959-962 was generated using homologous recombination-based ends-out gene targeting (Gong and Golic, 2003). Because these four miRNAs are located within a single intron of the CG31646 locus (Supplementary Figure 1), a targeting vector was prepared with homology regions contained within that intron. The DNA was amplified and cloned into the targeting vector pW25-Gal4-attB2 (Weng et al., 2009). It allows introduction of Gal4 and the mini-white marker in place of the miRNA following homologous recombination (Fig 3A). The reporter genes in this knock-in strain are flanked by loxP sites to permit subsequent excision of the Gal4 and mini-white reporter cassette. Flies carrying the targeted allele were crossed to flies carrying a heat-shock promoter driven Cre recombinase transgene, and progeny lacking the mini-white marker were selected on the basis of eye color. Homozygous knock-out lines were established and the absence of the miRNA cluster DNA confirmed by PCR. The KO lines contain a single loxP site in place of the four miRNAs within the CG31646 intron (Fig. 3A).
Figure 3. Generation of the cluster knock-out and characterization of cluster expression.
The miR-959-962 knock out mutant was generated using homologous recombination based ends-out gene targeting (Gong and Golick, 2003). Upstream and downstream homology arms flanking miR-959-962 cluster were amplified and cloned into a targeting vector. A) The targeting vector pW25-Gal4-attB2 would allow Gal4 and mini-white gene knock-in at the targeted locus. Therefore miR-959-962 cluster-Gal4 KO is a Gal4 knock-in mutant and can be used as a Gal4 driver to report the cluster expression pattern. miR-959-962 cluster KOw- was generated from Gal4 and mini-white knock-in mutant by using Cre/loxP recombination to excise both Gal4 and mini-white genes. B) Total RNA was extracted from w118 and the KO flies, and all cluster members were assayed by miRNA RT-PCR. The miR 959-962 KO flies lacked the expression of the miR-959, miR-960, miR-961 and miR-962 where as the expression of miR-963 and miR-964 was unaffected. C) The Gal4 knock-in mutant was crossed to UAS-mCD8GFP line and intact brains were dissected to check for expression of the reporter gene. The brains were not cleaned thoroughly to assay the peri-cerebral tissue. Gal4 under the control of the cluster miRNA promoter shows circadian oscillation with higher levels at ZT12 as manifested by the expression of the reporter gene GFP. Moreover GFP showed strong localization in the peri-cerebral fat-body. Lsp2-Gal4 was used as a control for peri-cerebral fat-body localization.
The miR-959-962 KO mutant was homozygous viable and fertile. Quantitative real-time PCR analysis using RNA isolated from the central nervous systems of homozygous miR-959-962 mutant third instar larvae showed no detectable miR-962 miRNA. In contrast, the levels of miR-963 were not reduced (data not shown). Similar results were observed by assaying all six miRNAs with miRNA-specific RT-PCR using adult head RNA from the KO strain as a template: miR-963-964 were detected, whereas miR-959-962 were not (Fig. 3B).
The Gal4 gene present in the knock-in strain was used to assess head tissue expression of the cluster. The strain was crossed to UAS-mCD8GFP, and “brains” stained for GFP expression at two time points, ZT12 and ZT0. The brains were not well-cleaned so that adjacent head tissue could be assayed. Staining was prominent in the peri-cerebral fat body, which was assessed by comparison to the well-characterized Lsp2-Gal4 driver. Although this tissue is probably not the sole site of cluster expression (see below), the pattern suggests that the fat body is a major site of cluster miRNA expression. Moreover, staining was much stronger at ZT12 than at ZT0, consistent with the cluster miRNA analyses. This suggests that Gal4 is under the circadian transcriptional regulation that normally governs cluster expression.
Possible Functions of the Cluster miRNAs from affected mRNAs
To address possible functions of the cluster miRNAs, the knock-out strain as well as a cluster over-expression strain was compared with companion WT strains. The over-expression strain was a stable line containing the broad circadian driver tim-gal4 and a UAS-cluster genomic DNA transgene. This was favored over a fat body driver because of the possibility of non-fat body expression. Indeed, cluster miRNA levels in RNA from purified LNvs (Nagoshi et al 2010; Kula-Eversole et al 2010) were similar to those from heads relative to bantam miRNA (data not shown). Over-expression of the six cluster miRNAs at all circadian times was apparent by RTPCR of head RNA (Supplementary Figure 2).
We first assayed circadian behavior in the two strains relative to companion WT strains. The period of the knock-out strain period is short by about 30 min, whereas the period of the overexpression strain is long by more than an hour (Supplementary Figure 3). Although this period alteration could reflect effects from the fat body, we favor the interpretation that it derives from circadian neuron expression.
mRNA changes in the knock-out and cluster overexpression strains relative to their WT counterparts were then assayed on Affymetrix expression arrays. The strategy was based on the observation that miRNAs often cause a decrease in the steady-state levels of their target mRNAs (Guo et al., 2010; Lim et al., 2005). To accommodate the possibility that important mRNA changes only appear at certain circadian times, RNA was assayed from heads collected at two different times, ZT4 and ZT16, and a criterion of 1.5-fold was used to identify mRNAs altered in the over-expression and the KO strains.
Many individual mRNAs were altered in the two strains and some in opposite directions, i.e., up in the KO strain and down in the over-expression strain; this makes them candidate direct target mRNAs (Table S1). Moreover, many putative target 3’ UTRs have predicted miRNA seed-complementary sequences, and some of these 3’ UTRs are cluster miRNA targets in S2 cell reporter gene assays (Supplementary Figure 4).
To address possible biological functions, we searched for GO terms that were enriched in the putative target mRNAs. The most consistent categories were metabolism (oxidation-reduction) and immune function, both of which appeared in the over-expression as well as the KO strain data (Table 1). However, “immunity” was only apparent in the ZT4 data. Proteolysis or related terms were enriched in the ZT16 data (Table 1); they might also reflect immune function as many immunity-relevant proteins are cleaved for activation. Indeed, Ance-4 is related to metaloproteases, and Drosomycin (Drs) is an innate immunity peptide; both mRNAs are cluster targets by S2 cell criteria (Supplementary Figure 4). Both of these functions, metabolism/feeding and innate immunity, are consistent with the relatively prominent fat body expression of the cluster miRNAs (Fig. 3C). It is however possible that the proteolysis term is unrelated to immune function and that the difference between the ZT4 and ZT16 terms just reflects circadian regulation (see below).
Table 1. DAVID GO analysis enriched categories for immune function and metabolism.
Total RNA was extracted from wild type, null and cluster over-expression flies at ZT4 and ZT16 time points and microarrays were performed as described by the supplier (Affymetrix). We looked for mRNAs whose levels went up in the null and down in the over-expression flies by at least 1.5 fold and Gene Ontology (GO) analysis was performed using DAVID.
| GO TERM | Number of Genes | Benjamini Value | |
|---|---|---|---|
| Immune Response | 15 | 3.3 E−04 | ZT4 – Null |
| Oxidation Reduction | 26 | 6.4 E−03 | |
| Antimicrobial Humoral Response | 8 | 2.1 E−02 | |
| GO TERM | Number of Genes | Benjamini value | ZT4 – Over-expression |
| Oxidation Reduction | 68 | 2.3 E−07 | |
| Innate Immune Response | 17 | 5.2 E−03 | |
| Peptidase M13, Neprilysin | 10 | 1.0 E−03 | |
| Enzyme Inhibitor Activity | 19 | 3.2 E−03 | |
| GO TERM | Number of Genes | Benjamini value | ZT16 – Null |
| Oxidation Reduction | 86 | 3.7 E−11 | |
| Proteolysis | 96 | 4.3 E−11 | |
| Post-mating Behavior | 15 | 6.8 E−11 | |
| Behavior | 18 | 2.6 E−08 | |
| Peptidase Activity | 93 | 5.0 E−08 | |
| GO TERM | Number of Genes | Benjamini value | ZT16 – Over-expression |
| Oxidation Reduction | 108 | 8.5 E−16 | |
| Extracellular Region | 87 | 6.5 E−12 | |
| Proteolysis | 104 | 1.4 E−09 | |
| Post-mating Behavior | 14 | 8.2 E−09 | |
| Peptidase Activity | 106 | 3.7 E−08 |
The cluster miRNAs modulate immune function and feeding
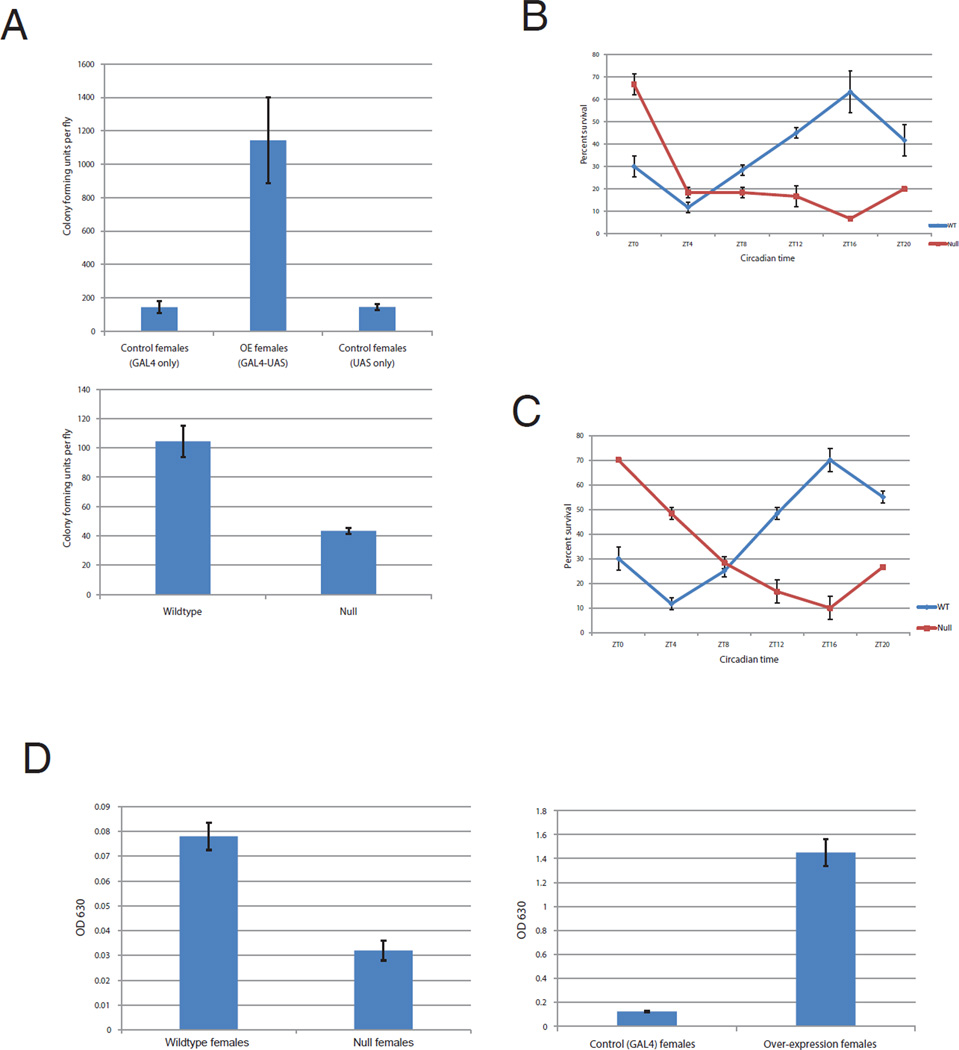
To assay immune function, flies were infected with an attenuated strain of Pseudomonas aeruginosa (PA14 plcs) at the reported peak survival time of ZT17 and assayed for colony forming units per fly (cfu/fly) 24 hr post-infection (Lee and Edery, 2008). Cluster miRNA over-expression caused substantially higher bacterial titers (~ 7 fold) compared to the control strains, whereas the KO strain had lower titers compared to its control strain (Figures 4A). The data suggest that the miRNAs inhibit immune function against this specific pathogen and/or change the peak survival time.
Figure 4. The cluster miRNAs modulate feeding and immunity.
A) To investigate whether miRNA cluster affects immunity, we compared immune response of WT, knock-out and miRNA over-expression flies and assayed the levels of bacteria from the peak survival time point ZT17 (Lee and Edery, 2008). Female flies were entrained under LD conditions for three days and used for injections on the 4th day. 20 flies were infected with P.aeruginosa at the peak survival time point, ~ ZT17 and fly extract was prepared after 24 hr. Serial dilutions of the extract were plated on LB plates with gentamycin and number of colony forming units per fly were calculated (cfu/fly). Null flies showed lower bacterial titer in response to the immune challenge in comparison to the wild type flies. The cluster over-expression flies showed 7 fold more bacterial titers in comparison to the two control flies (tim-gal4 and uas lines). B) Wild type (w1118) and null flies were infected with PA14 plcs at different times of the day during LD cycles as described in Lee and Edery, 2008, and % survival was calculated using values from two different experiments. Error bars indicate standard deviation. C) As in B above except the bacterial stock was diluted to 0.5X of what was used in B. D) Flies over-expressing the cluster eat more in response to starvation where as the null flies eat less under the same conditions. Female flies were entrained for three days under LD conditions and the feeding assay was performed on the 4th day. Null flies eat less food in response to starvation whereas female flies over-expressing the cluster eat ~ 11 times more after starvation in comparison to the control strain (tim-gal4 flies). The error bars indicate standard deviation.
To test the second possibility, the KO and its companion WT strain were infected at different ZTs (zeitgeber time; circadian time but in an LD cycle). The survival curve of the WT strain at 48 hours post-infection was indistinguishable from that originally reported by Lee and Edery, 2008 (Figure 4B and 4C). In contrast, the cluster KO strain had a dramatically shifted curve. The fact that there is still a survival curve in this strain suggests that there are other circadian modulators of immune function and/or that the light cycle has a prominent impact. In either case, the data indicate that the cycling miRNAs contribute to the temporal regulation of immune function, at least against Pseudomonas aeruginosa (Figure 4B, 4C). and see Discussion).
To address food/metabolism, the strain over-expressing the cluster and the KO strain were assayed compared to control strains in a typical starvation-feeding paradigm: flies were assayed for food intake in the 15 min after transfer to normal food following 16 hr of starvation. Flies over-expressing the cluster showed a dramatic increase in food consumption after starvation compared to control flies, whereas KO flies ate less than control flies. This was especially apparent in females (Fig. 4D).
To gain insight into this feeding behavior phenotype, we watched and videotaped flies after the 16 hr of starvation, immediately after transfer from starvation vials to food vials (Supplementary Video 1). A phenotype was particularly apparent in KO female flies, which were hyperactive compared to control flies. It was as if they could not "focus" on feeding. One interpretation is that the miRNAs suppress a foraging response, which is normally induced by 16 hr of starvation. The role of the miRNAs in suppression of foraging could lead to an apparently paradoxical feeding response in a vial: in the KO strain, enhanced foraging would lead to reduced food consumption. In the wild however, low cluster miRNAs levels in the late night-early morning would not only lead to increased foraging but also to increased feeding.
Feeding regulates cluster miRNA expression
This interpretation suggests that the cluster miRNA expression itself might be regulated by food availability, i.e., successful feeding regulates the timing of cluster miRNA expression, which then suppresses the timing of foraging and food intake as well as the extent and timing of immune function and metabolism. To test this prediction, we first assayed miRNA expression under the conditions used for the feeding assays, i.e., flies were starved and then assayed for cluster miRNA expression and cycling.
Under these conditions, cluster miRNAs remain at the low levels characteristic of late night or very early morning, i.e., ZT20 and ZT0, and fail to achieve the normal increase characteristic of ZT4, 8 and 12 (Figure 5A). Cluster pri-miRNA expression was affected similarly (Fig. 5B). Bantam miRNA levels, assayed in parallel, were unaffected by starvation (Fig. 5A), and there was little or no effect on per mRNA levels or cycling (Fig. 5B). The observations suggest that feeding leads to transcription of the cluster and that this regulation occurs downstream of the central clock (see Discussion). Although the tissue heterogeneity of Drosophila heads complicates this interpretation, RNA analysis after dissection of different tissues suggests that it is correct: starvation inhibited cluster miRNA expression in the brain and abdominal fat body as well as the head with little or no effect on bantam or per mRNA cycling (Supplementary Fig 5).
Figure 5. Transcription of the cluster is under food control.
Total RNA was extracted from fly heads across six different circadian time points under ad lib (AL) and starvation (S) conditions. Canton S flies were starved for at least 8 hr before lights on (starting no later than ZT16) and then collected for RNA extraction every four hr after that (e.g. ZT0 flies were starved for 8 hr whereas ZT20 flies were starved for 28 hr. RNA levels were then assayed using RT-PCR and compared to the flies fed (AL). A) The levels of mature miRNAs were assayed under AL and S conditions. The levels showed robust oscillation under AL conditions in WT but remained low under S conditions. Bantam was used as a control and did not show any significant difference under AL and S conditions. B) The levels of the pri-miRNA also did not show any oscillation under S conditions and remained low indicating that food is controlling the expression of the miRNA cluster. The effect of starvation on circadian clock was assayed by measuring the levels of one of the core clock gene transcripts, per whose levels and cycling did not show any significant difference under AL and S conditions. rpl32 was used as a control and its levels were unchanged under the experimental conditions. C) Feeding time determines the phase of the cluster expression. Flies were subjected to a restricted feeding (RF) paradigm and fed for only two hours from ZT10 to ZT12. The levels of mature and pri-miRNA were assayed across circadian time. As shown earlier under AL conditions the levels of the mature and the pri-miRNA peak at around ZT12 and this peak shifts to around ZT0-4 under RF paradigm strongly suggesting that the peak expression is determined by the time of feeding. Circadian clock was assayed by measuring the levels of per mRNA and we did not observe any significant difference under AL and RF conditions. rpl32 was used a control and its levels were unchanged under the experimental conditions. D) Flies were entrained for 3 days under AL conditions and on the 4th day were shifted to RF paradigm as described in materials and methods. Fat-body from the abdomen was purified on the 5th day of RF paradigm and per mRNA levels were assayed from fat-body and fly heads. Restricted feeding had no effect on the phase and the levels of per mRNA. The mRNA for rpl32 was assayed as a control. E) per01 flies were subjected to the RF paradigm as described in materials and methods and the oscillation of the cluster miRNAs was assayed using RT-PCR. The levels of the mature miRNA do not show any oscillation in per01 flies under AL and RF conditions.
The strong effect of starvation suggests that the phase of cluster miRNA transcription might be determined by the phase of feeding. This is because flies feed predominantly in the early morning (Seay and Thummel, 2011; Xu et al., 2008), a few hours before cluster miRNA and pri-miRNA levels begin to rise (Fig. 2A, 2B and 2D). We tested this prediction explicitly, by subjecting flies to a restricted feeding regimen. They were fed for only two hours, between ZT10 and ZT12, which is almost 12 hours out of phase from their normal feeding time peak of ZT0 to ZT2 under ad-libitum conditions (Xu et al 2008).
The results indeed show that the phase of cluster miRNA expression as well as pri-miRNA expression in heads is shifted by almost 12 hours under these restricted feeding conditions, with little or no effect on per mRNA expression (Fig. 5C). There was similarly little effect of restricted feeding on per mRNA isolated from dissected abdominal fat body (Fig 5D), certainly nothing comparable to the dramatic effect on the phase of cluster miRNA expression. This suggests that restricted feeding impacts cluster miRNA expression downstream of the central clock. However, the clock is necessary for this effect of restricted feeding, as it cannot restore cluster miRNA cycling to the arrhythmic per01 strain (Fig 5E; see Discussion).
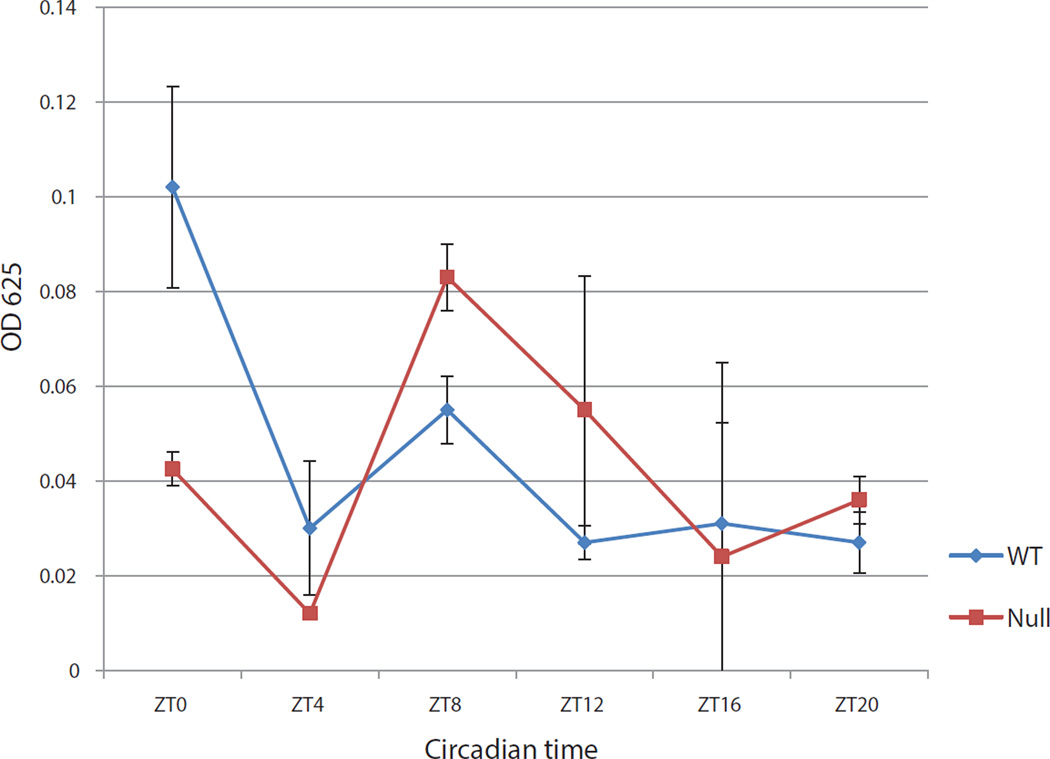
Is this relationship between feeding time and cluster expression reciprocal, i.e., do the cluster miRNAs regulate feeding time under normal ad lib conditions? To address this issue, we used the assay ofXu et al. (2008) to compare feeding time between the cluster knock out strain and its wild-type parental strain under ad-lib conditions. The assay with the wild-type strain recapitulates published observations: there is a quite strong feeding peak at about dawn or ZT0 with a secondary peak during the late day/early night (Fig. 6). The KO strain shows a reduced dawn peak and a larger secondary peak (Fig. 6). This alteration in the feeding curve is very similar to that characteristic of the fully arrhythmic clkjrk strain (Xu et al 2008), indicating that the cluster miRNAs make a substantial contribution to the circadian control of feeding time.
Figure 6. Cluster miRNAs regulate feeding time in Drosophila melanogaster.
Feeding rhythms of wild type and cluster null flies under LD conditions. Following entrainment in LD conditions for 3 days flies were fed blue dye food for 2 hr at six different time points over a 24 hr cycle. Fly bodies were separated from heads and homogenized in PBS and the absorbance was measured at 625 nm. Flies shifted to normal food were used as controls and their absorbance at 625 nm was subtracted from the flies that ate blue-dye food. The experiment was repeated two times and relative food consumption was double plotted across circadian time. The error bars indicate standard deviation.
The data taken together suggest that feeding regulates the circadian cycling of the cluster miRNAs, predominantly at the transcriptional level and downstream of the core clock. The cycling miRNAs then regulate numerous physiological processes including metabolism and immune function as well as foraging and feeding time. A successful feeding event will lead to up-regulation of the cluster miRNAs a few hours later, which then feeds back to down-regulate metabolic and immune functions as well as foraging and feeding functions characteristic of the late-night and early morning. Near the end of the night, miRNA levels fall to minimal values, which lead to an up-regulation of these functions. This miRNA-mediated post-transcriptional circuit allows the fly to properly regulate the daily cycle of feeding and other physiological functions (graphical abstract).
DISCUSSION
Using Illumina deep-sequencing of small RNAs from Drosophila heads, we identified a small set of miRNAs that undergo robust circadian cycling. We focused on six of them, which appear to be synthesized together on a single pri-miRNA and peak at the end of the day. Although the data implicate the activity of a circadian transcriptional activator or repressor in the regulation of the six cluster miRNAs, the clock probably also contributes to the short half lives required for robust miRNA cycling. RNA profiling after manipulation of feeding indicates that the levels and cycling of the cluster miRNAs are sensitive to nutrition and/or starvation and that target functions include metabolism and innate immunity. Indeed, over-expression and especially deletion phenotypes indicates a robust miRNA effect on immunity and feeding. The data taken together suggest that the circadian clock regulates the timing of food intake, which regulates cluster miRNA cycling. These oscillations in turn regulate the daily cycling of multiple physiological processes. As feeding up-regulates the cluster miRNAs with a likely down-regulation of physiologically relevant mRNAs, we suggest that a successful feeding bout up-regulates transcription of the cluster miRNAs to modulate mRNA translation associated with starvation, feeding and stress. As feeding resumes when miRNA levels are low, the data suggest that the cluster miRNAs and their timing contribute to a feedback circuit that regulates daily feeding.
It may be relevant that the all 6 cluster miRNAs have low read numbers, explaining perhaps why they escaped detection in previous less sensitive attempts to identify cycling miRNAs (Yang et al., 2008). There are 22 other miRNAs with similar low read numbers; most of these appear non-cycling, making the relationship between the low read numbers and cycling uncertain. Bantam in contrast has an average read number approximately 3500 greater than the peak values of the individual cluster miRNAs, and the previously identified cycling miRNAs, 263a and 263b (Yang et al., 2008), also have much higher read numbers than the cluster miRNAs (Supplementary Figure 6). It is notable that we did not detect cycling of these two miRNAs (all time points were +/− 20% of each other) as compared to an amplitude of at least 5-fold for the cluster miRNAs in the same assay (Fig. 1A and Supplementary Figure 6).
The sequencing indicates low cluster miRNA levels around ZT0 (Figure 1A), and this conclusion is reinforced by the PCR assays (Figure 2A). Nonetheless, the miRNAs could be cyclically processed by U addition to their 3’ ends, precluding an assignment to the genome at these times (Ameres et al., 2010; Ibrahim et al., 2010; Kim et al., 2010). However, we searched the RNA sequencing data without success for partial miRNA sequences with the addition of non-coded U residues (Ameres et al., 2010). This suggests transcriptional regulation, which is also consistent with the very similar phase and amplitude of pri-miRNA cycling.
Although the location of the primers used to assay pri-miRNA (Supplementary Figure 1) suggests a single transcription unit, it is still possible that there are two primary transcripts, one of which includes miRNAs 959-962 and the other miRNAs 963-964. This would explain the presence of miRNAs 963-964 in the KO strain (Fig. 3B). However, these observations can be equally well accommodated by a single transcription start site and regulatory region unaffected by the deletions. In either case, we assume that this regulatory region still functions properly despite the internal additions/deletions created by homologous recombination. It presumably controls the expression of Gal4 in the knock-in strain, which cycles robustly and similarly to cluster miRNA cycling (Fig. 3C). Fat body expression is consistent with the immune and metabolism functions suggested by the mRNA analyses; the latter might include the response to noxious food components including genotoxic substances (Table 1). Feeding and immunity are also indicated by the phenotypic characterization of the KO and over-expression strains. Interestingly, it is not known whether peri-cerebral or abdominal fat body is more important for the expression of immunity-relevant molecules (N. Silverman, personal communication).
The peak pri-miRNA phase implicates a cycling transcriptional activator, with peak activity at about ZT12. Although this timing is compatible with a direct role for the master positive transcription factor CLK-CYC, CLK tiling and ChIP-seq data show no indication of binding to DNA within or near the cluster, which contrasts with other miRNA genes (Abruzzi et al., 2011); K. Abruzzi, personal communication). An activator downstream of the core clock is therefore indicated. Equally possible is cycling of a downstream repressor with minimal activity at about ZT12 (Sancar et al., 2011).
Both of these possibilities are compatible with the starvation response, which reinforces this transcription-centric view. The absence of cycling and low levels of the pri-miRNA as well as the cluster miRNAs (Fig. 5A, B) indicate that starvation freezes the normally cycling activity of the key activator at a low level, or it freezes the key repressor at a high level. Restricted feeding provides a similar indication: activity of the putative activator must increase or the putative repressor decrease a few hours after feeding. Intriguingly, all of this appears to occur downstream of the core oscillator, as per mRNA cycling -- phase as well as amplitude – is largely unaffected by starvation (Fig. 5C).
These results are reminiscent of restricted feeding experiments on mammalian liver (Damiola et al., 2000; Hara et al., 2001). Importantly, they had prominent effects on the core clock within the liver. The prominent difference with mammals may reflect the strong effect of light and cryptochrome on clock gene mRNA oscillations in flies, especially in peripheral tissues (Plautz et al., 1997; Stanewsky et al., 1998); these presumably include the fat body. Consistent with our reasoning are results of a starvation experiment in which RNA from abdominal fat body was assayed. It indeed indicates that the food effects on cluster miRNA transcription occur downstream of the central clock (Supplementary Figure 5). Although this conclusion differs from recent results indicating a major effect of the feeding on the fat body central clock (Xu et al. 2011), those experiments were done in constant darkness. We therefore prefer the interpretation that the central clock “wins” over food under standard light-dark entrainment conditions and that the observed feeding effects on the cluster miRNAs shown here occur downstream of the central clock. However, we cannot rule out the possibility that the peri-cerebral fat body, the major source of the cluster miRNAs in our experiments, is different from abdominal fat body.
The data also point to important post-transcriptional regulation of the cluster miRNAs by the clock. For example, cluster miRNA levels are not only non-cycling in the arrhythmic per01 strain background, but pri-miRNA levels are also relatively high and mature miRNA levels low. One interpretation is that Drosha processing of the cluster pri-miRNA is inefficient in per01 flies. Given the direct role of PER in transcriptional regulation (Menet et al., 2010), cluster miRNA processing efficiency may be coupled to circadian transcription (Obernosterer et al., 2007; Thomson et al., 2006).
In a similar vein, even the short half lives of these six miRNAs may be linked to circadian transcription. The similar pri-miRNA and miRNA circadian profiles suggest cytoplasmic miRNA half-lives of no longer than an hour or two; much longer would give rise to dampened and delayed mature miRNA profiles relative to that of pri-miRNA (Gatfield et al., 2009). Although not unprecedented (Chatterjee and Grosshans 2009; Bail et al. 2010; Krol et al. 2010; Burns et al. 2011; Rissland et al. 2011), rapid miRNA turnover is unusual. Perhaps their common transcription is relevant to their short half-lives. In addition, a relationship of this nature might help explain the miRNA response to starvation in per01 flies. Starvation dramatically increases mature miRNA levels in this strain without a comparable effect on pri-miRNA levels (Supplementary Figure 7), suggesting that the aberrant transcriptional regulation during starvation of the per01 strain may be unable to maintain the coupling of short miRNA half-life to transcription; stable miRNA expression may result as a default state. Consistent with coupling to transcriptional regulation, the cluster miRNAs have long half-lives when expressed in S2 cells under actin promoter control (data not shown).
However, we have no insight into the mechanism responsible for the normal instability of the cluster miRNAs in vivo. 3’ -> 5’ exoribonucleases have been shown to play a role in miRNA degradation in plants and animals (Ramachandran and Chen, 2008), whereas the 5’ -> 3’ exonuclease Xrn2 has been implicated in C. elegans (Chatterjee and Grosshans, 2009). Two recent studies indicate that extensive base pairing between miRNAs and their targets leads to miRNA degradation (Ameres et al., 2010; Cazalla et al., 2010). Although we do not find runs of untemplated nucleotides in our sequencing of the cluster miRNAs (data not shown), it is still possible that target mRNA cycling is necessary for the cluster miRNA short half lives and cycling rather than vice versa.
We also have no particular insight into cluster miRNA mechanism of action. Current dogma dictates that the alterations in miRNA levels should change target mRNA translation and levels. This view is consistent with our microarray assays from the KO and over-expression strains (Table S1). Although these two strains are not complete opposites (the KO strain is missing four of the six miRNAs, whereas the over-expression strain increases levels of all six), there are many mRNAs that move in opposite directions in the two strains: up in the deletion strain and down in the over-expression strain. Despite the likelihood of indirect effects in the two strains, some mRNAs are candidate direct targets of miRNAs 959–962, with good target sequences in their 3’UTRs. Some of these respond in traditional S2 cell reporter gene assays to cluster miRNA addition (Supplementary Figure 4). Taken together with the miRNA fat body expression and the functions of metabolism and immunity as indicated by the coding sequences of putative target mRNAs (Table 1), they may contribute to the feeding and immunity-related phenotypes affected in the deletion and the over-expression strains. However, we have not identified specific mRNA targets that contribute to these phenotypes.
These effects on innate immunity functions, feeding and foraging (Figure 4A, Figure 4B, Figure 4C, Figure 4D, Figure 6 and Supplementary Video 1) indicate that these cycling processes (Lee and Edery, 2008; Seay and Thummel, 2011; Wang et al., 2011; Xu et al., 2008) are influenced by these miRNAs. Given the effect of the cluster knock out on ad libitum feeding (Fig. 6), it is even possible that there is no direct effect of the circadian clock on the timing of feeding; all clock regulation may occur via the cluster miRNAs. As their expression is strongly regulated by feeding (Fig. 5A and 5C), a feedback circuit is indicated (graphical abstract). Its timing must also reflect the temporal parameters that govern the synthesis and turnover of the cluster miRNAs. Although this circuit integrates circadian and environmental timing (graphical abstract), its mechanism is unknown. We speculate that two transcription factors function together on the promoter of the cluster miRNAs: one is a circadian factor required for transcription or perhaps even for cycling transcription, whereas the other is a nutrient-regulated factor that determines the phase of cycling. A major future challenge will be to understand the molecular mechanism that underlies this integration of circadian and environmental regulation.
MATERIALS AND METHODS
Fly strains
clkJRK, tim-GAL4 (yw;tim-GAL4/CyO) and clkAR flies were described previously (Allada et al., 2003; Allada et al., 1998; Kaneko and Hall, 2000). The plasmid pUAST-CLU was generated by cloning a 2.4 kb genomic DNA fragment, 500 bases upstream and downstream of the Dme-miR-959 and Dme-miR-964 respectively, into the XhoI and XbaI sites of pUAST vector. The cloning was confirmed by restriction digestion and conventional sequencing. The transgenic yw;UASCLU/CyO fly line was generated by The Best Gene (thebestgene.com). The transgenic fly line was then crossed to tim-GAL4/CyO to generate timgal4; UAS-CLU stable lines. The miR-959-962 knock out mutant was generated using homologous recombination based ends-out gene targeting (Gong, 2003 Genetics). Upstream and downstream homology arms flanking miR-959-962 cluster were amplified and cloned into the targeting vector pW25-Gal4-attB2 (Weng 2009) Genetics), using primer pairs 5’GCGGCCGCCGCTCGACTATTCTGCACTT and 5'GCGGCCGCTTAATTAATGCACTGCTTTAGCATCCAC for upstream homology arm, and 5’GGCCGGCCTCGTTGGACCAGACAATACACT and 5’GGCCGGCCGGCCAGACAAAAA form downstream homology arm. Donor transgene was obtained by integrating targeting vector into landing strain, ZH-attP-86Fb (Bischof 2007) PNAS) as described in Chen et al., Methods Mol Biol 2011. The targeting vector pW25-Gal4-attB2 would allow Gal4 knock-in at targeted locus, therefore miR-959-962 cluster-Gal4 KO is a Gal4 knock-in mutant and can be used as a Gal4 driver to report the cluster expression pattern. miR-959-962 cluster KOw- was generated from Gal4 knock-in mutant by using Cre/loxP recombination to excise both Gal4 and mini-white genes (Weng 2009) Genetics).
RNA extraction and RT-PCR
Total RNA from fly heads was extracted using TRIzol reagent (Invitrogen) as suggested by the supplier. For RT-PCR analysis 5 µg of total RNA was treated with RQ1 DNase (Promega) as described by the supplier. From this 2 µg was used for reverse transcription reaction using Superscript II and random hexamers. Where required, transcript specific reverse transcription was performed using a gene-specific primer. Cycling parameters for the PCR reaction were 94 °C for 2 min followed by 28 cycles of 94 °C for 30 sec, 55 °C for 45 sec and 72 °C for 1 min. miRNA RT-PCR was essentially performed as described by (Chen et al., 2005). RP49 and Bantam were used as controls for mRNA and miRNA RT-PCRs respectively. Primers used for various gene-expression analyses are described in Table S2.
Preparation of small RNA sequencing libraries
Small RNA sequencing libraries were essentially prepared as described in (Seitz et al., 2008) with one exception. The 2S rRNA was depleted by hybridizing to a DNA oligonucleotide followed by RNaseH treatment. Following depletion of 2S rRNA, 19 to 29 nucleotide long RNAs were gel purified and adapters were ligated at 3’ and 5’ end using Rnl2[(1-249) Ho et al., 2004] and T4 RNA ligase (Ambion) respectively. RNA was PCR amplified using primerscorresponding to the adapters and the PCR products were gel purified on a 2% agarose gel. The libraries were sequenced using the Illumina deep sequencing platform. Reads were aligned to the genome with Seqmap (Jiang and Wong, 2008) using the first 20 bases and allowing at most 1 mismatch. Mature miRNA sequences downloaded from miRBase were then aligned to this subset of reads. Only mature sequences completely contained within the read with at most 2 mismatches were kept. The miRNA reads were then normalized to total reads. Sequencing data are deposited on the Gene Expression Omnibus database under the accession number GSE40981 (GEO, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40981).
Microarrays and Expression Analysis
Total RNA was extracted as described above and the probes for the arrays were prepared as described by the supplier (Affymetrix). Hybridization of the probes, staining and washing was according to the manufacturer’s protocol. The normalized log2 values were obtained from CEL files using the GCRMA algorithm in the Bioconductor analysis package for the R software. Microarray data are deposited on the Gene Expression Omnibus database under the accession number GSE40981 (GEO, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40981).
Feeding assays
The feeding assay was as described in Xu et al., 2008. Briefly, 2 to 5 day old female flies were entrained at 25 °C in LD under ad lib (AL) conditions for three days. On the 4th day in LD, flies were shifted from normal food to either blue-color food (15 % sucrose/1 % agar diets containing 1% FD&C Blue no. 1 McCormick) or non-dye containing food for 2 hours. The flies were homogenized in PBS and centrifuged at 14,000 rpm for 30 minutes. The absorbance of the supernatant was measured at 625 nm. The flies shifted to no-dye containing food served as controls and their absorbance was subtracted from the supernatant of the flies fed with food containing blue dye. Feeding assays in response to starvation were done by starving the flies for 24 h. Female flies were entrained for three days in LD cycle under ad lib conditions and were shifted to starvation vials at ZT0 on the 4th day. Following 24 h starvation flies were shifted to either blue-color food (as above) or non-dye containing food for 15 min. Samples were processed as above and the absorbance was measured at 625 nm.
Pseudomonas aeruginosa infections and survival rates
PA14 plcs strain of P.aeruginosa was a kind gift from Laurence Rahme. P.aeruginosa was cultured as described in Lee and Edery, 2008. Female flies were entrained for three days in LD and infected on the 4th day. Flies were anesthetized using CO2 and a 10 µµm tungsten needle (Ted Pella, Inc.) was used for pricking. Tungsten needle was dipped into the concentrated bacterial culture and pricked gently on the surface of the abdomen to prevent injury to the internal organs. Following infections flies were returned to the same LD conditions and their survival was counted 48 hr post-infection.
Bacterial growth assay
24 hr following infections flies were rinsed once with 70 % ethanol to eliminate bacteria from the surface. Fly extract was prepared by homogenizing flies in ice cold LB media and serial dilutions were plated on LB agar plates containing gentamycin. The plates were incubated at 37 °C overnight and visible colonies on the plates were counted manually. Colony forming units per fly were averaged (Cfu/fly) and plotted for comparison.
S2 cell Transfection Assays
S2 cells were cultured in insect tissue culture media (HyClone) containing 10 % FBS (GIBCO). Cells were seeded in a six-well plate and transfections were performed at ~ 80 % confluence using Cellfectin (Invitrogen) as suggested by the supplier. 50 ng of luciferase reporter constructs were co-transfected with 50 ng of either the wild type or mutated seed sequence pAc-CLU plasmid. 48 hr post-transfection luciferase assays were performed using the Dual-Luciferase Reporter Assay system (Promega).
Statistical analysis
Data from two independent experiments were averaged and plotted in Microsoft Excel. Statistical significance was calculated using standard deviation.
Supplementary Material
01
02
03
04
HIGHLIGHTS.
The miRNA cluster 959-964 shows circadian oscillations.
The cluster pri-miRNA is under circadian transcriptional control.
Cluster transcription is also under feeding control.
The cluster regulates immunity and feeding.
ACKNOWLEDGEMENTS
We thank C. Li and P. Zamore for the small RNA cloning protocol, L. Rahme for the PA14 plcs strain and the fly injection protocol. We thank P. Chilakamarri for help with fly dissections. We are also grateful to S. Kadener, P. Nawathean, M. Marr, N. Lau and members of Rosbash laboratory for critical comments and suggestions. This work was supported in part by NIH grants GM023549 and NS045713 to M.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Kadener S, Nandakumar N, Rosbash M. A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. Embo J. 2003;22:3367–3375. doi: 10.1093/emboj/cdg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, White N, So W, Hall J, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci U S A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, Shibata S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson S, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. The Journal of Comparative Neurology. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One. 2010;5:e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Edery I. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr Biol. 2008;18:195–199. doi: 10.1016/j.cub.2007.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: noncanonical pathways for microRNA production. Mol Genet Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brügger B, Ha N, Sachsenheimer T, Gin E, Wdowik S, Lohmann I, Wieland F, Höfer T, et al. A Global Circadian Repressor Controls Antiphasic Expression of Metabolic Genes in Neurospora. Molecular Cell. 2011;44:687–697. doi: 10.1016/j.molcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Seay DJ, Thummel CS. The circadian clock, light, and cryptochrome regulate feeding and metabolism in Drosophila. J Biol Rhythms. 2011;26:497–506. doi: 10.1177/0748730411420080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5' precision of both MicroRNAs and their miRNA* strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Ko ML, Ko GY. Rhythmic expression of microRNA-26a regulates the L-type voltagegated calcium channel alpha1C subunit in chicken cone photoreceptors. J Biol Chem. 2009;284:25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta M, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, Lee DU, Fu XD, Dong X. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R, Chen YW, Bushati N, Cliffe A, Cohen SM. Recombinase-mediated cassette exchange provides a versatile platform for gene targeting: knockout of miR-31b. Genetics. 2009;183:399–402. doi: 10.1534/genetics.109.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol. 2010;30:197–205. doi: 10.1128/MCB.01154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04