Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity (original) (raw)
Abstract
A consistent clinical feature of amyotrophic lateral sclerosis (ALS) is the sparing of eye movements and the function of external sphincters, with corresponding preservation of motor neurons in the brainstem oculomotor nuclei, and of Onuf’s nucleus in the sacral spinal cord. Studying the differences in properties of neurons that are vulnerable and resistant to the disease process in ALS may provide insights into the mechanisms of neuronal degeneration, and identify targets for therapeutic manipulation. We used microarray analysis to determine the differences in gene expression between oculomotor and spinal motor neurons, isolated by laser capture microdissection from the midbrain and spinal cord of neurologically normal human controls. We compared these to transcriptional profiles of oculomotor nuclei and spinal cord from rat and mouse, obtained from the GEO omnibus database. We show that oculomotor neurons have a distinct transcriptional profile, with significant differential expression of 1,757 named genes (q < 0.001). Differentially expressed genes are enriched for the functional categories of synaptic transmission, ubiquitin-dependent proteolysis, mitochondrial function, transcriptional regulation, immune system functions, and the extracellular matrix. Marked differences are seen, across the three species, in genes with a function in synaptic transmission, including several glutamate and GABA receptor subunits. Using patch clamp recording in acute spinal and brainstem slices, we show that resistant oculomotor neurons show a reduced AMPA-mediated inward calcium current, and a higher GABA-mediated chloride current, than vulnerable spinal motor neurons. The findings suggest that reduced susceptibility to excitotoxicity, mediated in part through enhanced GABAergic transmission, is an important determinant of the relative resistance of oculomotor neurons to degeneration in ALS.
Electronic supplementary material
The online version of this article (doi:10.1007/s00401-012-1058-5) contains supplementary material, which is available to authorized users.
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disorder, in which there is selective loss of motor neurons in the cerebral cortex, brainstem and spinal cord. Symptoms often begin focally but with progression of disease, there is spread of weakness to the limb, bulbar and respiratory muscles. Mean survival from onset is 3 years. On pathological examination, there is loss of lower motor neurons in the ventral horn of the spinal cord, with astrocytic gliosis, and the presence of ubiquitinated inclusions in surviving motor neurons. An upper motor neuron axonopathy is demonstrated by myelin pallor and gliosis of the corticospinal tracts, which may be accompanied by a reduction in cortical pyramidal neurons [17]. Approximately 5 % of ALS cases are familial, and 70 % of these have mutations in C9ORF72, SOD1, TDP43 or FUS. In the majority of ALS cases, the cause of motor neuron degeneration is unknown, although a number of pathogenic processes, including excitotoxicity, oxidative stress, protein aggregation, mitochondrial dysfunction, dysregulation of the cytoskeleton and axonal transport, and inflammation are considered to play important roles [9].
A consistent clinical feature of the disease is the sparing of eye movements and the function of external sphincters, so that patients retain continence. Pathological studies have confirmed that there is relative sparing of the cranial motor nuclei of the oculomotor, trochlear and abducens nerves, and of Onuf’s nucleus in the sacral spinal cord, which innervates the external sphincters of the pelvic floor [36]. Although neuronal numbers are relatively well-preserved in these resistant motor nuclei, pathological changes resembling those seen in anterior horn cells are present to a lesser degree [40, 41]. The earliest and most severe changes in ALS affect motor neurons, but there is increasing recognition of variable clinical and pathological involvement outside the motor system. Fronto-temporal dementia (FTD), or more subtle cognitive deficits may affect patients with ALS [27], with ubiquitinated inclusions in the dentate granule cells and neocortex [33]. Other long tracts, including the dorsal column and spinocerebellar tracts [17] show degenerative changes in some patients. Patients who are treated with ventilation for a prolonged period after the onset of respiratory failure may develop more widespread CNS degeneration including, in some cases, loss of oculomotor neurons on neuropathological examination [13].
Thus ALS is a degenerative disease, in which motor neurons are relatively selectively vulnerable, but other neuronal subgroups are affected in some patients, particularly after a prolonged disease course. Two groups of lower motor neurons: the oculomotor nuclei, and the sacral nucleus of Onuf, are relatively resistant to the disease process. Rodents which overexpress mutant human SOD1 develop motor neuron degeneration, and in this animal model of ALS, oculomotor nuclei are also relatively spared [38]. Studying the differences in properties of neurons that are vulnerable and those that are resistant to the disease process in ALS may provide insights into the mechanisms of neuronal degeneration, and identify targets for therapeutic manipulation.
The oculomotor nucleus innervates four of the six extraocular muscles (EOMs), which display a distinct phenotype, gene expression profile [49] and disease responsiveness [47]. They have a unique composition of six fibre types, distinct from other skeletal muscles, some with very high mitochondrial content and marked fatigue resistance. Such differences may be determined by their distinct embryonic origin, or by demands imposed by the relative complexity of oculomotor control systems, and the specific discharge patterns of oculomotor neurons [46]. Motor unit discharge patterns are a key determinant of skeletal muscle properties, and EOMs and oculomotor neurons interact in a highly specific manner: explants of neonatal extraocular muscle grown in co-culture with the incorrect spinal motor neurons die faster than those grown with oculomotor neurons [48]. The pattern of innervation of EOMs is different from other skeletal muscles. Neuromuscular junctions are distributed throughout the fibre length at a higher density [12], and show some structural differences [23]. 20 % of EOM fibres are innervated by multiple neuromuscular junctions [44]. Oculomotor motor units are amongst the smallest seen in any skeletal muscle, [47], with higher maximum motor neuron discharge rates. Even in the primary position of gaze, 70 % of oculomotor neurons are active, commonly discharging at 100 Hz [50]. This level of activity would be predicted to place a significant metabolic demand on the neuron.
We have used microarray analysis to compare the gene expression profiles of isolated motor neurons from the ALS-resistant oculomotor nucleus and ALS-vulnerable spinal cord in post-mortem tissue from four neurologically normal control subjects. We show that motor neuron subtypes with different susceptibility to the disease process in ALS also have markedly different transcriptional profiles. Oculomotor (OM) and lumbar spinal cord (LSC) motor neurons show differential expression of genes with a function in synaptic transmission, ubiquitin-dependent proteolysis, mitochondrial function, transcriptional regulation, immune system functions, and the extracellular matrix. The most striking differences are seen in genes with a function in synaptic transmission, in particular GABA and glutamate receptor subunits. We have validated these observed changes, using unpublished transcriptional data from the oculomotor nucleus and spinal cord of two other species, obtained from the public repository GEO Omnibus. Furthermore, we demonstrate the functional impact of altered expression of GABA and glutamate receptors on whole-cell currents induced by agonists at GABA and glutamate receptors.
Materials and methods
Case selection
Brain and spinal cord tissue from four neurologically normal human control subjects (Table 1) was obtained from the Sheffield Brain Tissue Bank. Tissue donated for research was obtained with informed consent from the next of kin, and in accordance with the Human Tissue Authority guidelines on tissue donation. Midbrain and lumbar spinal cord sections were collected postmortem, processed according to standard protocols [18], and stored at −80 °C until required. No abnormality of the midbrain or spinal cord was found at postmortem on full neuropathological examination.
Table 1.
Characteristics of cases
| Case number | Sex | Age at death | Cause of death | Postmortem delay time (h) |
|---|---|---|---|---|
| 1 | M | 64 | Ischaemic heart disease | 12 |
| 2 | M | 64 | Empyema | 30 |
| 3 | F | 59 | Pneumonia | 5 |
| 4 | M | 67 | Hepatocellular carcinoma | 63 |
Tissue preparation and laser capture microdissection
Frozen tissue sections were mounted in Cryo-M-Bed embedding compound (Bright UK), and 10 μm sections cut in a cryostat at −22 °C. Frozen sections were thaw mounted onto slides at room temperature, fixed in 70 % ethanol, washed in DEPC-treated water, and stained for 1 min in a solution of 0.1 % w/v Toluidine Blue in 0.1 M sodium phosphate. They were then washed and dehydrated through graded ethanol concentrations (70, 90 and 100 %), and xylene. OM and LSC motor neurons, identified by staining, anatomical location, size and morphology, were isolated on Capsure Macro LCM caps using the Arcturus PixCell II laser capture microdissection system (Arcturus Bioscience). Approximately 800 successful laser capture firings were made from the spinal cord and 800 from the OM nucleus of each case (supplementary figure 1). The procedure of laser capture microdissection does not prevent the collection of small amounts of adjacent neuropil and glial cells, but significantly enriches the extracted RNA for motor neuron-expressed transcripts.
RNA isolation and amplification, and Affymetrix GeneChip processing
From each sample >50 ng RNA was extracted using the PicoPure™ RNA isolation kit (Arcturus), and amplified using a 2-cycle linear amplification process, with the GeneChip two-cycle target labelling and control kit (Affymetrix), and MEGAscript® T7 kit (Ambion). The linear amplification technique has been shown to generate highly reproducible gene expression profiles from small starting quantities of RNA [32]. This procedure produced 50–100 μg of biotin-labelled antisense RNA for each sample. 15 μg amplified cRNA was fragmented by heating to 94 °C for 20 min, and spiked hybridization controls were added. Each sample was hybridized to one GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix) and scanned in the GeneChip Scanner 3000 to detect fluorescent hybridization signals. We carried out quality control (QC) measures, according to Affymetrix protocols, to ensure that RNA was of sufficient quality and was matched between samples. CEL files generated by the Affymetrix GeneChip Operating System were taken forward for microarray data analysis.
Microarray data analysis
We wanted to detect differentially expressed transcripts between the group of oculomotor samples and the group of spinal cord samples. The bioconductor1 [10] package PUMA [45] was used to carry out the normalization and differential expression analysis. PUMA is a Bayesian probabilistic model for probe-level analysis that takes into account the probe-level measurement error while estimating the gene expression levels. This measurement error is propagated downstream into the analysis, for instance when detecting differentially expressed transcripts This has been shown to increase the accuracy in detecting differential expression, making the model more resistant to outliers even at small sample sizes [29]. The output of this procedure is the posterior probability (PPLR value), estimated using Bayesian inference, of a transcript’s differential expression in the group of oculomotor samples and the group of spinal cord samples. Since this computation is essentially a univariate test, care must be taken to account for multiple hypotheses testing. For this reason, we computed q values from the posterior probabilities as suggested in [52]. Genes that were significantly (q value less than 0.001) differentially expressed were assigned Gene Ontology (Gene Ontology project; http://www.geneontology.org/ [1]) and Kegg Pathway (Kyoto Encyclopaedia of Genes and Genomes; http://www.genome.jp/kegg/ [20]) annotations, and gene ontology (GO) enrichment analysis was performed using DAVID software (NIAID/NIH; http://david.abcc.ncifcrf.gov/summary.jsp, [6]).
Analysis of differential expression of oculomotor nucleus and spinal cord tissue of rat and mouse
Two datasets were obtained from the Gene Expression Omnibus public functional genomics data repository (http://www.ncbi.nlm.nih.gov/geo/). Dataset GSE3305 analysed total RNA extracted from OM nucleus and spinal cord of rats at 6, 18 and 30 months of age, using TRIzol. Tissues from four animals were combined in each RNA sample. Biotinylated RNA samples were hybridized to rat RA230 Affymetrix microarray chips (n = 18). Dataset GSE3343 analysed RNA extracted from the oculomotor nucleus and spinal cord of mice at 10 weeks of age, hybridized to mouse 430A Affymetrix microarray chips (n = 6). The data were normalized, and probe-level analysis conducted using MAS5 software, and made freely available to the scientific community. For the purposes of the present study, as there were relatively few significant differences in the gene expression profile between the three rat age groups, the data from 6, 18 and 30 months were combined. As the .CEL files were unavailable in the repository for PUMA analysis, differential expression between oculomotor and spinal tissue was determined using an unpaired t test.
Quantitative PCR
RNA extracted from laser-captured OM and LSC motor neurons as above, which was not required for hybridization to microarray chips, was taken forward for use in quantitative PCR to verify expression levels of GABRA1. cDNA was synthesized using Superscript II reverse transcriptase, according to manufacturer’s protocol (Invitrogen). QPCR was performed using 12.5 ng cDNA, 1× SYBR Green PCR master mix (Applied Biosystems), 900 nM forward primer (CCTTCCAGACTTCTCATGGCTAAC) and 600 nM reverse primer (TAGCAGGAAGCAGACTAATAAGAAATATTC), to a total volume of 20 μl. After an initial denaturation at 95 °C for 10 min, templates were amplified by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, on an MX3000P Real-Time PCR system (Stratagene). Gene expression values, calculated using the ΔΔ_C_ t calculation (ABI PRISM 7700 Sequence Detection System protocol; Applied Biosystems), were normalized to β-actin expression, and statistical analysis performed using an unpaired t test.
Preparation of acute spinal cord and midbrain slices for patch clamp recording
Adult male Sprague-Dawley rats were anaesthetized with sodium pentobarbital (50 mg kg−1) and decapitated according to the UK Animal (Scientific Procedures) Act 1986 guidelines. Brainstem was isolated, glued on its rostral end to the stage of a vibroslicer, and sliced from the caudal end to the midbrain region of OM nucleus, identified by anatomical landmarks. Two or three 300-μm-thick transverse slices through the OM nucleus were obtained per preparation. The spinal cord was isolated, and 300 μm transverse sections prepared from the lumbar limb expansion, using a McIlwain tissue chopper. Slices were maintained in continuously bubbled (95 % O2/5 % CO2) bicarbonate buffered saline for at least 1 h prior to recordings.
Electrophysiology
Whole-cell electrophysiological experiments were recorded as previously described [39]. The location of electrode placement for OM neuron recording is shown in supplementary figure 2. The constituents of all buffers used are detailed in the supplementary experimental procedures. Voltage clamp recordings were performed using an Axon Multi-Clamp 700B amplifier (Axon Instruments) using unpolished borosilicate pipettes placed at the cell soma. Pipettes had a resistance of 2–4 MΩ when filled with intracellular solution. Pipettes filled with high concentrations of Cl− for GABA-induced current recordings were used to maintain the Cl− equilibrium potential close to 0 mV, thereby facilitating the observation of GABAR-mediated whole-cell currents at resting potentials. Cs+ in the pipette solution would block K+-dependent membrane conductance. Cells were accepted for study if a stable seal formed with a whole-cell resistance of at least 120 MΩ and a series resistance of <10 MΩ. Receptors were activated by focal perfusion of agonists from a micropipette with its tip located 30–50 μm from the cell. Three cells were used for dose–response recordings for AMPA (5 μM to 5 mM) or kainate (50 μM to 50 mM)-induced whole-cell currents in OM and LSC motor neurons. Currents were recorded in extracellular perfusion buffer with 20 mM extracellular Na+ at −60 mV. Na+ was reduced from 125 mM in normal extracellular solution to 20 mM to reduce the driving force for agonist-evoked current. 100 μM AMPA and 1 mM kainate, which were close to the EC50 from the dose–response recordings, were used to measure Ca2+ permeability of AMPA receptors in six cells per agonist, in Na+-free extracellular solutions containing 50 mM Ca2+. Three cells were used for dose–response recording for GABA (0.1–100 mM)-induced whole-cell currents in OM and LSC motor neurons. 10 mM GABA, which was close to the EC50 of the dose–response recordings, was used for repeat recording in six cells. All extracellular solutions were supplemented with MK-801 (10 μM), tetrodotoxin (1.0 μM), and Cd2+ (100 μM) to block NMDA receptors, voltage-gated Na+ channels, and Ca2+ channels, respectively. Cells were held at a membrane potential of −60 mV. All recordings were performed at room temperature 21 °C. Current recordings were sampled onto an IBM-PC-compatible computer using pClamp10 software (Axon). Data were filtered at 3 kHz and sampled at 20–40 kHz.
Results
Overall patterns of differential gene expression
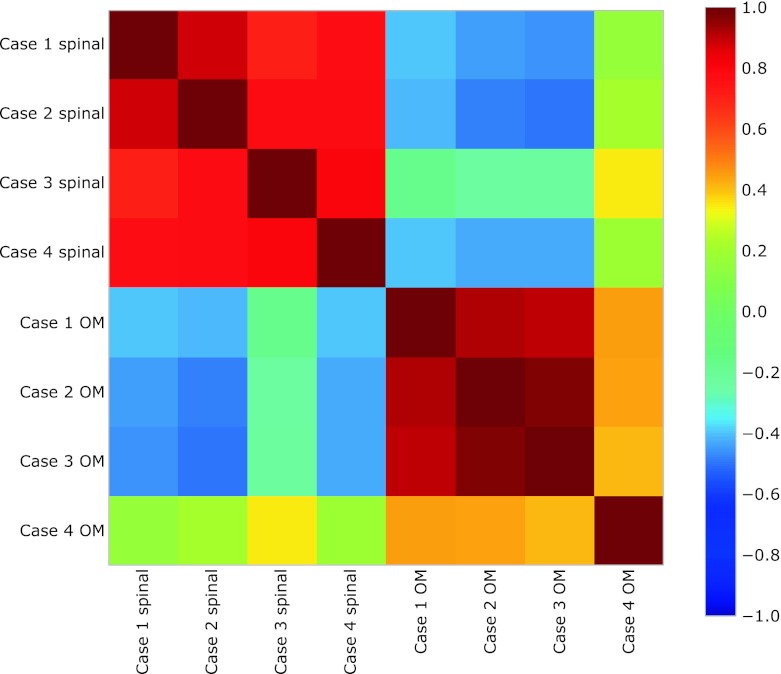
A total of 1,521 unique named genes were upregulated in OM neurons, and 236 in LSC motor neurons, at a significance level of q < 0.001, which was the threshold used for downstream analysis (Table 2). OM and LSC motor neurons cluster according to their gene expression profiles, as shown in the covariance plot (Fig. 1).
Table 2.
Numbers of differentially expressed probe sets and PPLR values at different thresholds of q
| q value threshold | Upregulated in oculomotor neurons | Upregulated in spinal motor neurons | ||
|---|---|---|---|---|
| No. of probe sets | PPLR values | No. of probe sets | PPLR values | |
| 1.0e−5 | 957 | <7.66e−5 | 209 | <1.17e−4 |
| 1.0e−4 | 1,417 | <0.00060 | 265 | <0.000899 |
| 1.0e−3 | 2,196 | <0.0059 | 340 | <0.011 |
Fig. 1.
Covariance matrix for the gene expression dataset, evaluated using only the top 100 differentially expressed genes. The samples on both axes are ordered according to the tissue type. The two visible blocks correspond to the two groups, indicating that the most differentially expressed genes are able to capture the difference between oculomotor and spinal cord samples. Each square in the plot shows the correlation between each pair of samples. Warm colours correspond to a high positive correlation, while cold colours correspond to a high negative correlation
Gene ontology analysis
Gene ontology (GO) analysis was performed to determine the functional categories over-represented amongst significantly differentially expressed genes. A significant proportion of genes expressed at higher levels in OM neurons belonged to the functional clusters of synaptic transmission, ubiquitin-mediated protein degradation, and mitochondrial oxidative phosphorylation (Fig. 2a; Table 3). Genes involved in all stages of ubiquitin-mediated proteolysis, and genes with mitochondrial functions, including the respiratory chain complex were upregulated in OM neurons (Supplementary tables 1 and 2). Three KEGG pathways relevant to neurodegenerative diseases (Huntington’s disease, Alzheimer’s disease and Parkinson’s disease) were also enriched amongst genes upregulated in OM neurons. Although ALS was not an enriched functional category, there was upregulation in OM neurons of the expression of a number of genes in which mutations are known to cause motor neuron degeneration, including C9ORF72 (FC 2.56, q = 4.0E−6), BSCL2 (FC 3.09, q = 0.003), optineurin (FC 2.0, q = 0.001), spastin (FC 2.4, q = 2.95E−6), and VAPB (FC 1.8, q = 1.0E−5).
Fig. 2.
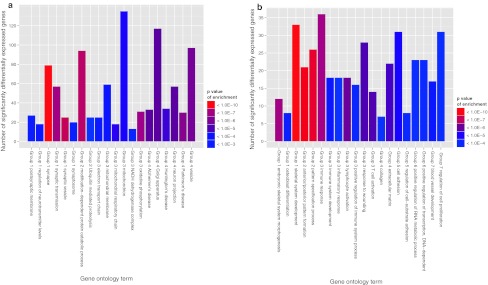
Numbers of significantly differentially expressed genes assigned to enriched gene ontology terms. The 20 most enriched KEGG pathways and GO terms in the categories of biological process and cell component are shown. Redundant terms were removed for clarity. a Genes upregulated in oculomotor neurons (q value for enrichment <0.05). Group 1 synaptic function, Group 2 ubiquitin-mediated proteolysis, Group 3 oxidative phosphorylation and mitochondrial function, Group 4 other. b Genes upregulated in spinal motor neurons (q value for enrichment <0.005). Group 1 skeletal development, Group 2 embryonic pattern formation, Group 3 immune system processes, Group 4 the extracellular matrix, Group 5 cell adhesion, Group 6 regulation of transcription, Group 7 other
Table 3.
Gene ontology enrichment analysis
| Gene ontology term | Count | P value | q value | Fold enriched | |
|---|---|---|---|---|---|
| GO terms enriched in genes upregulated in oculomotor neurons | |||||
| 1. Synaptic function | GO:0045202—synapse | 79 | 2.60E−14 | 1.32E−10 | 2.50 |
| GO:0008021—synaptic vesicle | 25 | 1.91E−08 | 2.42E−05 | 3.70 | |
| GO:0007268—synaptic transmission | 57 | 1.98E−07 | 0.00012 | 2.07 | |
| GO:0045211—postsynaptic membrane | 27 | 0.000125 | 0.015 | 2.25 | |
| GO:0019717—synaptosome | 20 | 0.00013 | 0.015 | 2.65 | |
| GO:0001505—regulation of neurotransmitter levels | 18 | 9.25E−05 | 0.012 | 2.90 | |
| 2. Ubiquitin-dependent proteolysis | GO:0019941—modification-dependent protein catabolic process | 94 | 4.57E−08 | 3.86E−05 | 1.77 |
| hsa04120: ubiquitin-mediated proteolysis | 25 | 0.00048 | 0.043 | 2.13 | |
| 3. Mitochondrial function | hsa00190: oxidative phosphorylation | 31 | 2.85E−07 | 0.00014 | 2.78 |
| GO:0005746—mitochondrial respiratory chain | 18 | 2.94E−05 | 0.0055 | 3.16 | |
| GO:0005739—mitochondrion | 135 | 4.15E−05 | 0.0075 | 1.40 | |
| GO:0031966—mitochondrial membrane | 59 | 8.51E−05 | 0.012 | 1.68 | |
| GO:0022900—electron transport chain | 25 | 9.91E−05 | 0.013 | 2.37 | |
| GO:0030964—NADH dehydrogenase complex | 13 | 0.00020 | 0.020 | 3.48 | |
| 4. Other | hsa05012: Parkinson’s disease | 30 | 6.85E−07 | 0.00031 | 2.73 |
| GO:0031982—vesicle | 97 | 1.39E−06 | 0.00050 | 1.63 | |
| GO:0005794—Golgi apparatus | 117 | 4.68E−06 | 0.0013 | 1.51 | |
| GO:0043005—neuron projection | 57 | 5.05E−06 | 0.0013 | 1.87 | |
| hsa05010: Alzheimer’s disease | 33 | 5.14E−06 | 0.0013 | 2.36 | |
| hsa05016: Huntington’s disease | 34 | 1.68E−05 | 0.0034 | 2.20 | |
| GO terms enriched in genes upregulated in spinal motor neurons | |||||
| 1. Skeletal system | GO:0001501—skeletal system development | 33 | 1.09E−16 | 3.17E−13 | 6.30 |
| GO:0048704—embryonic skeletal system morphogenesis | 12 | 1.65E−09 | 6.01E−07 | 12.83 | |
| GO:0001649—osteoblast differentiation | 8 | 4.69E−06 | 0.00031 | 11.61 | |
| 2. Ant/post specification | GO:0009952—anterior/posterior pattern formation | 21 | 1.14E−13 | 1.66E−10 | 9.14 |
| GO:0007389—pattern specification process | 26 | 1.62E−12 | 1.57E−09 | 5.93 | |
| 3. Immune response | GO:0006955—immune response | 36 | 1.97E−09 | 6.37E−07 | 3.18 |
| GO:0046649—lymphocyte activation | 18 | 2.76E−08 | 7.31E−06 | 5.51 | |
| GO:0042110—T cell activation | 14 | 1.39E−07 | 2.18E−05 | 6.77 | |
| GO:0009611—response to wounding | 28 | 1.52E−07 | 2.21E−05 | 3.22 | |
| GO:0002684—positive regulation of immune system process | 16 | 8.57E−06 | 0.000509 | 4.10 | |
| GO:0002520—immune system development | 18 | 2.88E−06 | 0.000215 | 3.97 | |
| GO:0006954—inflammatory response | 18 | 2.46E−05 | 0.00116 | 3.37 | |
| 4. Extracellular matrix | GO:0031012—extracellular matrix | 22 | 2.42E−07 | 3.36E−05 | 3.86 |
| GO:0005581—collagen | 7 | 2.00E−05 | 0.000971 | 12.12 | |
| 5. Cell adhesion | GO:0007155—cell adhesion | 31 | 1.21E−06 | 0.000113 | 2.70 |
| GO:0010810—regulation of cell-substrate adhesion | 8 | 8.79E−06 | 0.000512 | 10.60 | |
| 6. Transcription | GO:0045893—positive regulation of transcription, DNA-dependent | 23 | 1.15E−05 | 0.000598 | 2.94 |
| GO:0051254—positive regulation of RNA metabolic process | 23 | 1.32E−05 | 0.000663 | 2.91 | |
| 7. Other | GO:0001568—blood vessel development | 17 | 2.68E−06 | 0.000205 | 4.23 |
| GO:0042127—regulation of cell proliferation | 31 | 1.26E−05 | 0.000644 | 2.40 |
In LSC motor neurons, upregulated genes were enriched for the functions of skeletal system development, anterior/posterior pattern formation, immune system processes, the extracellular matrix, cell adhesion, and transcription (Fig. 2b; Table 3). Transcriptional regulators expressed at a higher level in LSC motor neurons included 17 of the 39 homeobox (Hox) genes which operate distinct genetic programs along the anterior/posterior axis (Supplementary table 3), and MALAT1, a non-coding RNA recently identified as a principle binding target of TDP43, and thought to play a role in the recruitment of splicing factors [55], which is 8.7-fold upregulated in LSC motor neurons relative to OM neurons (q value <1.0E−17). Homeobox genes engrailed 1 and 2, which are known molecular markers of midbrain tissue [5] were upregulated in OM neurons. Extracellular matrix components expressed at higher levels in LSC motor neurons included seven different collagen subunits (Supplementary table 4).
The majority of gene ontology categories enriched in OM and LSC-upregulated genes identify functions which are known to play a role in the pathogenesis of ALS. It was not possible within the scope of this study to explore the potential role that each functional category may play in the relative resistance of OM neurons to the degenerative process. We focussed on the differences observed in the most highly enriched functional category of synaptic transmission.
Differential expression of genes with a function in synaptic transmission
Glutamate-mediated neurotransmission
A number of glutamate receptor subunits were upregulated in OM neurons (Table 4). The AMPA glutamate receptor subtype consists of four subunits, GluR1–GluR4, and the presence of the GluR2 subunit determines the calcium permeability of the receptor. In the absence of GluR2, the AMPA receptor–ion channel complex becomes permeable to calcium. In this study, we showed upregulation of the GluR2 subunit in resistant OM neurons, relative to the vulnerable lumbar motor neuron population. The GluR1 subunit was also expressed at higher levels, as were the synaptic scaffolding proteins, GRIP1 and 2, which interact with GluR2 and GluR3 to stabilize these receptor subunits at the synaptic membrane [7]. There was no differential expression of NMDA receptors.
Table 4.
Upregulated glutamate receptor subunits and transporters in human oculomotor neurons
| Gene name | Symbol | Probe id | Fold change | PPLR value | q value |
|---|---|---|---|---|---|
| Glutamate receptor, ionotropic, AMPA 1 | GLUR1 | 209793_at | 4.82 | 3.49E−15 | 0 |
| 211520_at | 3.20 | 5.60E−06 | 6.59E−07 | ||
| Glutamate receptor, ionotropic, AMPA 2 | GLUR2 | 205358_at | 2.31 | 1.14E−05 | 1.36E−06 |
| 236538_at | 3.31 | 0.002008 | 0.00031 | ||
| Glutamate receptor, ionotropic, kainate 2 | GRIK2 | 213845_at | 5.34 | 5.09E−07 | 4.74E−08 |
| Glutamate receptor, metabotropic 3 | GRM3 | 205814_at | 2.00 | 3.58E−08 | 3.42E−09 |
| Glutamate receptor, metabotropic 7 | GRM7 | 207548_at | 2.50 | 2.30E−06 | 2.72E−07 |
| Glutamate receptor interacting protein 1 | GRIP1 | 235957_at | 2.57 | 6.37E−06 | 7.33E−07 |
| Glutamate receptor interacting protein 2 | GRIP2 | 216481_at | 3.80 | 3.19E−09 | 2.23E−10 |
GABA-mediated neurotransmission
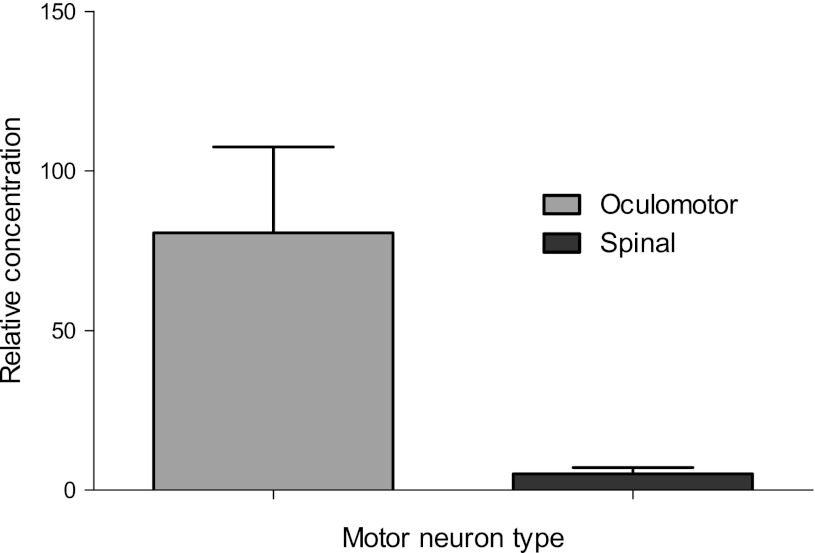
GABA is the most widely distributed inhibitory neurotransmitter in the mammalian CNS. It signals through GABAA ligand-gated chloride channels, composed of different combinations of 16 subunits, and GABAB metabotropic receptors, which are heterodimers of subunits 1 and 2. In OM neurons, there was upregulation relative to LSC motor neurons, of six GABAA receptor subunits and of GABAB receptor subunit 2 (Table 5). The differential expression of GABRA1 was confirmed using quantitative PCR (Fig. 3). Relative concentration of GABRA1 was 16-fold higher in OM neurons compared to LSC motor neurons, when determined by QPCR.
Table 5.
Upregulated GABA receptor subunits and genes in human oculomotor neurons
| Gene name | Symbol | Probe id | Fold change | PPLR value | q value |
|---|---|---|---|---|---|
| Gamma-aminobutyric acid (GABA) A receptor, alpha 1 | GABRA1 | 244118_at | 7.56 | 1.16E−07 | 1.08E−08 |
| Gamma-aminobutyric acid (GABA) A receptor, beta 1 | GABRB1 | 207010_at | 5.21 | 5.35E−08 | 4.57E−09 |
| Gamma-aminobutyric acid (GABA) A receptor, beta 2 | GABRB2 | 155712 _at | 8.76 | 3.78E−20 | 0 |
| 242344_at | 9.21 | 2.61E−05 | 3.21E−06 | ||
| Gamma-aminobutyric acid (GABA) A receptor, epsilon | GABRE | 204537_at | 8.18 | 1.54E−08 | 1.62E−09 |
| Gamma-aminobutyric acid (GABA) A receptor, gamma 1 | GABRG1 | 241805_at | 2.79 | 7.27E−05 | 9.25E−06 |
| Gamma-aminobutyric acid (GABA) receptor, theta | GABRQ | 238123_at | 26.3 | 1.13E−08 | 9.84E−10 |
| Gamma-aminobutyric acid (GABA) B receptor, 2 | GABBR2 | 209990_at | 3.10 | 4.93E−09 | 4.30E−10 |
| 211679_at | 2.33 | 4.83E−06 | 5.75E−07 |
Fig. 3.
Concentration of GABRA1 relative to β-actin in laser-captured motor oculomotor and spinal motor neurons determined by QPCR (2 cases, n = 6 replicates). Error bars represent SEM
Verification of microarray findings by QPCR and in other species
Limited motor neuron numbers in the oculomotor nucleus precluded the verification of differential expression by QPCR in a large panel of genes. We therefore aimed to confirm the changes found in human tissue, using microarray datasets from two other species—rat and mouse, which were obtained from a public repository. These studies were performed on whole oculomotor nucleus and whole spinal cord, and the gene expression profiles obtained are therefore from a more heterogeneous cell population than that of our human study, which was highly enriched for motor neurons, due to the laser capture process. Nevertheless, the differential gene expression profiles obtained from mouse and rat oculomotor nucleus versus spinal cord, and those from human OM neurons versus LSC motor neurons were highly correlated. On gene ontology analysis, only 2 of the top 20 GO terms enriched amongst genes upregulated in OM neurons and LSC motor neurons, respectively, were unique to the human study, the others all being significantly enriched in the mouse and/or rat study. In both other species, functional categories relating to synaptic transmission were the most highly enriched in differentially expressed genes (Supplementary table 5). Homologues of 12 of the 16 GABA and glutamate receptor subunits that were significantly differentially expressed between human OM neurons and LSC motor neurons were also significantly differentially expressed between oculomotor nucleus and spinal cord in mouse and/or rat tissue (Table 6).
Table 6.
Significantly upregulated glutamate and GABA receptor subunits and genes in mouse and rat oculomotor nucleus
| Gene name | Symbol | Mouse | Rat | ||||
|---|---|---|---|---|---|---|---|
| Probe id | Fold change | P value | Probe id | Fold change | P value | ||
| Glutamate receptor, ionotropic, AMPA 1 | GLUR1 | 1435239_at | 2.49 | 0.0024 | 1371013_at | 2.05 | 4.79E−06 |
| 1448972_at | 1.83 | 0.0379 | |||||
| Glutamate receptor, ionotropic, AMPA 2 | GLUR2 | 1387171_at | 2.19 | 4.86E−09 | |||
| 1368401_at | 2.02 | 4.23E−08 | |||||
| Glutamate receptor, metabotropic 7 | GRM7 | 1369781_at | 1.29 | 0.0011 | |||
| Glutamate receptor interacting protein 1 | GRIP1 | 1376988_at | 1.48 | 0.0101 | |||
| Glutamate receptor interacting protein 2 | GRIP2 | 1387954_at | 1.29 | 0.0011 | |||
| 1388016_at | 4.95 | 2.38E−06 | |||||
| Gamma-aminobutyric acid (GABA) A receptor, alpha 1 | GABRA1 | 1421281_at | 8.24 | 1.98E−04 | |||
| 1436889_at | 5.62 | 1.85E−05 | |||||
| Gamma-aminobutyric acid (GABA) A receptor, beta 1 | GABRB1 | 1369904_at | 2.30 | 3.82E−04 | |||
| 1369371_at | 1.96 | 1.42E−08 | |||||
| 1388030_at | 1.71 | 5.67E−04 | |||||
| 1388039_at | 1.42 | 9.54E−07 | |||||
| 1375720_at | 1.39 | 6.49E−07 | |||||
| Gamma-aminobutyric acid (GABA) A receptor, beta 2 | GABRB2 | 1450319_at | 2.39 | 0.0059 | 1369818_at | 2.51 | 1.93E−06 |
| 1387383_at | 1.64 | 3.11E−05 | |||||
| 1368952_at | 1.38 | 0.00285 | |||||
| Gamma-aminobutyric acid (GABA) A receptor, epsilon | GABRE | 1388049_at | 1.84 | 3.19E−05 | |||
| Gamma-aminobutyric acid (GABA) A receptor, gamma 1 | GABRG1 | 1387706_at | 1.25 | 0.0300 | |||
| Gamma-aminobutyric acid (GABA) receptor, theta | GABRQ | 1387706_at | 1.25 | 0.0300 | |||
| Gamma-aminobutyric acid (GABA) B receptor, 2 | GABBR2 | 1370701_at | 1.63 | 0.0279 |
Glutamate and GABA receptor-mediated currents in lumbar spinal cord and oculomotor neurons in slice preparations from rat
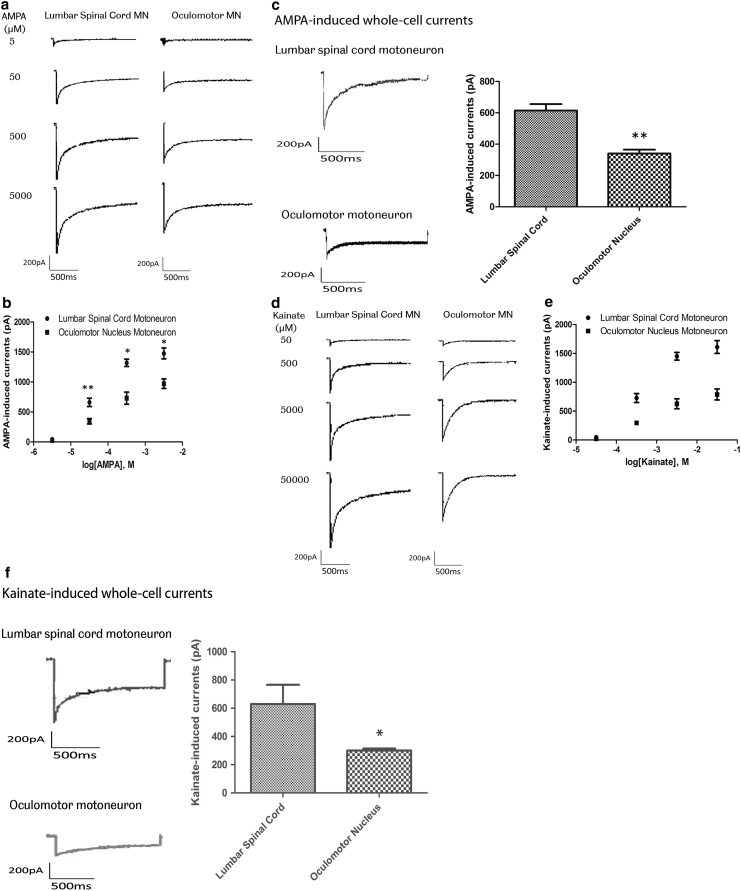
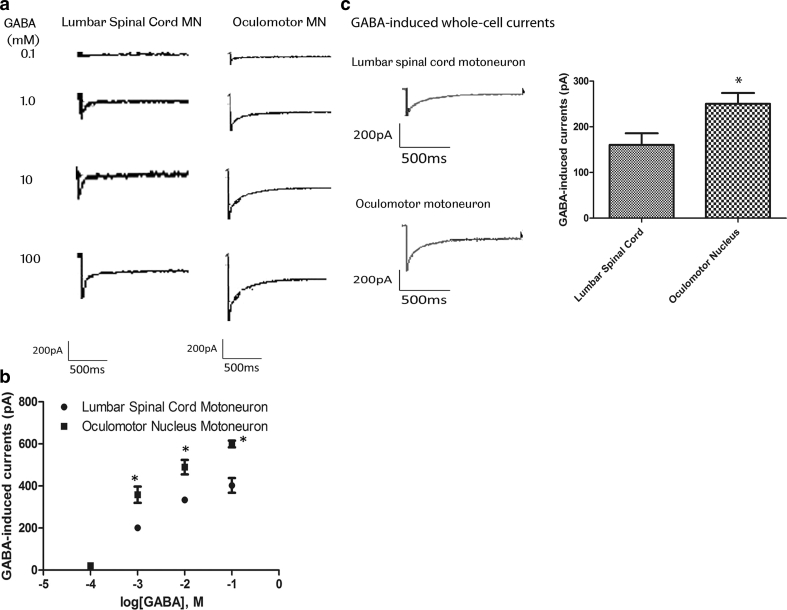
The functional significance of the observed changes in glutamate and GABA receptor subunits was determined using patch clamp recording of whole-cell currents in slice preparations of the rat lumbar spinal cord and midbrain. To assess AMPA receptor-mediated whole-cell currents, three cells were used for dose–response recordings for AMPA- and kainate-induced whole-cell currents in OM and LSC motor neurons. 100 μM AMPA and 1 mM kainate were used to measure Ca2+ permeability of AMPA receptors in six cells per agonist. Both AMPA- and kainate-induced currents were significantly larger in LSC motor neurons than in OM neurons. AMPA-induced currents were 614.00 ± 102.21 pA in LSC motor neurons and 340.67 ± 58.80 pA in OM neurons. Kainate-induced currents were 629.50 ± 334.57 in LSC motor neurons and 300.50 ± 34.22 in OM neurons (Fig. 4). To assess GABA receptor-mediated whole-cell currents, three cells were used for dose–response recording for GABA. 10 mM GABA was used for repeat recording in six cells. Amplitudes of whole-cell currents evoked by 10 mM GABA were significantly smaller in LSC motor neurons than in OM neurons. GABA-induced currents were 160.33 ± 62.43 pA in LSC motor neurons and 250.33 ± 58.03 pA in OM neurons (Fig. 5).
Fig. 4.
a and b Concentration–response relation for AMPA-induced whole-cell currents in lumbar spinal cord and oculomotor neurons. Currents were recorded in 20 mM extracellular Na+ at −60 mV, in response to AMPA concentrations ranging from 5 μM to 5 mM. a Representative current traces elicited by AMPA. b Each point represents mean ± SEM from three cells (*P < 0.05, **P < 0.01, Student’s t test). EC50 values estimated from fits to pooled data were 118.6 μM for lumbar spinal cord motor neurons and 123.2 μM for oculomotor neurons. c Whole-cell currents recorded in Na+-free extracellular solution containing 50 mM Ca2+ at −60 mV in lumbar spinal cord and oculomotor neurons, evoked by AMPA 100 μM (n = 6). Columns represent the peak amplitudes of agonist-induced whole-cell currents (mean ± SEM, **P < 0.001, P < 0.05, Student’s t test). d and e Concentration–response relation for kainate-induced whole-cell currents in lumbar spinal cord and oculomotor neurons. Currents were recorded in 20 mM extracellular Na+ at −60 mV, in response to kainate concentrations ranging from 50 μM to 50 mM. d Representative current traces elicited by kainate. e Each point represents mean ± SEM from three cells (*P < 0.05, **P < 0.01, Student’s t test). EC50 values estimated from fits to pooled data were 1.12 mM for lumbar spinal cord motor neurons and 1.26 mM for oculomotor motor neurons, respectively. f Whole-cell currents recorded in Na+-free extracellular solution containing 50 mM Ca2+ at −60 mV in lumbar spinal cord and oculomotor neurons, evoked by kainate 1 mM (n = 6). Columns represent the peak amplitudes of agonist-induced whole-cell currents (mean ± SEM, **P < 0.001, P < 0.05, Student’s t test)
Fig. 5.
a and b Concentration–response relation for GABA-induced whole-cell currents in lumbar spinal cord and oculomotor motor neurons. Currents were recorded at −60 mV, in response to GABA concentrations ranging from 0.1 mM to 100 mM. Each point represents mean ± SEM from three cells (*P < 0.05, Student’s t test). EC50 values estimated from fits to pooled data were 6.1 mM for lumbar spinal cord motor neurons and 5.7 mM for oculomotor motor neurons, respectively. c GABA-induced whole-cell currents in lumbar spinal cord and oculomotor neurons. Whole-cell currents evoked by 6 mM GABA were recorded at −60 mV. Columns represent the peak amplitudes of GABA-induced whole-cell currents (mean ± SEM, *P < 0.05, n = 6, Student’s t test)
Discussion
We have identified marked differences in the transcriptional profiles of human spinal motor neurons, which are susceptible, and oculomotor neurons, which are resistant to the degenerative process in ALS. Nearly 2,000 genes are significantly differentially expressed by the two motor neuron subtypes, with a conservative false discovery rate of 0.001. This is the first study of differences in the gene expression characteristics between ALS-resistant and ALS-vulnerable motor neuron subtypes in human tissue. A recent paper by Hedlund et al. [14] used microarray analysis of motor neurons of normal mice, to compare the transcriptional profiles of OM, hypoglossal and cervical spinal motor neurons. They also found differential expression between genes in the functional categories of mitochondrial function, ubiquitination, cell adhesion and RNA metabolism. Upregulation of Hox genes was seen in cervical spinal motor neurons and hypoglossal neurons, compared to oculomotor neurons. However, three genes highlighted in the study, IGF-1 and guanine deaminase (upregulated in oculomotor neurons), and peripherin (upregulated in spinal motor neurons), showed no differential expression in our study of human motor neurons, which may reflect species-specific differences in expression between the motor neuron subtypes.
Functional categories of genes differentially expressed between oculomotor and spinal motor neurons
Using Gene Ontology enrichment analysis, we have shown that motor neuron subtypes with different susceptibility to degeneration in ALS show differential expression of genes with a function in synaptic transmission, ubiquitin-dependent proteolysis, mitochondrial function, transcriptional regulation, immune system functions and the extracellular matrix. These differences may reflect their diverse embryological origin, the different milieu in the brainstem, or differences in the structure and function of oculomotor units compared to motor units of other skeletal muscles. For example, upregulation of genes with mitochondrial functions, including the respiratory chain complex, is likely to reflect the high metabolic demand placed on oculomotor neurons, due to their high firing rate. Hox genes are key controllers of rostro-caudal patterning, and of motor neuron pools within each level of the hindbrain and spinal cord [3, 4, 11]. The patterns of differential expression of Hox genes and of engrailed 1 and 2 therefore reflect the positions of these neurons on the rostro-caudal axis. The functional implications of differential expression of collagen subunits between OM and LSC motor neurons are unknown, but are intriguing in the light of findings of altered composition of the extracellular matrix in patients with ALS, and the clinical observation that they are resistant to bedsores [24, 42, 43].
Amongst the differences in gene expression observed, are transcriptional characteristics that render oculomotor neurons resistant to the degenerative process in ALS. For example, there is evidence that the capacity of the proteasome system to degrade proteins may be a limiting factor in the vulnerability of neurons to the degenerative process [19]. Our observation of an increase in the expression of genes encoding components of the ubiquitin–proteasome system in OM neurons compared to LSC motor neurons is a novel finding, which may contribute to their relative resistance to degeneration. We have made this array data freely available to the scientific community (http://www.ncbi.nlm.nih.gov/geo/; GSE 40438) to explore other gene expression changes which may lead to a functional difference between OM and LSC motor neurons.
We used datasets from two different species, obtained from the GEO Omnibus repository, to confirm the differential gene expression seen on microarray analysis in human OM and LSC motor neurons. There was marked concordance in the functional categories of genes differentially expressed between the three species analysed, with the most significant enrichment occurring in all three species, in genes with a function in synaptic transmission. GABA and glutamate receptor subunits differentially expressed between the motor neuron subtypes in human tissue showed concordant changes in mouse and/or rat tissue in the majority of cases.
Relative resistance of oculomotor neurons to excitotoxicity
There is substantial evidence that excitotoxicity plays a role in the motor neuronal death of ALS, and that motor neurons are selectively vulnerable to excitotoxic cell injury. Motor neurons possess atypical AMPA receptors that express proportionately fewer GluR2 receptor subunits, relative to other GluR subunits [22, 53]. This low expression of GluR2 would render them more permeable to calcium, and thus more vulnerable to excitotoxic injury. Spinal motor neurons show a higher AMPA receptor current density than ALS-resistant dorsal horn neurons [57]. SOD1G93A transgenic mice crossed with ChAT-GluR2 mice that overexpress the GluR2 subunit in cholinergic neurons have reduced calcium permeability of the motor neuronal AMPA receptors, and show prolonged survival compared to the single heterozygote SOD1G93A mice [54]. GluR2 pre-mRNA is edited by the enzyme ADAR2 (adenosine deaminase acting on RNA2), to convert adenosine to inosine at the Q/R site. Failure of this editing function has been described in motor neurons of patients with sporadic ALS [21]. Furthermore, conditional knockdown of ADAR2 in motor neurons of mice [15], or expression of a functionally modified GluR2 subunit with increased calcium permeability [25] causes a slow decline in motor neurons of the spinal cord and brainstem. In ADAR2 knockout mice, this motor neuron degeneration spares the oculomotor nucleus, suggesting that excitotoxicity is an important determinant of the selective vulnerability of spinal motor neurons, a finding that is further supported by the present study.
We observed upregulation in OM neurons relative to LSC motor neurons of several GABA and glutamate receptor subunits. A previous study showed no difference between ALS-resistant and -vulnerable brainstem motor nuclei in the expression of mRNA or protein of GluR2 and GluR4 [26]. However, we found that OM neurons express higher levels of GluR2, which may render them less susceptible to excitotoxic injury. Although the proportional expression of GluR2 relative to other GluR receptor subunits was not determined in this study, our functional data confirm that the expression changes observed lead to a higher level of calcium influx in response to AMPA/kainate in spinal motor neurons than in oculomotor neurons. The higher level of expression of GABA receptor subunits in OM neurons may reflect increased expression of GABA receptors, which may be neuroprotective through increasing GABA transmission to protect against excessive neuronal excitability. Alternatively, the upregulation of some GABA receptor subunits in OM neurons may reflect alteration in subunit composition, which is thought to determine functional diversity of the receptor [28]. This finding is supported by an earlier study showing lower expression of GABAR subunits in vulnerable than resistant motor neurons in brainstem motor nuclei of normal rats [31].
We carried out patch clamp recording of AMPA and GABA-induced whole-cell currents in OM and LSC motor neurons from rat, in order to determine the functional effect of the observed differences in expression of AMPA and GABA receptor subunits. Amplitudes of whole-cell currents evoked by AMPA receptor agonists were dramatically larger in LSC motor neurons than OM neurons. This difference in AMPA receptor current between the two types of motor neuron, observed with both AMPA and kainate as agonists, was detected over the full range of the agonist concentration–response curves. These whole-cell currents were generated by inward influx of calcium ions, through AMPA receptor activation, as they were observed in extracellular solutions free of Na+, and in the presence of MK-801 to block NMDA receptors, tetrodotoxin to block voltage-gated Na+ channels and Cd2+ to block voltage-gated Ca2+ channels. The higher amplitude of the receptor-mediated current in LSC motor neurons could be explained by a higher density of AMPA receptors, but this is not supported by the microarray data, which showed reduced expression of AMPA receptor subunits in LSC motor neurons. Alternatively, the differential expression of GluR1 and GluR2 subunits may reflect an alteration in subunit composition, which could result in an increase in single-channel conductance or in open probability of calcium channels. A relatively higher somatic size in spinal motor neurons could also result in higher amplitude of the receptor-mediated current, but this is unlikely to explain the differences observed, as the GABAA-induced current was decreased in LSC motor neurons relative to OM neurons. Amplitudes of GABAA receptor-mediated inward chloride currents were significantly higher in OM neurons than in LSC motor neurons. This higher amplitude could be explained by a higher expression of GABA receptors in OM neurons, reflected by the observed increase in GABRA1, B1 and B2 receptor subunit expression, or by altered subunit composition of the receptors, resulting in a change in the physiological channel properties.
The difference in AMPA and GABA-mediated inward currents, with an increase in AMPA-mediated inward calcium current, and decrease in GABA-mediated inward current in LSC motor neurons, relative to OM neurons, would be predicted to increase the susceptibility of spinal motor neurons to excitotoxicity. Interest in excitotoxicity in ALS has largely focussed on the role of excess glutamate activity; however, insufficient synaptic inhibition may also play an important role. Spinal motor neurons and oculomotor neurons are innervated by GABAergic inhibitory neurons [35, 51], which generate fast inhibitory potentials to regulate the amplitude and duration of excitatory postsynaptic potentials. GABAergic transmission has been shown to be neuroprotective in motor neuron cultures exposed to glutamate [58], and in other experimental paradigms where excitotoxicity contributes to neuronal death [34]. Loss of cortical inhibitory signals [8], and depletion of cortical GABAergic interneurons [37] have been demonstrated in patients with ALS. PET studies have also shown reduction in binding of the GABAA receptor ligand, flumazenil, in the extramotor cortex of ALS patients, correlating with the extent of cognitive dysfunction [30]. Furthermore, patients with rapidly progressive disease were shown to have loss of cortical inhibitory GABAergic mechanisms, compared to those with a more slowly progressive ALS variant [56]. There has been increasing interest recently in the potential role of degeneration of inhibitory interneurons in the pathogenesis of ALS. Reduced expression of GABAergic and glycinergic markers has been demonstrated in the ventral horns, and reduced glycine-mediated currents in cultured motor neurons from mutant SOD1 mice [2, 16]. The present study contributes to a growing body of evidence that a reduction in GABA-mediated synaptic transmission plays a role in the pathogenesis of selective degeneration of spinal motor neurons in ALS.
Conclusion
We have used genome-wide transcriptome analysis to determine systematically the differences between two populations of motor neurons that show differential susceptibility to degeneration in ALS. The transcriptional profiles of oculomotor and spinal motor neurons are distinct, with differences in nearly 2,000 genes, at a conservative false discovery rate. Significant differences might be expected due to the different embryological origin, location, and structure and function of oculomotor units. However, amongst the changes observed are those that determine the disease-resistant properties of oculomotor neurons. Analysis of the transcriptional profiles of oculomotor nucleus and spinal cord tissue of two further species showed that differential expression of genes with a function in synaptic transmission, including GABA and glutamate receptor subunits, is conserved across the three species. We carried out functional studies, using patch clamp recording to measure whole-cell currents induced by AMPA and GABA receptor agonists, and found significant upregulation of AMPA-induced calcium currents, and downregulation of GABA-induced chloride currents in the disease-susceptible spinal motor neurons. These microarray and electrophysiology data provide strong evidence that reduced susceptibility to excitotoxicity, mediated in part through increased GABAergic transmission, is an important determinant of the relative resistance of oculomotor neurons to degeneration in ALS.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by a Clinical Lecturer Starter Grant, from the Academy of Medical Sciences and the Wellcome Trust to A.B., a Wellcome Programme grant to P.J.S. and a grant from the Motor Neurone Disease Association to J.K.
Conflict of interest
There are no conflicts of interest to declare.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Footnotes
1
Bioconductor is an open-source computational framework for the analysis of high-throughput genomic data implemented in the R programming language.
Contributor Information
Alice Brockington, Email: alicebrockington@yahoo.co.uk.
Pamela J. Shaw, Phone: +0114-2222260, FAX: +0114-2222290, Email: pamela.shaw@sheffield.ac.uk
References
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Q, Martin LJ. Glycine receptor channels in spinal motoneurons are abnormal in a transgenic mouse model of amyotrophic lateral sclerosis. J Neurosci. 2011;31(8):2815–2827. doi: 10.1523/JNEUROSCI.2475-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalla Torre di Sanguinetto SA, Dasen JS, Arber S. Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr Opin Neurobiol. 2008;18(1):36–43. doi: 10.1016/j.conb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123(3):477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Davidson D, Graham E, Sime C, Hill R. A gene with sequence similarity to Drosophila engrailed is expressed during the development of the neural tube and vertebrae in the mouse. Development. 1988;104(2):305–316. doi: 10.1242/dev.104.2.305. [DOI] [PubMed] [Google Scholar]
- 6.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 7.Dong H, O’Brien R, Fung E, Lanahan A, Worley P, Huganir R. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386(6622):279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 8.Enterzari-Taher M, Eisen A, Stewart H, Nakajima M. Abnormalities of cortical inhibitory neurons in amyotrophic lateral sclerosis. Muscle Nerve. 1997;20(1):65–71. doi: 10.1002/(SICI)1097-4598(199701)20:1<65::AID-MUS9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Ferraiuolo L, Kirby J, Grierson AJ, Sendtner M, Shaw PJ. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7(11):616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 10.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guthrie S. Patterning and axon guidance of cranial motor neurons. Nat Rev Neurosci. 2007;8(11):859–871. doi: 10.1038/nrn2254. [DOI] [PubMed] [Google Scholar]
- 12.Harrison AR, Anderson BC, Thompson LV, McLoon LK. Myofiber length and three-dimensional localization of NMJs in normal and botulinum toxin treated adult extraocular muscles. Invest Ophthalmol Vis Sci. 2007;48(8):3594–3601. doi: 10.1167/iovs.06-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi H, Kato S. Total manifestations of amyotrophic lateral sclerosis. ALS in the totally locked-in state. J Neurol Sci. 1989;93(1):19–35. doi: 10.1016/0022-510X(89)90158-5. [DOI] [PubMed] [Google Scholar]
- 14.Hedlund E, Karlsson M, Osborn T, Ludwig W, Isacson O. Global gene expression profiling of somatic motor neuron populations with different vulnerability identify molecules and pathways of degeneration and protection. Brain. 2010;133(Pt 8):2313–2330. doi: 10.1093/brain/awq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hideyama T, Yamashita T, Suzuki T, Tsuji S, Higuchi M, Seeburg PH, Takahashi R, Misawa H, Kwak S. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J Neurosci Off J Soc Neurosci. 2010;30(36):11917–11925. doi: 10.1523/JNEUROSCI.2021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hossaini M, Cardona Cano S, van Dis V, Haasdijk ED, Hoogenraad CC, Holstege JC, Jaarsma D. Spinal inhibitory interneuron pathology follows motor neuron degeneration independent of glial mutant superoxide dismutase 1 expression in SOD1-ALS mice. J Neuropathol Exp Neurol. 2011;70(8):662–677. doi: 10.1097/NEN.0b013e31822581ac. [DOI] [PubMed] [Google Scholar]
- 17.Ince P. Neuropathology. In: Brown RJ, Meininger V, Swash M, editors. Amyotrophic lateral sclerosis. London: Martin Dunitz; 2000. pp. 83–112. [Google Scholar]
- 18.Ince P, McArthur F, Bjertness E, Torvik A, Candy J, Edwardson J. Neuropathological diagnoses in elderly patients in Oslo: Alzheimer’s disease, Lewy body disease, vascular lesions. Dementia. 1995;6(3):162–168. doi: 10.1159/000106940. [DOI] [PubMed] [Google Scholar]
- 19.Kabashi E, Agar J, Taylor D, Minotti S, Durham H. Focal dysfunction of the proteasome: a pathogenic factor in a mouse model of amyotrophic lateral sclerosis. J Neurochem. 2004;89(6):1325–1335. doi: 10.1111/j.1471-4159.2004.02453.x. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427(6977):801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 22.Kawahara Y, Kwak S, Sun H, Ito K, Hashida H, Aizawa H, Jeong SY, Kanazawa I. Human spinal motoneurons express low relative abundance of GluR2 mRNA: an implication for excitotoxicity in ALS. J Neurochem. 2003;85(3):680–689. doi: 10.1046/j.1471-4159.2003.01703.x. [DOI] [PubMed] [Google Scholar]
- 23.Khanna S, Richmonds CR, Kaminski HJ, Porter JD. Molecular organization of the extraocular muscle neuromuscular junction: partial conservation of and divergence from the skeletal muscle prototype. Invest Ophthalmol Vis Sci. 2003;44(5):1918–1926. doi: 10.1167/iovs.02-0890. [DOI] [PubMed] [Google Scholar]
- 24.Kolde G, Bachus R, Ludolph AC. Skin involvement in amyotrophic lateral sclerosis. Lancet. 1996;347(9010):1226–1227. doi: 10.1016/S0140-6736(96)90737-0. [DOI] [PubMed] [Google Scholar]
- 25.Kuner R, Groom AJ, Bresink I, Kornau HC, Stefovska V, Muller G, Hartmann B, Tschauner K, Waibel S, Ludolph AC, Ikonomidou C, Seeburg PH, Turski L. Late-onset motoneuron disease caused by a functionally modified AMPA receptor subunit. Proc Natl Acad Sci USA. 2005;102(16):5826–5831. doi: 10.1073/pnas.0501316102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laslo P, Lipski J, Nicholson L, Miles G, Funk G. GluR2 AMPA receptor subunit expression in motoneurons at low and high risk for degeneration in amyotrophic lateral sclerosis. Exp Neurol. 2001;169(2):461–471. doi: 10.1006/exnr.2001.7653. [DOI] [PubMed] [Google Scholar]
- 27.Leigh PN, Ray-Chaudhuri K. Motor neuron disease. J Neurol Neurosurg Psychiatry. 1994;57(8):886–896. doi: 10.1136/jnnp.57.8.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levitan E, Schofield P, Burt D, Rhee L, Wisden W, Köhler M, Fujita N, Rodriguez H, Stephenson A, Darlison M. Structural and functional basis for GABAA receptor heterogeneity. Nature. 1988;335(6185):76–79. doi: 10.1038/335076a0. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Milo M, Lawrence ND, Rattray M. Probe-level measurement error improves accuracy in detecting differential gene expression. Bioinformatics. 2006;22(17):2107–2113. doi: 10.1093/bioinformatics/btl361. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd CM, Richardson MP, Brooks DJ, Al-Chalabi A, Leigh PN. Extramotor involvement in ALS: PET studies with the GABA(A) ligand [(11)C]flumazenil. Brain J Neurol. 2000;123(Pt 11):2289–2296. doi: 10.1093/brain/123.11.2289. [DOI] [PubMed] [Google Scholar]
- 31.Lorenzo LE, Barbe A, Portalier P, Fritschy JM, Bras H. Differential expression of GABAA and glycine receptors in ALS-resistant vs. ALS-vulnerable motoneurons: possible implications for selective vulnerability of motoneurons. Eur J Neurosci. 2006;23(12):3161–3170. doi: 10.1111/j.1460-9568.2006.04863.x. [DOI] [PubMed] [Google Scholar]
- 32.Luzzi V, Mahadevappa M, Raja R, Warrington JA, Watson MA. Accurate and reproducible gene expression profiles from laser capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J Mol Diagn JMD. 2003;5(1):9–14. doi: 10.1016/S1525-1578(10)60445-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackenzie IR, Feldman HH. Ubiquitin immunohistochemistry suggests classic motor neuron disease, motor neuron disease with dementia, and frontotemporal dementia of the motor neuron disease type represent a clinicopathologic spectrum. J Neuropathol Exp Neurol. 2005;64(8):730–739. doi: 10.1097/01.jnen.0000174335.27708.0a. [DOI] [PubMed] [Google Scholar]
- 34.Madden K. Effect of gamma-aminobutyric acid modulation on neuronal ischemia in rabbits. Stroke. 1994;25(11):2271–2274. doi: 10.1161/01.STR.25.11.2271. [DOI] [PubMed] [Google Scholar]
- 35.Magoul R, Onteniente B, Geffard M, Calas A. Anatomical distribution and ultrastructural organization of the GABAergic system in the rat spinal cord. An immunocytochemical study using anti-GABA antibodies. Neuroscience. 1987;20(3):1001–1009. doi: 10.1016/0306-4522(87)90258-2. [DOI] [PubMed] [Google Scholar]
- 36.Mannen T, Iwata M, Toyokura Y, Nagashima K. Preservation of a certain motoneurone group of the sacral cord in amyotrophic lateral sclerosis: its clinical significance. J Neurol Neurosurg Psychiatry. 1977;40(5):464–469. doi: 10.1136/jnnp.40.5.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nihei K, McKee A, Kowall N. Patterns of neuronal degeneration in the motor cortex of amyotrophic lateral sclerosis patients. Acta Neuropathol. 1993;86(1):55–64. doi: 10.1007/BF00454899. [DOI] [PubMed] [Google Scholar]
- 38.Nimchinsky E, Young W, Yeung G, Shah R, Gordon J, Bloom F, Morrison J, Hof P. Differential vulnerability of oculomotor, facial, and hypoglossal nuclei in G86R superoxide dismutase transgenic mice. J Comp Neurol. 2000;416(1):112–125. doi: 10.1002/(SICI)1096-9861(20000103)416:1<112::AID-CNE9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 39.Ning K, Li L, Liao M, Liu B, Mielke JG, Chen Y, Duan Y, El-Hayek YH, Wan Q. Circadian regulation of GABAA receptor function by CKI epsilon-CKI delta in the rat suprachiasmatic nuclei. Nat Neurosci. 2004;7(5):489–490. doi: 10.1038/nn1236. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto K, Hirai S, Amari M, Iizuka T, Watanabe M, Murakami N, Takatama M. Oculomotor nuclear pathology in amyotrophic lateral sclerosis. Acta Neuropathol. 1993;85(5):458–462. doi: 10.1007/BF00230482. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto K, Hirai S, Ishiguro K, Kawarabayashi T, Takatama M. Light and electron microscopic and immunohistochemical observations of the Onuf’s nucleus of amyotrophic lateral sclerosis. Acta Neuropathol. 1991;81(6):610–614. doi: 10.1007/BF00296370. [DOI] [PubMed] [Google Scholar]
- 42.Ono S, Imai T, Takahashi K, Jinnai K, Yamano T, Nagao K, Shimizu N, Yamauchi M. Decreased type IV collagen of skin and serum in patients with amyotrophic lateral sclerosis. Neurology. 1998;51(1):114–120. doi: 10.1212/WNL.51.1.114. [DOI] [PubMed] [Google Scholar]
- 43.Ono S, Mechanic GL, Yamauchi M. Amyotrophic lateral sclerosis: unusually low content of collagen in skin. J Neurol Sci. 1990;100(1–2):234–237. doi: 10.1016/0022-510X(90)90038-O. [DOI] [PubMed] [Google Scholar]
- 44.Pachter BR. Rat extraocular muscle. 1. Three dimensional cytoarchitecture, component fibre populations and innervation. J Anat. 1983;137(Pt 1):143–159. [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson RD, Liu X, Sanguinetti G, Milo M, Lawrence ND, Rattray M. puma: a Bioconductor package for propagating uncertainty in microarray analysis. BMC Bioinforma. 2009;10:211. doi: 10.1186/1471-2105-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porter JD. Extraocular muscle: cellular adaptations for a diverse functional repertoire. Ann NY Acad Sci. 2002;956:7–16. doi: 10.1111/j.1749-6632.2002.tb02804.x. [DOI] [PubMed] [Google Scholar]
- 47.Porter JD, Baker RS. Muscles of a different ‘color’: the unusual properties of the extraocular muscles may predispose or protect them in neurogenic and myogenic disease. Neurology. 1996;46(1):30–37. doi: 10.1212/WNL.46.1.30. [DOI] [PubMed] [Google Scholar]
- 48.Porter JD, Hauser KF. Survival of extraocular muscle in long-term organotypic culture: differential influence of appropriate and inappropriate motoneurons. Dev Biol. 1993;160(1):39–50. doi: 10.1006/dbio.1993.1284. [DOI] [PubMed] [Google Scholar]
- 49.Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Andrade FH. Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc Natl Acad Sci USA. 2001;98(21):12062–12067. doi: 10.1073/pnas.211257298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson DA. Oculomotor unit behavior in the monkey. J Neurophysiol. 1970;33(3):393–403. doi: 10.1152/jn.1970.33.3.393. [DOI] [PubMed] [Google Scholar]
- 51.Soghomonian J, Pinard R, Lanoir J. GABA innervation in adult rat oculomotor nucleus: a radioautographic and immunocytochemical study. J Neurocytol. 1989;18(3):319–331. doi: 10.1007/BF01190835. [DOI] [PubMed] [Google Scholar]
- 52.Storey J. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat. 2003;31(6):2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 53.Sun H, Kawahara Y, Ito K, Kanazawa I, Kwak S. Expression profile of AMPA receptor subunit mRNA in single adult rat brain and spinal cord neurons in situ. Neurosci Res. 2005;52(3):228–234. doi: 10.1016/j.neures.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Tateno M, Sadakata H, Tanaka M, Itohara S, Shin RM, Miura M, Masuda M, Aosaki T, Urushitani M, Misawa H, Takahashi R. Calcium-permeable AMPA receptors promote misfolding of mutant SOD1 protein and development of amyotrophic lateral sclerosis in a transgenic mouse model. Hum Mol Genet. 2004;13(19):2183–2196. doi: 10.1093/hmg/ddh246. [DOI] [PubMed] [Google Scholar]
- 55.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14(4):452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner MR, Osei-Lah AD, Hammers A, Al-Chalabi A, Shaw CE, Andersen PM, Brooks DJ, Leigh PN, Mills KR. Abnormal cortical excitability in sporadic but not homozygous D90A SOD1 ALS. J Neurol Neurosurg Psychiatry. 2005;76(9):1279–1285. doi: 10.1136/jnnp.2004.054429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandenberghe W, Ihle EC, Patneau DK, Robberecht W, Brorson JR. AMPA receptor current density, not desensitization, predicts selective motoneuron vulnerability. J Neurosci Off J Soc Neurosci. 2000;20(19):7158–7166. doi: 10.1523/JNEUROSCI.20-19-07158.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent A, Backus C, Taubman A, Feldman E. Identification of candidate drugs for the treatment of ALS. Amyotroph Lateral Scler Other Motor Neuron Disord. 2005;6(1):29–36. doi: 10.1080/14660820510026171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.