Differential Expression of Exosomal microRNAs in Prefrontal Cortices of Schizophrenia and Bipolar Disorder Patients (original) (raw)
Abstract
Exosomes are cellular secretory vesicles containing microRNAs (miRNAs). Once secreted, exosomes are able to attach to recipient cells and release miRNAs potentially modulating the function of the recipient cell. We hypothesized that exosomal miRNA expression in brains of patients diagnosed with schizophrenia (SZ) and bipolar disorder (BD) might differ from controls, reflecting either disease-specific or common aberrations in SZ and BD patients. The sources of the analyzed samples included McLean 66 Cohort Collection (Harvard Brain Tissue Resource Center), BrainNet Europe II (BNE, a consortium of 18 brain banks across Europe) and Boston Medical Center (BMC). Exosomal miRNAs from frozen postmortem prefrontal cortices with well-preserved RNA were isolated and submitted to profiling by Luminex FLEXMAP 3D microfluidic device. Multiple statistical analyses of microarray data suggested that certain exosomal miRNAs were differentially expressed in SZ and BD subjects in comparison to controls. RT-PCR validation confirmed that two miRNAs, miR-497 in SZ samples and miR-29c in BD samples, have significantly increased expression when compared to control samples. These results warrant future studies to evaluate the potential of exosome-derived miRNAs to serve as biomarkers of SZ and BD.
Introduction
Knowing which molecules are specifically altered in neuropsychiatric patients would represent a crucial step towards uncovering mechanisms of the development of neuropshychiatric diseases and generating more successful therapeutic strategies. An immediate impact of identifying such molecules would be access to a set of biomarkers to help define and monitor populations at risk.
MicroRNAs (miRNAs, miRs) may up- or down-regulate the translation of messenger RNA (mRNA) or render it unstable, and have recently been proposed as biomarkers for brain neoplasms, degenerative diseases, autism, and schizophrenia [1]. Dysregulation of miRNAs in brains of patients diagnosed with schizophrenia (SZ) [2] and other neuropsychiatric disorders is plausible considering that many miRNAs are expressed in human brain [3] where they regulate neuronal development [4] and differentiation [5] including dendritic spine development [6] and plasticity [7],[8], as well as cognitive functions [9]. A group of miRNAs has been recently designated as “misexpressed” in the prefrontal cortices (PFCs) of both SZ and bipolar disorder (BD) patients [10]. The target analysis of another reported set of differentially expressed miRNAs in the PFCs of SZ patients revealed many genes implicated in signaling pathways [11]. While a specific BD miRNA profile has not yet been established, alterations in neurochemical regulation including an excess in signaling activity have been associated with BD [12].
Signaling activity in neurons commonly requires the release of signaling molecules from their specific secretory vesicles [13]. Exosomes are secretory vesicles defined by size (30–90 nm), buoyant density (∼1.1–1.2 g/ml), lipid composition, and the presence as well as the absence of specific marker proteins [14]. Exosomes represent the alternative route for the content of multivesicular bodies (MVBs, exosomal precursor organelles) destined for degradation in lyzosomes. Rather then being degraded, MVBs’ intraluminal vesicles are fused with the plasma membrane and secreted into the extracellular space as exosomes [15]. Because of the specific molecules on their surface, including cell-adhesion proteins, exosomes can be incorporated by specific recipient cells [15],[16]. The exosomal involvement in neuronal signaling was suggested by the presence of MVBs in dendritic spines [17],[18] and by the resemblance of synaptic spinules (evaginations of the postsynaptic membrane that bud into presynaptic axon) to exosomes [19],[20]. The role of exosomes in synaptic activity is further corroborated by the dependence of long term potentiation (LTP)-induced structural spine plasticity on exocytic trafficking from recycling endosomes [21].
Cultured primary cortical neurons and astrocytes do release exosomes [22]. Studies of exosomal cargo in neurons and microglia have revealed proteins important for the development of neurodegenerative diseases such as Alzheimer’s disease [23]–[26] and Parkinson’s disease [27]. Exosomes in neuronal cell lines have also been shown to transfer prions [28] as well as wild-type and mutant superoxide dismutase, thus propagating cell-to-cell mutant toxicity [29]. Microarray analysis of exosomal content from mouse bone marrow mast cells and human and mouse mast cell lines has established the presence of mRNA and miRNA [30]. A new name for this mRNA - “exosomal shuttle RNA” [30] - has been proposed to underscore the ability of exosomes to mediate the exchange of genetic material between cells. Changes in exosomal miRNAs have been reported in patients diagnosed with Alzheimer’s disease [31], while RNA content from the glioblastoma microvesicles was shown to provide diagnostic biomarkers [32]. Postmortem human PFC samples have been used to successfully generate miRNA profiles [11]. Here we report the results of exosomal miRNA expression analysis in PFCs from patients diagnosed with SZ, BD, and mental illness-free controls. We have observed aberrations in SZ and BD samples in comparison to controls, suggesting both common and differential pathobiology. These results may forecast reproducible, disease-associated changes of miRNA expression serving as biomarkers in exosomes, cellular particles potentially obtainable from the cerebrospinal fluid (CSF) of living patients.
Materials and Methods
Human Brain Tissue
Written informed consent was obtained from the next of kin for the use of the postmortem brain tissue by the following sources of frozen PFC (Brodmann area 9, BA9): BrainNet Europe II, a consortium of 18 brain banks across Europe (BNE; cases SZ 1–6, BD 5–9, and C 6–10), McLean 66 Cohort Collection at Harvard Brain Tissue Resource Center (McLean; cases SZ 7 and 8, BD 1–4, and C 1–5), and autopsy service at Boston Medical Center (BMC; cases C 11–13) (Table 1).
Table 1. Analyzed human prefrontal cortices (BA9).
| Case andDisease | Age | Gender/Hemisphere | PMI | Neuropathology and pertinentclinical history | Source | RIN | Experiment |
|---|---|---|---|---|---|---|---|
| SZ1 | 74 | M/left | 6 | BB II, alpha-synuclein immunoreactivityin few SN neurons | BNE | 8.3 | L, P |
| SZ2 | 65 | M/left | 3.5 | BB I, lacunar infarct in the caudate | BNE | 8.3 | L, P |
| SZ3 | 82 | M/left | 4.5 | BB VI, amyloid angiopathy, lacunarinfarct in putamen | BNE | 9.1 | L, P |
| SZ4 | 77 | M/left | 6 | BB II, old infarct in the frontal lobe | BNE | 8.3 | L, P |
| SZ5 | 84 | M/left | 2.5 | BB II, amyloid angiopathy, argirofilicgrain disease | BNE | 8.3 | L, P |
| SZ6 | 79 | M/left | 2.5 | multiple infarcts in the teritories of middleand posterior cerebral artery circulation | BNE | 7.8 | L, P |
| SZ7 | 63 | M/right | 22.3 | BB I | McLean | 7.6 | L |
| SZ8 | 35 | M/left | 28 | BB I, non-specific fronto-temporo-parietaland thalamic atrophy | McLean | 6.1 | L |
| BD1 | 40 | M/left | 30.8 | acute hypoxic changes, focalarteriolosclerosis | McLean | 6.7 | L |
| BD2 | 38 | M/left | 22 | parietal cortex infarct, focal acute hypoxicchanges, focal arteriolosclerosis | McLean | 7.9 | L |
| BD3 | 50 | M/right | 30.5 | no neuropathology reported | McLean | 7.1 | L |
| BD4 | 74 | M/right | 25 | no neuropathology reported | McLean | 6.6 | L |
| BD5 | 73 | M/right | 5.3 | BB II, temporal SPs, small old corticaland striatal infarcts | BNE | 6.2 | L, P |
| BD6 | 70 | M/left | 6.4 | BB III, many SP, few NP, contusions (dueto fall); alcohol abuse | BNE | 6.2 | L, P |
| BD7 | 68 | M/left | NA | BB I | BNE | 5.5 | P |
| BD8 | 93 | M/left | 6 | BB I | BNE | 5.1 | P |
| BD9 | 90 | F/left | 6.5 | BB I-II, rare SP | BNE | 7.7 | P |
| C1 | 67 | M/right | 22.3 | BB I, lacunar infarcts in putamen, focalarteriolosclerosis | McLean | 8.0 | L |
| C2 | 37 | M/right | 18.7 | minimal acute hypoxic-ischemic changein hippocampus | McLean | 6.4 | L |
| C3 | 38 | M/left | 28.8 | no neuropathology reported | McLean | 7.7 | L |
| C4 | 50 | M/left | 24.1 | BB I | McLean | 6.2 | L |
| C5 | 40 | M/right | 28 | no neuropathology reported | McLean | 7.1 | L |
| C6 | 68 | M/left | 10.2 | no neuropathology reported | BNE | 6.4 | L, P |
| C7 | 71 | M/left | 7.6 | BB I, lacunar infarct in internal capsule | BNE | 6.1 | P |
| C8 | 82 | F/left | 7 | severe atherosclerosis in the circle ofWillis, focal gliosis in striatum | BNE | 6.1 | P |
| C9 | 96 | M/right | 5.4 | BB I, rare SP, rare NP, hypoxic-ischemicchanges in striatum and hippocampus | BNE | 6.0 | P |
| C10 | 65 | F/left | 12.8 | no neuropathology | BNE | 8.1 | P |
| C11 | 80 | M/left | 17 | no neuropathology | BMC | 5.9 | P |
| C12 | 72 | F/left | 23 | no neuropathology | BMC | 5.1 | P |
| C13 | 68 | F/left | 25 | no neuropathology | BMC | 7.0 | P |
Extractions of Exosome-containing Pellets
Exosome extraction protocol from primary and cultured cells [30] was modified to obtain exosome-containing pellets from frozen brain tissue that was previously evaluated for RNA integrity (Figure S1). Only the brain tissue samples yielding an RNA integrity number (RIN) of ≥5.1 (range 5.1–9.1, Table 1) and thus well within accepted standards [33] were used to obtain exosome-containing pellets. From the original frozen prefrontal cortex stored at −80°C procured by each brain bank, we cut approximately 600 mg of grey matter using a razor blade. The frozen tissue was put in 1 ml phosphate-buffered saline, pH 7.4 (PBS, 4°C), gently teased with a small spatula, and vortexed (VWR mini-vortexer) on a medium power. Each tube was centrifuged at 300×g for 10 min, decanted, centrifuged twice at 1,200×g for 10 min with decanting in between, and finally filtered through a 0.2 um syringe filter (Millipore, Carrigtowhill, Cork, Ireland). The filtrate was centrifuged twice at 10,000×g for 30 min with decanting in between. The final centrifugation to obtain exosome-containing pellets was performed at 22,000×g for 22 hrs. All centrifugations were performed at 2°C.
As a control exosomal extraction procedure, exosomes from cultured human H4 cells (HTB 148 ATCC, Manassas, VA, USA) were prepared as previously published [16]–[34] with minor modifications. Briefly, H4 cells were cultured in OPTIMEM medium (Life Technologies) without serum or antibiotics. After 48 hours, conditioned medium was collected and centrifuged for 5 min at 500×g at 4°C to remove cell debris. The supernatants were then sequentially centrifuged at 300×g (10 min) and twice at 2,000×g (10 min), 4°C. The supernatant was filtered through a 0.45 um (Whatman, Florham Park, NJ) and 0.22 um (Millipore, Carrigtowhill, Cork, Ireland) filter, and centrifuged for 1 hr at 10,000×g at 4°C. The supernatant was removed and then subjected to ultracentrifugation at 100,000×g for 2 hours at 4°C. The supernatant was collected and the exosome-containing pellet was re-suspended in PBS for Western Blotting.
Transmission Electron Microscopy (TEM) and Immuno-gold Labeling
For morphological identification of exosomes, the pellets were either embedded in a hydrophilic resin upon fixation in TEM fixative (4% paraformaldehyde, 0.2% glutaraldehyde, in 0.2 M cacodylate buffer, pH 7.2.) or directly adsorbed to a carbon-coated grid that has been made hydrophilic by an exposure to a glow discharge (30 sec). Upon blocking with 1% Bovine Serum Albumin (BSA), grids with pellets were incubated in primary antibody (CD63, BD Pharmingen, and GAPDH, Ambion) solutions in 1% BSA, rinsed in PBS, exposed to either bridging antibody first or directly to protein A-gold (5 nM) solution in 1% BSA, rinsed in PBS, and finally in water. Excess liquid was removed with a filter-paper (Whatman #1). Optional negative staining was achieved by incubation in 0.75% uranyl formate for 30 seconds. The examination was carried out in a JEOL 1200EX TEM or a TecnaiG2 Spirit BioTWIN and images were recorded with an AMT 2k CCD camera.
Western Blotting
The expression of an exosomal marker flotillin-2 [35] in the exosomal preparations from frozen postmortem PFC was compared to the flotillin-2 expression in the cell-culture-derived exosomal preparations described above. Equal protein amounts (2.5 mg/ml) from each exosome-containing pellet reconstituted in PBS and respective supernatant were blotted with anti-flotillin-2 antibody (BD Transduction Laboratories, 1∶1,000).
Isolation, Purification, and Linear Amplification of miRNA from Exosome-containing Pellets
Exosome-containing pellets re-suspended in 20 ul of PBS were incubated with 0.4 mg/ml of RNase A/T1 (Fementas) for 10 min at 37°C. RNase was inactivated by adding 20 units/ul of SuperRase-in RNase inhibitor (Ambion) for 10 min at 25°C. The RNase treatment destroyed the higher molecular weight extra-exosomal cellular RNAs and preserved the small RNAs contained in the exosomes, i.e. ribosomal RNA and miRNA (Figure S2). Upon RNase inactivation, 60 ul of miRNA extraction buffer (Arcturus) was added, followed by an incubation at 42°C for 30 min and purification using PicoPure RNA Isolation Kit (Lifetech). The expected size range of miRNAs (∼19–28 nucleotides, nt) and pre-miRNAs (45–60 nt) were well-represented in the purified preparations (Figure S2). Because Luminex profiling system operates with inputs of 2.5–5 ug of total RNA per sample, we needed to ensure that our samples are sufficiently enriched for miRNA. To that end, we used Invitrogen NCode miRNA amplification system according to the manufacturer’s protocol to linearly amplify miRNA in our material (Table S2). We found that this amplification step upon above described RNA treatment did not induce changes in the RNA profile (Figure S3).
Luminex miRNA Assay
We performed miRNA expression analysis using a FlexMAP3D instrument by (Luminex Corporation, Austin, TX) and a manufacturer’s assay for 312 miRNA (Table S1). This assay uses 5.6 um polystyrene micro-beads each of which contains a mixture of two fluorescent dyes that enable the beads to be identified as one of a specific set. Oligonucleotides, specific for each of our 312 miRNA assayed, were attached to the beads according to the manufacturer’s protocol. The beads, upon incubation with miRNA containing samples, were passed through a fluidic tube that causes the micro-spheres to line up in single file before they pass through the detection chamber that contains two lasers. One of the laser beams classifies each bead into the appropriate bead set, while the other scans the beads for the presence of fluorescent reporter molecules and quantifies the number of reporter molecules on each bead. All 312 miRNA were assayed simultaneously in each sample. Intra-normalization of the obtained expression values was performed according to the Luminex protocol. Negative values were considered “0” because they indicate the expression below the background, i.e. the absence of a specific miRNA signal (Table S1).
Statistical Analysis of Luminex miRNA Expression Data
Student’s t-tests and statistical software packages including an Excel plug-in named Significance Analysis of Microarrays (SAM) [36],[37] and an R package named Prediction Analysis of Microarrays (PAM) [38] were used to analyze Luminex expression data on 312 miRNAs from the prefrontal cortex of 8 SZ, 6 BD, and 6 control samples (Table 1 and Table S1). Because the statistical analysis of these data involves multiple comparisons we applied Bonferroni Step-down Holm Correction [39] to Student’s t-test outcomes [0.05/(312–number of miRNAs ranked higher)], resulting in a stricter threshold for significance (Table S1). Next, we used SAM and PAM because they provide multiclass testing [36]–[38] while a simple t-test only enables analysis between two groups. Multiclass testing was a necessary tool to identify a set of miRNAs that could be used to differentiate between all three groups. Both SAM and PAM use a modified z-score statistic [36]–[38] to produce a ranked list of candidate biomarker miRNAs. The adjustment for codependency of expression was performed as local False Discovery Rate (FDR) [37], [40] presented in Tables 2 and 3. FDR is a method of permutation testing [41] that evaluates the risk of Type 1 error. Local FDR is an FDR assessment of not only a single miRNA, but also its “neighbors”, i.e. other closely ranked miRNAs. In this way, a co-dependency is accounted for. Cluster analysis [38] was applied to those miRNAs that had “0″ q-values in SAM (Tables 2 and 3). Clustering was carried out through use of 2*(1-cc) where cc equals a correlation between the cubed root of a single miRNA expression value for a sample and the average of cube-rooted values for all samples in the cluster it joins at that time point. The highest possible correlation between samples is 1 and the lowest is 0, resulting in a dendrogram that presents the clustered samples on a scale of 0 to 2. The dendrogram begins clustering miRNA with most similar expression patterns at the bottom of the graph and then additional miRNA are added to existing clusters as the graph is assembled vertically. In order to assess the predictive power of SAM-ranked miRNAs, the misclassification rate [38]–[42] was established as a means to classify clinical groups. A low misclassification rate indicates that a miRNA or a set of miRNAs is a reliable predictor of a group phenotype (a biomarker). Because of the limited number of cases available for analysis, we performed Wilcoxon test to supplement our findings (Table S3).
Table 2. SAM test of Luminex miRNA expression data for all 3 examined groups of cases: SZ, BD, and C.
| miRNA | z-score:C | z-score:BD | z-score: SZ | q-value(%) | local FDR(%) |
|---|---|---|---|---|---|
| hsa-miR-31 | −2.19709 | −2.54002 | 3.55283 | 0 | 1.553882119 |
| hsa-miR-33 | −2.17637 | −1.95758 | 3.10046 | 0 | 2.433615922 |
| hsa-miR-96 | −1.99363 | −1.9829 | 2.9824 | 0 | 2.493468788 |
| hsa-miR-28 | −1.40723 | −1.68092 | 2.31611 | 0 | 2.300536436 |
| hsa-miR-30e-5p | −1.01353 | −2.15639 | 2.37744 | 0 | 2.438595672 |
| hsa-miR-199a* | −1.52064 | −1.43723 | 2.21841 | 0 | 2.580013124 |
| hsa-miR-501 | 2.162398 | −0.31372 | −1.38651 | 0 | 2.585758655 |
| hsa-miR-504 | 1.9952 | −0.30377 | −1.26857 | 0 | 2.662187029 |
| hsa-miR-15b | −0.83294 | −1.75496 | 1.94092 | 0 | 2.659473233 |
| hsa-miR-29c | 2.068676 | −0.39402 | −1.25599 | 0 | 2.649467758 |
| hsa-miR-455 | −1.49405 | −1.08385 | 1.93342 | 0 | 2.611617439 |
| hsa-miR-380-3p | 1.878938 | −0.83204 | −0.78517 | 0 | 2.621226767 |
| hsa-miR-323 | −1.39982 | −0.99834 | 1.79862 | 0 | 2.626487216 |
| hsa-miR-527 | 1.535186 | 0.010721 | −1.15943 | 0 | 2.807651873 |
| hsa-miR-93 | −1.31706 | −0.81606 | 1.59984 | 0 | 2.851798957 |
| hsa-miR-32 | −1.01365 | −1.26054 | 1.70564 | 0 | 2.86415899 |
| hsa-miR-20b | −1.24865 | −1.25458 | 1.87742 | 0 | 2.873400151 |
| hsa-miR-516-5p | 1.441361 | −0.31318 | −0.84614 | 0 | 2.997241768 |
| hsa-miR-92 | −1.12108 | −0.96419 | 1.56396 | 0 | 3.013501643 |
| hsa-miR-30a-3p | −1.18983 | −1.04924 | 1.67931 | 0 | 3.063908167 |
| hsa-miR-497 | 1.63309 | −0.87672 | −0.56728 | 0 | 3.16010056 |
| hsa-miR-498 | 1.256994 | −0.11349 | −0.85762 | 2.17948718 | 4.322678597 |
| hsa-miR-133b | −1.34128 | −0.7324 | 1.55526 | 2.17948718 | 4.639798252 |
| hsa-miR-499 | 1.736963 | −0.88259 | −0.64078 | 2.17948718 | 4.738725328 |
| hsa-miR-10b | −0.72213 | −1.17673 | 1.42415 | 2.17948718 | 4.878212949 |
| hsa-miR-202* | −0.9101 | −1.73414 | 1.98318 | 2.17948718 | 4.898293562 |
| hsa-miR-202 | −0.14303 | 1.238359 | −0.82149 | 2.17948718 | 5.065305767 |
| hsa-miR-149 | 1.490285 | −0.74638 | −0.55793 | 2.17948718 | 7.055913859 |
| hsa-miR-523 | 0.88386 | 0.910065 | −1.34544 | 2.17948718 | 7.62760309 |
| hsa-miR-199b | −0.87254 | −0.91723 | 1.34233 | 2.17948718 | 7.857837003 |
| hsa-miR-377 | −0.97987 | −1.24239 | 1.6667 | 3.73626374 | 9.314491623 |
| hsa-miR-518b | 0.322664 | 0.980094 | −0.97707 | 3.73626374 | 9.367786992 |
| hsa-miR-26b | 1.336505 | −0.36918 | −0.72549 | 3.73626374 | 9.819393964 |
| hsa-miR-190 | −1.44232 | −1.32539 | 2.07578 | 3.73626374 | 10.24724163 |
| hsa-miR-326 | −0.1038 | 1.199379 | −0.82169 | 3.73626374 | 10.34765218 |
| hsa-miR-494 | 0.293894 | 0.910858 | −0.90356 | 3.73626374 | 11.67285654 |
| hsa-miR-30e-3p | 1.314221 | −0.87488 | −0.32951 | 3.73626374 | 12.18718064 |
| hsa-miR-512-3p | −0.17555 | 1.286875 | −0.83349 | 3.73626374 | 12.89683212 |
| hsa-miR-302a* | −0.47638 | −1.30373 | 1.33509 | 3.73626374 | 13.04097715 |
| hsa-miR-19b | −0.28889 | 1.482134 | −0.89494 | 3.73626374 | 13.11908036 |
| hsa-miR-302c | −0.12265 | 1.445789 | −0.99236 | 4.84330484 | 14.25507383 |
| hsa-miR-218 | 0.521858 | 1.207283 | −1.29686 | 4.84330484 | 14.36727552 |
| hsa-miR-338 | −1.33167 | −0.78432 | 1.58699 | 4.84330484 | 14.76902994 |
| hsa-miR-423 | 0.477963 | 0.812936 | −0.96817 | 4.84330484 | 14.83356578 |
| hsa-miR-200c | −1.36615 | −1.22514 | 1.94347 | 4.84330484 | 15.64595147 |
| hsa-miR-325 | 0.109128 | 1.083419 | −0.89441 | 4.84330484 | 15.97503816 |
| hsa-miR-376b | 1.029142 | −0.00167 | −0.7706 | 4.84330484 | 16.33334873 |
| hsa-miR-518c | −1.34489 | −0.8751 | 1.66499 | 4.84330484 | 17.02063785 |
| hsa-miR-525* | −0.93878 | −1.06791 | 1.50502 | 4.84330484 | 17.14036115 |
| hsa-miR-125b | −0.86013 | −0.85856 | 1.28902 | 4.84330484 | 17.17411684 |
| hsa-miR-299-3p | 1.138288 | −0.02057 | −0.83829 | 4.84330484 | 18.19059244 |
| hsa-miR-363* | 0.02007 | 0.987996 | −0.75605 | 4.84330484 | 18.33451874 |
| hsa-miR-542-3p | 0.990978 | 0.007651 | −0.74897 | 4.84330484 | 18.33773541 |
| hsa-miR-371 | 0.798332 | 0.593946 | −1.04421 | 4.84330484 | 18.36737645 |
| hsa-miR-105 | 1.116132 | 0.542534 | −1.244 | 7.00549451 | 22.95334699 |
| hsa-miR-449 | 0.914034 | 0.511142 | −1.06888 | 7.00549451 | 23.04808523 |
| hsa-miR-370 | 1.088913 | −0.34469 | −0.55817 | 8.02968961 | 25.80170896 |
| hsa-miR-302d | −0.82471 | −1.17696 | 1.50126 | 8.02968961 | 27.03187735 |
| hsa-miR-22 | −1.01777 | −1.05168 | 1.55209 | 8.02968961 | 27.64688383 |
| hsa-miR-17-5p | 0.337338 | 0.922478 | −0.94486 | 8.02968961 | 27.75618225 |
| hsa-miR-518a-2* | 1.201014 | −0.2914 | −0.68221 | 8.02968961 | 28.15873897 |
| hsa-miR-103 | −0.48953 | −0.73475 | 0.91821 | 8.02968961 | 28.42825986 |
| hsa-miR-520d* | 1.460088 | −0.4613 | −0.74909 | 9.90675991 | 29.0140902 |
| hsa-miR-18a* | −0.89462 | −0.66084 | 1.1666 | 9.90675991 | 30.34475207 |
| hsa-miR-210 | −0.05258 | 1.249962 | −0.89804 | 9.90675991 | 31.08028153 |
| hsa-miR-520e | −0.47633 | −1.04849 | 1.14362 | 9.90675991 | 31.70287333 |
| hsa-miR-516-3p | 1.187797 | 0.108884 | −0.97251 | 9.90675991 | 32.5639352 |
| hsa-miR-329 | −0.9053 | 1.414948 | −0.38224 | 9.90675991 | 33.43751026 |
| hsa-miR-182 | −1.07408 | −0.71552 | 1.3422 | 9.90675991 | 33.54506373 |
| hsa-miR-380-5p | 0.31583 | 0.983345 | −0.97438 | 9.90675991 | 34.0190772 |
| hsa-miR-30c | −1.18267 | 0.402311 | 0.58527 | 9.90675991 | 35.0288034 |
| hsa-miR-106b | −0.08408 | 1.027975 | −0.70792 | 11.587147 | 35.57300849 |
| hsa-miR-219 | 1.517496 | −0.96688 | −0.41296 | 11.587147 | 35.95711034 |
| hsa-miR-16 | −0.92068 | −0.50831 | 1.07174 | 11.587147 | 36.48103455 |
| hsa-miR-490 | 0.150144 | 0.835278 | −0.73907 | 11.587147 | 37.01897704 |
| hsa-miR-367 | 0.797077 | 0.41705 | −0.9106 | 11.587147 | 37.31177035 |
| hsa-miR-320 | −0.25449 | 0.937805 | −0.51249 | 11.587147 | 37.998358 |
| hsa-miR-135b | 0.337006 | 0.857104 | −0.89558 | 11.587147 | 38.84789583 |
| hsa-miR-206 | −0.3697 | 0.94394 | −0.43068 | 11.587147 | 39.10460163 |
| hsa-miR-526b* | 1.208306 | −0.78313 | −0.31888 | 11.587147 | 40.34787385 |
| hsa-miR-425 | −0.85102 | 1.009775 | −0.11907 | 11.587147 | 40.77866903 |
| hsa-miR-376a | 0.920047 | −0.1274 | −0.59448 | 11.587147 | 40.92694087 |
| hsa-miR-503 | 0.888295 | −0.13607 | −0.56417 | 14.1171329 | 42.85245703 |
| hsa-miR-487a | 0.162841 | 0.82095 | −0.73784 | 14.1171329 | 43.30927731 |
| hsa-miR-106a | 0.521766 | 0.595635 | −0.83805 | 14.1171329 | 43.48858371 |
| hsa-miR-100 | 0.056388 | −1.20747 | 0.86331 | 14.1171329 | 44.10398892 |
| hsa-let-7f | −0.93439 | −0.05581 | 0.74264 | 14.1171329 | 44.67888809 |
| hsa-miR-511 | 0.310344 | −1.0703 | 0.56996 | 14.1171329 | 44.90173075 |
| hsa-miR-544 | 1.131482 | −0.56382 | −0.42574 | 14.1171329 | 47.12197196 |
| hsa-miR-487b | −0.82112 | −0.41659 | 0.92829 | 14.1171329 | 47.26591312 |
| hsa-miR-363 | −1.27178 | −0.56825 | 1.38003 | 14.1171329 | 47.48233294 |
| hsa-miR-376a* | 0.390564 | 0.732719 | −0.84246 | 14.1171329 | 48.56245152 |
| hsa-miR-375 | 0.243853 | 0.644572 | −0.66632 | 16.5048544 | 49.01224431 |
| hsa-miR-153 | −0.9969 | −1.14696 | 1.60789 | 16.5048544 | 49.40211523 |
| hsa-miR-431 | −0.82106 | −0.91197 | 1.29978 | 16.5048544 | 49.44548869 |
| hsa-miR-217 | −0.99016 | −0.31608 | 0.97968 | 16.5048544 | 49.52370084 |
| hsa-miR-362 | −0.28573 | −1.02521 | 0.98321 | 16.5048544 | 49.87006572 |
| hsa-miR-500 | 0.980354 | −0.56183 | −0.31389 | 16.5048544 | 50.74516842 |
| hsa-miR-372 | −0.96458 | −0.95534 | 1.43994 | 16.5048544 | 50.99446275 |
| hsa-miR-148a | 0.828874 | −1.04752 | 0.16398 | 16.5048544 | 51.40114981 |
| hsa-miR-10a | −0.64047 | −0.52695 | 0.87556 | 16.5048544 | 52.40792839 |
| hsa-miR-433 | −0.22833 | 0.946138 | −0.53835 | 16.5048544 | 52.46791499 |
| hsa-miR-29b | 0.620123 | −1.14248 | 0.39177 | 16.5048544 | 52.72930619 |
| hsa-miR-382 | −0.23578 | 0.894415 | −0.49398 | 16.5048544 | 53.59705171 |
| hsa-miR-335 | −0.3399 | 0.856229 | −0.38725 | 16.5048544 | 53.75101383 |
| hsa-miR-381 | −0.81724 | −0.5501 | 1.02551 | 16.5048544 | 54.38769193 |
| hsa-miR-513 | 0.767999 | 0.015731 | −0.5878 | 18.2605683 | 54.93952212 |
| hsa-miR-328 | 0.233808 | 0.700291 | −0.70057 | 18.2605683 | 55.70006535 |
| hsa-miR-518e | −0.73833 | −0.62201 | 1.02026 | 18.2605683 | 56.00690632 |
| hsa-miR-301 | 0.120635 | 0.905541 | −0.76963 | 18.2605683 | 56.02980543 |
| hsa-miR-25 | −0.66639 | −0.6356 | 0.97649 | 18.2605683 | 56.32551846 |
| hsa-miR-518d | 0.155299 | 0.838491 | −0.74534 | 19.8489011 | 58.23570838 |
| hsa-miR-342 | 0.551634 | −1.20248 | 0.48813 | 22.5464191 | 59.93768974 |
| hsa-miR-521 | −0.96506 | −0.45523 | 1.06522 | 22.5464191 | 60.4907877 |
| hsa-miR-520a | 0.84235 | −1.11148 | 0.20185 | 22.5464191 | 60.95727772 |
| hsa-miR-409-5p | 0.882507 | −0.76108 | −0.09107 | 22.5464191 | 61.30748083 |
| hsa-miR-485-3p | 0.386518 | 0.492904 | −0.65957 | 24.0302433 | 62.62241634 |
| hsa-miR-200a | 0.092787 | −0.91805 | 0.61894 | 25.0641026 | 63.41566145 |
| hsa-miR-29a | 0.482776 | 0.519518 | −0.75172 | 25.0641026 | 63.78211013 |
| hsa-miR-429 | 0.055713 | 0.792367 | −0.63606 | 25.0641026 | 63.82820222 |
| hsa-miR-125a | 0.805608 | −0.59199 | −0.16021 | 28.4739454 | 65.48468447 |
| hsa-miR-502 | 0.754098 | −0.67512 | −0.05923 | 28.4739454 | 65.82932368 |
| hsa-miR-17-3p | −0.43522 | −0.7476 | 0.88711 | 28.4739454 | 66.19614625 |
| hsa-miR-148b | 0.174623 | −0.94181 | 0.57539 | 28.4739454 | 66.43951644 |
| hsa-miR-368 | 0.743991 | −0.88733 | 0.10751 | 28.4739454 | 67.12123684 |
| hsa-miR-142-3p | −0.20516 | −1.124 | 0.99687 | 31.1598558 | 67.73036369 |
| hsa-miR-205 | 0.249306 | 0.845495 | −0.8211 | 31.1598558 | 67.86858747 |
| hsa-miR-373* | 0.052207 | 0.60096 | −0.48988 | 31.1598558 | 68.0705944 |
| hsa-miR-517* | −0.63383 | −0.14053 | 0.58077 | 32.4408284 | 68.51734889 |
| hsa-miR-186 | −0.35485 | 0.80983 | −0.34124 | 32.4408284 | 68.75244349 |
| hsa-miR-452* | −0.27916 | −0.91071 | 0.8924 | 32.4408284 | 70.00183058 |
| hsa-miR-181d | 0.830156 | −0.52455 | −0.2292 | 32.4408284 | 70.12549652 |
| hsa-miR-489 | −0.56225 | −0.14396 | 0.52966 | 32.4408284 | 70.13050269 |
| hsa-miR-183 | 0.159174 | 0.686645 | −0.63436 | 32.4408284 | 70.19307622 |
| hsa-miR-30d | −0.43691 | 0.770014 | −0.24983 | 32.4408284 | 70.23265424 |
| hsa-miR-483 | 0.815477 | −0.84234 | 0.02015 | 32.4408284 | 70.60907344 |
| hsa-let-7a | −0.70813 | 0.463107 | 0.18377 | 32.4408284 | 70.71775515 |
| hsa-miR-451 | −1.23771 | 0.247949 | 0.74232 | 32.4408284 | 70.91507517 |
| hsa-miR-221 | −0.87556 | −0.58425 | 1.09486 | 32.4408284 | 71.4883905 |
| hsa-miR-507 | −0.35197 | −0.58369 | 0.70175 | 32.4408284 | 71.49526037 |
| hsa-miR-526b | 0.030514 | 0.581534 | −0.45904 | 32.4408284 | 71.53582005 |
| hsa-miR-496 | 0.850841 | −0.05299 | −0.59839 | 32.4408284 | 71.54645244 |
| hsa-miR-185 | 0.031791 | 0.573309 | −0.45383 | 32.4408284 | 71.56623333 |
| hsa-miR-510 | 0.742406 | −0.56788 | −0.13089 | 32.4408284 | 71.60493642 |
| hsa-miR-452 | 0.02098 | 0.573449 | −0.44582 | 32.4408284 | 71.81598617 |
| hsa-miR-127 | −0.70529 | −0.18458 | 0.6674 | 32.4408284 | 71.9211001 |
| hsa-miR-212 | −0.61587 | 0.689795 | −0.05545 | 32.4408284 | 72.13105619 |
| hsa-miR-26a | 0.832095 | -0.49388 | −0.25366 | 32.4408284 | 72.20091674 |
| hsa-miR-383 | −0.45492 | 0.701135 | −0.18466 | 32.4408284 | 72.32614427 |
| hsa-miR-422a | −0.26395 | 0.806814 | −0.40714 | 32.4408284 | 72.3764922 |
| hsa-miR-518c* | 0.001353 | 0.564767 | −0.42459 | 41.3047974 | 72.53390646 |
| hsa-miR-196b | −0.67575 | 0.967093 | −0.2185 | 41.3047974 | 72.53717424 |
| hsa-miR-150 | 0.050418 | 0.574385 | −0.4686 | 41.3047974 | 72.67544996 |
| hsa-miR-302b* | 0.246155 | −0.65687 | 0.30804 | 41.3047974 | 72.68497047 |
| hsa-miR-134 | 0.023544 | 0.583001 | −0.45491 | 41.3047974 | 72.76553336 |
| hsa-miR-9* | 0.6188 | −0.93082 | 0.23401 | 41.3047974 | 72.77217111 |
| hsa-miR-453 | −0.74878 | 0.408178 | 0.25545 | 41.3047974 | 72.87972425 |
| hsa-miR-34a | −0.58065 | −0.27546 | 0.64208 | 41.3047974 | 72.96315149 |
| hsa-miR-199a | −0.04344 | 0.698393 | −0.49121 | 41.3047974 | 73.19135993 |
| hsa-miR-526c | 0.000846 | 0.548959 | −0.41235 | 41.3047974 | 73.26518389 |
| hsa-miR-515-3p | −0.73256 | 0.530075 | 0.15187 | 41.3047974 | 73.34759333 |
| hsa-miR-215 | −0.03675 | 0.717886 | −0.51085 | 41.3047974 | 73.34812817 |
| hsa-miR-181c | 0.87805 | −0.73324 | −0.1086 | 41.3047974 | 73.58552846 |
| hsa-miR-302c* | 0.01393 | 0.540437 | −0.41577 | 41.3047974 | 73.63892656 |
| hsa-miR-211 | −0.0302 | 0.714761 | −0.51342 | 41.3047974 | 73.73914691 |
| hsa-miR-515-5p | 0.051513 | 0.532635 | −0.43811 | 41.3047974 | 73.75924315 |
| hsa-miR-520g | −0.95069 | −0.92546 | 1.40711 | 41.3047974 | 74.09075907 |
| hsa-miR-142-5p | −0.0799 | 0.677113 | −0.44791 | 41.3047974 | 74.12613458 |
| hsa-miR-524* | −0.00157 | 0.565716 | −0.42311 | 41.3047974 | 74.15676194 |
| hsa-miR-324-5p | −0.21206 | 0.663358 | −0.33847 | 41.3047974 | 74.23525626 |
| hsa-miR-432* | −0.13292 | 0.678476 | −0.40916 | 41.3047974 | 74.28848132 |
| hsa-miR-518f* | −0.03101 | 0.5454 | −0.38579 | 41.3047974 | 74.30610926 |
| hsa-miR-346 | 0.224671 | 0.480879 | −0.52916 | 41.3047974 | 74.36735065 |
| hsa-miR-216 | −0.45903 | 0.7177 | −0.194 | 41.3047974 | 74.44567177 |
| hsa-miR-361 | 0.03535 | 0.671744 | −0.53032 | 41.3047974 | 74.55343752 |
| hsa-miR-98 | −0.60393 | 0.234364 | 0.27717 | 41.3047974 | 74.60460599 |
| hsa-miR-484 | 0.209461 | 0.738378 | −0.71088 | 41.3047974 | 74.64404077 |
| hsa-miR-189 | −0.80392 | −0.68433 | 1.11619 | 41.3047974 | 75.05309477 |
| hsa-miR-126* | 0.027436 | 0.621051 | −0.48637 | 41.3047974 | 75.09259849 |
| hsa-miR-545 | 0.351442 | 0.506327 | −0.64333 | 41.3047974 | 75.16836638 |
| hsa-miR-302a | −0.0864 | 0.685513 | −0.44934 | 41.3047974 | 75.20719598 |
| hsa-miR-432 | −0.81081 | 0.255479 | 0.4165 | 41.3047974 | 75.25585407 |
| hsa-miR-224 | −0.65873 | −0.04749 | 0.52966 | 41.3047974 | 75.292015 |
| hsa-let-7i | −0.54415 | −0.16195 | 0.52957 | 41.3047974 | 75.71872619 |
| hsa-miR-137 | 0.070381 | 0.650415 | −0.5406 | 41.3047974 | 75.76579176 |
| hsa-miR-369-5p | 0.749466 | −0.27253 | −0.3577 | 41.3047974 | 75.83285292 |
| hsa-miR-526a | −0.04561 | 0.52662 | −0.36076 | 43.4773989 | 76.60067761 |
| hsa-miR-181b | 0.487456 | 0.434359 | −0.69136 | 43.4773989 | 76.6991424 |
| hsa-miR-365 | −0.08579 | 0.600065 | −0.38571 | 43.4773989 | 76.70911636 |
| hsa-miR-512-5p | 0.584274 | −0.53636 | −0.03594 | 43.4773989 | 76.84890105 |
| hsa-miR-493-5p | −0.39502 | −0.55077 | 0.70934 | 43.4773989 | 77.04222504 |
| hsa-miR-200a* | −0.41277 | −0.31389 | 0.545 | 43.4773989 | 77.05673199 |
| hsa-miR-509 | 0.610327 | −0.27544 | −0.25117 | 43.4773989 | 77.06179362 |
| hsa-miR-187 | −0.53083 | −0.10011 | 0.4732 | 43.4773989 | 77.28591448 |
| hsa-miR-214 | −0.55444 | 0.28481 | 0.20223 | 43.4773989 | 77.6270228 |
| hsa-miR-193b | −0.5695 | −0.27523 | 0.63355 | 44.7455322 | 77.84340704 |
| hsa-miR-18b | 0.158782 | −0.67872 | 0.38996 | 44.7455322 | 78.06910367 |
| hsa-miR-27a | −0.67425 | −0.14205 | 0.61222 | 44.7455322 | 78.15702062 |
Table 3. SAM test of Luminex miRNA expression data for BD and C only.
| miRNA | q-value(%) | local FDR(%) |
|---|---|---|
| hsa-miR-219 | 0 | 27.05850719 |
| hsa-miR-380-3p | 0 | 28.5520989 |
| hsa-miR-499 | 0 | 29.39061044 |
| hsa-miR-497 | 0 | 29.86048144 |
| hsa-miR-149 | 0 | 30.16935542 |
| hsa-miR-501 | 0 | 32.7806583 |
| hsa-miR-29c | 0 | 34.68749497 |
| hsa-miR-30e-3p | 12.10826211 | 36.00723604 |
| hsa-miR-504 | 12.10826211 | 37.42550117 |
| hsa-miR-148a | 12.10826211 | 39.79413767 |
| hsa-miR-520a | 12.10826211 | 41.10693353 |
| hsa-miR-526b* | 12.10826211 | 43.98048662 |
| hsa-miR-15b | 17.20647773 | 47.26654182 |
| hsa-miR-342 | 17.20647773 | 47.55108918 |
| hsa-miR-29b | 17.20647773 | 49.13804596 |
| hsa-miR-409-5p | 17.20647773 | 50.09111912 |
| hsa-miR-516-5p | 17.20647773 | 50.61439857 |
| hsa-miR-520d* | 17.20647773 | 50.79672442 |
| hsa-miR-500 | 17.20647773 | 50.82589406 |
| hsa-miR-368 | 18.95206243 | 53.22108834 |
| hsa-miR-30e-5p | 18.95206243 | 54.66652218 |
| hsa-miR-181c | 18.95206243 | 55.4595906 |
| hsa-miR-26b | 18.95206243 | 55.51339027 |
| hsa-miR-511 | 22.70299145 | 58.90676274 |
| hsa-miR-527 | 27.24358974 | 63.91589056 |
| hsa-miR-483 | 27.24358974 | 64.697843 |
| hsa-miR-544 | 27.24358974 | 65.0689104 |
| hsa-miR-518a-2* | 27.24358974 | 65.91311804 |
| hsa-miR-370 | 30.06189213 | 67.72976336 |
| hsa-miR-100 | 69.07664285 | 70.77603166 |
| hsa-miR-498 | 69.07664285 | 71.22258291 |
| hsa-miR-9* | 71.07023411 | 74.68821123 |
| hsa-miR-125a | 71.07023411 | 76.54730008 |
| hsa-miR-502 | 72.64957265 | 78.54613247 |
| hsa-miR-26a | 75.17482517 | 84.87201631 |
| hsa-miR-181d | 77.26224393 | 94.90250066 |
| hsa-miR-510 | 77.26224393 | 95.66638517 |
| hsa-miR-512-5p | 78.16485434 | 100 |
| hsa-miR-299-3p | 78.16485434 | 100 |
| hsa-miR-142-3p | 78.16485434 | 100 |
| hsa-miR-148b | 82.00595701 | 100 |
| hsa-miR-376a | 82.00595701 | 100 |
| hsa-miR-302a* | 82.00595701 | 100 |
| hsa-miR-503 | 82.00595701 | 100 |
| hsa-miR-202* | 82.00595701 | 100 |
| hsa-miR-376b | 82.00595701 | 100 |
| hsa-miR-200a | 84.15841584 | 100 |
| hsa-miR-542-3p | 84.15841584 | 100 |
| hsa-miR-520e | 84.15841584 | 100 |
| hsa-miR-516-3p | 84.15841584 | 100 |
| hsa-miR-302b* | 84.54907162 | 100 |
qPCR Analysis
Upon isolation and purification of RNA from exosome-containing pellets as above, the RNA concentration of each sample tested (Table 1) was calculated using Agilent Bioanlyzer 2100 data. 20 ng of RNA was used to synthesize cDNA in a 20 ul reaction volume employing Exiqon mIRCURY LNA microRNA PCR Universal cDNA Synthesis Kit (Denmark). Quantitative PCR (qPCR) reactions for miRNAs of interest were performed according to the same manufacturer’s protocol. Briefly, cDNA was diluted to a final concentration of 0.1 ng/ul, added to a primer set generated by Exiqon and PCR SYBR Green Master Mix by the same manufacturer, and run for 40 cycles on a Bio-Rad iCycler. To obtain a change in CT (Delta-CT) for each miRNA of interest we needed to have an internal control in every sample. Luminex data as well as preliminary qPCRs established that miR-423 had a stable and robust expression in the samples tested (Table S1). Thus, all the analyzed samples were run in duplicates for each miRNA of interest and for miR-423 as a reference [43]–[45], (http://www.exiqon.com/ls/Documents/Scientific/miRNA-qPCR-guidelines.pdf) in a minimum of three separate plates. In each plate, the average CT value for miR-423 was subtracted from average CT value for a given miRNA of interest to obtain a value of change in CT (Delta-CT) for every sample. Student’s t-tests on control and BD and on control and SZ Delta-CT values were performed using Graph Pad Prism 5.
Results
Exosomal Marker Identification in Exosome-containing Pellets from Postmortem Human Prefrontal Cortex
To confirm the presence of exosomes in our pellet-preparations from postmortem human frozen BA9 cortices, we examined the morphology and antigenicity of the pellet content (Figure 1). On electron microscopy, membrane-bound vesicles with diameters of 70–90 nm were immunoreactive for antigens commonly found in exosomes, CD63 and GAPDH (Figure 1A and B). Next, we evaluated the efficiency of our exosomal extractions from brain tissue. Western blot analysis revealed the presence of flotillin-2, protein commonly associated with exosomal membrane [35], in both BA9 pellets and in exosomes-containing pellets from the medium of cultured H4 cells (Figure 1C). The exosomal extraction procedures depleted flotillin-2 from the supernatants (Figure 1C).
Figure 1. Characterization of exosome-containing pellets from human brain tissue.
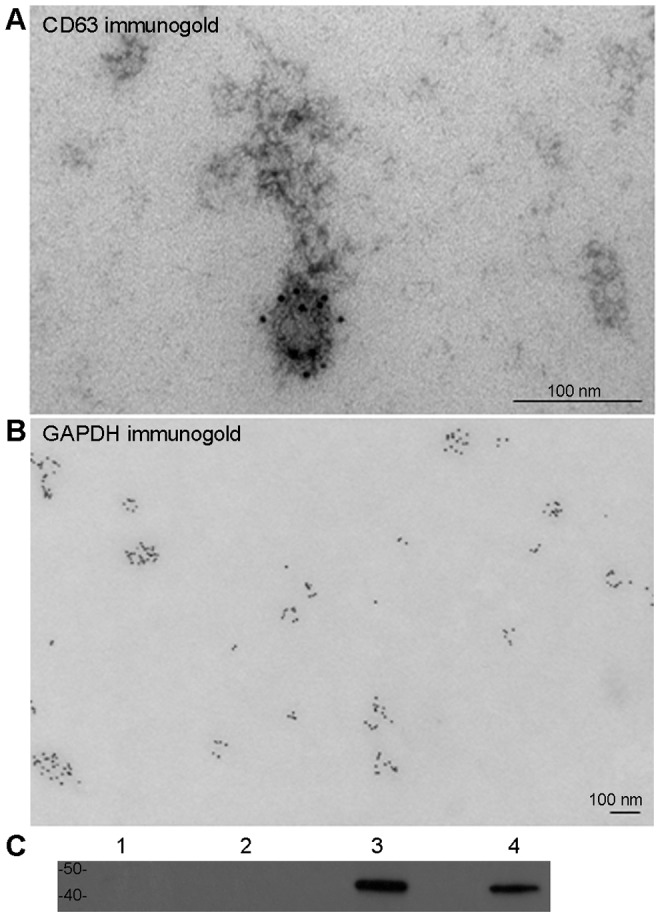
Electron microscopy of exosomal extractions from BA9 cortices demonstrates the presence of microvesicles (∼70–100 nm in diameter). Upon immuno-gold labeling procedure with antibodies against CD63 (A; additional negative staining highlights membrane of the vesicle) and GAPDH (B), the microvesicles reveal the presence of exosome-associated antigens. Bars indicate 100 nm. Comparison of exosomal extraction procedure products from BA9 cortices and H4 cell-culture reveals similar outcomes in Western blot. Supernatant of BA9 exosome-containing pellets (lane 1), supernatant of H4 exosome-containing pellets (lane 2), BA9 exosome-containing pellets reconstituted in PBS (lane 3), and of H4 exosome-containing pellets reconstituted in PBS (lane 4), show robust presence of exosomal marker flotillin-2 in the pellets, but not in the supernatants (C).
Luminex miRNA Expression Analysis in Exosome-containing Pellets from SZ and BD Patients Suggest Differential Expression of a Subset of miRNAs in Comparison to Controls
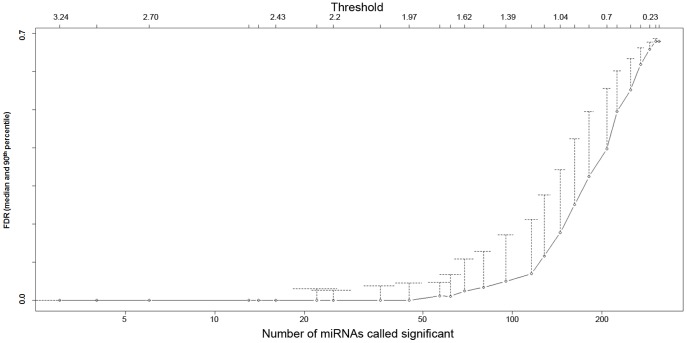
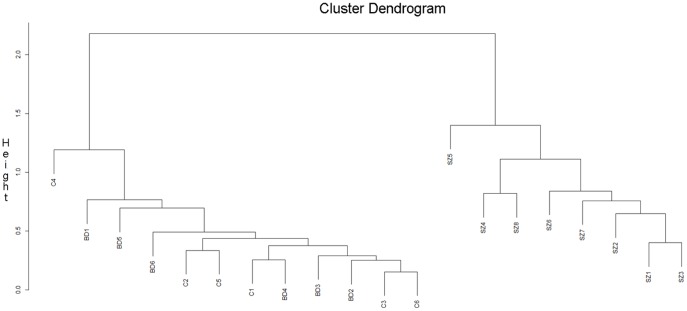
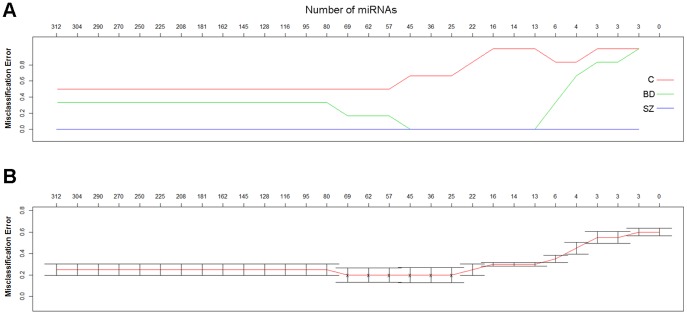
Luminex miRNA expression data in exosome-containing pellets from prefrontal (BA9) gray matter of 8 SZ patients (SZ 1–8), 6 BD patients (BD 1–6), and 6 controls (C 1–6) (Table 1) were submitted to Student’s t-test. These data reveal several significantly differentially expressed miRNAs in either BD or SZ, or both in comparison to controls. Bonferroni Step-down Holm Correction was then applied to correct for multiple comparisons [39]. This correction resulted in the absence of significantly differentially expressed miRNAs in BD samples, while the expression of only three miRNAs (miR- 31, -33, and -96) was significantly enhanced in SZ samples in comparison to controls and to BD (Table S1). In an alternative statistical approach towards identification of miRNAs differentially expressed in the three groups examined, we submitted raw Luminex miRNA expression data to Significance Analysis of Microarrays (SAM) program [36],[37] (Table 2). SAM ranked the expressions of examined miRNA expression using z-scores (Table 2). For the 21 top-ranked miRNAs, Prediction Analysis of Microarrays (PAM) [38]– derived q-value was 0% (Figure 2). We next examined how the expression of these 21 top-ranked miRNAs might influence the clustering of our samples. Surprisingly, the dendrogram from the clustering analysis [38] (Figure 3) suggests that the expression the 21 top-ranked miRNAs segregated SZ samples from BD samples and controls, leaving BD samples and controls undifferentiated from one another. In order to remove the influence of SZ group on the ranking system, the SAM program was run again for BD and control groups only (Table 3) and yielded 7 miRNAs with q- value of 0% (Table 3). This group of miRNAs, however, had a rather limited ability to segregate BD samples from controls (cluster dendrogram not shown). These results prompted us to submit the entire Luminex data to a PAM-derived misclassification rate analysis to determine if the miRNA expression values of each sample in a group have a reliable predictive power, i.e. if they can serve as biomarkers for a given group [42]–[38]. The misclassification rate was 0 for the SZ group (Figure 4), indicating that: a) the examined miRNA expressions were tightly grouped in SZ as previously indicated by the cluster dendrogram (see Figure 3); and b) the expression values of a very small number of miRNAs can assign an SZ sample to its correct group. The control samples had most variable miRNA expression levels associated with high misclassification rate regardless of how many miRNAs were evaluated (Figure 4). Finally, Figure 4 shows that the misclassification rate for BD samples is dependent on the number of miRNAs evaluated: if ∼13–16 miRNAs are taken into account, BD samples separate well from controls and reach zero misclassification rate that is characteristic of SZ samples (Figure 4).
Figure 2. Median and 90th percentile of False Discovery Rates (FDR) in order left-to-right of threshold value and number of statistically significant results for all three groups (C, BD, and SZ).
The Y-axis is an estimate of the percentage of false positives. High-ranked miRNA (at the left) have a low rate of false positives, while lower-ranked miRNA (moving toward the right) have higher rates of false positives.
Figure 3. Hierarchical clustering analysis of top 21 ranked miRNAs from FDR analysis.
Correlation coefficient (cc) was generated to assess the relationship between the expression values of each sample and the rest of the samples (see Methods). The coefficient is 1 if their expression profiles are highly similar and 0 if their expression profiles are highly divergent. The clustering graph is built so that the samples with similar expression patterns are clustered at the bottom while more differential patterns are at the top of the graph.
Figure 4. Misclassification rate analysis.
(A) Controls (red) have the most variable miRNA expression. The expression data have less variability in BD (green) and the least in SZ (blue; misclassification rate = 0), resulting in stronger predictive power within their respective clinical groups.
Covariate Analysis: the Effect of Medications on SAM Results
The clinical information accompanying our samples failed to provide sufficient data to control for tobacco, alcohol or recreational drug consumption (Table S4), while the information of prescribed medications (Table 4) was well defined. The cases were marked if the patients took at least one medication from the seven drug classes constructed on the basis of the common mechanism(s) underlying drug effect (A-G; Table 5). Since a standard error is used to generate a covariate effect, only drug classes with at least three case-subjects taking a drug and three case-subjects not taking a drug could be evaluated as covariates. Thus, only drug classes A (antipsychotics used in SZ only), B (neurotransmitter receptor-site binders used in both BD and SZ), and C (sedatives, hypnotics, anticonvulsants and analgesics used in both BD and SZ) could be controlled for.
Table 4. Medications (grouped in classes A-G) prescribed to subject cases for psychiatric and neurological symptoms.
| Case | Medications prescribed for psychiatric and neurological symptoms, daily mg | A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|---|---|
| SZ1 | Levomepromazine 100, Haloperidol 10, Diazepam 5 | X | X | |||||
| SZ2 | Amisulpride 200, Quetiapine 200, Clonazepam 0.5, Oxcarbazepine 300 | X | X | |||||
| SZ3 | Haloperidol 5, Olanzapine 10, Lorazepam 1, Clomethiazole 192, Imipramine 50 | X | X | X | ||||
| SZ4 | Lormetazepam 1, Olanzapine 10, Lorazepam 1, Clomethiazole 192 | X | X | |||||
| SZ5 | Olanzapine 15 | X | ||||||
| SZ6 | Phenitoine 100 | X | ||||||
| SZ7 | Clozapine 400, Trazadone 50, Haloperidol 5, Lorazepam 1 | X | X | X | X | |||
| SZ8 | None | |||||||
| BD1 | Lithium 400, Methylphenidate 20, Diazepam 10, Oxazepam 20 | X | X | X | ||||
| BD2 | Lithium 400, Quetiapine 25, Midazolam (amount?) | X | X | X | ||||
| BD3 | Risperidone 1.5, Gabapentin 600, Nefazodone 600, Topiramate 75, Ziprasidone 20 | X | X | X | ||||
| BD4 | Valproic acid 1500, Paroxetine 20, Clonazepam 4, Olanzapine 10 | X | X | X | ||||
| BD5 | Lithium (amount?) | X | ||||||
| BD6 | Valproic acid 1000, Quetiapine 125 | X | X | |||||
| BD7 | Moclobemide 300 | X | ||||||
| BD8 | Dexamphetamine sulphate 5 | X | ||||||
| BD9 | Tranylcypromine 60 | X | ||||||
| C1–C13 | None |
Table 5. Classes (A–G) encompassing the medications in Table 4.
| Class | Description of medication class | Prescribed medicationfrom the class | Effects |
|---|---|---|---|
| A | Antipsychotics specifically for SZ | Levomepromazine | Phenothiazine Group; dopamine, adrenalin, histamine, acetylcholine and serotonin antagonist; antipsychotic effects |
| A | Antipsychotics specifically for SZ | Haloperidol | dopamine antagonist; effects similar to Phenothiazine group; antipsychotic effects |
| B | Neurotransmitter receptor- site binders usedin BD and SZ | Olanzapine | dopamine, serotonin, adrenergic antagonist with antihistaminic effect; antipsychotic properties |
| B | Neurotransmitter receptor- site binders usedin BD and SZ | Clozapine | dopamine, serotonin, adrenergic antagonist with antihistaminic effect; antipsychotic properties |
| B | Neurotransmitter receptor- site binders usedin BD and SZ | Quetiapine | dopamine, serotonin, adrenergic antagonist with antihistaminic effect; antipsychotic properties |
| B | Neurotransmitter receptor- site binders usedin BD and SZ | Amisulpride | dopamine and serotonin antagonist; antipsychotic properties |
| B | Neurotransmitter receptor- site binders usedin BD and SZ | Risperidone | dopamine, serotonin, adrenergic antagonist with antihistaminic effect; antipsychotic properties |
| B | Neurotransmitter receptor- site binders usedin BD and SZ | Ziprasidone | dopamine, serotonin, adrenergic antagonist with antihistaminic effect; antipsychotic properties |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Diazepam | Benzodiazipines, i.e. GABA agonists; sedative, hypnotic, with anticonvulsant effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Clonazepam | Benzodiazipines, i.e. GABA agonists; sedative, hypnotic, with anticonvulsant effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Lorazepam | Benzodiazipines, i.e. GABA agonists; sedative, hypnotic, with anticonvulsant effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Oxazepam | Benzodiazipines, i.e. GABA agonists; sedative, hypnotic, with anticonvulsant effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Midazolam | Benzodiazipines, i.e. GABA agonists; sedative, hypnotic, with anticonvulsant effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Clomethiazole | GABA agonist; sedative, hypnotic, with anticonvulsant effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Oxcarbazepin | ion, mostly sodium, channel stabilizer; anticonvulsant and mood stabilizing effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Phenitoin | sodium channel stabilizer, anticonvulsant effect |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Gabapentin | GABA agonist; anticonvulsant and analgesic effects |
| C | Sedatives, hypnotics, anticonvulsants and analgesicsin BD and SZ | Topiramate | mechanism unclear; anticonvulsant effect |
| D | Antipsychotics specifically for BD | Valproic acid | GABA effects enhancer; anticonvulsant and mood-stabilizing effects |
| D | Antipsychotics specifically for BD | Lithium | mechanism unclear; mood-stabilizing effects |
| E | Psycho-stimulants used in BD | Moclobemide | monoamine oxidase inhibitors; antidepressant effects |
| E | Psycho-stimulants used in BD | Tranylcypromine | monoamine oxidase inhibitors; antidepressant effects |
| E | Psycho-stimulants used in BD | Methylphenidate | dopamine enhancer, psychostimulant |
| E | Psycho-stimulants used in BD | Dexamphetamine | psychostimulant |
| F | Serotonin and adrenergic antagonists for BD | Nefazodone | serotonin and adrenergic antagonist; antidepressant effects |
| F | Serotonin and adrenergic antagonists for BD | Paroxetine | serotonin re-uptake inhibitor; antidepressant and anxiolytic effects |
| G | Serotonin antagonist for SZ | Trazodone | serotonin antagonist and reuptake inhibitor, antidepressant, anxiolytic, and sedative effects |
In a simple format of covariate adjustment [46] the effects of multiple independent variables are added together in a linear regression model. To guard against Type I errors, we designed a model to adjust for covariates using the SAM program. SAM was rerun to compare case subjects taking a specific drug class medication against case subjects not taking it, and a list of covariate effects for drug classes A–C was derived for top-ranked miRNA in the study (Table 6). Those effects were then subtracted from the original z-scores (Tables 2 and 3) and p-values were added to demonstrate the effects of the covariate on significance (Tables 7 and 8).
Table 6. Covariate effects of medications on highly-ranked miRNAs in SAM (top-21 in Table 2 and top-12 in Table 3).
| miRNA | Class A | Class B | Class C |
|---|---|---|---|
| hsa-miR-31 | −0.5474735 | 0.0027362 | |
| hsa-miR-33 | 0.6342707 | −0.3112329 | 0.9637398 |
| hsa-miR-96 | 0.040926 | 0.5554684 | |
| hsa-miR-28 | 1.1396825 | 1.0107414 | |
| hsa-miR-30e-5p | 0.3379079 | ||
| hsa-miR-199a* | −0.8762626 | ||
| hsa-miR-501 | 0.0812317 | −0.167766 | −0.6013733 |
| hsa-miR-504 | 0.2946595 | −0.1356475 | −0.3858902 |
| hsa-miR-15b | −0.1307195 | ||
| hsa-miR-29c | −0.1678 | 0.3820155 | 0.1279756 |
| hsa-miR-455 | −0.2162923 | −0.2365768 | |
| hsa-miR-380-3p | −0.1295169 | −0.2513642 | −0.2837673 |
| hsa-miR-323 | 0.0956179 | −0.3292096 | |
| hsa-miR-527 | 0.3199353 | −0.3980191 | −0.4156484 |
| hsa-miR-93 | 0.8926459 | 0.8342627 | 1.4247242 |
| hsa-miR-32 | −0.3081563 | ||
| hsa-miR-20b | −0.3025591 | ||
| hsa-miR-516-5p | 0.1006308 | −0.2462411 | −0.4384718 |
| hsa-miR-92 | 1.5916982 | 1.9080176 | |
| hsa-miR-30a-3p | 0.2072069 | ||
| hsa-miR-497 | 0.1045716 | −0.2486034 | 0.036478 |
| hsa-miR-219 | 0.3630727 | −0.2988121 | |
| hsa-miR-499 | −0.0609767 | −0.3046793 | −0.2561058 |
| hsa-miR-149 | 0.654253 | −0.2538624 | 0.04254 |
| hsa-miR-30e-3p | 0.4068861 | −0.0672197 | 0.11598 |
| hsa-miR-148a | |||
| hsa-miR-520a | −0.0963095 | −1.5626633 | |
| hsa-miR-526b* | 0.3595811 | 0.594503 |
Table 7. Top-ranked miRNAs from Table 2, adjusted for covariate effects of medications (classes A, B, C, and B+C).
| miRNA | SZ score (z-score from SAM) | p-value | Class A covariate adjust | p-value | Class B covariate adjust | p-value | Class C covariate adjust | p-value | Classes B+C covariate adjust | p-value |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-31 | 3.55283 | 0.0007 | 4.10031 | 0.00009 | 3.55010 | 0.00073 | 3.55283 | 0.00072 | 3.5501 | 0.0007 |
| hsa-miR-33 | 3.10046 | 0.0032 | 2.46619 | 0.01906 | 3.41170 | 0.00118 | 2.13672 | 0.04069 | 2.4480 | 0.0199 |
| hsa-miR-96 | 2.98240 | 0.0046 | 2.94147 | 0.00527 | 2.42693 | 0.02099 | 2.98240 | 0.00467 | 2.4269 | 0.0210 |
| hsa-miR-28 | 2.31611 | 0.0272 | 1.17643 | 0.19970 | 1.30537 | 0.17017 | 2.31611 | 0.02729 | 1.3054 | 0.1702 |
| hsa-miR-30e-5p | 2.37744 | 0.0236 | 2.03954 | 0.04985 | 2.37744 | 0.02363 | 2.37744 | 0.02363 | 2.3774 | 0.0236 |
| hsa-miR-199a* | 2.21841 | 0.0340 | 3.09467 | 0.00332 | 2.21841 | 0.03406 | 2.21841 | 0.03406 | 2.2184 | 0.0341 |
| hsa-miR-501 | −1.38651 | 0.1525 | −1.46774 | 0.13587 | −1.21874 | 0.18983 | −0.78514 | 0.29312 | −0.6174 | 0.3297 |
| hsa-miR-504 | −1.26857 | 0.1784 | −1.56323 | 0.11756 | −1.13292 | 0.20999 | −0.88268 | 0.27022 | −0.7470 | 0.3018 |
| hsa-miR-15b | 1.94092 | 0.0606 | 2.07164 | 0.04666 | 1.94092 | 0.06066 | 1.94092 | 0.06066 | 1.9409 | 0.0607 |
| hsa-miR-29c | −1.25599 | 0.1812 | −1.08819 | 0.22068 | −1.63801 | 0.10430 | −1.38397 | 0.15311 | −1.7660 | 0.0839 |
| hsa-miR-455 | 1.93342 | 0.0615 | 2.14972 | 0.03957 | 2.17000 | 0.03788 | 1.93342 | 0.06154 | 2.1700 | 0.0379 |
| hsa-miR-380-3p | −0.78517 | 0.2931 | −0.65565 | 0.32178 | −0.53381 | 0.34597 | −0.50140 | 0.35182 | −0.2500 | 0.3867 |
| hsa-miR-323 | 1.79862 | 0.0791 | 1.70300 | 0.09357 | 2.12783 | 0.04147 | 1.79862 | 0.07915 | 2.1278 | 0.0415 |
| hsa-miR-527 | −1.15943 | 0.2037 | −1.47937 | 0.13356 | −0.76141 | 0.29855 | −0.74378 | 0.30254 | −0.3458 | 0.3758 |
| hsa-miR-93 | 1.59984 | 0.1109 | 0.70719 | 0.31068 | 0.76558 | 0.29760 | 0.17512 | 0.39287 | −0.6591 | 0.3210 |
| hsa-miR-32 | 1.70564 | 0.09315 | 2.01380 | 0.05252 | 1.70564 | 0.09315 | 1.70564 | 0.09315 | 1.7056 | 0.0931 |
| hsa-miR-20b | 1.87742 | 0.06847 | 2.17998 | 0.03706 | 1.87742 | 0.06847 | 1.87742 | 0.06847 | 1.8774 | 0.0685 |
| hsa-miR-516-5p | −0.84614 | 0.27890 | −0.94677 | 0.25484 | −0.59990 | 0.33325 | −0.40767 | 0.36713 | −0.1614 | 0.3938 |
| hsa-miR-92 | 1.56396 | 0.11743 | −0.02774 | 0.39879 | 1.56396 | 0.11743 | −0.34406 | 0.37601 | −0.3441 | 0.3760 |
| hsa-miR-30a-3p | 1.67931 | 0.09740 | 1.47210 | 0.13500 | 1.67931 | 0.09740 | 1.67931 | 0.09740 | 1.6793 | 0.0974 |
| hsa-miR-497 | −0.56728 | 0.33965 | −0.67185 | 0.31834 | −0.31867 | 0.37919 | −0.60376 | 0.33247 | −0.3551 | 0.3746 |
Table 8. Top-ranked miRNAs from Table 3, adjusted for covariate effects of medications (classes B, C, and B+C).
| miRNA | BD score (z-score from SAM) | p-value | Class B covariate adjust | p-value | Class C covariate adjust | p-value | Classes B+C covariate adjust | p-value |
|---|---|---|---|---|---|---|---|---|
| hsa-miR-219 | −2.8237 | 0.0074 | −2.8237 | 0.0074 | −3.1225 | 0.0030 | −3.1225 | 0.0030 |
| hsa-miR-380-3p | −2.2933 | 0.0287 | −2.0419 | 0.0496 | −2.0095 | 0.0529 | −1.7581 | 0.0850 |
| hsa-miR-499 | −2.2226 | 0.0337 | −1.9179 | 0.0634 | −1.9665 | 0.0576 | −1.6618 | 0.1002 |
| hsa-miR-497 | −2.1947 | 0.0358 | −1.9460 | 0.0600 | −2.2311 | 0.0331 | −1.9825 | 0.0558 |
| hsa-miR-149 | −2.1785 | 0.0371 | −1.924 | 0.0625 | −2.2211 | 0.0338 | −1.9672 | 0.0576 |
| hsa-miR-501 | −2.0751 | 0.0463 | −1.9073 | 0.0647 | −1.4737 | 0.1346 | −1.3059 | 0.1700 |
| hsa-miR-29c | −2.0161 | 0.0522 | −2.3981 | 0.0224 | −2.1440 | 0.0400 | −2.5260 | 0.0164 |
| hsa-miR-30e-3p | −1.9787 | 0.0563 | −2.0274 | 0.0444 | −2.0947 | −1.9115 | 0.0641 | 0.0510 |
| hsa-miR-504 | −1.9404 | 0.0607 | −1.4189 | 0.1191 | −1.5545 | −1.8048 | 0.0782 | 0.1457 |
| hsa-miR-148a | −1.8790 | 0.0682 | −1.8790 | 0.0682 | −1.8790 | −1.8790 | 0.0682 | 0.0682 |
| hsa-miR-520a | −1.8457 | 0.0726 | −0.2830 | 0.0726 | −1.8457 | −0.2830 | 0.3832 | 0.3832 |
| hsa-miR-526b* | −1.7745 | 0.0826 | −2.3691 | 0.0826 | −1.7745 | −2.3691 | 0.0241 | 0.0241 |
When adjusted for drug class A (antipsychotics used in SZ only), the significance of results for miRNAs 31, 33, 96, 30e-5p, and 199a* was preserved, while miRNAs 15b, 455, 32, and 20b acquire significance (Table 7, see also Table 2). After the adjustment for B (neurotransmitters receptor-site binders) and C (sedatives, hypnotics, anticonvulsants and analgesics) drug classes in SZ subjects, the significance for miRNAs 31, 33, 96, 30e-5p, and 199a* was still preserved while miR-455 (again) acquired significance together with miR-323 (Table 7, see also Table 2). In BD subjects, the consideration of combined effects of B and C drug classes heightened the significance of the results for miRNAs 219 and 29c while miRNA 30e-3p and 526b* acquired significance (Table 8, see also Table 3). Importantly, Wilcoxon test also indicated the expression of miRNAs 219 and 526b to be significantly different in BD cases in comparison to controls, but failed to discover miRNA 29c (Table S3; see Discussion).
Finally, while effects related to gender were not relevant to this study due to a male dominated cohort, we did consider the ages of our cases (Table 1). Although the SZ group of cases is on average older than BD and control groups, no significant difference in age between groups exists (Table S5).
qPCR Analysis Confirms Differential Expression of miR-497 in SZ, and miR-29c in BD Samples in Comparison to Controls
To verify microarray data for miRNAs that may be differentially expressed in our three analyzed groups, controls, BD, and SZ, we performed a series of qPCR experiments. The primers for the following miRNAs worked in reactions with cDNA derived from each of our samples available for analysis: miR-31, -15b, -29c, -497, -219, and -149. These miRNAs were among the highest ranked according to the likelihood to be differentially expressed in both SZ and BD (miR-31, -15b, -29c, and -497; Table 2), or just BD (miR-219 and -149; Table 3). To further test the reproducibility of our data we used additional BD and additional control cases that became available through brain banks and BMC autopsy service (Table 1): SZ 1–6 (same as in Luminex experiment), BD 5–9 (three new samples), and C 6–13 (seven new samples). As shown in Table 9 and Figure 5, qPCR analyses confirmed miR-29c to be significantly differentially expressed (2.77 fold increased) in the examined BD samples, and miR-497 in SZ samples (2.35 fold increased) in comparison to controls.
Table 9. Student’s t-test comparison of qPCR Delta-CT.
| miRNA | p-value, BD | Significant fold change | p-value, SZ | Significant fold change |
|---|---|---|---|---|
| hsa-miR-15b | 0.6971 | 0.7906 | ||
| hsa-miR-29c | 0.0237 | 2.77 | 0.501 | |
| hsa-miR-31 | 0.426 | 0.3699 | ||
| hsa-miR-149 | 0.591 | 0.5011 | ||
| hsa-miR-219 | 0.8723 | 0.4059 | ||
| hsa-miR-497 | 0.1969 | 0.0026 | 2.35 |
Figure 5. In comparison to controls, the expression of miR-29c and miR-497 is significantly increased in BD and SZ samples, respectively.
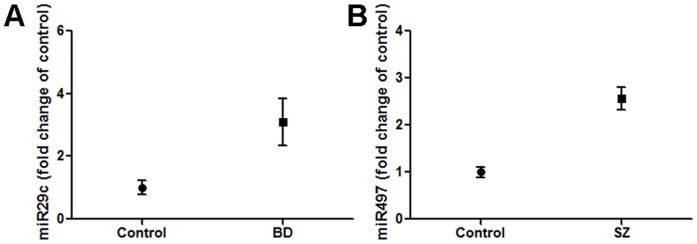
Average exosomal RNA extracted from BA9 cortices of BD samples show a 2.77 fold increase of miR-29c in comparison to controls (A). SZ samples reveal 2.35 fold increase of miR-497 when compared to controls (B). Error bars indicate S.E.M.
Discussion
While genetic component has been extensively researched in BD and SZ patients, the biological markers for these diseases have proven remarkably elusive. The putative susceptibility genes in both BD and SZ regulate cell-signaling [47]–[48]. This study analyzed the expression of miRNAs found in exosomes (Figure 1A and B), signaling vesicles capable of cell-to-cell transfer of miRNAs, in the prefrontal cortices of patients diagnosed with either BD or SZ and of individuals with no mental illnesses. Standard statistical analysis (Student’s t-test with Bonferroni Step Down/Holm correction) of the results of Luminex oligonucleotide- based screen for the expression of 312 miRNAs revealed only three miRNAs differentially expressed in SZ in comparison to BD and control samples (Table S1). While these three miRNAs (miR-31, -33, and -96) were also most highly ranked in a z-score based SAM, an additional 18 miRNAs were suggested to have truly differential expression (Table 2). Accordingly, the expression values of about 21 miRNAs were not false discoveries in a three-way comparison (Figure 2). These 21 miRNAs distinguished SZ but not BD samples from controls relatively well (Figure 3). Upon elimination of the distinctive influence of SZ samples in the three-way comparison (Table 2), the short list of most differentially expressed miRNA in BD compared to controls (Table 3) still featured several of the original top 21-ranked miRNAs. The covariate adjustment for the medications used by some of the subject cases (Tables 4 and 5), had nominal effects on significance of differential miRNA expression in the SZ group, while in the BD group the difference in the expression of miR-30e-3p, -219 and -29c attained heightened significance (Tables 6–8). To verify the microarray data, we performed qPCR analysis of the selected miRNAs in a partially new set of samples. Two miRNAs were confirmed to have significantly altered expression: in comparison to controls, the expression of miR-497 and miR-29c were significantly higher in SZ samples and BD samples, respectively (Table 9).
Our study has several novel and important findings. While exosomal content has been intensively studied in human diseases ever since the report of Valadi et al. in 2007 [30], we demonstrated here for the first time that exosomal miRNAs could be obtained from frozen postmortem brain tissue. On electron microscopy our PFC- derived exosome-containing pellets showed membrane bound vesicles within the range of the reported size of exosomes [49] (Figure 1A and B). In addition to the identification by size, shape, and miRNA content (intracellular micro-vesicles of similar size, e.g. lysosomes, do not contain miRNAs), we demonstrated a robust signal for CD63 and GAPDH in the micro-vesicles extracted from the human PFC (Figure 1A and B). According to Mathivanan and Simpson (2009) [50] exosomes from different sources have varying sets of protein markers on their surface, with CD63 being the second most, and GAPDH being the fourth most commonly reported exosomal antigen. Finally, our exosome-containing pellets were enriched with exosomal marker flotillin-2 [35] on Western blots, similar to the pellets from the medium of cultured cells (Figure 1C). Having insight into the cortical exosomal miRNA content may be of particular importance because exosomes are released into the CSF, and thus accessible for evaluation in living patients [51]. A recent study identified a substantial number of miRNAs exclusively or predominantly expressed in CSF [52]. Comparing the data from brain tissue with CSF will indeed require clinical investigation since postmortem brain depositories do not currently have sufficient number of samples and/or amount of CSF from neuropsychiatric patients. So far, the search for specifically altered miRNA expression in peripheral blood mononuclear cells (PBMCs) in SZ yielded diverse results [53], [54]. Three miRNAs were found to be differentially expressed in PBMCs as well as in PFC (BA46) of SZ patients [53]–[55].
The second novel and important feature of our investigation lies in the fact that our statistical analyses pointed to both differences and similarities between the miRNA expression changes in SZ and BD samples in comparison to control samples. The SAM top 21- ranked miRNAs (Table 2, Figure 2) had the ability to separate SZ samples, but not BD samples from the controls (Figure 3). This finding is directly connected to the suggested inherent differences in the variability of miRNA expression values in control samples in the three analyzed groups of samples, illustrated by the misclassification error rate analysis (Figure 4). The high variability of miRNA expression values in control samples represents perhaps a defining feature - the agility of the cortical transcriptional regulation in the absence of mental disease.
Initial statistical analysis revealed three miRNAs (miR-31, -33, and -96) differentially regulated in SZ samples (Table S1). Of those three, miR-33 was previously reported among misexpressed miRNAs in PFCs of both SZ and BD individuals [10]. Since we performed qPCR for additional miRNAs that were likely to be differentially expressed in SZ and BD samples according to SAM (Tables 2 and 3), we found significantly increased miR-497 expression in exosome-containing pellets from PFCs of SZ patients (Table 9). Perkins et al. (2007) [11] reported 16 miRNAs to be differentially expressed in PFCs of SZ subjects, one being miR-195. Both miR-195 and -497 belong to the well-studied miR-15/107 gene family. miR-15/107 miRNAs share most of their targets and are implicated in the pathogenesis of neoplasms, neurodegenerative diseases and heart disease [56]. Importantly, the up-regulation of these miRNAs expected to affect cortical gene expressions, has been reported in SZ [57]. Interestingly, miR-497 was also found to promote ischemic neuronal death by negatively regulating anti-apoptotic proteins, bcl-2 and bcl-w [58]. Three more miRNAs from our SAM top 21-ranked miRNAs were on the list of significantly down-regulated miRNAs in SZ by Perkins and colleagues [11]: miR-92, -20b, and -30e. miR-92 and -20b belong to miR-17-92 cluster known to be a potent oncogene [59]. Potentially pertinent for SZ pathobiology, miR-92 has also been suggested to developmentally regulate neuronal K(+)Cl(−) co-transporter 2 (KCC2) that modulates effects of GABA [60]. Finally, miR-30e may regulate neuronal death as down-regulation of miR-30e expression promoted neuronal survival in long-lived calorie-restricted mice [61]. However, we were not able to validate differential expression of miRNA-30e in the exosome-containing samples of PFCs in SZ (data not shown). We found miR-29c to be significantly increased in the exosome-enriched preparations from PFC of individuals diagnosed with BD in comparison to controls according to SAM and qPCR analysis. Interestingly, the expression of miR-29c was not significantly different in BD cases compared to controls according to Wilcoxon test applied to our Luminex data (p-value 0.0649; Table S3). The fact that miR-29c was significantly differentially expressed in SAM (Tables 2 and 3), further confirmed by qPCR analysis in BD vs. controls (Table 9), suggests that FDR testing through the use of SAM might be a superior strategy for discovery of true positives.
Like miR-497, the miRNA confirmed to be up-regulated in SZ samples, miR-29c was among the top-ranked in both SAM analyses (Table 2 and 3). In other words, both of these miRNAs were considered highly differentially regulated in both diseases according to SAM. Moreover, miR-29c was also reported by Perkins et al. to be differentially expressed miRNAs in PFC of SZ patients [11]. Interestingly, miR-29 together with miR-31, the top-ranked differentially expressed miRNA by SAM but not by qPCR (Table S1, Table 8), was proposed to regulate several cell-adhesion machinery components that are affected in the pathogenesis of many diseases [62]. The up-regulation of miR-29c in the cortical exosomes of BD patients is particularly intriguing because miR-29c is induced by canonical Wnt signaling [63] that is antagonized by GSK-3, a know substrate of inhibition by lithium, a first line of therapy for BD [64].
The interpretation of our findings has the following limitations. First, the relationship between exosomal and cellular miRNAs is not understood: is the exosomal miRNA aberration a part of pathogenesis, or a corrective attempt? Second, age-associated common brain pathology, may complicate the interpretation of our results, although vascular and early Alzheimer’s disease-associated pathologies were relatively evenly distributed in our samples (Table 1). Third, we were limited by a finite number of the samples available that fulfilled the RNA quality standards. However, the statistical methods we applied were developed specifically as a “non-parametric” test for miRNA expression studies. This test has a built-in mechanism for evaluating small sample sizes, and makes use of a “positive constant” standard error value as one of its formulaic variables. This ensures that miRNAs with small values are not elevated high in the rankings simply because they have a small standard error. By comparison t-tests often assign strong significance to miRNAs with small expression levels due to their small standard errors. The PAM and SAM programs we used are therefore optimal in dealing with irregular data with small sample size such as ours.
Fourth, our microarray examined the expression of 312 miRNA, while hundreds more are known to exist and thus may be important for the pathobiology of SZ and BD. Finally, due to the variable efficiency of the primers in our cDNA samples, we could carry qPCRs for only six miRNAs (Table 8, 9) featured among our top-ranked in SAM analyses (Tables 2 and 3). Nevertheless, the results of qPCR validation are remarkable considering that we examined different samples from the ones analyzed in the microarray, in particular controls - most likely to exhibit highly variable miRNA expressions (Figure 4). Interestingly, our qPCR analysis did not validate SZ-associated highly significant up-regulation of miR-31 by Luminex assay (Table 2, Table S1). Considering, however, that Gardiner et al. (2012) [54] confirmed miR-31 down-regulation in PBMCs of SZ patients, miR-31 dysregulation in SZ cannot be ruled-out. Our search for exosome-derived biomarkers yielded two candidates, miR-497 and miR-29c, in the PFC of SZ and BD patients respectively, potentially offering a novel insight into the pathogenesis of these diseases. In addition, rapid development of exosome nanotechnology [65] adds the promise of alternative therapeutics delivery [66]–[67] to the emerging diagnostic potential of brain-derived exosomes in neuropsychiatric diseases.
Acknowledgments
We thank George Tejada and David Ennulat of Harvard Brain Tissue Resource Center and Michiel Kooreman of The Netherlands Brain Bank for tissue and clinical histories procurement, Maria Ericsson and Howard Mulhern for expert electron microscopy processing, Ozge Cagsal-Getkin for help with Luminex assay, and Zhigang Xie for critical reading of the manuscript.
Supporting Information
Figure S1
RNA quality control. Representative RNA (sample C13, RIN = 7.0; the sample closest to the average (6.96) and median (6.85) value for the set) yields two strong bands at 40 s and 45 s representing the 18 S and 28 S ribosomal RNA (A). The electropherogram shows a marker peak and the two ribosomal peaks corresponding to 18 S and 28 S subunits. RIN value on a scale of 1–10 is computed based on the presence of correct ribosomal peaks, the ratio between those peaks, and the extent of RNA fragmentation (B). Relationship between RIN and mRNA and miRNA profiles (C): Next to the ladder (left), total and small RNA profiles of three degradation stages of a single RNA sample are shown (RIN 8.4 = not degraded, RIN 3.1 = partially degraded, RIN 2.5 = severely degraded). Note that both mRNA (RT PCR, middle) and miRNA (Luminex, right) profiles obtained from a sample with RIN 3.1 (low in comparison to the average RIN of 6.96 in this study) are still relatively similar to the profile obtained from a sample with high (8.4) RIN.
(TIF)
Figure S2
RNase treatment effect on total RNA profile. Agilent RNA 6000 Pico chips have the ability to resolve RNAs in the size range of 25 nt to 6000 nt (top row). Agilent Small RNA chips have superior resolving power for RNAs in the range of 4 nt to 150 nt only (bottom row). Electropherogram from Pico chip shows that in addition to miRNAs, our exosome preparations also contain higher molecular weight cellular RNAs of sizes up to 4000 nt. These higher molecular weight species cannot be seen with the small RNA chips. Digesting exosome preparations with RNase A or a combination of RNase A and RNase T1 destroys the higher molecular weight extra-exosomal cellular RNAs and preserves only the small RNAs contained in the exosome itself. Note, also, that the fluorescent units scale [FU] on the Y-axis of all of the electropherograms represents the quantity of RNA. As such, RNase treated preps contain much less RNA than untreated preps as the exosomal RNA represents a small portion of the cellular RNA. Similarly, RNase digestion reduces both the size and amount of small RNAs present in exosomal preparations. Exosomal RNA preparations not treated with RNase contain more and larger RNAs in the 4–150+ nt range than do preparations treated with RNase A alone or RNases A and T1. The expected size ranges of miRNAs (∼19–28 nt, yellow) and pre-miRNAs (45–60 nt, blue) are indicated in the small RNA assay panels (bottom row). RNAs in these size ranges are well represented in our purified exosomal RNA preparations.
(TIF)
Figure S3
NCode amplification of exosome-derived RNA. Amplification of miRNAs (which have no poly-A tails) requires the addition of a 3′ Oligo(dT) 24-mer as well as a 5′ T7 promoter template, which increases the size of each miRNA by ∼40 nt. In this case, the expected size range of amplified miRNAs is ∼60 nt (arrow) as opposed to that of the native ∼20 nt species. Profiles of amplified miRNA upon treatment with RNase A (green), without RNase (blue), and with RNase A/T1 (red – optimal for exosome-derived RNA used in Luminex assay) are similar.
(TIF)
Table S1
Student’s T tests of Luminex miRNA expression data with corrected p-values.
(DOCX)
Table S2
Quantities of exosome-derived miRNA from the analyzed cases for Luminex assay, before and after NCode amplification.
(DOCX)
Table S3
Wilcoxon non-parametric test and fold change for top BD hits as means for statistical verification. 12 top-ranked miRNAs according to SAM in Table 3, with q-values lower than 15%, ending with miR-526b* - the last miRNA to have significantly changed expression in BD according to this test. miRNAs 219, 380–3p, 148a, 520a, and 526b* (p-values in bold) have significantly changed expression in BD cases in comparison to controls by this analysis. The expression of miR-29c is not significantly different in BD cases according to Wilcoxon test (p-value 0.0649; see Discussion).
(DOCX)
Table S4
Phenotype traits (tobacco, alcohol, and recreational drug use) of analyzed cases.
(DOCX)
Table S5
An ANOVA analysis of differences in age between C, BD, and SZ groups of cases. No significant difference in age between the groups exists.
(DOCX)
Funding Statement
This work was supported by the National Institute of Mental Health (R21 MH086079; ID and CRV) and National Institute of Aging (T32 AG00015-21; PFK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.De Smaele E, Ferretti E, Gulino A (2010) MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res 1338: 100–111. [DOI] [PubMed] [Google Scholar]
- 2.Beveridge NJ, Cairns MJ (2011) MicroRNA dysregulation in schizophrenia. Neurobiol Dis 46: 263–71. [DOI] [PubMed] [Google Scholar]
- 3.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, et al. (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113: 673–676. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, et al. (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317: 1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, et al. (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289. [DOI] [PubMed] [Google Scholar]
- 7.Presutti C, Rosati J, Vincenti S, Nasi S (2006) Non coding RNA and brain. BMC Neurosci 7 Suppl 1S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiore R, Siegel G, Schratt G (2008) MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta 1779: 471–8. [DOI] [PubMed] [Google Scholar]
- 9.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S (2006) Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124: 191–205. [DOI] [PubMed] [Google Scholar]
- 10.Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM (2011) Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry 69: 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, et al. (2007) microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol 8: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vawter MP, Freed WJ, Kleinman JE (2000) Neuropathology of bipolar disorder. Biol Psychiatry 48: 486–504. [DOI] [PubMed] [Google Scholar]
- 13.Suudhof TC (2008) Neurotransmitter release. Handb Exp Pharmacol: 1–21. [DOI] [PubMed]
- 14.Smalheiser NR (2007) Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct 2: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Niel G, Porto-Carreiro I, Simoes S, Raposo G (2006) Exosomes: a common pathway for a specialized function. J Biochem 140: 13–21. [DOI] [PubMed] [Google Scholar]
- 16.Fevrier B, Raposo G (2004) Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol 16: 415–421. [DOI] [PubMed] [Google Scholar]
- 17.Blanpied TA, Scott DB, Ehlers MD (2002) Dynamics and regulation of clathrin coats at specialized endocytic zones of dendrites and spines. Neuron 36: 435–449. [DOI] [PubMed] [Google Scholar]
- 18.Cooney JR, Hurlburt JL, Selig DK, Harris KM, Fiala JC (2002) Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci 22: 2215–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarrant SB, Routtenberg A (1977) The synaptic spinule in the dendritic spine: electron microscopic study of the hippocampal dentate gyrus. Tissue Cell 9: 461–473. [DOI] [PubMed] [Google Scholar]
- 20.Spacek J, Harris KM (2004) Trans-endocytosis via spinules in adult rat hippocampus. J Neurosci 24: 4233–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park M, Salgado JM, Ostroff L, Helton TD, Robinson CG, et al. (2006) Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron 52: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faure J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, et al. (2006) Exosomes are released by cultured cortical neurones. Mol Cell Neurosci 31: 642–648. [DOI] [PubMed] [Google Scholar]
- 23.Okabayashi S, Kimura N (2010) LGI3 interacts with flotillin-1 to mediate APP trafficking and exosome formation. Neuroreport 21: 606–610. [DOI] [PubMed] [Google Scholar]
- 24.Bulloj A, Leal MC, Xu H, Castano EM, Morelli L (2010) Insulin-degrading enzyme sorting in exosomes: a secretory pathway for a key brain amyloid-beta degrading protease. J Alzheimers Dis 19: 79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharples RA, Vella LJ, Nisbet RM, Naylor R, Perez K, et al. (2008) Inhibition of gamma-secretase causes increased secretion of amyloid precursor protein C-terminal fragments in association with exosomes. FASEB J 22: 1469–1478. [DOI] [PubMed] [Google Scholar]
- 26.Yuyama K, Sun H, Mitsutake S, Igarashi Y (2012) Sphingolipid-modulated exosome secretion promotes the clearance of amyloid-beta by microglia. J Biol Chem 287: 10977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, et al. (2010) Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci 30: 6838–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, et al. (2007) Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol 211: 582–590. [DOI] [PubMed] [Google Scholar]
- 29.Gomes C, Keller S, Altevogt P, Costa J (2007) Evidence for secretion of Cu,Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci Lett 428: 43–46. [DOI] [PubMed] [Google Scholar]
- 30.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, et al. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659. [DOI] [PubMed] [Google Scholar]
- 31.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, et al. (2008) Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis 14: 27–41. [DOI] [PubMed] [Google Scholar]
- 32.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, et al. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10: 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durrenberger PF, Fernando S, Kashefi SN, Ferrer I, Hauw JJ, et al. (2010) Effects of antemortem and postmortem variables on human brain mRNA quality: a BrainNet Europe study. J Neuropathol Exp Neurol 69: 70–81. [DOI] [PubMed] [Google Scholar]
- 34.Thery C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3: Unit 3.22. [DOI] [PubMed]
- 35.Strauss K, Goebel C, Runz H, Mobius W, Weiss S, et al. (2010) Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J Biol Chem 285: 26279–26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Efron B, Tibshirani R (2002) Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol 23: 70–86. [DOI] [PubMed] [Google Scholar]
- 38.Tibshirani R, Hastie T, Narasimhan B, Chu G (2002) Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 99: 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm S (1979) A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics 6: 65–70. [Google Scholar]
- 40.Storey JD (2003) The positive false discovery rate: A Bayesian interpretation and the q-value. Annals of Statistics 31: 2013–2035. [Google Scholar]
- 41.Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 29: 1165–1188. [Google Scholar]
- 42.Tibshirani R, Hastie T, Narasimhan B, Chu G (2003) Class prediction by nearest shrunken centroids, with applications to DNA microarrays. Statistical Science 18: 104–117. [Google Scholar]
- 43.Liang Y, Ridzon D, Wong L, Chen C (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer SU, Pfaffl MW, Ulbrich SE (2010) Normalization strategies for microRNA profiling experiments: a 'normal' way to a hidden layer of complexity? Biotechnol Lett 32: 1777–1788. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh D, Chinnaiyan AM (2005) Covariate adjustment in the analysis of microarray data from clinical studies. Funct Integr Genomics 5: 18–27. [DOI] [PubMed] [Google Scholar]
- 47.Harrison PJ, Weinberger DR (2005) Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry 10: 40–68. [DOI] [PubMed] [Google Scholar]
- 48.Owen MJ, O'Donovan MC, Harrison PJ (2005) Schizophrenia: a genetic disorder of the synapse? BMJ 330: 158–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, et al. (2009) Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 100: 1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathivanan S, Simpson RJ (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9: 4997–5000. [DOI] [PubMed] [Google Scholar]
- 51.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, et al. (2012) Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med 10: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallego JA, Gordon ML, Claycomb K, Bhatt M, Lencz T, et al. (2012) In vivo microRNA detection and quantitation in cerebrospinal fluid. J Mol Neurosci 47: 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, et al. (2011) MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One 6: e21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, et al. (2012) Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry 17: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ (2011) Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry 69: 180–187. [DOI] [PubMed] [Google Scholar]
- 56.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, et al. (2010) The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol 402: 491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ (2010) Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry 15: 1176–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, et al. (2010) miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis 38: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, et al. (2011) miR-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev 25: 1734–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbato C, Ruberti F, Pieri M, Vilardo E, Costanzo M, et al. (2010) MicroRNA-92 modulates K(+) Cl(-) co-transporter KCC2 expression in cerebellar granule neurons. J Neurochem 113: 591–600. [DOI] [PubMed] [Google Scholar]
- 61.Khanna A, Muthusamy S, Liang R, Sarojini H, Wang E (2011) Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging (Albany NY) 3: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valastyan S, Weinberg RA (2011) Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci 124: 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kapinas K, Kessler CB, Delany AM (2009) miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem 108: 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valvezan AJ, Klein PS (2012) GSK-3 and Wnt Signaling in Neurogenesis and Bipolar Disorder. Front Mol Neurosci 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lakhal S, Wood MJ (2011) Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. Bioessays 33: 737–741. [DOI] [PubMed] [Google Scholar]
- 66.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, et al. (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29: 341–345. [DOI] [PubMed] [Google Scholar]
- 67.Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, et al. (2011) Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther 19: 1769–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
RNA quality control. Representative RNA (sample C13, RIN = 7.0; the sample closest to the average (6.96) and median (6.85) value for the set) yields two strong bands at 40 s and 45 s representing the 18 S and 28 S ribosomal RNA (A). The electropherogram shows a marker peak and the two ribosomal peaks corresponding to 18 S and 28 S subunits. RIN value on a scale of 1–10 is computed based on the presence of correct ribosomal peaks, the ratio between those peaks, and the extent of RNA fragmentation (B). Relationship between RIN and mRNA and miRNA profiles (C): Next to the ladder (left), total and small RNA profiles of three degradation stages of a single RNA sample are shown (RIN 8.4 = not degraded, RIN 3.1 = partially degraded, RIN 2.5 = severely degraded). Note that both mRNA (RT PCR, middle) and miRNA (Luminex, right) profiles obtained from a sample with RIN 3.1 (low in comparison to the average RIN of 6.96 in this study) are still relatively similar to the profile obtained from a sample with high (8.4) RIN.
(TIF)
Figure S2
RNase treatment effect on total RNA profile. Agilent RNA 6000 Pico chips have the ability to resolve RNAs in the size range of 25 nt to 6000 nt (top row). Agilent Small RNA chips have superior resolving power for RNAs in the range of 4 nt to 150 nt only (bottom row). Electropherogram from Pico chip shows that in addition to miRNAs, our exosome preparations also contain higher molecular weight cellular RNAs of sizes up to 4000 nt. These higher molecular weight species cannot be seen with the small RNA chips. Digesting exosome preparations with RNase A or a combination of RNase A and RNase T1 destroys the higher molecular weight extra-exosomal cellular RNAs and preserves only the small RNAs contained in the exosome itself. Note, also, that the fluorescent units scale [FU] on the Y-axis of all of the electropherograms represents the quantity of RNA. As such, RNase treated preps contain much less RNA than untreated preps as the exosomal RNA represents a small portion of the cellular RNA. Similarly, RNase digestion reduces both the size and amount of small RNAs present in exosomal preparations. Exosomal RNA preparations not treated with RNase contain more and larger RNAs in the 4–150+ nt range than do preparations treated with RNase A alone or RNases A and T1. The expected size ranges of miRNAs (∼19–28 nt, yellow) and pre-miRNAs (45–60 nt, blue) are indicated in the small RNA assay panels (bottom row). RNAs in these size ranges are well represented in our purified exosomal RNA preparations.
(TIF)
Figure S3
NCode amplification of exosome-derived RNA. Amplification of miRNAs (which have no poly-A tails) requires the addition of a 3′ Oligo(dT) 24-mer as well as a 5′ T7 promoter template, which increases the size of each miRNA by ∼40 nt. In this case, the expected size range of amplified miRNAs is ∼60 nt (arrow) as opposed to that of the native ∼20 nt species. Profiles of amplified miRNA upon treatment with RNase A (green), without RNase (blue), and with RNase A/T1 (red – optimal for exosome-derived RNA used in Luminex assay) are similar.
(TIF)
Table S1
Student’s T tests of Luminex miRNA expression data with corrected p-values.
(DOCX)
Table S2
Quantities of exosome-derived miRNA from the analyzed cases for Luminex assay, before and after NCode amplification.
(DOCX)
Table S3
Wilcoxon non-parametric test and fold change for top BD hits as means for statistical verification. 12 top-ranked miRNAs according to SAM in Table 3, with q-values lower than 15%, ending with miR-526b* - the last miRNA to have significantly changed expression in BD according to this test. miRNAs 219, 380–3p, 148a, 520a, and 526b* (p-values in bold) have significantly changed expression in BD cases in comparison to controls by this analysis. The expression of miR-29c is not significantly different in BD cases according to Wilcoxon test (p-value 0.0649; see Discussion).
(DOCX)
Table S4
Phenotype traits (tobacco, alcohol, and recreational drug use) of analyzed cases.
(DOCX)
Table S5
An ANOVA analysis of differences in age between C, BD, and SZ groups of cases. No significant difference in age between the groups exists.
(DOCX)