Hypoxia-regulated microRNAs in human cancer (original) (raw)
Abstract
Hypoxia plays an important role in the tumor microenvironment by allowing the development and maintenance of cancer cells, but the regulatory mechanisms by which tumor cells adapt to hypoxic conditions are not yet well understood. MicroRNAs are recognized as a new class of master regulators that control gene expression and are responsible for many normal and pathological cellular processes. Studies have shown that hypoxia inducible factor 1 (HIF1) regulates a panel of microRNAs, whereas some of microRNAs target HIF1. The interaction between microRNAs and HIF1 can account for many vital events relevant to tumorigenesis, such as angiogenesis, metabolism, apoptosis, cell cycle regulation, proliferation, metastasis, and resistance to anticancer therapy. This review will summarize recent findings on the roles of hypoxia and microRNAs in human cancer and illustrate the machinery by which microRNAs interact with hypoxia in tumor cells. It is expected to update our knowledge about the regulatory roles of microRNAs in regulating tumor microenvironments and thus benefit the development of new anticancer drugs.
Keywords: microRNA, hypoxia, HIF1, human cancer, angiogenesis, apoptosis, cell cycle, cancer metastasis, chemoresistance, radioresistance
Introduction
As the oxygen (O2) concentration is significantly lower in tumor tissue than in the surrounding normal tissue, hypoxia has been a focus of attention because of its important role in maintaining tumor microenvironments1. For instance, the average O2 pressure in breast tumors is approximately 10 mmHg, whereas normal breast tissue has an O2 pressure of more than 60 mmHg2. In general, hypoxia regulates tumor-cell phenotypes by altering genes that are sensitive to O2 pressure. Hypoxia-inducible factor-1 (HIF1) is one of the most studied genes that play a vital role in the process of hypoxia3. As a transcription factor, HIF1 can be induced by low O2 pressure and can subsequently influence the expression of a number of genes via transcriptional regulation. Thus, aside from the disruption of normal cellular metabolic processes, low O2 results in dysregulation of hypoxia-related genes, such as HIF1. This dysregulation can account for many oncogenic phenotypes, including tumor cell transformation, invasion, metastasis, and resistance to chemotherapy and radiation therapy3.
MicroRNAs (miRNAs) are a class of endogenous, 18–24 nucleotide, non-protein-coding small RNA molecules that regulate eukaryotic gene expression at the post-transcriptional and translational levels. They are involved in a wide variety of normal and pathological cellular processes, such as cell death, neuronal patterning, development, metabolism, and oncogenesis4. Numerous studies show that miRNAs with oncogenic and tumor suppressive signatures are correlated with various types of human cancer; however, the mechanistic role of miRNAs in malignant diseases is not yet completely understood. In particular, we lack a clear understanding of how miRNAs interact with conditions in the tumor microenvironment, such as hypoxia. Recently, a few reports revealed that miRNAs were associated with several key signaling pathways that respond to hypoxia and played important roles in hypoxic adaptation. Here, we will summarize these novel discoveries to illustrate the interaction between miRNAs and hypoxia in order to better understand the role of this interaction in tumorigenesis.
Hypoxia regulates the expression of miRNAs
Previous studies have discovered a number of miRNAs that are differentially expressed in response to hypoxia (hypoxia- regulated miRNAs; HRMs) by using the microarray method. For instance, miR-210, -155, -372/373, and -10b were found to be up-regulated5,6,7,8,9,10,11, whereas miR-20b and miR-200b were found to be down-regulated, in response to hypoxia12,13. Surprisingly, most of the HRMs apparently lack consistent phenotypes across studies. The exception to this rule appears to be a “master” HRM, miR-210, a finding that has been confirmed by various groups14. This lack of consistency may be attributable to a combination of technical variables, including the sensitivity of screening methods, the duration and severity of oxygen deprivation, and the cellular context15. However, in addition to the large variety of miRNA profiling platforms, we think that the lack of appropriate normalization methods specific to miRNAs is also of significance when calibrating variations in measurements of expression16,17,18,19. Most of the methods currently used for miRNA analysis were derived from established normalization methods based on the assumption that the total number of arrayed probes is large enough (>5000). To date, only approximately 2000 human miRNAs have been named; obviously, these established methods cannot well meet the needs of miRNA profiling analysis. Therefore, in addition to calling for novel specific normalization methods, it is extremely important to functionally validate candidate HRMs that are identified from array data.
Similar to protein encoding genes, miRNAs are transcribed from miRNA genes via classical transcription machinery that involves RNA polymerase. Thus, transcription factors play crucial roles in regulating the expression of miRNAs. For instance, c-Myc and E2F can transcriptionally up-regulate miR-17-92 and significantly influence tumor transformation mediated by miR-17-9220,21. Since Kulshreshtha et al first reported in 2007 that the expression of an miRNA panel could be induced by hypoxia22, numerous additional studies of HRMs have emerged. Table 1 summarizes most of the HRMs that have been reported to date. Hypoxia response elements (HRE) contained in the promoter regions of HRM genes can be bound by the α and β subunits of HIF1, and hypoxia can improve the affinity of such a complex thereby promoting the transcription of HRMs. Many HRMs, such as miR-210, -155, and -373, have been demonstrated to contain HREs by which HIF1 regulates the expression of these HRMs5,6,14.
Table 1. Differentially expressed miRNAs in response to hypoxia.
| Upregulated miRNAs | Downregulated miRNAs |
|---|---|
| miR-10b, miR-103, miR-107, miR-125a, miR-152, miR-155, miR-181b, miR-188, miR-191, miR-193b, miR-203, miR-205, miR-206, miR-21, miR-210, miR-213, miR-224, miR-23, miR-24, miR-26, miR-27, miR-30a-5p, miR-30c, miR-30d, miR-322, miR-333, miR-335, miR-339, miR-373, miR-451, miR-491, miR-497, miR-512-5p, miR-562, miR-93 | let-7f, miR-128b, miR-150, miR-159, miR-17-92, miR-181d, miR-196a, miR-196b, miR-199a, miR-199b, miR-200a, miR-200b, miR-20b, miR-22, miR-25, miR-30, miR-424, miR-449, miR-489, |
Many transcription factors, such as TWIST, PPARγ, and GATA1, can be regulated by HIF1 at the transcriptional level23,24,25. As a result, HIF1 can be involved in the regulation of miRNA expression by influencing these transcription factors. For example, when HIF1 is stabilized by insufficient oxygen levels, TWIST can be induced to up-regulate miR-10b8. MiR-10b is a well-documented oncogenic miRNA that mediate the metastasis of various human cancers26,27,28. Therefore, HRM expression induced by HIF1 may account for the mechanism by which hypoxia facilities tumor progression. Intriguingly, studies also reported that HIF1 had been associated with down-regulation of miRNAs. The miR-17-92 cluster was reported to be reduced in hypoxia-treated cells containing wild-type p53, but not in p53-deficient cells29. As shown in this study, p53 directly repressed the expression of miR-17-92 through the c-Myc independent transcriptional regulation, which was thought to be responsible for hypoxia-induced apoptosis29. In contrast, Lei et al found that the knockdown of HIF1 resulting in the increase of miR-20b13, while Chan et al reported that hypoxia led to the reduction of miR-200b12.
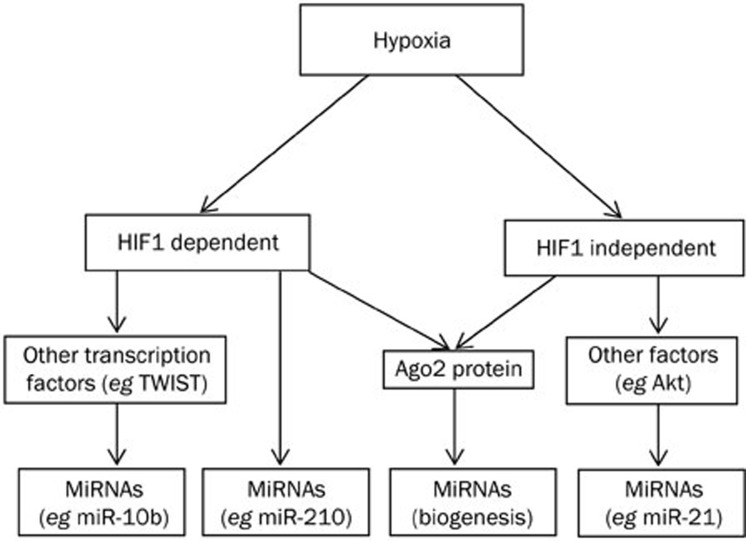
In addition to HIF1, other genes and signaling pathways may also contribute to the adaptation of tumor cells to hypoxia. For example, oxygen deprivation could promote the induction of miR-21 via an Akt2-dependent process. The hypoxia-generated signals transduced by Akt2 have been reported to increase the activity of NF-κB and CREB, which were able to transcriptionally up-regulate the expression of miR-2130. Hypoxia was also involved in the biogenesis of miRNA. The protein Ago2 is a crucial component of the RNA-induced silencing complex (RISC), and the hydroxylation of Ago2 is a key step for the assembly of Ago2 to heat shock protein 90 (Hsp90) in RISC31. Previous studies have shown that hypoxia was able to increase the level of type 1 collagen prolyl-4-hydroxylase[C-P4H(I)], which could lead to prolyl-hydroxylation and accumulation of Ago2, thus increasing the endonuclease activity of Ago2 through either the HIF1 independent or dependent pathways31,32. These mechanisms are summarized in Figure 1.
Figure 1.
The machinery that hypoxia regulates miRNA expression.
miRNAs regulate HIF1
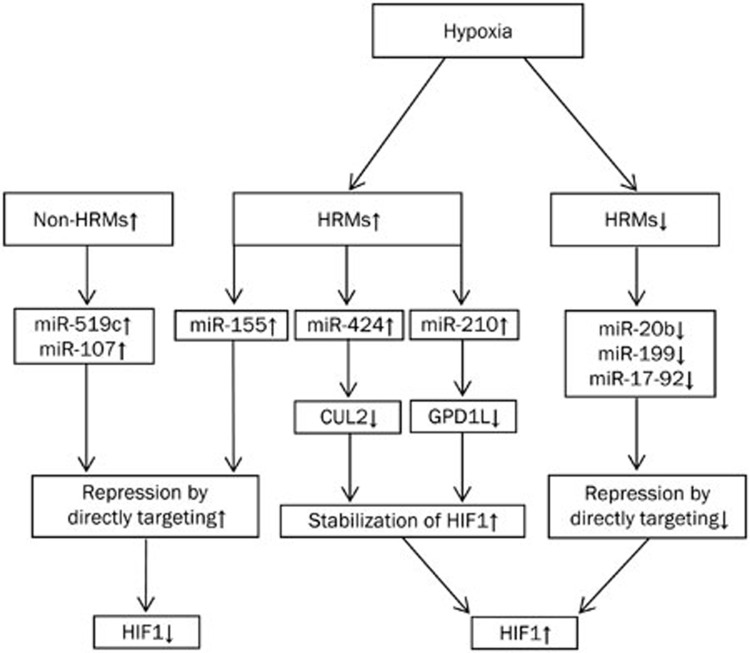
Thanks to the signature of regulating hundreds of target genes simultaneously, miRNAs are capable of repressing the expression of genes associated with hypoxia. For example, Kelly et al found that hypoxia-induced miR-210 could repress glycerol-3-phosphate dehydrogenase 1-like (GPD1L), which, in turn, stabilized HIF1α by reducing hyperhydroxylation33. Similarly, cullin 2 (CUL2), a scaffolding protein critical to the assembly of the ubiquitin ligase system, was able to be suppressed by miR-42434. Given that hypoxia can induce miR-424 in human endothelial cells, the decline of CUL2 potentially stabilized HIF1α34. Therefore, when low O2 induces the expression of HRMs, genes targeted by these miRNAs stabilize HIF1 by forming positive-feedback loops. Moreover, some HRMs are also involved in the destabilization of HIF1. The down-regulation of miR-20b, miR-199, and miR-17-92 by hypoxia stabilized HIF1 because these HRMs were able to repress the expression of HIF1 through direct targeting13,29,35,36. In addition, Brunning et al reported that the hypoxic induction of miR-155 could negatively influence the stability and activity of HIF1α in in vitro and in vivo models6. Some non-HRMs, such as miR-519c and miR-107, were reported to target HIF1α37 and HIF1β38, respectively. Although HIF2 is another important isoform of HIF that has been intensively studied in hypoxia, there are few studies reporting associations between HIF2 and miRNAs. As such, we only summarize the regulation of HIF1 by miRNAs in Figure 2.
Figure 2.
The machinery that miRNA regulates HIF1.
The roles of HRMs in human cancer
Angiogenesis
Angiogenesis is a highly coordinated process of tissue remodeling that leads to the formation of new blood vessels39. Hypoxic regions modulate the induction of angiogenesis via the regulation of pro- and anti-angiogenic factors40,41. When cells are subjected to hypoxia, HIF1 up-regulates a variety of angiogeneic growth factors via transcriptional modulation, including vascular endothelial growth factor (VEGF)42, angiopoietin 243, stromal-derived factor 144, and stem cell factor45. When these factors bind to specific receptors that are expressed on the surface of vascular endothelial and smooth muscle cells, angiogenic budding of new capillaries from existing vessels is initiated. As angiogenesis is important for tumor growth and metastasis, we will summarize the recent studies that have revealed an additional layer of angiogenic regulation via the action of specific HRMs.
The miRNA miR-210 was reported to target the receptor tyrosine kinase ligand Ephrin-A3 (EFNA3) and enhance the differentiation of human umbilical vein endothelial cells (HUVEC)46. When induced by hypoxia, miR-424 was able to promote angiogenesis through targeting CUL2, a scaffolding protein critical to the assembly of the ubiquitin ligase system described earlier. This process stabilizes HIF1α and allows it to transcriptionally activate VEGF34. MiR-20b has been shown to negatively regulate angiogenesis by targeting VEGF and HIF1α13,47. In low-oxygen conditions, miR-20b could be down-regulated by HIF1α and attenuate the inhibitory effect on VEGF and HIF1α. The regulatory interactions among miR-20b, HIF1α, and VEGF thereby enabled tumor cells to adapt to different oxygen concentrations13,47. In addition to the inhibitory effects of miR-519c on angiogenesis through the direct targeting of HIF1α37, miR-21 was demonstrated to induce tumor angiogenesis by targeting PTEN and activating the Akt and ERK1/2 signaling pathways, which led to elevation of HIF1α and VEGF48. Delivery of the miR-200b mimics in human microvascular endothelial cells (HMECs) suppressed the angiogenic response, whereas miR-200b-depleted HMECs exhibited elevated angiogenesis in vitro, as evidenced by the matrigel tube formation and cell migration assays. Both oxygen deprivation and HIF1 stabilization could inhibit miR-200b expression12. In addition, down-regulation of miR-107 was found to promote tumor angiogenesis in hypoxic conditions, which may be attributed to the reduced inhibition of HIF1β by miR-10738.
Metabolism
When oxygen levels are insufficient, a cell's metabolism shifts from mitochondrial oxidative phosphorylation to glycolysis. As such, HIF1 may be involved in the induction of kinases and enzymes that are necessary for this adaptation49. Recently, several groups have demonstrated that miR-210 contributes to this metabolic shift by repressing several steps that occur during mitochondrial metabolism, particularly the electron transport chain (ETC) complexes50,51,52,53. The iron-sulfur cluster scaffold homolog and cytochrome c oxidase assembly factor (COX10) proteins that repress mitochondrial respiration are both targeted by miR-210. Additionally, miR-210 can target NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (NDUFA4)54, succinate dehydrogenase complex55, and GPD1-L52, which play important roles in cell metabolism.
Apoptosis
Fasanaro et al reported that inhibition of miR-210 induces endothelial cell apoptosis in both normoxic and hypoxic conditions by targeting Ephrin-A347. Additionally, Kim et al showed that ischemic preconditioning could augment survival of mesenchymal stem cells through miR-210 expression by targeting the apoptotic component CASP8AP256. Recently, miR-21 was found to be an anti-apoptotic miRNA under hypoxic conditions57, where it exhibited a protective effect against hypoxia/reoxygenation-induced cell apoptosis through targeting programmed cell death 4 (PDCD4) in cultured cardiac myocytes. This protective effect was further confirmed in rat hearts after ischemia/reperfusion injury in vivo57. Another study also suggested that miR-21 might regulate apoptosis though targeting PTEN and Fas ligand (FasL)58.
Cell cycle and proliferation
The HRM miR-210 was recently demonstrated to inhibit the expression of E2F3 and MNT54,59. E2F3 belongs to the E2F family that control cell cycle progression via influencing genes required for DNA synthesis at G1/S phase54. MNT is a known antagonist of c-Myc and involved in regulation of cell cycle and proliferation. Induction of miR-210 and subsequent MNT repression could accelerate the G1/S transition and promote tumor cell proliferation59. Therefore, induction of miR-210 by hypoxia may be involved in these essential cellular processes in tumor cells.
Cancer metastasis
Ying et al reported that miR-210 was induced by hypoxia in hepatocellular carcinoma (HCC) cells and could promote hypoxia-induced HCC cell metastasis60. Vacuole membrane protein 1 (VMP1) was identified as the direct target of miR-210, and down-regulation of VMP1 by hypoxia was involved the mobility of HCC cells60. Chen et al found that miR-103/107 expression was elevated by hypoxia in colon tumor cells and substantially repressed the tumor metastasis suppressor death-associated protein kinase (DAPK) and Kruppel-like factor 4 (KLF4)61. Loayza-Puch et al showed that miR-372/373 could be up-regulated by hypoxia through the transcriptional regulation of HIF1α and TWIST, while miR-21 was up-regulated by RAS/ERK signaling11. The induction of these HRMs sequentially decreased the expression of the membrane-anchored metalloproteinase regulator RECK, which was recognized as a suppressor of tumor metastasis11.
Cancer therapy
Hypoxia has been reported to be correlated with chemoresistance and radioresistance in anticancer therapy62,63. Although it has been studied for decades, the mechanism by which cancer cells are resistant to anticancer therapy during hypoxic conditions is still not completely understood. Zhu et al reported that the multidrug resistance gene MDR1 could be induced in HIF1 transfected human hepatoma HepG2 cells, which consequently became more resistant to 5-FU treatment than the control cells64. A recent study revealed that suppression of the “master” HRM, miR-210, by the antisense method, was able to significantly sensitize human hepatoma cells to radiation by inhibiting proliferation and promoting apoptosis65. Therefore, HRMs could be responsible for tumor cell chemoresistance and radioresistance and have the potential to serve as novel biomarkers and therapeutic targets for future anticancer therapy.
Summary and perspective
Even though a number of studies on hypoxia and human cancer have been published, the physiological and pathophysiological regulation of hypoxia is still poorly understood. Research on miRNAs promises to shed light on the mechanism that underlies the regulation of hypoxia, for two reasons. First, miRNAs can rapidly respond to stress caused by hypoxia by post-transcriptional/translational modulation at the cellular level. Second, miRNAs are capable of regulating numerous genes and influencing multiple components of a signaling pathway simultaneously. As such, it is important to understand the function and regulatory basis of HRMs in response to hypoxia. In our opinion, future studies on HRMs will continue to focus on the following directions. (1) The discovery of novel HRMs; (2) The validation of newly discovered HRMs and their functions during hypoxic conditions; (3) The identification of HRM target genes; (4) The development of novel therapeutic and preventive drugs that target HRMs; and (5) The study of the translational potential of HRMs in treating cancer patients. To complement previously published review articles, here we summarize the latest findings on hypoxia-regulated miRNAs in human cancers. We expect that this review will be able to further our understanding of the regulatory role played by miRNAs in the tumor microenvironment and will benefit the development of new anticancer drugs.
Acknowledgments
This study is supported by the NIH/NCI grant 1R21CA160280-01A1 (Yaguang XI). We sincerely convey our apology to the colleagues who have greatly contributed to this field but whose publications were not cited in this review due to limitations of time and space.
References
- Noman MZ, Buart S, Romero P, Ketari S, Janji B, Mari B, et al. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res. 2012;72:4629–41. doi: 10.1158/0008-5472.CAN-12-1383. [DOI] [PubMed] [Google Scholar]
- Vaupel P, Mayer A, Hockel M. Tumor hypoxia and malignant progression. Methods Enzymol. 2004;381:335–54. doi: 10.1016/S0076-6879(04)81023-1. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med. 2002;8:S62–7. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- Pocock R. Invited review: decoding the microRNA response to hypoxia. Pflugers Arch. 2011;461:307–15. doi: 10.1007/s00424-010-0910-5. [DOI] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–67. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, et al. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31:4087–96. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–9. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque I, Banerjee S, Mehta S, De A, Majumder M, Mayo MS, et al. Cysteine-rich 61-connective tissue growth factor-nephroblastoma-overexpressed 5 (CCN5)/Wnt-1-induced signaling protein-2 (WISP-2) regulates microRNA-10b via hypoxia-inducible factor-1alpha-TWIST signaling networks in human breast cancer cells. J Biol Chem. 2011;286:43475–85. doi: 10.1074/jbc.M111.284158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–52. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal CS, Michael MZ, Rawlings LH, Van der Hoek MB, Gleadle JM. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med. 2010;8:64. doi: 10.1186/1741-7015-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Yoshida Y, Matsuzaki T, Takahashi C, Kitayama H, Noda M. Hypoxia and RAS-signaling pathways converge on, and cooperatively downregulate, the RECK tumor-suppressor protein through microRNAs. Oncogene. 2010;29:2638–48. doi: 10.1038/onc.2010.23. [DOI] [PubMed] [Google Scholar]
- Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286:2047–56. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Li B, Yang Z, Fang H, Zhang GM, Feng ZH, et al. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One. 2009;4:e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Le QT, Giaccia AJ. MiR-210 — micromanager of the hypoxia pathway. Trends Mol Med. 2010;16:230–7. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–83. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Howel P, Bruheim S, Ju J, Owen LB, Fodstad O, et al. Systematic evaluation of three microRNA profiling platforms: microarray, beads array, and quantitative real-time PCR array. PLoS One. 2011;6:e17167. doi: 10.1371/journal.pone.0017167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang XF, Howell P, Qian X, Huang K, Riker AI, et al. A personalized microRNA microarray normalization method using a logistic regression model. Bioinformatics. 2010;26:228–34. doi: 10.1093/bioinformatics/btp655. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang XF, Xi Y. Normalizing bead-based microRNA expression data: a measurement error model-based approach. Bioinformatics. 2011;27:1506–12. doi: 10.1093/bioinformatics/btr180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhang SG, Wang XF, Tan M, Xi Y. Testing for differentially-expressed microRNAs with errors-in-variables nonparametric regression. PLoS One. 2012;7:e37537. doi: 10.1371/journal.pone.0037537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- Krishnan J, Suter M, Windak R, Krebs T, Felley A, Montessuit C, et al. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9:512–24. doi: 10.1016/j.cmet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Shen GM, Liu XL, Wang F, Zhao YZ, Zhang JW. Hypoxia-inducible factor 1-mediated human GATA1 induction promotes erythroid differentiation under hypoxic conditions. J Cell Mol Med. 2012;16:1889–99. doi: 10.1111/j.1582-4934.2011.01484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao L, Cui Q, Gary BD, Dyess DL, Taylor W, et al. Sulindac inhibits tumor cell invasion by suppressing NF-kappaB-mediated transcription of microRNAs. Oncogene. 2012;31:4979–86. doi: 10.1038/onc.2011.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010;12:210. doi: 10.1186/bcr2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature Biotechnol. 2010;28:341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, et al. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009;28:2719–32. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polytarchou C, Iliopoulos D, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, et al. Akt2 regulates all Akt isoforms and promotes resistance to hypoxia through induction of miR-21 upon oxygen deprivation. Cancer Res. 2011;71:4720–31. doi: 10.1158/0008-5472.CAN-11-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, So J, Davis-Dusenbery BN, Qi HH, Bloch DB, Shi Y, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011;31:4760–74. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer KH, Gess B, Lohaus C, Meyer HE, Katschinski D, Kurtz A. Oxygen tension regulates the expression of a group of procollagen hydroxylases. Eur J Biochem. 2003;270:4515–22. doi: 10.1046/j.1432-1033.2003.03846.x. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31:2696–706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–54. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540–5. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70:2675–85. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6334–9. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. 2000;6:389–95. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–49. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia–inducible factor 1. Mol Cell Biol. 1996;16:4604–13. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MP, Tournaire R, Pouyssegur J. The angiopoietin-2 gene of endothelial cells is up-regulated in hypoxia by a HIF binding site located in its first intron and by the central factors GATA-2 and Ets-1. J Cell Physiol. 2008;217:809–18. doi: 10.1002/jcp.21558. [DOI] [PubMed] [Google Scholar]
- Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–64. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- Kelly BD, Hackett SF, Hirota K, Oshima Y, Cai Z, Berg-Dixon S, et al. Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circ Res. 2003;93:1074–81. doi: 10.1161/01.RES.0000102937.50486.1B. [DOI] [PubMed] [Google Scholar]
- Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–83. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio S, D'Andrea A, Ferla R, Surmacz E, Gulotta E, Amodeo V, et al. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224:242–9. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- Liu LZ, Li C, Chen Q, Jing Y, Carpenter R, Jiang Y, et al. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS One. 2011;6:e19139. doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–84. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–43. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–8. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–64. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–78. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HW, Haider HK, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem. 2009;284:33161–8. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, et al. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;87:431–9. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, et al. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem. 2010;285:20281–90. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–68. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, et al. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology. 2011;54:2064–75. doi: 10.1002/hep.24614. [DOI] [PubMed] [Google Scholar]
- Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–41. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- Wu XZ, Xie GR, Chen D. Hypoxia and hepatocellular carcinoma: The therapeutic target for hepatocellular carcinoma. J Gastroenterol Hepatol. 2007;22:1178–82. doi: 10.1111/j.1440-1746.2007.04997.x. [DOI] [PubMed] [Google Scholar]
- De Milito A, Fais S. Tumor acidity, chemoresistance and proton pump inhibitors. Future Oncol. 2005;1:779–86. doi: 10.2217/14796694.1.6.779. [DOI] [PubMed] [Google Scholar]
- Zhu H, Chen XP, Luo SF, Guan J, Zhang WG, Zhang BX. Involvement of hypoxia-inducible factor-1-alpha in multidrug resistance induced by hypoxia in HepG2 cells. J Exp Clin Cancer Res. 2005;24:565–74. [PubMed] [Google Scholar]
- Yang W, Sun T, Cao J, Liu F, Tian Y, Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318:944–54. doi: 10.1016/j.yexcr.2012.02.010. [DOI] [PubMed] [Google Scholar]