Efficient Clinical Scale Gene Modification via Zinc Finger Nuclease–Targeted Disruption of the HIV Co-receptor CCR5 (original) (raw)
Abstract
Since HIV requires CD4 and a co-receptor, most commonly C-C chemokine receptor 5 (CCR5), for cellular entry, targeting CCR5 expression is an attractive approach for therapy of HIV infection. Treatment of CD4+ T cells with zinc-finger protein nucleases (ZFNs) specifically disrupting chemokine receptor CCR5 coding sequences induces resistance to HIV infection in vitro and in vivo. A chimeric Ad5/F35 adenoviral vector encoding CCR5-ZFNs permitted efficient delivery and transient expression following anti-CD3/anti-CD28 costimulation of T lymphocytes. We present data showing CD3/CD28 costimulation substantially improved transduction efficiency over reported methods for Ad5/F35 transduction of T lymphocytes. Modifications to the laboratory scale process, incorporating clinically compatible reagents and methods, resulted in a robust ex vivo manufacturing process capable of generating >1010 CCR5 gene-edited CD4+ T cells from healthy and HIV+ donors. CD4+ T-cell phenotype, cytokine production, and repertoire were comparable between ZFN-modified and control cells. Following consultation with regulatory authorities, we conducted in vivo toxicity studies that showed no detectable ZFN-specific toxicity or T-cell transformation. Based on these findings, we initiated a clinical trial testing the safety and feasibility of CCR5 gene-edited CD4+ T-cell transfer in study subjects with HIV-1 infection.
Maier and colleagues show that costimulation with anti-CD3 and anti-CD28 substantially improves T cell transduction efficiency by a chimeric Ad5/F35 adenoviral vector encoding a zinc finger protein nuclease (ZFN) targeting the chemokine receptor CCR5. Incorporation of clinically compatible reagents and methods resulted in a robust ex vivo manufacturing process capable of generating >1010 CCR5 gene-edited CD4+ T cells from both healthy as well as HIV+ donors. T cell phenotype, cytokine production, and repertoire were comparable between ZFN-modified and control cells. Further in vivo toxicity studies showed no detectable ZFN-specific toxicity or transformation.
Introduction
Highly active antiretroviral therapy (HAART) controls HIV replication and generally improves immune status in people who are HIV+, significantly prolonging survival. HAART is a lifelong drug therapy, with difficulties in medication adherence and long-term toxicities. However, many patients still present late, which is associated with diminished immune restoration and shorter survival durations. These patients would benefit from immune restoration in addition to antiretroviral therapy to address the immune activation and incomplete immune restoration that persists during HAART.
Immune-based therapies are attractive since there is evidence that control of HIV-1 infection is associated with strong virus-specific polyfunctional CD4+ T cells that support antiviral CD8+ T cells (Pantaleo and Koup, 2004). We have shown that reconstituting CD4+ helper T-cell activity through adoptive transfer of costimulated CD4+ T cells can improve CD4 counts and may augment natural immunity to HIV-1 infection (Levine et al., 2002). A treatment modality that protects CD4+ T cells from HIV infection in vivo may result in improved antiviral immunity as well as overall immune function and reduction in disease-related morbidity.
Gene therapy for HIV-1 infection, including antisense RNA, transdominant proteins, ribozymes, RNA decoys, single chain antibodies, and RNAi (RNA-interference), has long been proposed as an alternative to antiretroviral drug regimens (Sarver and Rossi, 1993; Dropulic and June, 2006). Payloads targeting entry of HIV have also been investigated both in preclinical studies and in human trials (Li et al., 2005; van Lunzen et al., 2007). Blocking HIV at an early step in the replication cycle may be important for conferring a selective advantage to genetically modified cells in the body and hence allow outgrowth of HIV-resistant cells in vivo (von Laer et al., 2006).
A naturally occurring mutation of the HIV entry co-receptor C-C chemokine receptor 5 (CCR5; Δ32) that explains why certain high exposure-risk patients remained HIV-resistant was described in 1996 (Liu et al., 1996). The homozygous 32-base-pair (bp) deletion in CCR5 results in a dysfunctional receptor (Quillent et al., 1998; Carrington et al., 1999). CD4 T cells from individual's homozygous for the CCR5Δ32 mutation required a 100-fold higher level of HIV to be infected (Paxton et al., 1996). In HIV+ long-term nonprogressors (defined by the criteria of HIV positivity for >7 years, CD4 cell counts >600, no antiretroviral therapy, and no symptoms), the frequency of CCR5Δ32 heterozygotes is significantly higher than in the general population, indicative of a protective effect in heterozygotes (Cohen et al., 1997; Eugen-Olsen et al., 1997). The functional eradication of HIV-1 in a 40-year-old HIV-positive man in Berlin who underwent hematopoietic stem and progenitor cell (HSPC) transplantation from a homozygous CCR5Δ32 (CCR5−/CCR5−) donor and achieved sustained virologic suppression without antiretroviral therapy has increased the rationale for immune-based therapies of HIV-1 infection that target CCR5 (Hutter et al., 2009; Hutter and Thiel, 2011; June and Levine, 2012).
Designed zinc-finger protein (ZFP) domains have been successfully used as engineered transcription factors to modulate endogenous gene expression by fusion to well-characterized transcriptional activation or repression domains (Liu et al., 2001; Rebar et al., 2002; Snowden et al., 2003). Importantly, engineered ZFPs can bind with singular specificity (Tan et al., 2003) and fusion of an engineered ZFP to the catalytic domain of the type IIS restriction enzyme FokI creates a designer zinc finger nuclease capable of cleaving DNA specifically at a unique and predetermined site in the human genome (Kim et al., 1996; Smith et al., 1999; Urnov et al., 2005; Rahman et al., 2011). We previously showed that engineered ZFNs targeting human CCR5 efficiently generate a double strand break at a predetermined site in the CCR5 coding region upstream of the natural CCR5Δ32 mutation. Transient expression of the _CCR5_-targeted ZFNs was sufficient to selectively, efficiently, and stably modify the CCR5 locus in both primary T cells and T-cell lines. In addition, ZFN-modified T cells show a marked growth advantage when challenged both in vitro and in vivo with CCR5-tropic HIV (Perez et al., 2008).
The studies presented here have moved the technology of disruption of CCR5 by engineered ZFNs from the research bench to clinical scale using good manufacturing practice (GMP)-compliant reagents, supplies, and procedures. Following CD3/CD28 stimulation and Ad5/F35 adenoviral vector transduction, more than 1×1010 CCR5 gene-edited CD4+ T cells from healthy and HIV-1 infected donors can be generated. CD4+ T cell phenotype, function as assayed by cytokine production and repertoire were comparable between ZFN-modified and control cells. In vivo toxicity studies showed no detectable ZFN-specific toxicity or T-cell transformation. Based on these data and following regulatory approval by the National Institutes of Health (NIH) Recombinant DNA Advisory Committee, University of Pennsylvania Institutional Review Board (IRB) and Institutional Biosafety Committee (IBC), and Food and Drug Administration Center for Biologics Evaluation and Research (FDA-CBER), we initiated a Phase I clinical trial testing this first use of ZFNs in HIV-1 infected subjects (www.clinicaltrials.gov NCT00842634).
Material and Methods
Leukapheresis or whole blood collection and cell separation
Leukapheresis was performed on donors consented on institutional IRB-approved protocols using a Baxter CS3000 (Baxter, Deerfield, IL) or a COBE Spectra (CaridianBCT, Lakewood, CO) in the apheresis unit at the Hospital of the University of Pennsylvania. Leukapheresis cell products were elutriated within 24 hr of collection using the Elutra® Cell Separation System (CaridianBCT) using a protocol developed to maximize lymphocyte recovery and purity (Powell Jr. et al., 2009). For the experiments shown in Figure 1 and Table 1, whole blood from HIV-1+ or healthy donors was collected via venipuncture, and peripheral blood mononuclear cells (PBMC) were isolated using Lymphocyte Separation Media (Lonza, Walkersville, MD). PBMC, peripheral blood lymphocytes (PBL), or elutriated lymphocytes next underwent CD8 depletion to enrich for CD4+ T cells. For smaller scale experiments, CD8 T-cell depletion was performed using the Miltenyi MidiMACS® System, LD columns, and research grade CD8 microbeads (Miltenyi, Auburn, CA) according to the manufacturer's instructions. For clinical scale validation and engineering runs, CD8 depletion was performed on the Miltenyi CliniMACS® system following CD8 enrichment 1.1 protocol and the negative fraction retained for ex vivo stimulation and transduction as described below. Thirteen unique human HIV-1+ and healthy donors were used for the studies shown in figures and tables (Supplementary Table 2; Supplementary Data available online at www.liebertonline.com/hum); altogether >20 unique donors were used in preclinical process development, validation, and engineering studies.
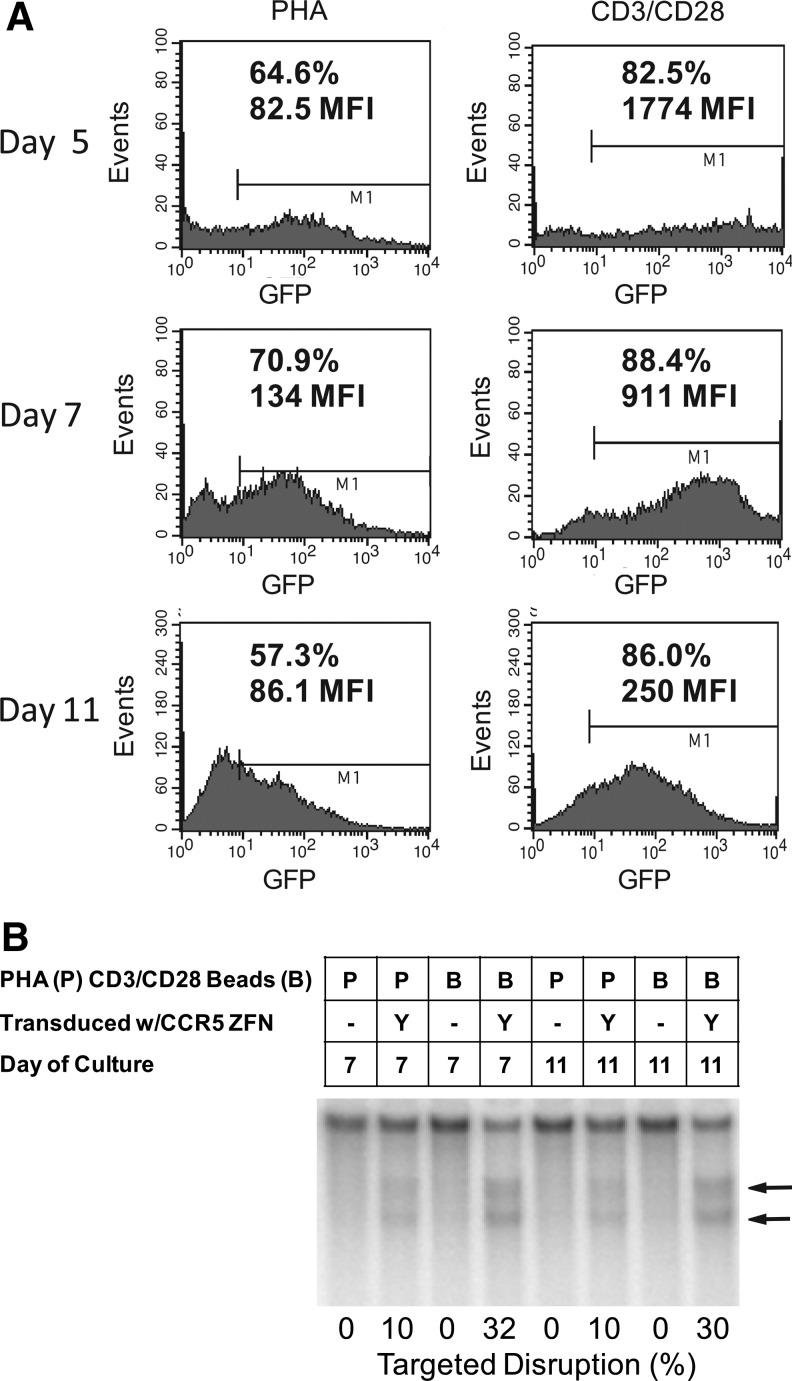
FIG. 1.
Effects of the method of T-cell activation on Ad5/F35 vector transduction and expression efficiency. Primary CD4 T cells were activated overnight with 5 μg/ml phytohaemagglutinin (PHA) or anti-CD3/28 antibody-conjugated beads at a 3:1 bead:cell ratio and then infected at a multiplicity of infection (MOI) of 600 with Ad5/F35 vector. (A) Ad5/F35 vector-expressing green fluorescent protein (GFP) was used to measure transduction efficiency by fluorescence activity at Days 5 to 11 post-stimulation. The percent of cells expressing GFP are shown, as well as the mean fluorescence intensity (MFI), which reflects the level of GFP expression in each cell. (B) Ad5/F35 vector expressing the _CCR5_-specific ZFN, SB-728, was used to measure nuclease activity after transduction. Boxes above the gel show the method of T-cell stimulation, ZFN treatment, and the day of culture. Nuclease activity was measured by using the surveyor nuclease assay on CCR5. Cleavage fragments are indicated by the arrows to the right of the gel. Percent disruption was determined by image analysis as described in the methods and is indicated at the bottom of the gel.
Table 1.
Zinc Finger Nuclease–Mediated Genome Disruption of CCR5 at Five Days Following PHA or CD3/CD28 Stimulation (Percent of Alleles Disrupted)
| | PHA | CD3/CD28 | | | ----------------- | ---------- | ---- | | Donor 1 (healthy) | 4.1 | 16.1 | | Donor 2 (healthy) | 7.3 | 24.6 | | Donor 3 (HIV-1+) | 10.4 | 37.4 |
Media and culture conditions
CD4-enriched lymphocytes were initially seeded into XVIVO™ 15 media formulated without Phenol Red or Gentamicin (Lonza, Walkersville, MD) and supplemented to a final concentration of 20 m_M_ HEPES (Lonza), 2 m_M_ L-Glutamax (Invitrogen, Carlsbad, CA), 0.5% human serum albumin (CSL Behring, Kanakakee, IL), 10 m_M_ N-acetylcysteine (Roxane Laboratories, Columbus, OH), 1 m_M_ sodium pyruvate, 1% minimal essential vitamin mix (both from Lonza), 500 n_M_ Norvir® (ritonavir; Abbott Laboratories, North Chicago, IL), 1 μ_M_ Retrovir® (zidovudine; GlaxoSmithKline, Research Triangle Park, NC), and 100 IU/mL of Interleukin-2 (Chiron Corp, Emeryville, CA). This media formulation also served as Ad5/F35 vector transduction medium. At Day 3 of culture and thereafter, cells were fed with XVIVO™-15 supplemented as above, except that 5% pooled human AB serum (Valley Biomedical, Winchester, VA) was substituted for 0.5% human serum albumin, with 100 IU to 500 IU/mL of Interleukin-2 added, depending on the culture cell concentration. For clinical scale experiments, on Day 5, once cells were seeded into the WAVE™ Bioreactor, 0.02% Pluronic (CellGro®, Manassas, VA) was added to the media. Cell counts and size determinations were performed on a Coulter Multisizer™ 3 (Beckman Coulter Particle Characterization, Miami, FL). Cell viability was assessed by Trypan blue dye exclusion.
T-cell activation and adenoviral transduction
CD4-enriched lymphocytes were seeded at 1×106 per mL in transduction media and stimulated with anti-CD3 and anti-CD28 mAb-conjugated beads as described (Levine et al., 1997) or phytohaemagglutanin (PHA; Sigma, St. Louis, MO) at 5 ug/mL and incubated in a 37°C, 5% CO2 incubator. Following overnight incubation, cells were transduced with either Ad5/F35 CCR5-ZFN or Ad5/F35 green flourescent protein (GFP) vector. Cells were cultured in either T25 plastic flasks for research scale or Baxter Lifecell flasks (Baxter, Deerfield, IL) for clinical scale experiments. At culture Day 3 and after, cells were counted and fed with fresh media containing human serum. For clinical-scale experiments, the cells were transferred to the WAVE Bioreactor™ 2/10 (GE Healthcare Bio-Sciences, Piscataway, NJ). Seeding concentration for the WAVE 2/10 was 0.5×106/mL, and media perfusion was initiated once the culture had reached maximum bag volume. When the cell culture reached 5×106 cells per mL, perfusion was increased to one culture volume per 24 hr. For both research scale and clinical scale experiments, cells were cultured for 9–12 days before harvest. For clinical scale, anti-CD3/anti-CD28 beads were removed with a MaxSep Magnetic Cell Separator (Baxter), and cells were concentrated and washed with a Baxter Fenwal Harvester (Levine et al., 1998) and subsequently cryopreserved using a controlled rate freezer.
Ad5/F35-GFP and Ad5/F35-CCR5-ZFN vectors
The ZFNs targeting the human CCR5 gene were designed and assembled as previously described (Perez et al., 2008). The CCR5-ZFNs were inserted into the pAdEasy-1/F35 vector linked together via a 2A peptide sequence and under control of the tet-regulated cytomegalovirus internal promoter (Invitrogen) to regulate transgene expression during vector production. The GFP reporter gene was independently inserted into the pAdEasy-1/F35 vector to create the Ad5/F35-GFP vector. The Ad5/F35 vectors were packaged in TREx 293 cells (Invitrogen), essentially as previously described (Nilsson et al., 2004).
Flow cytometry
Cells for analysis were resuspended in flow cytometry staining medium (3% heat-inactivated pooled human AB serum and 0.05% sodium azide along with 2m_M_ EDTA in PBS w/o Ca or Mg) prior to the addition of staining antibody. Cell samples were incubated in the dark for 30 min at 4°C, washed with staining buffer, and resuspended in either staining buffer or 1% paraformaldehyde. Cell acquisition was performed using a Becton Dickinson Biosciences (San Jose, CA) FACSCalibur flow cytometer and accompanying BD CELLQuest Pro software. Antibodies were purchased from BD Biosciences. T cells were defined as CD3+ gated on CD45+ cells.
Cytokine analysis
Following 9–12 days of culture, 2×106 cells from either transduced ZFN-modified or nontransduced control cultures were washed, resuspended in fresh media, and either stimulated with fresh anti-CD3/anti-CD28 mAb beads or left unstimulated (control), each at a final cell concentration of 1×106 per mL. Supernatants were harvested 24 hr later. Cytometric bead array reagents in Th1/Th2 kits or Flex Sets designed for multiplex analysis (Becton Dickinson) were used to measure concentrations of IL-2, TNF-α, Interferon-γ, MIP-1α, MIP-1β, GM-CSF, and RANTES. Approximately 300 beads per cytokine were collected and analyzed using the FCAP Array™ software.
Surveyor nuclease assay
The Surveyor nuclease assay to detect genetic modification of sites in the genome by the CCR5-ZFNs was performed as previously described (Perez et al., 2008). Briefly, genomic DNA was extracted from modified and control cells using the MasterPure™ DNA purification kit (Epicentre Biotechnologies, Madison, WI). After radioactive polymerase chain reaction (PCR) amplification of the region encompassing the CCR5 ZFN target region, the Surveyor nuclease (Surveyor mutation detection kit; Transgenomic, Omaha, NE) was used according to the manufacturer. The following primers were used in the PCR: CCR5: 5′-AAGATGGATTATCAAGTGTCAAGTCC-3′ and 5′-CAAAGTCCCACTGGGCG-3′; CCR2: 5′-GGCTCTACTCGCTGGTGTTC-ATC-3′ and 5′-GGCAGAGTAATAAGAAAAAGCAGATC-3′; and ABLIM2: 5′-GGGTTCTCTTGTTTGTTCCATACC-3′ and 5′-GGGTCAGTCTCTTAGCCGTGTG-3′. Products were resolved by PAGE and bands quantified by phosphorimaging.
T-cell repertoire by T-cell receptor Vβ analysis
Approximately 1×106 cells from Day 0 (before transduction) and various time points post-transduction and expansion were collected, and cells were pelleted and frozen. The pellets were analyzed by TCLand Expression (Nantes, France) for the expression of 26 TCR Vβ chain genes. Complementary determining region 3 length distribution (CDR3-LD) alteration was performed on a AB3730 capillary sequencer and analyzed using GeneMapper® and Immunoscope® software (Pannetier et al., 1995). Global CDR3-LD alterations were measured according to Gorochov et al. (1998). T-cell receptor (TCR) variable chain Vβ to hypoxanthine-guanine phosphoribosyltransferase transcript (HPRT) ratios were measured by real-time PCR (Gagne et al., 2000). Data were displayed as a bidimensional TopView or as a tridimensional TCLandscape® (Sebille et al., 2001; Guillet et al., 2002) where the x-axis displays the 26 Vβ families, the z-axis shows the Vβ transcript/HPRT transcript ratios, and the y-axis gives the 13 possible CDR3 lengths per Vβ family. Percentages of alterations compared to healthy donors as controls are represented as a color code, ranging from deep blue (values −30%) to dark red (30%) in the integrated landscapes (Sebille et al., 2001; Guillet et al., 2002). MatLab® 5.3 software (The MathWorks Inc, Natick, MA) was used to compute and display the data.
Karyotyping
Karyotype analysis was performed by the University of Pennsylvania Clinical Cytogenetics Laboratory, which is accredited by the College of American Pathologists. Colcemid as a metaphase arresting agent was added to cell cultures for 2 hr. Cells were exposed to hypotonic solution for 40 min at 37°C, followed by three changes of 1:3 glacial acetic acid and methanol. Cell suspensions were dropped onto water-wet microscope slides. Dried slides were aged in a 60°C oven for 14 hr and stained with Wright Stain for G-banding and analyzed at 100x magnification on a bright field microscope. Images were captured and karyotypes prepared on the Applied Imaging Computer Karyotyping system. Three slide preparations were made from each condition, and a total of 10–15 cells were analyzed per slide.
In vivo toxicity studies in mice
NOD/scid/γcnull (NSG) mice (Shultz et al., 2005) were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in microisolator cages under sterile conditions. While xenogeneic graft-versus-host disease (xGVHD) drives activation and expansion of human T cells in this model (Garcia et al., 1997), it also leads to xGVHD-mediated death in host animals within 2–4 months, depending on infused cell dose and route of administration. T-cell expansion amplifies detection of any genotoxic event, which partially mitigates concern regarding the ability of this model to detect such events within the limited study duration. All animal-use protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Studies used 4–6-week-old male and female mice in equal proportions. Including a pilot study that served to validate the design for Study 1, four unique human donors overall were used for in vivo studies, human T cells were collected, and CCR5-ZFN modified, expanded, and cryopreserved as described above. Twenty million T cells in a cushion of 2 million autologous PBMC (to promote engraftment) were administered to each of the 24 mice per donor in the CCR5-ZFN-modified T-cell group and eight mice per donor in the unmodified T-cell control group. A dose of 20 million cells in a mouse corresponds to a human dose of ∼5×1010 cells. The total dose of CCR5-ZFN-modified T cells administered during preclinical studies from four separate human donors was >3×109 cells, which approaches a single human dose of 5–10×1010 total cells. Cells were injected via the tail vein to mimic the clinical route of administration. Animals were weighed at baseline, weekly post-treatment, and prior to necropsy. Peripheral blood was collected by retro-orbital bleeding at 2–4-week intervals. Daily cage-side observations were performed to assess gross animal health. Twice weekly, animals were physically examined and formally scored for signs of xGVHD, including but not limited to activity level, behavior, general grooming, and condition of fur and posture. Animals judged to be suffering from severe xGVHD were euthanized.
All animals were scheduled to undergo a full necropsy according to a schedule outlined in the experimental design. However, animals judged to be suffering from severe xGVHD were euthanized and necropsied before their scheduled sacrifice date. Peripheral blood collected by cardiac puncture was analyzed for the presence of human cells using the TruCount assay (BD Biosciences, Franklin Lakes, NJ). Samples were stained for human CD45, CD4, and CD8. Animals were scored as positively engrafted if there were greater than 20 CD45+ cells/uL of peripheral blood. Bone marrow harvested from femurs was also stained with antibodies to human CD45, CD4, and CD8, and analyzed by flow cytometry. A second study was conducted utilizing 10 million cells per animal administered intravenously, in the absence of a PBMC cushion with the aim of minimizing xGVHD and supporting longitudinal observation of the animals to detect potential delayed adverse effects. In this study, IL-2 intraperitoneal injections were given at 2000 IU biweekly at days 0, 4, 7, and 10. There was a single donor for this study and all 38 animals received CCR5-ZFN-modified T cells.
Multiple organs were subjected to gross macroscopic analysis at necropsy in order to detect major changes that may have resulted from tumor formation or xGVHD-induced gross tissue damage. Organs and tissues collected included brain, heart, kidney, liver, spleen, lungs, lymph nodes (typically absent), skin (ear), reproductive organs, eye, skeletal muscle, thymus (typically absent), and gut. Any unusual or unidentified organs or tissues were also collected. The brain, heart, kidney, liver, lung, testes (if present), and spleen were weighed. All collected organs were sectioned: one section was formalin fixed and the other was snap-frozen. Fixed tissues were used to prepare paraffin blocks for H&E analysis and immunohistochemistry. In rare cases where potential neoplasms were detected, sections of the affected tissue were stained for with anti-human-CD3 mAb to determine the T-cell content of the neoplasm. Lymphoid neoplasms characteristic of B-cell lymphoma were identified in a pilot study in both control and treatment groups. There was an absence of CD3 staining, and blinded independent histopathologic analysis, performed by Pathology Associates International (PAI, Frederick, Maryland), determined that the neoplasms were characteristic of B-cell lymphoma. Additional review of the histopathology findings in studies 1 and 2 was obtained by a board certified hematology/oncology pathologist at the Hospital at the University of Pennsylvania and also by WIL-Biotechnics LLC (Hillsborough, NC). Retrospective analysis of the donors showed both were Epstein-Barr-virus (EBV) positive. Therefore, the tumors may have arisen from B cells in the PBMC cushion. These cells are known to result in lymphoproliferation in immune-deficient mice (Mosier et al., 1992; Wagar et al., 2000), which illustrates the sensitivity of this model.
Engraftment of human CD4 T cells in skin, liver, testes, lung, ovaries, brain, bone marrow, kidneys, heart, spleen, and blood was assessed by duplex PCR involving coamplification of human and mouse repetitive sequences from snap-frozen tissue sections (Pelz et al., 2005). The limit of detection of the assay was 1 copy of human DNA per 2000 copies of mouse genomic DNA. Quantitation of CCR5-ZFN-modified T cells in blood, spleen, and lung tissues was performed using the Cel-1 Surveyor nuclease assay described above and previously (Perez et al., 2008).
Statistical analysis in vivo toxicity studies
The data from in vivo toxicity studies consisted of up to nine observations (baseline plus eight weekly follow-ups) on animals in two treatment arms (mock transfected and SB 728-T transfected). Two experiments were conducted, each with three separate T-cell donors (although one donor was used in both experiments). To account for this, we considered each donor-experiment combination a group, leading to six groups. We analyzed the weight data using mixed linear models incorporating group, treatment arm, week, and animal as explanatory factors. All analyses were conducted in Proc Mixed of SAS Version 9.2 (SAS Institute, Cary, NC) on a Sun server.
Results
Enhanced efficiency of zinc finger nuclease (ZFN) delivery by Ad5/F35 vector following CD3/CD28 stimulation
We have previously demonstrated that activation of T cells via immobilized anti-CD3 and anti-CD28 mAbs enabled highly efficient and potent stimulation of T cells compared with either phorbol 12-myristate 13-acetate (PMA) or PHA activation (Levine et al., 1995, 1997). Furthermore, since 1996 the CD3/CD28 activation method has been used to prepare several hundred activated T-cell products that have been infused into study subjects in clinical trials (Levine et al., 2002, 2006; Laport et al., 2003; Rapoport et al., 2004, 2005, 2009; Porter et al., 2006). In vitro studies performed with the ZFNs encoded by SB-728 demonstrated that transient ZFN expression post gene transfer using a chimeric Ad5/F35 adenoviral vector was sufficient to modify >30% of the CCR5 alleles (Perez et al., 2008; and data not shown). Ad5/F35 vector had previously been demonstrated to efficiently transduce hematopoietic cells, including T lymphocytes (Shayakhmetov et al., 2000; Chen et al., 2002; Cho et al., 2003; Nilsson et al., 2004; Schroers et al., 2004; Jung et al., 2005; Lu et al., 2006). Earlier studies by other groups in which T lymphocytes had been transduced with Ad5/F35 vectors used PHA or PMA for cell stimulation (Chen et al., 2002; Schroers et al., 2004). However, the use of either PMA or PHA, available in research grades only, is problematic from a regulatory and toxicity perspective. We directly compared the efficiency of Ad5/F35 transduction in experiments with two separate Ad5/F35 vectors encoding either GFP (Fig. 1A) or a ZFN targeted to CCR5 (Fig. 1B) following PHA stimulation or anti-CD3/CD28 stimulation of T cells. CD3/CD28-stimulated cells were transduced more efficiently, as measured by both percent GFP positive cells and GFP mean fluorescence intensity (Fig. 1A). Using the CCR5 ZFN previously selected for further clinical development (Perez et al., 2008), CD3/CD28 stimulation of T cells also facilitated superior genetic modification of CCR5 compared with PHA stimulation as shown in Figure 1B and Table 1. We have previously shown that during the first 5–7 days post-stimulation, T-cell expansion mediated by PHA, PMA, and CD3/CD28 stimulation is roughly equivalent. However, further expansion and restimulation demonstrated the superiority of CD3/CD28 stimulation in terms of cell yield and cell function (Levine et al., 1997). Taken together, these data, and the regulatory advantage of using a clinically validated method for T-lymphocyte stimulation, indicated that maximal ZFN-mediated CCR5 disruption and T-cell yield could be obtained by adapting the Ad5/F35 delivery of ZFNs to large-scale CD3/CD28 cell stimulation to generate sufficient numbers of _CCR5_-modified T cells for a clinical trial.
Development of clinical ex vivo transduction process
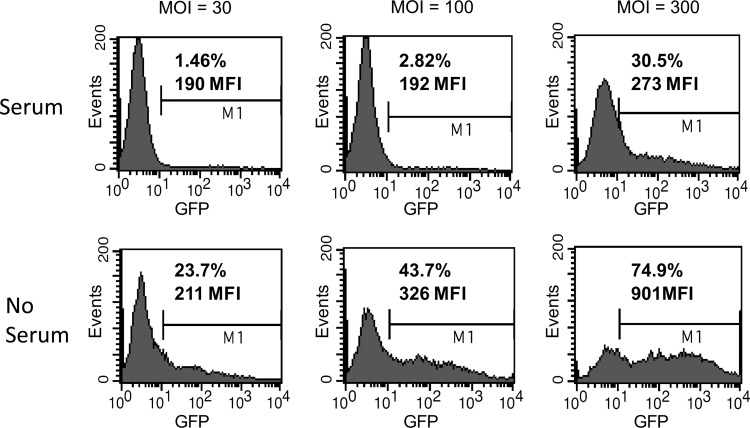
While early experiments showed that Ad5/F35-mediated transduction of CCR5-ZFNs was efficient, these studies were conducted with a media formulation containing fetal calf serum (Perez et al., 2008). This presented a regulatory obstacle for clinical process development due to the risk of bovine spongiform encephalopathy (BSE) contamination. Of more concern is the reported immune reactivity to adoptively transferred cells prepared with bovine serum, even in immunosuppressed subjects, ranging from anti–fetal calf serum (FCS) antibodies to Arthus reactions and anaphylaxis (Selvaggi et al., 1997; Mackensen et al., 2000; Horwitz et al., 2002; Tuschong et al., 2002). We first performed Ad5/F35 transduction in standard clinical media that we previously validated for T-cell expansion. This medium was based on XVIVO-15, developed for serum-free cultures, but it was supplemented with pooled human AB serum as used in several previous clinical trials, including gene transfer trials (Levine et al., 2002, 2006; Laport et al., 2003; Rapoport et al., 2004, 2005, 2009; Porter et al., 2006). This media was compared to the research-grade media consisting of RPMI 1640 supplemented with 10% FCS. While there was efficient transduction in RPMI 1640/10% FCS, there was minimal transduction in XVIVO-15 supplemented with 5% pooled human serum (data not shown), likely due to anti-adenovirus antibodies in lots of pooled human serum (Nwanegbo et al., 2004). We then tested a medium formulation of XVIVO-15 supplemented with 0.5% human serum albumin without addition of pooled human serum. To optimize T-cell expansion following transduction, medium supplemented with pooled human serum was added 3 days after the addition of Ad5/F35 CCR5-ZFN vector. As shown in Figure 2, efficient transduction was obtained when compared with pooled human serum–supplemented media.
FIG. 2.
Human serum inhibition of Ad5/F35 transduction in T cells. Primary CD4 T cells were activated overnight with anti-CD3/28 antibody-conjugated beads at a 3:1 bead:cell ratio and then transduced at an MOI of 30, 100, or 300 with Ad5/F35 vector-expressing GFP in the presence of 5% pooled human serum or in the absence of human serum. GFP was measured on Day 3 after initial activation. The percent of cells expressing GFP and the MFI of cells gated as positive are shown.
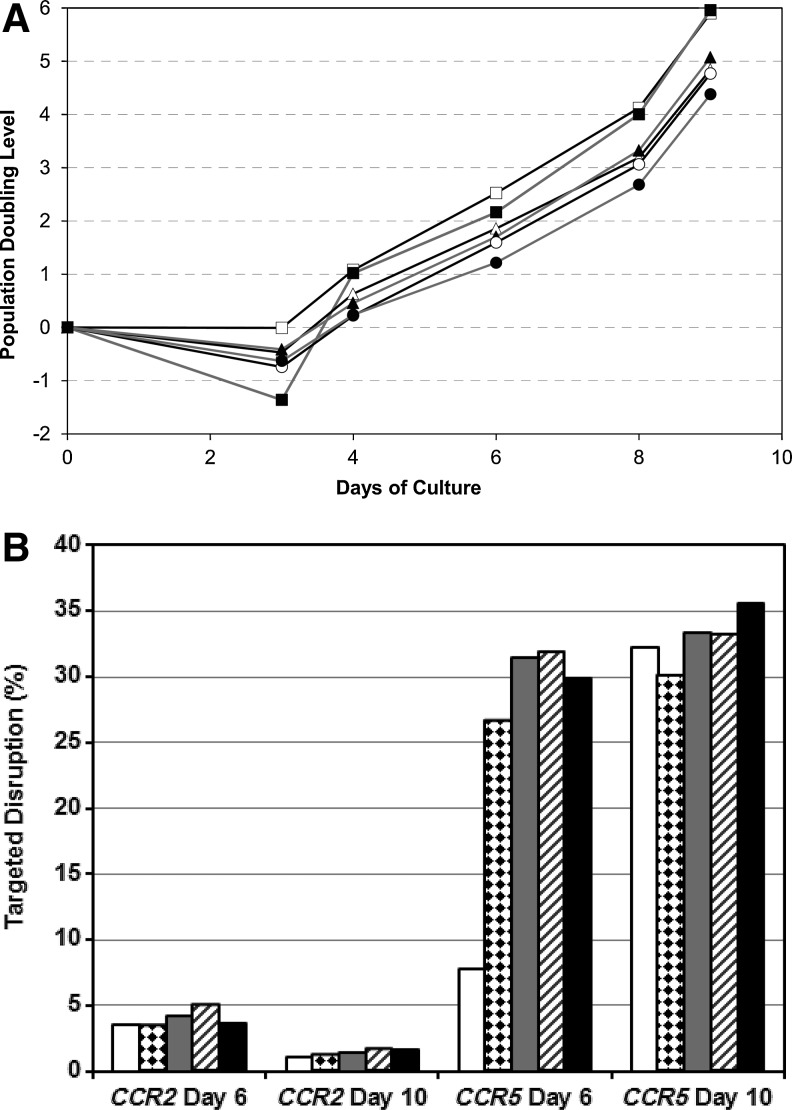
Using the transduction media and process described above, we next established the optimal Ad5/F35 CCR5-ZFN vector MOI to maximize ZFN-mediated CCR5 modification while minimizing the potential negative effects of higher levels of vector or ZFN on T-cell expansion. Additionally, it was important to establish whether off-target gene modification could be observed at higher MOI. Based on pilot studies, we focused on a range of MOI from 400 to 2,000 infectious units per cell, which at our observed particle to infectious unit ratio of 8–10 corresponds to 3,200 to 20,000 vector genomes per cell. As expected, Ad5/F35 CCR5-ZFN transduction has a negative impact on overall T-cell expansion capacity at higher MOIs in both healthy donors and HIV-1 infected donors. As shown in Figure 3A in an HIV-1 infected donor, a modest impairment of T-cell growth was observed at MOIs 800, 1200, and 2000, with no, or minimal, effect on T-cell growth at MOIs of 400 and 600. A similar effect was observed in an identical MOI titration with a healthy donor (data not shown). To establish the effect of Ad5/F35 CCR5-ZFN vector MOI on the frequency of both on- and off-target gene modification, samples from this experiment were analyzed for disruption of the CCR5 locus, as well as disruption at two potential off-target sites, CCR2 and ABLIM2, previously identified by a bioinformatic search of the human genome (Perez et al., 2008). On- and off-target modification was determined by Surveyor nuclease assay and deep sequencing of the modified cells as described (Perez et al., 2008). Figure 3B shows that at Day 6 post-transduction, there was a trend for increased CCR5 modification with increasing MOI from 400 to 800, with no added benefit from higher MOIs. By Day 10 of culture, only marginal increases in CCR5 disruption were observed at higher MOIs. For CCR2, low level disruption (<4%) was observed at an MOI of 400, but the frequency of disruption did not increase with higher MOI, and by Day 10 there was a relative decrease in the percentage of CCR2 disrupted cells. There was no observed disruption at ABLIM2 at a level of detection in the Surveyor nuclease assay of ∼1% (data not shown). Previous studies conducted in small scale cultures using ultra-deep sequencing showed only rare (∼1/20,000) events at ABLIM2 (Perez et al., 2008). Based on these results, and to minimize the effect of higher MOI on cell growth, an MOI of 600 was chosen for further clinical development.
FIG. 3.
Ad5/F35 CCR5 ZFN vector titration and T-cell expansion. To evaluate the best balance of MOI for efficient transduction and preservation of robust cell expansion, CD4 T cells were activated overnight with CD3/28 beads as previously described and then transduced with Ad5/F35 vector-encoding CCR5-ZFN at MOI's of 0 or mock transduced, 400, 600, 800, 1200, and 2000. (A) Cells were monitored for growth and population doubling level calculated based on starting number of cells and number of cells continued in culture at each time point after sampling: Mock transduced (black line, open square symbols), MOI 400 (gray line, filled square symbols), MOI 600 (black line, open triangle symbols), MOI 800 (gray line, closed triangle symbols), MOI 1200 (black line, open circle symbols), and MOI 2000 (gray line, closed circle symbols). (B) Gene disruption at CCR5 and the closely related CCR2 was measured by the SURVEYOR nuclease assay: MOI 400 (white), MOI 600 (diamond hatch), MOI 800 (gray), MOI 1200 (diagonal hatch), and MOI 2000 (black).
Functional and phenotypic comparability of CCR5-ZFN-modified and -unmodified T cells
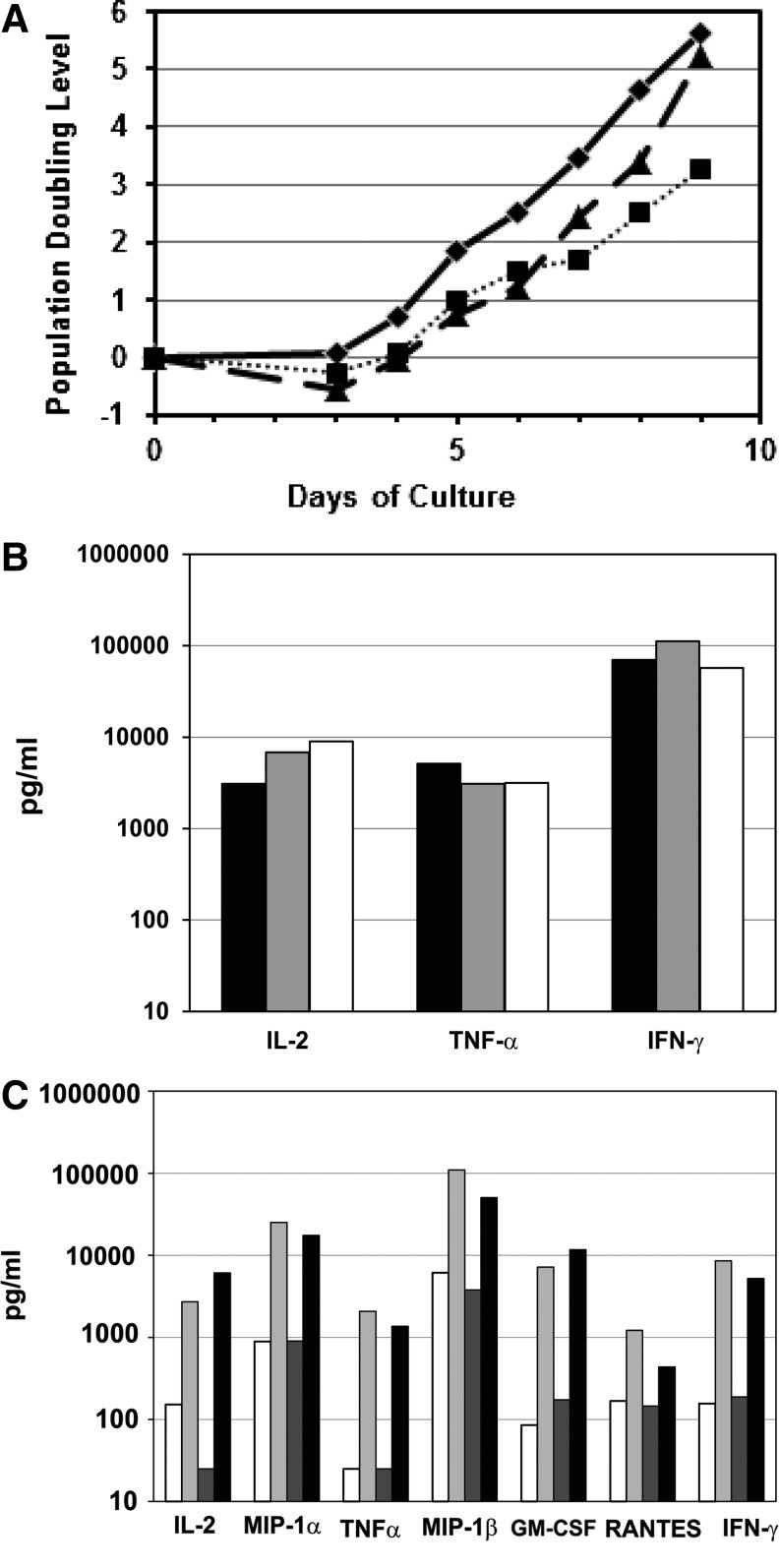
We previously developed a large-scale method of T-cell isolation and culture in static gas permeable bags, followed by washing, concentration, and formulation of the cell product sufficient to meet doses of expanded T cells of up to 3×1010 (Levine et al., 1998). More recently, we have integrated a dynamic bioreactor (Wave, GE Healthcare Life Sciences, Piscataway, NJ) that allows for media perfusion during the culture process (Hami et al., 2004; Hollyman et al., 2009). A possible advantage of the dynamic perfusion method is that it achieves further reduction of remaining adenoviral vectors that could contribute to the immunogenicity of the Ad5/F35-transduced T-cell product. Translating the research scale static process to a clinical scale dynamic perfusion culture required validation studies that addressed T-cell growth, possible decreases in transduction efficiency, or dilution of autocrine T-cell cytokine production by perfusion. Deficiencies in any of these processes could result in less potent T cells as the final product. To address these questions, we first compared growth rates in untransduced and transduced research scale T-cell cultures to growth rates in transduced clinical scale cultures employing the dynamic perfusion system. Shown in Figure 4A is a representative growth curve comparing these conditions using cells from an HIV-1 infected donor. These data indicate transduced cells were more efficiently expanded using the clinical scale dynamic perfusion procedure than the static small-scale procedure. In fact, growth of transduced cells using the clinical scale dynamic perfusion system was comparable to growth of untransduced cells using the static small-scale procedure. While it appears in Figure 4A that the growth rate accelerated in the clinical scale condition between Day 8 and Day 9, this likely reflects a sampling artifact resulting from the design of the bioreactor bag. On Day 9, the cell count was obtained from a sample after all cells had been removed from the bioreactor bag and thus more accurately reflects the total number of cells in the culture. In addition, the clinical scale process enabled targeted ZFN-mediated CCR5 disruption at 20.9% of alleles on Day 9, compared to 14.9% of alleles for the research scale process.
FIG. 4.
Validation of large-scale ex vivo clinical process for T-cell expansion and CCR5 modification incorporating a dynamic perfusion bioreactor. (A) Cells from an HIV-1 infected donor were stimulated ex vivo and transduced as described in the Methods section. Research scale untransduced, solid line and diamond symbols; research scale transduced, dotted line and square symbols; clinical scale transduced, dashed line and triangle symbols. (B) The production of IL-2, TNF-α, and Interferon-γ in supernatants are shown—research scale untransduced, research scale CCR5-ZFN transduced, and clinical scale CCR5-ZFN-transduced cultures. Expanded T cells from the culture in (A) were harvested, washed, resuspended in fresh media, and restimulated with fresh anti-CD3/CD28 mAb-coated beads. Supernatants were collected 24 hr later. Research scale untransduced, black bars; research scale transduced, gray bars; clinical scale transduced, white bars. An additional HIV-1 infected donor (C) was compared for production of cytokines as well as β-chemokines as in (B). Spontaneous secretion of cytokines and β-chemokines (production in the absence of restimulation) was also assayed as a measure potency of final product cells for infusion. Cells were harvested at the end of culture, and beads were removed and resuspended in fresh culture media. Supernatants were collected 24 hr later. Shown is the production of IL-2, TNF-α, MIP-1α, MIP-1β, GM-CSF, RANTES, and Interferon-γ. Research scale untransduced and unstimulated (spontaneous), white; research scale untransduced and restimulated with fresh anti-CD3/anti-CD28 mAb beads, light gray; clinical scale transduced and unstimulated (spontaneous), dark gray; clinical scale transduced and restimulated with fresh anti-CD3/anti-CD28 mAb beads, black.
As a gauge of T-cell function, the production of IL-2, TNF-α, and Interferon-γ was measured by the cytometric bead array assay in supernatants from research scale untransduced, research scale CCR5-ZFN transduced, and clinical scale CCR5-ZFN transduced cultures (Fig. 4B). Expanded T cells from the HIV-1 infected donor shown in Figure 4A were harvested, and the remaining anti-CD3/CD28 mAb-coated beads were removed. The cells were washed, resuspended in fresh media, and restimulated with fresh anti-CD3/CD28 mAb-coated beads. Supernatants were collected 24 hr later. All three culture methods resulted in comparable production of IL-2, TNF-α, and Interferon-γ, indicating that CCR5-ZFN-modified T cells transduced at research scale or at clinical scale following media perfusion in the wave bioreactor were not impaired in cytokine production compared to control untransduced T cells. These results were confirmed in a subsequent clinical scale run with a separate HIV-1+ donor in which production of granulocyte macrophage colony-stimulating factor (GM-CSF) and chemokines was also examined (Fig. 4C). Chemokine production, like cytokine production, is an important functional indicator in the setting of HIV infection (Verani and Lusso, 2002). Spontaneous secretion of cytokines and chemokines (production in the absence of restimulation) was also measured, as this might better reflect the functional status of the final cell product upon reinfusion to study subjects. Figure 4C depicts production of IL-2, TNF-α, MIP-1α, MIP-1β, GM-CSF, RANTES, and Interferon-γ by research scale untransduced control T cells and clinical scale CCR5-ZFN-transduced T cells. Spontaneous secretion of cytokines was comparable between the two conditions for all cytokines except for IL-2. Spontaneous IL-2 levels were lower in the clinical scale culture, which might reflect increased consumption in a rapidly growing culture. Perfusion culture media is supplemented with IL-2, but media used to assess spontaneous cytokine production was not supplemented with IL-2. For cells restimulated with fresh anti-CD3/CD28 mAb-coated beads, production of all cytokines and chemokines tested was comparable between the control research scale condition and the clinical scale transduced CCR5-ZFN-modified condition. Taken together, these results demonstrate functional comparability in control T cells and CCR5-ZFN-modified T cells expanded under clinical scale media perfusion conditions.
Control unmodified T cells and transduced CCR5-ZFN-modified T cells were also phenotyped by flow cytometry. Equivalent levels of CD3+CD4+, CD4+CD28+, activation (CD25), and homing (CD62L) receptors were observed on cells expanded by either method. CD3+CD4+, CD3+CD28+, and CD3+CD25+ percentages were routinely >90% (data not shown) and consistent with previous studies (Levine et al., 1996, 1998). To look for putative chromosomal abnormalities that might be induced by CCR5-ZFN modification of T cells, the FDA and the NIH Recombinant DNA Advisory Committee (RAC) requested that we perform karyotype analysis, in addition to the bioinformatic search, and ultra-deep sequencing of potential CCR5-ZFN off-target sites, and intranuclear staining for sites of DNA double-strand breaks conducted in our previous study (Perez et al., 2008). In both control expanded and CCR5-ZFN modified and expanded T cells, a normal karyotype was described and no clonal abnormalities were present (data not shown).
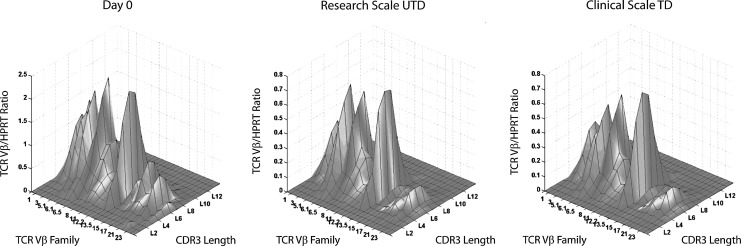
TCR repertoire analysis can provide information on the loss of diversity of CD4+ T cells during progressive HIV infection (Connors et al., 1997; Gea-Banacloche et al., 1998, 2000; Gorochov et al., 1998; Martinon et al., 1999). TCR analysis can detect aberrant clonal expansions or deletions (Pannetier et al., 1995) and is thus essential for safety monitoring in adoptive T-cell transfer therapy. For example, TCR repertoire analysis has been used to track the development of leukemic clones in children after gene therapy for x-linked severe combined immunodeficiency (SCID-X1) (Hacein-Bey-Abina et al., 2003). T cells from a total of three donors (one healthy, two HIV-1+) were analyzed for TCR Vβ chain repertoire before and after a control research scale expansion or after a clinical scale expansion of CCR5-ZFN-transduced cells. Figure 5 illustrates a three-dimensional plot of the complementary determining region 3 (CDR3) length distribution for the 26 TCR Vβ family genes analyzed as a ratio of the Vβ to HPRT transcripts. Similarity was seen between the research scale untransduced condition and the clinical scale CCR5-ZFN-modified condition in the average percent perturbation as measured by skewedness to healthy donor controls (Supplementary Table 1). Thus, by multiple in vitro measures, control, and transduced CCR5-ZFN-modified T cells in both healthy and HIV+ donors were equivalent in phenotype, expansion, cytokine and chemokine production, and TCR diversity.
FIG. 5.
T-cell receptor (TCR) Vβ repertoire of CCR5-ZFN T cells produced with large-scale ex vivo clinical process. TCR Vβ chain repertoire before and after a control research-scale expansion or after a clinical-scale expansion of CCR5-ZFN-transduced cells from a representative HIV+ donor. T cells from a total of three donors (one healthy, two HIV+) were analyzed. Three-dimensional plot of the complementary determining region 3 (CDR3) length distribution for the 26 TCR Vβ family of genes was analyzed as a ratio of the Vβ to hypoxanthine-guanine phosphoribosyltransferase transcript (HPRT). X-axis, TCR Vβ family; y-axis, TCR Vβ/HPRT ratio; z-axis, CDR3 length. Calculated TCR Vβ percentages of alteration are listed in Supplementary Table 1.
In vivo toxicity studies in mice
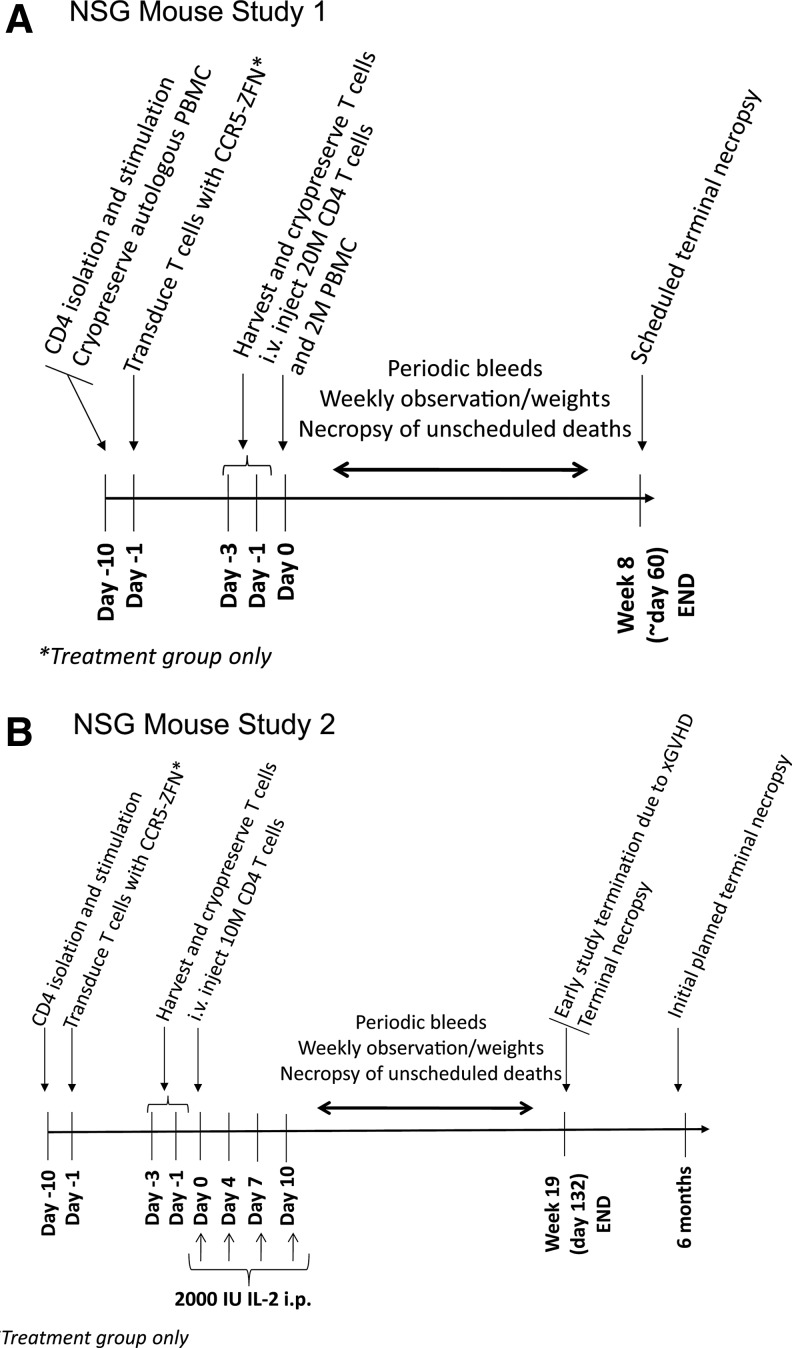
In pre–investigational new drug (IND) discussions, the FDA requested that animal studies be performed to evaluate the safety of CCR5-ZFN-modified T cells. Although there are limitations, immunodeficient mouse xenotransplant models can address some safety concerns by using in vivo engraftment to search for evidence of genotoxicity. The NOD/scid/γcnull (NSG) mouse (Shultz et al., 2005) supports high levels of engraftment of activated human T cells. In addition to lacking T-, B-, and natural killer cells, NSG mice also have impaired dendritic cell function (Ito et al., 2002). While xenoreactivity drives activation and pauci-clonal expansion of human T cells in NSG mice (Garcia et al., 1997), it also leads to xenogeneic graft-versus-host disease (xGVHD)-mediated death in host animals (Nervi et al., 2007). Consequently, we performed a series of pilot animal studies to establish the optimal cell dose, route of administration, and study duration in the NSG mouse model. FDA feedback following these pilot studies was incorporated into the design of full-scale studies 1 and 2 shown in Figure 6 and described below. These studies were conducted to evaluate whether ex vivo CCR5-ZFN modification of T cells led to in vivo T-cell proliferative abnormalities or tumor formation related to the CCR5-ZFN-modified T cells.
FIG. 6.
NOD/scid/γcnull (NSG) mouse–human xenograft biotoxicity study design. The days on study and the procedures at each timepoint are shown. See Methods for a detailed description of study procedures. (A) Study 1 utilized three unique human donors. CD4 T cells were transduced (24 mice/donor) or mock transduced (8 mice/donor) with Ad5/F35 vector-encoding CCR5-ZFN, and infused along with autologous peripheral blood mononuclear cells (PBMC) to support engraftment. Study 1 duration was restricted to 8 weeks due to onset of xenogeneic graft-versus-host disease (xGVHD). (B) Study 2 (38 mice) was performed using a single donor and without co-injection of autologous PBMC to delay xGVHD onset and allow a longer study observation period. Engraftment was supported by IL-2 injections concomitant with CCR5-ZFN-modified CD4+ T cells.
In the first study, 20 million CCR5-ZFN-modified CD4 T cells compared to mock control unmodified cells were administered along with 2 million autologous PBMCs (Table 2) for each of three unique human donors. Engraftment of human cells was detected overall across donors in 92–100% of animals. There was no adverse effect on CD4 T-cell engraftment of the CCR5-ZFN-modified cells compared to mock control unmodified cells (Supplementary Fig. S1). Peak engraftment levels were detected at 8 weeks post injection, corresponding with the development of xGVHD, which is endemic to this model (Nervi et al., 2007). There was also no adverse effect on body weight of the CCR5-ZFN-modified cells compared to mock control unmodified cells for each of three unique human donors, indicating no global adverse effects of the CCR5-ZFN-modified cells (Supplementary Fig. S2). Study 2 was performed with a single arm of CCR5-ZFN-modified cells and without a PBMC cushion to reduce xGVHD and thereby to extend the window for monitoring tumor formation. In this study, animals were evaluated for up to 18 weeks due to development of xGVHD across all animals prior to the planned 6-month duration.
Table 2.
In Vivo Studies—Unscheduled Deaths, CCR5 Modification in the Infused Cells and Recovered Cells, Change in Body Weight, Graft-Versus-Host Disease, and Pathology Findings
| Study | Donor | Group | n | Unscheduled deaths | CCR5 modification pre-infusion | CCR5 modification at necropsyamean±SD | Avg % weight change±SD | % GVHDb | Pathology findings |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | CCR5-ZFN | 24 | 18 (75%) | 27.5 | 0.7 (1.2) | −5.8 (6.2) | 33.3 | None detected |
| 1 | 10 | Mock | 8 | 8 (100%) | 0 | 0 | NA | 37.8 | None detected |
| 1 | 11 | CCR5-ZFN | 24 | 0 | 24.8 | 2.3 (4.0) | −3.7 (3.9) | 4.2 | None detected |
| 1 | 11 | Mock | 8 | 0 | 0 | 0 | −4.6 (4.0) | 12.5 | None detected |
| 1 | 12 | CCR5-ZFN | 24 | 3 (13%) | 20 | 0 | −5.4 (6.7) | 0 | 1) Hepatic adenoma2) Liver and kidney nodules, no abnormal histopathology |
| 1 | 12 | Mock | 8 | 1 (13%) | 0 | 0 | −6.1 (3.7) | 0 | Tubular degeneration in testes with epididymis |
| 2 | 13 | CCR5-ZFN | 38 | 38 (100%)c | 36.1 | 25.9 (6.9) | −8.5 (1.0) | 100 | None detected |
In each of the two animal studies, comparison of CCR5 gene modification levels in the initial cell product compared to the cells isolated from the spleen at necropsy indicated no selective outgrowth of CCR5-ZFN-modified cells in vivo (Table 2), confirming prior observations (Perez et al., 2008). Microscopic histopathology in each of the studies and groups was consistent with xGVHD. No CCR5-ZFN- or T-cell-related tumor formation was observed in any animals. All unscheduled deaths where necropsy could be performed were also confirmed to be xGVHD-associated. The number of unscheduled deaths was similar between the mice receiving CCR5-ZFN-modified T cells and control mice (Table 2). Therefore, by all criteria agreed upon in consultation with the FDA (clinical observations, gross observations upon necropsy, molecular studies, and histopathology) no CCR5-ZFN-related toxicity was detected in any of the studies. The in vivo studies complemented in vitro studies performed in both healthy and HIV-1+ donors (Supplementary Table 2). Data from these in vivo and in vitro studies were submitted to the FDA to support the first in human clinical trial evaluating CCR5-ZFN-modified T cells in HIV-1 study subjects (www.clinicaltrials.gov identifier number NCT00842634).
Discussion
We show here an integrated strategy for the scale-up, validation, and provision of safety data to regulatory authorities for the first trial of ex vivo zinc finger nuclease genome-edited cells. Sufficiently large numbers of modified cells can be generated using clinically compatible procedures and reagents, and these modified cells retain functional activity and phenotypic properties comparable to research scale methods. We have previously shown that highly efficient and potent activation of T lymphocytes is achieved following costimulation with anti-CD3/CD28 beads when compared to either PHA or PMA activation through protein kinase C (Levine et al., 1995, 1997). Moreover, while the T-cell expansion rate mediated by PHA, PMA, and anti-CD3/CD28 stimulation is roughly equivalent during the first 5–7 days poststimulation, only costimulation via anti-CD3/CD28 enables extensive expansion of functional T cells secreting high levels of cytokines and chemokines (Levine et al., 1997). We show that this anti-CD3/CD28 costimulatory activation method produces an expanded T-cell product with enhanced Ad5/F35 transduction efficiency and superior genetic modification, using either the GFP or ZFN-CCR5 Ad5/F35 vectors tested. In addition, while earlier work by others utilized PHA- or PMA-stimulated T cells for Ad5/F35 vector transduction (Chen et al., 2002; Schroers et al., 2004), both PHA and PMA pose regulatory and toxicity challenges for clinical use.
Chimeric Ad5/F35 adenoviral vector derived from swapping the knob region of the fiber protein results in a pseudotyped virus with modified tropism and allows for transduction of cells of the hematopoietic lineage (Shayakhmetov et al., 2000; Chen et al., 2002; Hurez et al., 2002; Cho et al., 2003; Leen et al., 2004; Nilsson et al., 2004; Schroers et al., 2004; Jung et al., 2005; Lu et al., 2006). While the efficiency of Ad5/F35 delivery of CCR5-ZFNs to T cells is attractive, a potential drawback is the presence of residual adenoviral vector at the end of production. Though our clinical process incorporates perfusion during culture and extensive washing at the end of culture and prior to formulation, adenoviral vector and residual ZFN antigens theoretically could contribute to generation of an adenovirus immune response, which could limit administration of multiple infusions of these cells (Nwanegbo et al., 2004). One way to avoid this issue is to use alternative methods of delivery such as RNA electroporation, recently been shown to effectively express functional gene constructs in vivo (Zhao et al., 2010).
Modification of culture media was necessary for efficient clinical scale ex vivo transduction with the adenoviral vector Ad5/F35. Due to the presence of neutralizing adenoviral antibodies in pooled human serum, reformulation of media supplements to contain human serum albumin in substitution for pooled human serum was shown to enhance transduction of Ad5/F35 CCR5-ZFN vector efficiency with minimal impact on ex vivo expansion. Ad5/F35 CCR5-ZFN transduction at higher MOIs resulted in reduced T-cell growth in both healthy donors and HIV+ donors compared to the overall T-cell expansion achieved at lower MOIs.
Two potential off-target sites, CCR2 and ABLIM2, previously identified by a bioinformatics search of the human genome (Perez et al., 2008), were monitored in this study. Importantly, the low level of disruption (<4%) observed at CCR2 did not increase with higher vector MOIs at Day 6 and decreased at Day 10. This could be due to deleterious effects of higher adenovirus vector exposure or higher levels of ZFNs. It may be that multiply transduced CCR5- and _CCR2-_modified T cells are at a growth disadvantage through vector-mediated or other effects. In humans, mutant alleles of CCR2 have been correlated with delayed progression to AIDS in HIV-infected individuals, suggesting a possible beneficial phenotype from CCR2 loss in HIV-infected individuals (Smith et al., 1997). The next off-target site identified (Perez et al., 2008), ABLIM2 is located in an intron, and we show here that there was no detected disruption at ABLIM2 using the clinical cell-processing methods described. A newer unbiased method for analysis of ZFN specificity has identified a restricted number of additional off target cleavage sites under conditions of ZFN overexpression in K562 cells (Gabriel et al., 2011). Improvements in designed ZFN specificity (either via modification of the nuclease or DNA binding domains) could further reduce the risk of off-target cleavage. In our clinical trial NCT00842634 to date, we have not observed any adverse events attributed to ZFN gene editing in patients.
In the present study, we directly compared Ad5/F35 ZFN gene editing of CCR5 in CD4+ T cells expanded via our modified clinical scale process of T-cell expansion, which incorporates a dynamic perfusion method (Hami et al., 2004; Hollyman et al., 2009) with our previously described research scale process for T-cell expansion (Levine et al., 1997). The results shown validate that our clinical scale process of T-cell expansion can produce an expanded Ad5/F35 ZFN-CCR5-modified CD4+ T-cell population that shows enhanced growth capacity, cytokine and chemokine production, and comparable phenotype and TCR diversity to previous research scale expansion methods.
Our previous in vivo study using a mouse model of acute HIV infection showed, for the first time, that ZFN-guided genomic editing is highly specific, well tolerated, and sufficiently robust to support engraftment and HIV resistance in that model (Perez et al., 2008). In consultation with the FDA, we expanded on these studies to evaluate possible toxicity caused by the CCR5-ZFN. In our toxicity studies, the predominant findings of microscopic histopathology were consistent with xGVHD in all studies. No treatment-related toxicity, including CCR5-ZFN or T-cell-related tumor formation, was observed in any animals. While the NSG model was the best available animal model for evaluation of in vivo toxicity and biodistribution of genetically modified human cells, it is not ideal due to xenogenic-mediated pauci-clonal expansion leading to TCR restriction and differences in the trafficking of human cells in mice (Lin et al., 2007). There is importance in using animals to establish the safety of a modified cell product as required by the FDA, but in the future we hope that more informative studies can be developed in concert with the FDA that are less expensive and time consuming while still serving to ameliorate the risk to human patients.
Recent work by Holt et al. (2010) has shown that the same ZFN-CCR5 construct used in our preclinical studies shown here and previously (Perez et al., 2008) can be adapted to an additional transient gene delivery system (nucleofection) to effectively knock out CCR5 in human hematopoietic stem/progenitor cells (HSPCs). This yielded a cell population that retains functional capacities for both engraftment and hematopoietic reconstitution in the NSG mouse, which has been used for modeling HIV-1 infection in vivo (Kumar et al., 2008) and human hematopoiesis (Ishikawa et al., 2005). Consistent with our findings in both the NSG and NOG murine models, Holt et al. showed that engrafted human CD34+ HSPCs containing a minority of _CCR5_-deleted cells were capable of producing a human polyclonal T-cell population that survived viral challenge with CCR5-tropic HIV-1.
The potential therapeutic benefit of a _CCR5_-specific genetic knockout could confer long term resistance to HIV-1 infection, at least in the modified cells. Hutter et al. (2009) reported an HIV-1+ patient who underwent an allogeneic stem cell transplant for treatment of his acute myeloid leukemia with donor cells homozygous for the CCR5 32-bp deletion (Δ32). Following chemotherapy, antiretroviral drugs were discontinued, and the patient remained off antiretroviral drugs following stem cell transplantation without return of viremia. The investigators recently reported reconstitution of CD4+ T cells in peripheral blood, gut, and tissue with donor-derived cells, and that HIV remains undetectable in bone marrow, gut, brain/CSF, and blood of this patient at 45 months posttransplant (Allers et al., 2011). This example provides further proof of concept for provision of CCR5 gene-edited T cells as a potential treatment for HIV infection.
Supplementary Material
Supplemental data
Supplemental data
Supplemental data
Supplemental data
Acknowledgments
The authors would like to thank Daniel F. Heitjan and Kay See Tan for statistical analysis, Ewa Tomczak for karyotyping analysis, Jianbin Wang, Kenneth Kim, Nhu Tran, Bao-Lu Chen, Martin Giedlin at Sangamo BioSciences, Anne Chew for advice and constructive comments, members of the Clinical Cell and Vaccine Product Facility for technical assistance, and Sandra Bridges and Frosso Voulgaropoulou of the NIH Division of AIDS for advice and support.
Supported by 1RO1AI104400 and NIAID Integrated Preclinical/Clinical Program, grants 5U19AI066290 and 5U19AI082628, and Sangamo BioSciences.
Author Disclosure Statement
Sangamo BioSciences provided research funding for these studies. Sangamo has not selected or edited the data presented. The University of Pennsylvania coauthors have no personal financial relationship with Sangamo. B.L. and C.J. have a financial interest due to intellectual property and patents in the field of cell and gene therapy. Conflict of interest is managed in accordance with University of Pennsylvania policy and oversight.
References
- Allers K. Hutter G. Hofmann J., et al. Evidence for the cure of HIV infection by CCR5{Delta}32/{Delta}32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- Carrington M. Dean M. Martin M.P. O'Brien S.J. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet. 1999;8:1939–1945. doi: 10.1093/hmg/8.10.1939. [DOI] [PubMed] [Google Scholar]
- Chen Z. Ahonen M. Hamalainen H., et al. High-efficiency gene transfer to primary T lymphocytes by recombinant adenovirus vectors. J. Immunol. Methods. 2002;260:79–89. doi: 10.1016/s0022-1759(01)00521-x. [DOI] [PubMed] [Google Scholar]
- Cho H.I. Kim H.J. Oh S.T. Kim T.G. In vitro induction of carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocytes by dendritic cells transduced with recombinant adenoviruses. Vaccine. 2003;22:224–236. doi: 10.1016/s0264-410x(03)00569-3. [DOI] [PubMed] [Google Scholar]
- Cohen O.J., et al. Heterozygosity for a defective gene for CC chemokine receptor 5 is not the sole determinant for the immunologic and virologic phenotype of HIV-infected long-term nonprogressors. J. Clin. Invest. 1997;100:1581–1589. doi: 10.1172/JCI119682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors M., et al. HIV infection induces changes in CD4+ T-cell phenotype and depletions within the CD4+ T-cell repertoire that are not immediately restored by antiviral or immune-based therapies. Nat. Med. 1997;3:533–540. doi: 10.1038/nm0597-533. [DOI] [PubMed] [Google Scholar]
- Dropulic B. June C.H. Gene-based immunotherapy for human immunodeficiency virus infection and acquired immunodeficiency syndrome. Hum. Gene Ther. 2006;17:577–588. doi: 10.1089/hum.2006.17.577. [DOI] [PubMed] [Google Scholar]
- Eugen-Olsen J., et al. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS. 1997;11:305–310. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- Gabriel R., et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Gagne K., et al. Highly altered V beta repertoire of T cells infiltrating long-term rejected kidney allografts. J. Immunol. 2000;164:1553–1563. doi: 10.4049/jimmunol.164.3.1553. [DOI] [PubMed] [Google Scholar]
- Garcia S. Dadaglio G. Gougeon M.L. Limits of the human-PBL-SCID mice model: severe restriction of the V beta T-cell repertoire of engrafted human T cells. Blood. 1997;89:329–336. [PubMed] [Google Scholar]
- Gea-Banacloche J.C., et al. Progression of human immunodeficiency virus disease is associated with increasing disruptions within the CD4+ T cell receptor repertoire. J. Infect. Dis. 1998;177:579–585. doi: 10.1086/514233. [DOI] [PubMed] [Google Scholar]
- Gea-Banacloche J.C., et al. Longitudinal changes in CD4+ T cell antigen receptor diversity and naive/memory cell phenotype during 9 to 26 months of antiretroviral therapy of HIV-infected patients. AIDS Res. Hum. Retroviruses. 2000;16:1877–1886. doi: 10.1089/08892220050195838. [DOI] [PubMed] [Google Scholar]
- Gorochov G., et al. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat. Med. 1998;4:215–221. doi: 10.1038/nm0298-215. [DOI] [PubMed] [Google Scholar]
- Guillet M. Brouard S. Gagne K., et al. Different qualitative and quantitative regulation of V beta TCR transcripts during early acute allograft rejection and tolerance induction. J. Immunol. 2002;168:5088–5095. doi: 10.4049/jimmunol.168.10.5088. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hami L.S. Green C. Leshinsky N., et al. GMP production and testing of Xcellerated T Cells for the treatment of patients with CLL. Cytotherapy. 2004;6:554–562. doi: 10.1080/14653240410005348. [DOI] [PubMed] [Google Scholar]
- Hollyman D., et al. Manufacturing validation of biologically functional T cells targeted to CD19 antigen for autologous adoptive cell therapy. J. Immunother. 2009;32:169–180. doi: 10.1097/CJI.0b013e318194a6e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N., et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz E.M. Gordon P.L. Koo W.K., et al. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurez V. Dzialo-Hatton R. Oliver J., et al. Efficient adenovirus-mediated gene transfer into primary T cells and thymocytes in a new coxsackie/adenovirus receptor transgenic model. BMC. Immunol. 2002;3:4. doi: 10.1186/1471-2172-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter G. Thiel E. Allogeneic transplantation of CCR5-deficient progenitor cells in a patient with HIV infection: an update after 3 years and the search for patient no. 2. AIDS. 2011;25:273–274. doi: 10.1097/QAD.0b013e328340fe28. [DOI] [PubMed] [Google Scholar]
- Hutter G., et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Ishikawa F., et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chainnull mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., et al. NOD/SCID/γ(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- June C. Levine B. Blocking HIV's attack. Sci. Am. 2012;306:54–59. doi: 10.1038/scientificamerican0312-54. [DOI] [PubMed] [Google Scholar]
- Jung D. Neron S. Drouin M. Jacques A. Efficient gene transfer into normal human B lymphocytes with the chimeric adenoviral vector Ad5/F35. J. Immunol. Methods. 2005;304:78–87. doi: 10.1016/j.jim.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Kim Y.G. Cha J. Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laport G.G., et al. Adoptive transfer of costimulated T cells induces lymphocytosis in patients with relapsed/refractory non-Hodgkin lymphoma following CD34+ -selected hematopoietic cell transplantation. Blood. 2003;102:2004–2013. doi: 10.1182/blood-2003-01-0095. [DOI] [PubMed] [Google Scholar]
- Leen A.M. Sili U. Savoldo B., et al. Fiber-modified adenoviruses generate subgroup cross-reactive, adenovirus-specific cytotoxic T lymphocytes for therapeutic applications. Blood. 2004;103:1011–1019. doi: 10.1182/blood-2003-07-2449. [DOI] [PubMed] [Google Scholar]
- Levine B.L. Ueda Y. Craighead N., et al. CD28 ligands CD80 (B7-1) and CD86 (B7-2) induce long-term autocrine growth of CD4+ T cells and induce similar patterns of cytokine secretion in vitro. Int. Immunol. 1995;7:891–904. doi: 10.1093/intimm/7.6.891. [DOI] [PubMed] [Google Scholar]
- Levine B. L., et al. Antiviral effect and ex vivo CD4+ T cell proliferation in HIV-positive patients as a result of CD28 costimulation. Science. 1996;272:1939–1943. doi: 10.1126/science.272.5270.1939. [DOI] [PubMed] [Google Scholar]
- Levine B.L. Bernstein W.B. Connors M., et al. Effects of CD28 costimulation on long-term proliferation of CD4+ T cells in the absence of exogenous feeder cells. Journal of Immunology. 1997;159:5921–5930. [PubMed] [Google Scholar]
- Levine B.L. Cotte J. Small C.C., et al. Large-scale production of CD4+ T cells from HIV-1-infected donors after CD3/CD28 costimulation. J. Hematother. 1998;7:437–448. doi: 10.1089/scd.1.1998.7.437. [DOI] [PubMed] [Google Scholar]
- Levine B.L., et al. Adoptive transfer of costimulated CD4+ T cells induces expansion of peripheral T cells and decreased CCR5 expression in HIV infection. Nat. Med. 2002;8:47–53. doi: 10.1038/nm0102-47. [DOI] [PubMed] [Google Scholar]
- Levine B. L., et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.J. Kim J. Li S., et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol. Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- Lin C. Chen S. Yang L., et al. Evaluation of TCR Vbeta subfamily T cell expansion in NOD/SCID mice transplanted with human cord blood hematopoietic stem cells. Hematology. 2007;12:325–330. doi: 10.1080/10245330701342342. [DOI] [PubMed] [Google Scholar]
- Liu R., et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Liu P.Q., et al. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J. Biol. Chem. 2001;276:11323–11334. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- Lu Z.Z. Ni F. Hu Z.B., et al. Efficient gene transfer into hematopoietic cells by a retargeting adenoviral vector system with a chimeric fiber of adenovirus serotype 5 and 11p. Exp. Hematol. 2006;34:1171–1182. doi: 10.1016/j.exphem.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Mackensen A. Drager R. Schlesier M., et al. Presence of IgE antibodies to bovine serum albumin in a patient developing anaphylaxis after vaccination with human peptide-pulsed dendritic cells. Cancer Immunol.Immunother. 2000;49:152–156. doi: 10.1007/s002620050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F. Michelet C. Peguillet I., et al. Persistent alterations in T-cell repertoire, cytokine and chemokine receptor gene expression after 1 year of highly active antiretroviral therapy. AIDS. 1999;13:185–194. doi: 10.1097/00002030-199902040-00006. [DOI] [PubMed] [Google Scholar]
- Mosier D.E. Picchio G.R. Kirven M.B., et al. EBV-induced human B cell lymphomas in hu-PBL-SCID mice. AIDS Res. Hum. Retroviruses. 1992;8:735–740. [PubMed] [Google Scholar]
- Nervi B., et al. Factors affecting human T cell engraftment, trafficking, and associated xenogeneic graft-vs-host disease in NOD/SCID beta2mnull mice. Exp. Hematol. 2007;35:1823–1838. doi: 10.1016/j.exphem.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M. Ljungberg J. Richter J., et al. Development of an adenoviral vector system with adenovirus serotype 35 tropism; efficient transient gene transfer into primary malignant hematopoietic cells. J. Gene Med. 2004;6:631–641. doi: 10.1002/jgm.543. [DOI] [PubMed] [Google Scholar]
- Nwanegbo E. Vardas E. Gao W., et al. Prevalence of neutralizing antibodies to adenoviral serotypes 5 and 35 in the adult populations of The Gambia, South Africa, and the United States. Clin. Diagn. Lab Immunol. 2004;11:351–357. doi: 10.1128/CDLI.11.2.351-357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannetier C. Even J. Kourilsky P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol. Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- Pantaleo G. Koup R.A. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat. Med. 2004;10:806–810. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- Paxton W.A., et al. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat. Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- Pelz O. Wu M. Nikolova T., et al. Duplex polymerase chain reaction quantification of human cells in a murine background. Stem Cells. 2005;23:828–833. doi: 10.1634/stemcells.2004-0206. [DOI] [PubMed] [Google Scholar]
- Perez E.E., et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D.L., et al. A phase 1 trial of donor lymphocyte infusions expanded and activated ex vivo via CD3/CD28 costimulation. Blood. 2006;107:1325–1331. doi: 10.1182/blood-2005-08-3373. [DOI] [PubMed] [Google Scholar]
- Powell D.J., Jr. Brennan A.L. Zheng Z., et al. Efficient clinical-scale enrichment of lymphocytes for use in adoptive immunotherapy using a modified counterflow centrifugal elutriation program. Cytotherapy. 2009;11:923–935. doi: 10.3109/14653240903188921. [DOI] [PubMed] [Google Scholar]
- Quillent C., et al. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet. 1998;351:14–18. doi: 10.1016/S0140-6736(97)09185-X. [DOI] [PubMed] [Google Scholar]
- Rahman S.H. Maeder M.L. Joung J.K. Cathomen T. Zinc-finger nucleases for somatic gene therapy: the next frontier. Hum. Gene Ther. 2011;22:925–933. doi: 10.1089/hum.2011.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport A.P., et al. Molecular remission of CML after autotransplantation followed by adoptive transfer of costimulated autologous T cells. Bone Marrow Transplant. 2004;33:53–60. doi: 10.1038/sj.bmt.1704317. [DOI] [PubMed] [Google Scholar]
- Rapoport A.P., et al. Restoration of immunity in lymphopenic individuals with cancer by vaccination and adoptive T-cell transfer. Nat. Med. 2005;11:1230–1237. doi: 10.1038/nm1310. [DOI] [PubMed] [Google Scholar]
- Rapoport A.P., et al. Rapid immune recovery and graft-versus-host disease-like engraftment syndrome following adoptive transfer of Costimulated autologous T cells. Clin. Cancer Res. 2009;15:4499–4507. doi: 10.1158/1078-0432.CCR-09-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebar E.J., et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat. Med. 2002;8:1427–1432. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- Sarver N. Rossi J. Gene therapy: a bold direction for HIV-1 treatment. AIDS Res. Hum. Retroviruses. 1993;9:483–487. doi: 10.1089/aid.1993.9.483. [DOI] [PubMed] [Google Scholar]
- Schroers R. Hildebrandt Y. Hasenkamp J., et al. Gene transfer into human T lymphocytes and natural killer cells by Ad5/F35 chimeric adenoviral vectors. Exp. Hematol. 2004;32:536–546. doi: 10.1016/j.exphem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Sebille F. Gagne K. Guillet M., et al. Direct recognition of foreign MHC determinants by naive T cells mobilizes specific Vbeta families without skewing of the complementarity-determining region 3 length distribution. J. Immunol. 2001;167:3082–3088. doi: 10.4049/jimmunol.167.6.3082. [DOI] [PubMed] [Google Scholar]
- Selvaggi T.A. Walker R.E. Fleisher T.A. Development of antibodies to fetal calf serum with arthus-like reactions in human immunodeficiency virus-infected patients given syngeneic lymphocyte infusions. Blood. 1997;89:776–779. [PubMed] [Google Scholar]
- Shayakhmetov D.M. Papayannopoulou T. Stamatoyannopoulos G. Lieber A. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J. Virol. 2000;74:2567–2583. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz L.D., et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- Smith M.W. Carrington M. Winkler C., et al. CCR2 chemokine receptor and AIDS progression. Nat. Med. 1997;3:1052–1053. doi: 10.1038/nm1097-1052c. [DOI] [PubMed] [Google Scholar]
- Smith J. Berg J.M. Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden A.W., et al. Repression of vascular endothelial growth factor A in glioblastoma cells using engineered zinc finger transcription factors. Cancer Res. 2003;63:8968–8976. [PubMed] [Google Scholar]
- Tan S., et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschong L. Soenen S.L. Blaese R.M., et al. Immune response to fetal calf serum by two adenosine deaminase-deficient patients after T cell gene therapy. Hum. Gene Ther. 2002;13:1605–1610. doi: 10.1089/10430340260201699. [DOI] [PubMed] [Google Scholar]
- Urnov F.D., et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- van Lunzen J., et al. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol. Ther. 2007;15:1024–1033. doi: 10.1038/mt.sj.6300124. [DOI] [PubMed] [Google Scholar]
- Verani A. Lusso P. Chemokines as natural HIV antagonists. Curr. Mol. Med. 2002;2:691–702. doi: 10.2174/1566524023361862. [DOI] [PubMed] [Google Scholar]
- von Laer D. Hasselmann S. Hasselmann K. Impact of gene-modified T cells on HIV infection dynamics. J. Theor. Biol. 2006;238:60–77. doi: 10.1016/j.jtbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Wagar E.J. Cromwell M.A. Shultz L.D. Regulation of human cell engraftment and development of EBV-related lymphoproliferative disorders in Hu-PBL-scid mice. J. Immunol. 2000;165:518–527. doi: 10.4049/jimmunol.165.1.518. [DOI] [PubMed] [Google Scholar]
- Zhao Y., et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res. 2010;70:9053–9061. doi: 10.1158/0008-5472.CAN-10-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data
Supplemental data
Supplemental data
Supplemental data