Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems (original) (raw)
Abstract
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) systems in bacteria and archaea use RNA-guided nuclease activity to provide adaptive immunity against invading foreign nucleic acids. Here, we report the use of type II bacterial CRISPR-Cas system in Saccharomyces cerevisiae for genome engineering. The CRISPR-Cas components, Cas9 gene and a designer genome targeting CRISPR guide RNA (gRNA), show robust and specific RNA-guided endonuclease activity at targeted endogenous genomic loci in yeast. Using constitutive Cas9 expression and a transient gRNA cassette, we show that targeted double-strand breaks can increase homologous recombination rates of single- and double-stranded oligonucleotide donors by 5-fold and 130-fold, respectively. In addition, co-transformation of a gRNA plasmid and a donor DNA in cells constitutively expressing Cas9 resulted in near 100% donor DNA recombination frequency. Our approach provides foundations for a simple and powerful genome engineering tool for site-specific mutagenesis and allelic replacement in yeast.
INTRODUCTION
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) immune systems in bacteria are of interest to the biotechnology community owing to RNA-guided endonuclease activity (1,2). The Cas9 gene, from the type II bacterial CRISPR system of Streptococcus pyogenes, complexes with a designer genome targeting CRISPR guide RNA (gRNA) to determine the site specificity of the DNA cutting activity (2,3) (Figure 1A). It has been shown that Cas9 can function as an RNA-guided endonuclease in heterologous organisms (4–8). In the future, engineered versions of the Cas9 gene could function as a RNA-guided DNA binding protein, lacking endonuclease activity. CRISPR systems offer an advantage to zinc finger and transcription activator-like effector DNA-binding proteins, as the site specificity in nucleotide binding CRISPR-Cas proteins is governed by a RNA molecule instead of the DNA-binding protein, which can be more challenging to design and synthesize. To express RNA without modifications added by the RNA polymerase II transcription system, RNA polymerase III regulatory elements have been used for transcription of functional gRNA in human cells (4,5).
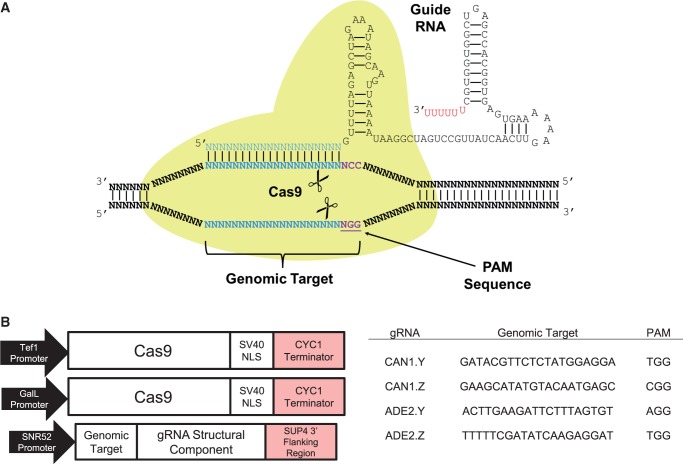
Figure 1.
Diagram of Cas9 complex and schematic of genetic constructs. (A) Illustration of Cas9 protein interacting with CRISPR gRNA to direct endonuclease activity proximal to the PAM sequence. (B) Design of the Cas9 and gRNA constructs. Cas9 gene contained a SV40 nuclear localization signal and was expressed under the Gal-L inducible promoter in CAN1 experiments and the TEF1 constitutive promoter in ADE2 experiments. The gRNA was expressed under the snoRNA SNR52 promoter and contained a terminator from the 3′ region of the yeast SUP4 gene. CAN1.Y and CAN1.Z were targeted to different loci in the CAN1 gene, whereas ADE2.Y and ADE2.Z were targeted to different loci in the ADE2 gene.
To examine RNA-guided Cas9 nuclease activity, we chose to design gRNAs to target the endogenous genomic negative selectable marker CAN1, a plasma membrane arginine permease, in haploid yeast cells and monitor mutation frequency at the locus. As this gene has a negative selection, its inactivation frequency can be observed experimentally. Nonsense mutations in CAN1 can be selected with media containing canavanine (a toxic arginine analogue), which is only imported into cells containing a functional CAN1 gene (9). Directed double-strand break at this locus increases mutation frequency owing to errors that occur in the repair pathway (9,10). The double-strand break can be resolved either by homologous recombination or through error-prone non-homologous end joining (11–13). To control for a potential genome-wide mutator phenotype, the mutation frequency of the non-targeted endogenous LYP1 gene, a lysine permease, was monitored by selecting for lyp1 mutants using a toxic lysine analogue, thialysine (14). The LYP1 and CAN1 genes are on separate chromosomes, and local mutation frequency in either locus should be independent, unless a global mutator phenotype is present.
We further examined the effects of genomic CRISPR-Cas activity on single- and double-stranded oligonucleotide transformation. It has previously been shown that induction of double-strand breaks near the oligonucleotide-targeting site can increase recombination efficiency by as high as 4000-fold (13). We first examined the effect of CRISPR-Cas on homologous recombination in Cas9 constitutively expressing cells by transforming a transient gRNA polymerase chain reaction (PCR) cassette containing a promoter, the gRNA sequence and a terminator, with an oligonucleotide donor DNA. In this experiment, a positive reporter system (in which gene correction could be assayed) was chosen to avoid ambiguity from the negative reporter system where the source of mutations could be from erroneous double-strand break repair or donor DNA.
Moreover, we examined the ability for CRISPR-Cas to stimulate recombination and select against wild-type sequences by co-transforming a gRNA plasmid with a donor DNA that mutates the genomically encoded protospacer-associated motif (PAM) sequence, a DNA motif required for cutting. A gRNA expression plasmid was co-transformed with a donor DNA in cells containing Cas9 constitutively expressed on plasmid. These cells were then selected for the gRNA and Cas9 plasmids, and the recombination frequency at the locus of integration was determined.
To ease the future use of CRISPR-Cas methods in for yeast genome engineering, we also calculated the frequency of gRNA target sites in yeast by calculating all 12 bpr ‘seed’ sequences crucial for gRNA genomic specificity proximal to a PAM sequence (2,6).
MATERIALS AND METHODS
Strains and media
The Saccharomyces cerevisiae strain used in the CAN1 mutagenesis analysis of the CRISPR system and the gRNA plasmid/donor DNA transformation in Cas9-expressing cells was BY4733 (MATa his3Δ200 trp1Δ63 leu2Δ0 met15Δ0 ura3Δ0), which was a kind gift from Fred Winston. Parental BY4733 was grown in YPAD before transformation and then propagated in the appropriate synthetic complete (SC) media minus the auxotrophic compound complemented by the plasmids. Strain VL6-48 (MATα, his3Δ200, trplΔ1, ura3-52, ade2-101, lys2, psio, cir°) was used for the homologous recombination experiments using the gRNA PCR product, owing to its native ade2-101 premature stop codon.VL6-48 was purchased from ATCC (MYA-3666). Plasmids p415-Gal-L and p426-Gal1 used in this study were a kind gift from Fred Winston (15).
Plasmid construction
The Cas9 gene was a codon-optimized version originally constructed for expression in human cells (4). This gene was C-terminally tagged with a SV40 nuclear localization signal. The p415 Gal-L and p414 TEF1p plasmids were each cut with XhoI and XmaI, and the backbone containing the promoter and terminator was gel purified. Cas9 was PCR amplified from a TOPO-TA vector with 20 base pair extended 5′ and 3′ regions identical to the promoter and terminator of the destined plasmid backbone (either p415 Gal-Lp or p414 TEF1p). The PCR amplified Cas9 was Gibson assembled into the vector using the NEB Gibson Assembly kit. For the gRNA expression plasmids, the p426-Gal1 plasmid was cut with XhoI and SacI to remove the Gal1 promoter, and the backbone was gel purified. The gRNA expression cassette containing the SNR52 promoter, the gRNA and SUP4 3′ flanking sequence were assembled by two rounds of PCR using Phusion 2X HF Master Mix. The outer two primers contained 20 base pair extended 5′ and 3′ regions identical to the p426-Gal1 plasmid backbone at the cut sites. In the first round of PCR contained all primers at 10 nM. The second round of PCR was a 50-μl reaction containing a 2 μl of 10-fold dilution of the first round product with the outer primers at concentrations of 10 nM. For the CAN1 experiments, the gRNA PCR products were Gibson assembled into the cut p426 plasmid. The KanMX sequence was PCR amplified with 50 bp homology arms to the CAN1 locus from the pFA6a-KanMX6 plasmid, commonly used for gene knockout in yeast. See Supplementary Material S1 for sequences of Cas9 gene and plasmid descriptions.
Transformation of plasmids for CAN1 mutagenesis
Transformation of plasmids (200 ng per transformation) was carried out using a standard lithium acetate transformation method (16). After transformation, cells were plated on selective media (SC-uracil and leucine, SC-tryptophan or SC-uracil and tryptophan) and allowed grow for 2 days until colonies appeared. All oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA). See Supplementary Material S1 for sequences of oligonucleotides. Double-stranded oligonucleotides were generated by annealing equimolar amounts of single-stranded oligonucleotides by first denaturing the mixture at 100°C for 5 min and then allowing cool to 25°C with a ramp of 0.1°C per second.
Electroporation of transient gRNA cassette and oligonucleotides
The gRNA cassette and donor single-stranded or double-stranded oligonucleotide were transformed into VL6-48 cells containing p414 TEFp Cas9 via electroporation as follows. Cultures were grown to saturation overnight in SC-tryptophan. The next morning, a 10 ml culture was inoculated in liquid SC-tryptophan to and OD600 = 0.3. Inoculated cells were grown in roller drum at 30°C until OD600 = 1.8 after 5 h. Cells were collected via centrifugation at 2250_g_ for 3 min and the media removed. The cell pellet was washed once by 10 ml ice-cold water and once by 10 ml of ice-cold electroporation buffer (1 M Sorbitol/1 mM CaCl2). The cells were conditioned by re-suspending the cell pellet in 2 ml 500 mM LiAc/10 mM Dithiothreitol (DTT) and placed in roller drum for 30 min at 30°C. Conditioned cells were collected by centrifugation and washed once by 10 ml ice-cold electroporation buffer. The cell pellet was re-suspended to a final volume of 1.6 ml in electroporation Buffer. In all, 400 μl of cells were used per electroporation with 1 nmol of oligonucleotide and 1 μg gRNA cassette (or 1 µg of salmon sperm DNA as a control). Cells were electroporated at 2.5 kV, 25 µF, 200Ω. Electroporated cells were transferred from each cuvette into 7 ml of 1:1 mix of 1 M sorbitol/YPAD media. The cells were incubated in a roller drum at 30°C for 12 h. Approximately 106–107 cells were plated on selective media, and cells were diluted appropriately on rich media. The ratio of colony count on selective plates over rich plates was used as a measure of correction frequency. Experiments were completed in quadruplicate.
Transformation of gRNA plasmid with donor DNA into Cas9-expressing cells
In all, 500 ng of either empty p426 or p426 containing a gRNA CAN1.Y expression cassette were transformed into BY4741 cells constitutively expressing Cas9 under the TEF1 promoter in a p414 plasmid backbone. The p426 plasmid was co-transformed with either 1 nmol of double stranded CAN1.Y oligonucleotide, 5 μg of KanMX cassette and 50 µg of salmon sperm DNA, or just 50 µg of salmon sperm DNA, using a standard lithium acetate transformation method (16). Cells were plated without dilution on SC without uracil and tryptophan and 10−5 dilutions onto Yeast Peptone Adenine Dextrose (YPAD) and allowed to recover for 2 days before the selective plates were replica plated to canavanine plates and YPAD plates with 100 μg/ml G418 antibiotic (Geneticin G418, purchased from Teknova). Transformation frequency was calculated by the ratio of number cells that recover on SC without uracil and tryptophan divided by the number on rich non-selective media. No donor DNA experiments were completed with three replicates, and the transformations containing donor DNA were completed with six replicates.
Galactose induction of Cas9
Cells were grown in 5 ml SC dropout without leucine and uracil media containing 2% glucose to saturation, washed twice in water and then inoculated to an OD = 0.3 in SC dropout without leucine and uracil media containing 2% galactose and 1% raffinose. Cells were allowed to grow for 16 hours before plating on YPAD, SC-arginine plates containing 60 μg/ml L-canavanine (Sigma) and SC-lysine containing 100 μg/ml thialysine (S-2-aminoethyl-l-cysteine, Sigma). In all, 107–108 cells were plated on canavanine and thialysine containing media, and cells were diluted appropriately on rich media. The ratio of colony count on canavanine or thialysine plates divided by the colony count on rich media plates for each culture was used as a measure of mutation frequency. Experiments were completed in quadruplicate. Sequence alignments of Sanger sequence files were completed using Lasergene Seqman Pro Software (17).
Toxicity of CRISPR system
To analyse the toxicity associated with of the Cas9 protein and gRNA, strains were grown in 5 ml SC dropout media containing 2% glucose to an OD = 2.0, washed twice in 5 ml water and then equal amounts of cells plated in on both YPAD and YPA Gal (2% galactose and 1% raffinose) and allowed to grow at 30°C for 2 days. Experiments were completed in quadruplicate.
Identification of CRISPR-Cas targets in yeast genome
Both strands of the complete the S. cerevisiae S288c genome (version R64-1-1, GenBank assembly GCA_000146045.2) were searched for sequences of the form N(21)GG that did not contain any string of six or more Ts, yielding 924 177 candidate Cas9 target sequences. Sequences containing six Ts in a row were excluded, as they can cause termination of RNA polymerase III transcripts (18,19). The yeast genome was then searched for other genomic occurrences of the S(12) ‘seed’ sequences (see text) within these targets followed by NGG using bowtie version one release 0.12.8 using parameters -l 15 -v 0 -k 2. Specifically, for each S(12), the four sequences S(12)AGG, S(12)CGG, S(12)GGG and S(12)TGG were considered input ‘reads’ to be mapped against the genome, and any candidate for whom more than one match was found among these four reads was rejected (20). All candidates passing this filter meet the Cas9 specificity conditions described in the text. A second round of seed checks was then performed using this same method looking for S(12)NAG matches. Here, targets that returned no matches for any of their four NAG reads were tagged as NoNAG targets to denote their greater specificity. The sequences of all targets have been provided in a .csv file in Supplementary Material S2. Sequence IDs in the header lines indicate the chromosome and location of the left endpoint of the sequences in the R64-1-1 version of the genome, followed by an ‘r’ if the sequence is presented as a reverse complement. NoNAG tags are provided in the header line after these IDs where applicable.
RESULTS
The gRNA expression and genomic target identification
The RNA polymerase III regulatory elements used in this study were based on constructs used to express bacterial tRNA genes in yeast (18,21). Specifically, the SNR52 snoRNA promoter and the yeast tRNA gene SUP4 3′ flanking sequence (as a terminator) were used to express gRNA (Figure 1B). This combination of promoter and terminator has been shown to produce transcripts with strings of U residues less than six ribonucelotides long, which is an important consideration, as the structural component of the gRNA contains a string of four U residues (4,19,22). The protospacer adjacent motif (PAM) sequence, a genomically encoded NGG nucleotide sequence directly 3′ of the 20 bp genome target, is a crucial design constraint to the site specificity. The design of the gRNA structural component used in this study was based on the sequence used by Mali et al. (4) for gRNA expression in human cells (Figure 1A and B). The 20 bp of genome sequence complementary in the gRNA mimics the processed CRISPR RNA (crRNA) found in the natural host, S. pyogenes. In this organism, a 39–42 base pair sequence, containing a 20 bp spacer-derived guide sequence and 19–22 base pair repeat-derived sequence, is processed by a trans-activating crRNA molecule and RNAse III enzyme to form functional gRNA. The functional gRNA designs in human cells build on the work of Jinek et al. (2) who demonstrated in vitro that Cas9 requires both a base paired activating trans-activating crRNA and the targeting crRNA, or a single chimeric gRNA with features of each molecule (4,5,23). It has been reported that mismatches in any of the last 12 nt of the 20 nt crRNA against a target dsDNA can ablate Cas9 activity, whereas mismatches in the first 8 nt have little effect. Therefore, 23 bp sequences of the form N(8)S(12)NGG for which the 12 bp ‘seed’ sequence S(12) followed by an NGG is found nowhere else in the genome have the greatest chance of being specifically targetable by Cas9. However, partial activity of a NAG PAM sequence has also been observed for the Cas9 system(6) so that targets that may be unique genomic S(12)NGG occurrences but have one or more S(12)NAG occurrences may be less specific. Using these constraints, we tabulated 645 392 genomic targets of maximal specificity (unique S(12)NGG with no S(12)NAG occurrences) and 108 493 genomic targets of lesser specificity (unique S(12)NGG but one or more S(12)NAG) (Supplementary Material S2).
CRISPR-Cas directed CAN1 mutagenesis
The CAN1 locus was targeted with gRNA expression constructs on high-copy 2 µ plasmids. The Cas9 gene was placed under the Gal-L promoter, an attenuated version of the strongly galactose-inducible Gal1 promoter, in a centromeric plasmid with ∼1 copy per cell (Figure 1B) and only induced with galactose during experiments (15). We chose to examine Cas9 under an inducible promoter, at first, to limit the potential toxicity of CRISPR-Cas activity.
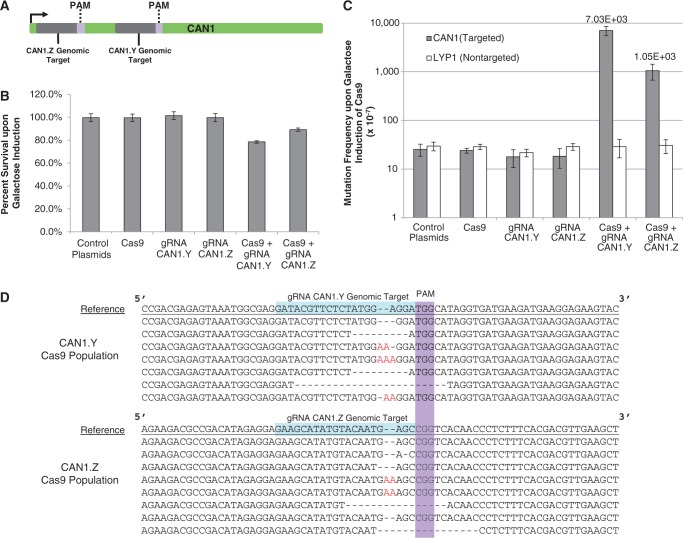
Two sites in the CAN1 gene were targeted. gRNA CAN1.Z was designed to direct endonuclease activity 58 bp downstream of the ATG start codon of the CAN1 gene, whereas the gRNA CAN1.Y genomic target site was located 207 bp downstream of the start codon (Figure 2A). On expression of Cas9 in strains also expressing gRNA, cell viability decreased to 78 and 89% with CAN1.Y and CAN1.Z, respectively, whereas strains containing only a single CRISPR-Cas component had viability near 100% (Figure 2B). The degree of toxicity also correlates with higher mutation frequency in the CAN1 locus, as CAN1.Y was the most toxic gRNA and had the highest CAN1 mutation frequency. Following galactose induction of Cas9, the mutation frequency in the CAN1 gene was 0.07 and 0.01% with CAN1.Y and CAN1.Z gRNAs, respectively (Figure 2C). Furthermore, the mutation rate in the LYP1 gene remained relatively constant across all strains, suggesting that CRISPR-Cas is site specific in yeast and does not induce random mutations genome-wide (Figure 2C). To further validate that mutations were caused by Cas9 and gRNA activity, the CAN1 gene from eight colonies were Sanger sequenced from the gRNA CAN1.Y/Cas9 and gRNA CAN1.Z/Cas9 canavanine resistant populations. Indeed, 8/8 colonies from the gRNA CAN1.Y/Cas9 canavanine resistant population and 7/8 colonies from the gRNA CAN1.Z/Cas9 canavanine resistant population were found to have frameshift CAN1 mutations directly upstream of their respective PAM sequences (Figure 2D). These mutations are proximal to the putative cleavage site of the Cas9 system, 3 bp upstream of the PAM sequence (2,4,5).
Figure 2.
CRISPR-Cas mediated genomic mutagenesis. (A) Schematic of the CAN1.Y and CAN1.Z genomic target sequences and respective PAM sequences in the CAN1 gene. (B) Percentage survival of yeast strains expressing different combinations of Cas9 and gRNA. Only combination of Cas9 and gRNA caused decrease in cell viability. Error bars represent standard deviation between four experiments. (C) CAN1 and LYP1 mutation frequency of yeast strains expressing different combinations of Cas9 and gRNA on galactose induction of Cas9. Mutation frequency of CAN1 (the gRNA-targeted gene) is elevated in strains expressing Cas9 and gRNA, whereas LYP1 (not targeted by gRNA) remained constant. Error bars represent standard deviation between four experiments. (D) Alignments of CAN1 gene from canavanine-resistant colonies post-galactose induction of Cas9 in gRNA-expressing strains. The underlined sequence is of the wild-type reference CAN1 gene, whereas the following eight sequences are from colonies from the population. The PAM sequence is highlighted in purple, whereas the gRNA guiding sequence is highlighted in blue. Nonsense mutations observed are largely deletions or insertions 5′ to the PAM sequence.
CRISPR-Cas stimulated homologous recombination with donor DNA and transient gRNA PCR cassette
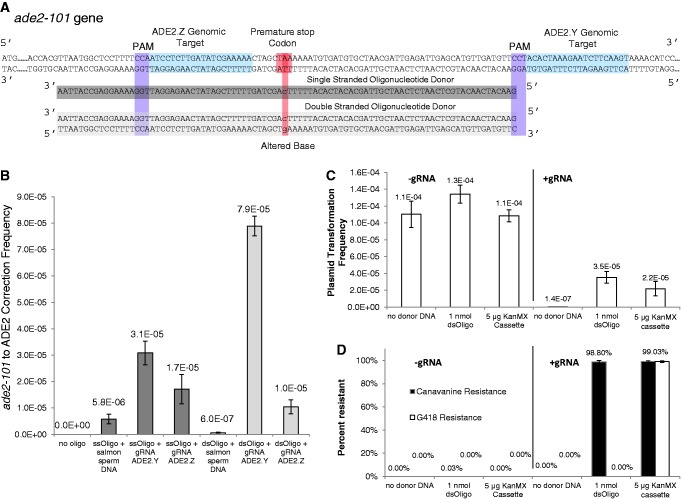
As a test system for donor DNA homologous recombination, an allele containing a nonsense mutation of the ADE2 gene, a phosphoribosylaminoimidazole carboxylase essential for adenine biosynthesis, was targeted for repair. The ade2-101 allele is common to many yeast laboratory strains and contains a premature stop codon at base 109 owing to a G to T transversion. In this positively selectable reporter, mutations causing correction of the mutation would be rare without donor DNA. We chose to target this mutation using a 90 mer oligonucleotide bearing the correct sequence of the ADE2 gene centred around the nonsense mutation. In yeast cells constitutively expressing Cas9 under the TEF1 promoter on a centromeric plasmid, either an ssDNA or dsDNA oligonucleotide containing these sequences were electroporated with either a transient PCR product of the gRNA cassette (containing the SNR52 promoter, gRNA sequence and SUP4 3′ flanking region) or salmon sperm DNA (as a control).
Two sites in the ADE2 gene were chosen to target with gRNAs based on the proximity of the ade2-101 mutation (Figure 3A). Cells electroporated with gRNA cassettes and oligonucleotides showed higher rates of homologous recombination than compared with cells with only oligonucleotides. The gRNA ADE2.Y had the largest effect on homologous recombination, improving single and double-stranded recombination rates 5-fold and 130-fold, respectively (Figure 3B). The activity of ADE2.Z was not as high as ADE2.Y, potentially for several reasons. One reason may be that the cut site of ADE2.Z was within the donor oligonucleotide; therefore, interactions between the gRNA and oligonucleotide may have decreased genomic cutting ability. Also, a stretch of five U residues within the gRNA genomic targeting region may have resulted in decreased full-length ADE2.Z transcription efficiency RNA polymerase III owing to premature transcription termination.
Figure 3.
The gRNA and donor DNA recombination in Cas9 constitutively expressing strains. (A) Diagram of ADE2 gene in the 5′–3′ direction displaying the gRNA genomic target regions and their respective PAM sequences. The sequence location of the single- or double-stranded oligonucleotide that will repair the premature stop codon is also displayed. The single-stranded oligonucleotide is from the non-coding strand of the ADE2 gene, whereas the double-stranded oligonucleotide is composed of annealed non-coding and coding strands. The oligonucleotide was designed centred around the mutant base pair. The gRNA sequences were designed based on the closest PAM sequences to the premature stop codon. (B) 90 mer single- and double-stranded oligonucleotides were electroporated into yeast expressing Cas9 under the TEF1 constitutive promoter, with and without a gRNA cassette. The gRNA cassette is a PCR product consisting of the SNR52 promoter, the genomic target, the guiding RNA scaffold and the SUP4 3′ flanking sequence. Electroporation of oligonucleotide and ADE.Y gRNA cassette increases oligonucleotide incorporation up to 5-fold for single-stranded oligonucleotide and 130-fold for double-stranded oligonucleotide. Approximately 106–107 cells were plated on selective media to monitor ADE2 correction frequency. Values are averages from four experiments, and error bars represent standard deviation from the experiments. (C) Transformation frequency of plasmid with and without gRNA CAN1.Y expression in cells containing Cas9 constitutively expressed on a second plasmid. Transformation frequency is defined by the ratio of number colonies that recover on defined media to select for both plasmids divided by the number of colonies on rich non-selective media. The plasmid transformation frequency decreases when gRNA expression is present on the plasmid, as expected owing to cell death associated with Cas9 DNA cleavage. (D) Percentage of cells containing gRNA and constitutive Cas9 plasmids resistant to Canavanine and G418 via replica plating. When a CAN1.Y double-stranded oligonucleotide or a KanMX resistance cassette targeted for disruption of the CAN1 and the CAN1.Y PAM sequence were co-transformed with the gRNA expression plasmid, nearly 100% of the cells selected for Cas9 and gRNA plasmids were resistant to canavanine. Furthermore, in the case of the KanMX resistance cassette donor DNA, the same percentage of cells resistant to canavanine was also resistant to G418, indicating all KanMX insertions integrated in the targeted locus. Values for Figure 3C and D represent an average of three experiments for the no donor DNA controls and six experiments for the transformations containing donor DNA. Error bars represent standard deviation of values.
CRISPR-Cas stimulated homologous recombination with donor DNA and gRNA expression plasmid
In cells containing a centromeric plasmid constitutively expressing Cas9 under the TEF1 promoter, donor DNA was co-transformed with a high-copy 2 µ plasmid with and without gRNA CAN1.Y expression elements. Donor DNA was designed to recombine within the gRNA CAN1.Y genomic target using homology arms to the site. A double-stranded 90 mer oligonucleotide donor centred around the PAM sequence contained 2 bp changes to mutate the PAM sequence and incorporate a premature TAG stop codon. A 1.4 kb KanMX cassette (conferring G418 resistance) was as amplified with 50 bp homology arms to the CAN1.Y target site and was also designed to disrupt the PAM sequence. On integration, both DNA donors would result in canavanine resistance. Selection of cells containing both gRNA.CAN1.Y and Cas9 plasmids resulted in a reduction of transformation frequency as compared with cells with a Cas9 plasmid and an empty vector (Figure 3C). This is likely due to toxicity of Cas9 DNA cleavage.
Furthermore, co-transformation of donor DNA with the gRNA.CAN1Y expression plasmid increased the transformation frequency as compared to a no donor DNA control (Figure 3C) Colonies containing both plasmids were then replica plated to canavanine media as well as rich media with G418 (to select for the KanMX integration event). Interestingly, the vast majority (near ∼100%) of the cells that received donor DNA selected for the gRNA CAN.1 Y and Cas9 plasmids were canavanine resistant and the same proportion were G418 resistant in the KanMX donor DNA co-transformation (Figure 3D). A small amount of transformants (an average of two colonies per replicate) resulted when gRNA CAN1.Y plasmid was transformed without donor DNA, all of which were canavanine sensitive (Supplementary Material S1). In the inducible Cas9 system, which lacked mutagenic donor DNA, canavanine resistant mutants arose with a frequency of 0.07%.Hence, in the no donor DNA control of this experiment there were too few transformants to likely observe such a low frequency. In cells co-transformed with an oligonucleotide containing a premature stop codon, to ensure that donor oligonucleotide recombination was the cause of canavanine resistance, we sequenced eight colonies of the canavanine resistant population. Indeed, all eight of these cells contained PAM sequence mutation and premature stop codon (Supplementary Material S1). These data suggest that under strong constitutive expression of Cas9, a genomically targeted gRNA on a plasmid can both stimulate recombination of donor DNA and select against wild type sequences with high frequency.
DISCUSSION
CRISPR-Cas has great potential as a foundational tool for genome engineering in S. cerevisiae owing to the user-designated site-specificity of Cas9 endonuclease activity and the simplicity of gRNA construction.
Yeast genome engineering methods using site-specific endonucleases could also benefit greatly from CRISPR systems. The delitto perfetto method for genomic oligonucleotide recombination in yeast, as described by Storici et al. (12,13), uses an induced double-strand break near the site of oligonucleotide recombination to obtain recombination frequencies of up to 20%. This technique requires initial targeted insertion of a CORE cassette containing a selectable marker with an I-SceI homing endonuclease site and a separate inducibile I-SceI gene. Using a similar approach, CRISPR-Cas could greatly simplify this method by removing the initial step requiring I-SceI endonuclease site integration. Furthermore, our experiments using a transient PCR product of the gRNA cassette show that the site specificity of the endonuclease could be directed easily using a non-integrating PCR product. More optimization, however, is required to attain these high frequencies of oligonucleotide recombination with a transient gRNA CRISPR system.
Transformation of donor DNA into cells selected for the presence of CRIPSR components on plasmids is a closer comparison with the delitto perfetto system, as site-specific DNA cleavage is more likely in all cells than on a transient PCR cassette. In this experiment, we saw recombination frequency near 100%, without selection for the integrated DNA. Removal of the PAM sequence in the donor DNA allowed recombinant cells to be protected from CRISPR-Cas activity and likely the associated toxicity. Although we did not test this hypothesis, mutations in the 12 bp ‘seed’ sequence, crucial for site specificity, may result in similar protection. Given the ease of designing site specificity for CRISPR-Cas targets with the high recombination frequency achieved for both a 90 mer double-stranded oligonucleotide and 1.4 kb double-stranded DNA cassette, we believe this method could be extremely valuable for engineering yeast genomes without the need of selectable markers attached to the integrated DNA.
In addition, the mutagenic capabilities of CRISPR-Cas, as demonstrated by the CAN1 mutagenesis experiments, highlight the potential for the use of CRISPR-Cas to make targeted knockouts. Further improvement in percentage mutagenized with an inducible system is needed before this would be a practical application, as the described induced Cas9 mutagenesis experiment only produced knockout rates of at most 0.07%. However, we have shown that if a mutagenic donor DNA to knockout the target gene is provided for homologous recombination, this frequency can be dramatically boosted.
One exciting finding of this study was the use of a transient gRNA cassette to stimulate homologous recombination. Because no plasmids or selectable markers were needed to produce functional gRNA in Cas9-expressing cells, many gRNAs can thus be easily synthesized and combinatorially transformed. This could allow for genomic targeting of many loci at once. Moreover, studies in several organisms have shown that multiple genomic targets are possible with the Cas9 system (4–6). Indeed, in the future, we plan to target multiple sites with gRNAs simultaneously, either on a plasmid or in a transient PCR product (potentially in an array form), and examine the possibility of CRISPR-Cas directed multiplex genome engineering towards engineering whole metabolic pathways and large gene networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials 1 and 2.
FUNDING
Department of Energy [DE-FG02-02ER63445]; Synthetic Biology Engineering Research Center from the National Science Foundation [SA5283-11210]; National Institutes of Health [P50 HG005550]. Funding for open access charge: Department of Energy [DE-FG02-02ER63445].
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
The authors are very grateful to Alejandro Chavez, Nikolai Eroshenko and Harris Wang for constructive and thought provoking conversations about the experiments described. Fred Winston and Dan Spatt have been extremely helpful with advice, plasmid donation and strain donation for this work, and they are very thankful to their patience and generosity. Two Cas9 expression plasmids and one gRNA expression plasmid described in the article are available on Addgene.org (plasmid #43802, #43803, and #43804).
REFERENCES
- 1.Carroll D. A CRISPR approach to gene targeting. Mol. Ther. 2012;20:1658–1660. doi: 10.1038/mt.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaya D, Davison M, Barrangou R. CRISPR-Cas Systems in bacteria and Archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 4.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–279. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whelan WL, Gocke E, Manney TR. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics. 1979;91:35–51. doi: 10.1093/genetics/91.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett CB, Westmoreland TJ, Snipe JR, Resnick MA. A double-strand break within a yeast artificial chromosome (YAC) containing human DNA can result in YAC loss, deletion or cell lethality. Mol. Cell. Biol. 1996;16:4414–4425. doi: 10.1128/mcb.16.8.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rattray AJ, McGill CB, Shafer BK, Strathern JN. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storici F, Snipe JR, Chan GK, Gordenin DA, Resnick MA. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol. Cell. Biol. 2006;26:7645–7657. doi: 10.1128/MCB.00672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl Acad. Sci. USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zwolshen JH, Bhattacharjee JK. Genetic and biochemical properties of thialysine-resistant mutants of Saccharomyces cerevisiae. J. Gen. Microbiol. 1981;122:281–287. doi: 10.1099/00221287-122-2-281. [DOI] [PubMed] [Google Scholar]
- 15.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 17.Burland TG. DNASTAR’s lasergene sequence analysis software. Methods Mol. Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J. Am. Chem. Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 19.Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dt) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- 20.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Wang L. Genetic incorporation of unnatural amino acids into proteins in yeast. Methods Mol. Biol. 2012;794:199–213. doi: 10.1007/978-1-61779-331-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acker J, Conesa C, Lefebvre O. Yeast RNA polymerase III transcription factors and effectors. Biochim. Biophys. Acta (BBA) 2012 doi: 10.1016/j.bbagrm.2012.10.002. pii, S1874–9399. [DOI] [PubMed] [Google Scholar]
- 23.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data