Division and subtraction by distinct cortical inhibitory networks in vivo (original) (raw)
. Author manuscript; available in PMC: 2013 May 14.
Published in final edited form as: Nature. 2012 Aug 16;488(7411):343–348. doi: 10.1038/nature11347
Abstract
Brain circuits process information through specialized neuronal subclasses interacting within a network. Revealing their interplay requires activating specific cells while monitoring others in a functioning circuit. Here we use a new platform for two-way light-based circuit interrogation in visual cortex in vivo to show the computational implications of modulating different subclasses of inhibitory neurons during sensory processing. We find that soma-targeting, parvalbumin-expressing (PV) neurons principally divide responses but preserve stimulus selectivity, whereas dendrite-targeting, somatostatin-expressing (SOM) neurons principally subtract from excitatory responses and sharpen selectivity. Visualized in vivo cell-attached recordings show that division by PV neurons alters response gain, whereas subtraction by SOM neurons shifts response levels. Finally, stimulating identified neurons while scanning many target cells reveals that single PV and SOM neurons functionally impact only specific subsets of neurons in their projection fields. These findings provide direct evidence that inhibitory neuronal subclasses have distinct and complementary roles in cortical computations.
Inhibition has fundamental and diverse roles in brain function, and is delivered by specialized cell types with distinct intrinsic properties and connectivity patterns1–3. This heterogeneity in cellular form and function suggests that different inhibitory subtypes may actually underpin distinct computational functions and even hold specific relevance to neurological disorders4 based on their unique morphologies and functional positions within the network. Previous pharmacological or intracellular studies in primary visual cortex (V1), which necessarily considered inhibition as a single entity, have produced diverse findings on the role of inhibition. On the one hand, inhibition has been proposed to sharpen neuronal responses by removing weak inputs5–7, though there have been conflicting reports on whether inhibition predominantly targets non-preferred responses or preferred ones8–10. On the other hand, inhibition has been posited to control response gain, a network mechanism by which cortical networks rapidly ‘divide’ or scale their dynamic range of responses11. This mechanism has been proposed as fundamental to processing across many brain systems, from primary sensory computations12 to attention13, multisensory integration14, and value estimation15.
Here we show that inhibition in the cerebral cortex can have either of these functions, depending on its cellular source. We propose that two key inhibitory neuron subclasses, soma-targeting PV neurons and dendrite-targeting SOM neurons, which together comprise a substantial proportion of cortical inhibitory neurons in mice16,17, drive different kinds of inhibition. We combined optogenetic activation of individual or populations of PV or SOM neurons with monitoring the effects in target cells using high-speed imaging of functional responses18 as well as cell-attached electrophysiological recordings19. These methods complement both static wiring diagrams20 and wiring patterns examined in tissue slices21–24 by revealing targeting specificity and functional consequences of inhibitory neuron activation in intact circuits processing visual information.
Optical dissection of network interactions
To measure the effects of distinct cell classes within a functioning network, we built a custom system combining optogenetic stimulation with in vivo two-photon imaging in the mammalian brain (Fig. 1a and Supplementary Fig. 1). Our imaging system (Supplementary Movie 1) sampled calcium responses from neurons loaded with a fluorescent reporter using a scan path customized for each image25, at high speed but also high dwell times within neurons, yielding highly repeatable measurements of orientation-selective responses and clear tuning curves (Fig. 1a–f, Supplementary Figs 2 and 3, and Supplementary Movie 2).
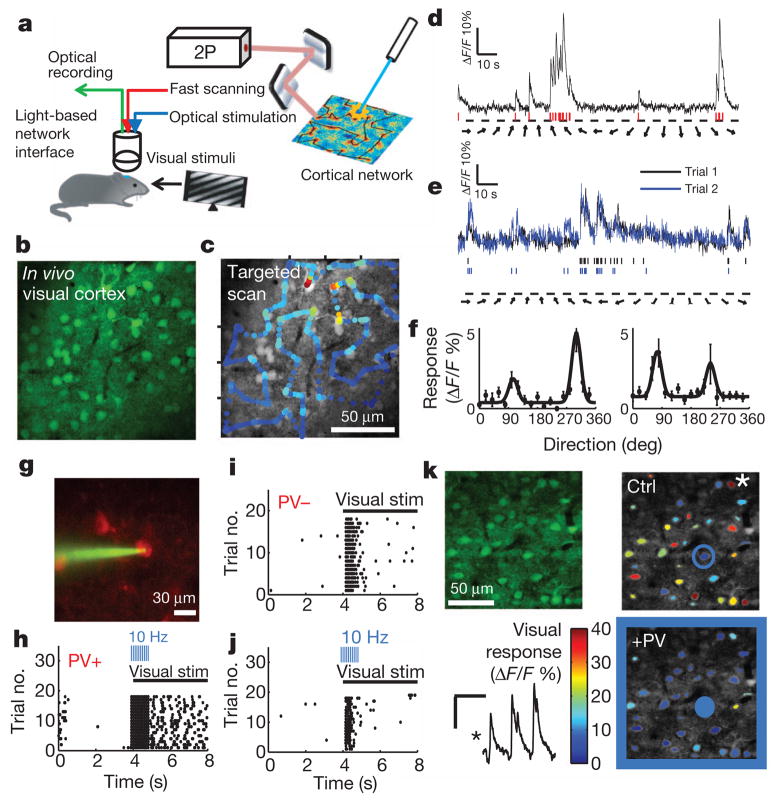
Figure 1. All-optical network dissection of cortical subclasses during visual computations.
a, Directed light, for optical recording and stimulation, was targeted to the V1 of an animal being shown visual stimuli. b, A traditional raster scan located cells in the network that were bulk-loaded with the calcium indicator dye OGB. c, Cells that were automatically identified were then imaged at high speed using targeted two-photon scanning along an arbitrary scan path. d, This enabled detection of robust cellular activity in response to episodically presented oriented drifting gratings, and this activity could be analysed as primary signals or deconvolved to estimate action potentials (red lines). e, Evoked optical traces were highly consistent over repeated presentations of visual stimuli. f, The clear, repeatable responses enabled the resolution of well-defined responses (dots), fit by dual Gaussian curves (lines). Data are shown as mean ± s.e.m. g, Image of an mCherry-ChR2+ PV+ (channelrhodopsin-2-and PV-positive) cell targeted in vivo for cell-attached recording. h, Evoked action potentials from an mCherry-ChR2+ PV+ cell. PV+ neurons were activated at the onset (3.9 s) of 4 s of visual stimulation, through 10-Hz stimulation of the PV+ neuron. i, j, Recording of the visual response of a PV− neuron in the control condition (i) and with PV activation (j). k, Left, cell population loaded with OGB dye (top), and responses of a cell marked by an asterisk in the network shown in the top right panel (bottom). Right, cells colour-coded by the magnitude of their visual response in the control condition (top) and when PV+ neurons were activated (bottom; colour bar, Δ_F_/F %). A ChR2+ PV neuron in the network is circled. +PV, optical PV activation; Ctrl, control; PV−, PV-negative; PV+, PV-positive.
To optically activate PV or SOM neurons, in parallel experiments, we used Cre/loxP recombination to express channelrhodopsin-2 (ChR2) in PV or SOM neurons in the mouse visual cortex (Supplementary Fig. 4). This led to highly specific and reliable on-demand activation of infected neurons in visual cortex that was verified both in slices and during visual stimulation in vivo (Fig. 1g and Supplementary Fig. 4). PV or SOM neurons were photo-activated for a 1-s interval at the onset of visual stimulation (Fig. 1h), enabling us to compare the control visual responses of neighbouring, non-infected neurons during episodically presented drifting oriented gratings (Fig. 1i) to responses in interleaved trials in which the cells were inhibited through PV or SOM activation (Fig. 1j and Supplementary Fig. 5). Concurrent calcium imaging and optogenetic stimulation enabled us to quantify interneuron suppression of neighbouring cells across the network (Fig. 1k).
Distinct functions of inhibitory cell classes
Using this system, we activated PV or SOM cells while recording the visual responses of non-infected cells to oriented drifting gratings (Supplementary Movie 3, Fig. 2a–d and Supplementary Fig. 5). Control responses of target cells were similar in PV and SOM experiments (Supplementary Fig. 6), and suppression by ChR2 was calibrated to a similar moderate range for all experiments (Supplementary Fig. 6i). Interestingly, PV activation caused a larger suppression when control responses were higher (Fig. 2e), whereas SOM activation caused a relatively uniform suppression of the full response profile (Fig. 2f), particularly when the control responses or baseline levels were high relative to the suppression thereby avoiding a ‘floor effect’. Indeed, the slope of the relationship between relative suppression and control response was significantly greater when PV cells were activated (Fig. 2g, i) than when SOM cells were activated (Fig. 2h, i; PV, n = 150 cells; SOM, n = 77 cells; P < 0.001, Kolmogorov–Smirnov test). Comparing the suppression at different response strengths for pooled cells further showed the asymmetric relationship between response and suppression when PV cells were activated (Fig. 2j; responses at 40, 60 and 80% of the maximum were less suppressed than those at maximum response strength; _P_ < 0.05–0.001 for all pair-wise comparisons with 100%). In contrast, SOM suppression affected weak and strong responses similarly (Fig. 2k; responses at 40, 60 and 80% compared to responses at 100%; _P_ > 0.2). Thus, suppression by SOM cells is relatively uniform across responses of different strengths, whereas suppression by PV cells is non-uniform and proportional to the response level of the target neuron.
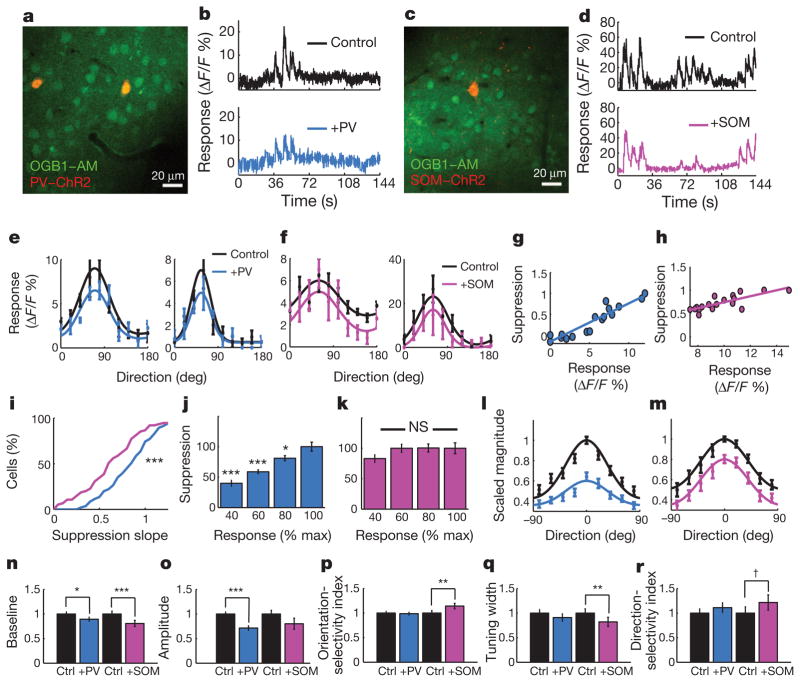
Figure 2. Impact of PV- and SOM-driven inhibition on the tuning of neuronal responses.
a, An imaging site showing neurons loaded with calcium indicator (OGB1-AM, green) and two PV+ neurons expressing mCherry-ChR2 (PV-ChR2, red) in visual cortex in vivo. b, Optical responses to visual stimuli, either without (Control) or with (+PV) simultaneous optical PV activation in interleaved trials, recorded during episodically presented oriented drifting gratings (see Fig. 1d). The photo-artefact from ChR2 stimulation has been removed from these trials. c, d, Same as (a and b), but from an experiment with mCherry-ChR2 expression in SOM neurons. e, f, Control tuning curves (black) were suppressed with ChR2 activation of PV neurons (blue) or SOM neurons (pink) in four example cells. g, h, The normalized suppression plotted as a function of control response strength in two example cells (blue, PV; pink, SOM). i, Cumulative density functions of the distributions of suppression versus response slopes for all cells suppressed by PV (blue) and SOM (pink) activation. j, k, Population data showing the amount of PV- and SOM-mediated suppression at different response levels. l, m, Average tuning curves showing control responses and effects of PV (blue) or SOM (pink) activation. n–r, Effects of PV and SOM activation on tuning, including baseline, peak-baseline amplitude, OSI, tuning width and DSI. NS, not significant. †P < 0.10; *P < 0.05; **P < 0.01; ***P < 0.001. Data are shown as mean ± s.e.m.
Proportionate suppression would ‘scale’ responses, reducing the tuning curve’s peak more strongly than its spontaneous response level or baseline, whereas uniform suppression would ‘shift’ the entire tuning curve downwards, including the baseline (Supplementary Fig. 7). Indeed, averaging the tuning curves of cells recorded during PV-cell ChR2 activation revealed a scaled down version of the control tuning curve (Fig. 2l), whereas activating SOM-cell ChR2 yielded a more uniform downwards shift of the control curve (Fig. 2m).
The presence of a response ‘floor’ or threshold influences how different forms of suppression impact the orientation tuning curve (Supplementary Fig. 7). Examining cells in which post-ChR2 responses were largely above the ‘floor’, to accurately quantify the full distribution of suppression across the whole curve (Supplementary Fig. 7a), showed that PV and SOM activation both reduced the baseline responses at non-preferred orientations (Fig. 2n; +PV (PV neurons activated optically), 11 ± 4% decrease in baseline, P < 0.05, n = 52 cells; versus +SOM, 19 ± 6%, P < 0.001, n = 25 cells). PV but not SOM activation decreased the peak-baseline amplitude of cells’ tuning functions (Fig. 2o; +PV, 28.5 ± 4.5% reduction, P < 0.001; +SOM, 19.8 ± 11.2%, P = 0.10), consistent with a larger reduction at the peak relative to the baseline. PV activation did not affect the orientation selectivity index (OSI) of target cells (Fig. 2p; control, 0.29 ± 0.01; versus +PV, 0.29 ± 0.01; P = 0.71; see also Supplementary Fig. 8), whereas SOM activation increased the OSI (Fig. 2p; control, 0.26 ± 0.01; versus +SOM, 0.30 ± 0.01; P < 0.01). Similarly, PV activation did not affect the tuning width of target cells (Fig. 2q; control, half-width at half-height, 38.7 ± 3.0 degrees; versus +PV, 35.2 ±3.0 deg; P = 0.31), whereas SOM activation narrowed the tuning width (Fig. 2q; control, 45.7 ± 4.1 deg; versus +SOM, 37.5 ± 4.3 deg; P < 0.01). The effects on the direction selectivity index (DSI) were similar in trend (Fig. 2r; PV: 0.33 ± 0.03 control DSI versus 0.36 ± 0.03 +PV DSI, P = 0.44; SOM: 0.28 ± 0.04 control DSI versus 0.34 ± 0.05 +SOM DSI, P = 0.10). Thus, the relatively uniform suppression by SOM cells leads to a sharpening in response selectivity of target neurons, whereas the non-uniform but proportional suppression by PV cells reduces response magnitude but does not change response selectivity.
PV and SOM effects measured electrophysiologically
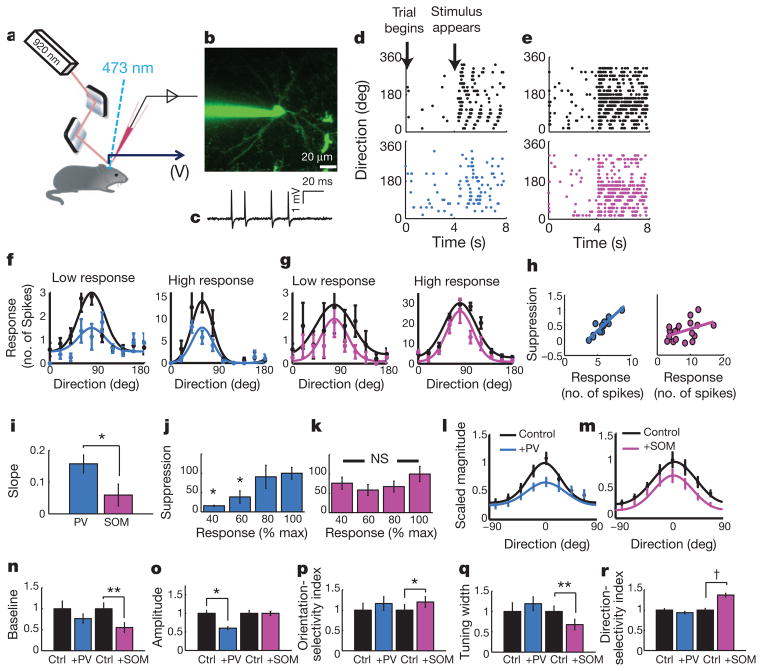
We examined further the different effects of PV and SOM activation using electrophysiological cell-attached recordings in vivo (Fig. 3a). Putative pyramidal neurons (Fig. 3b) were identified by their regular spiking properties (Fig. 3c); the peak:trough ratio of individual spikes was larger for all recorded cells than in identified fast-spiking PV-positive neurons (2.82 ± 0.20 for all recorded cells, n = 21; 1.41 ± 0.18 in PV-positive neurons, n = 53; P < 0.001). Moderate levels of PV- and SOM-mediated suppression (Supplementary Fig. 6i) had clearly different effects on spike responses of target cells and resultant orientation tuning curves (Fig. 3d, e), even in cells with very different response levels (Fig. 3f, g). PV suppression depended on the level of control response, whereas SOM suppression shifted tuning curves downwards more uniformly (Fig. 3h–k; PV, n = 21 cells, SOM, n = 17 cells). The changes in average tuning curves (Fig. 3l, m), and their parameters (Fig. 3n–r; Supplementary Fig. 8), showed that SOM but not PV suppression sharpened response selectivity, consistent with a model in which PV activation leads to a division of target cell responses but in which SOM activation leads to a subtraction (Supplementary Fig. 9).
Figure 3. Electrophysiological analysis of PV-and SOM-driven inhibition.
a, PV or SOM neurons were optogenetically activated while recording visual responses (measured in volts (V)) using targeted in vivo cell-attached recordings. b, A pyramidal neuron was patched under two-photon guidance and filled with Alexa 488 dye. c, Representative spikes recorded. d, e, Spike rasters depict responses of target cells in time over 18 directions, pooled over multiple trials in control conditions (black) and during activation of PV (blue) or SOM (pink) neurons. f, g, Control tuning curves (black) were suppressed with ChR2 activation of PV neurons (blue) or SOM neurons (pink) in four example cells with very different response levels. h, Representative examples of the relationship between PV-mediated (blue) and SOM-mediated (pink) suppression and control response strength. i, Bar graph of mean suppression versus response slopes during PV (blue) and SOM (pink) activation. j, k, Population data for the amount of PV- and SOM-mediated suppression seen at different response levels. l, m, Average tuning as in Fig. 2l, m, before (black) and during PV (blue) or SOM (pink) activation. n–r, Effects of PV and SOM activation on tuning-curve parameters, as in Fig. 2n–r. †P < 0.10; *P < 0.05; **P < 0.01; NS, not significant. Data are shown as mean ± s.e.m.
Differential inhibitory impact on target-cell gain
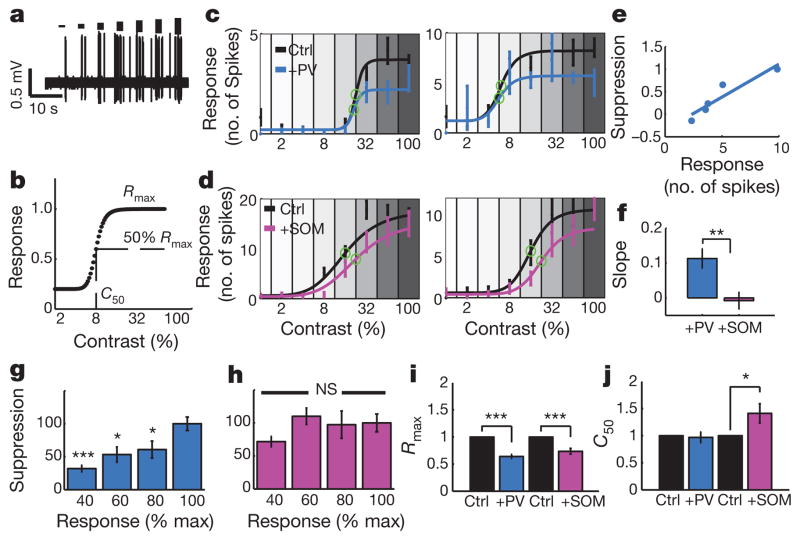
The divisive impact of PV activation suggests that PV neurons implement dynamic response gain control in cortex, which has been previously attributed to intracortical inhibition26,27. We carried out cell-attached recordings and examined the effects of PV and SOM activation on a canonical measure of response gain, the modulation of responses with increasing contrast (Fig. 4a, b). PV activation (Fig. 4c) led to contrast response curves with reduced gain (slope), whereas SOM activation (Fig. 4d) decreased responses relatively uniformly (with a floor effect at low response levels). The PV activation curves were better fit by a divisive scaling model than a subtractive model, whereas SOM activation curves were better fit by the subtractive model (Supplementary Fig. 10). PV- but not SOM-mediated suppression was dependent on the response level, as shown by both suppression-response slopes (Fig. 4e, f; PV, 0.11 ± 0.03, n = 17 cells; versus SOM, −0.008 ± 0.02, n = 16 cells; P < 0.01; Supplementary Fig. 11a, b) and suppression-response strength comparisons (PV, _P_ < 0.05–0.001 comparing suppression at 40%, 60% and 80% response to that at 100% (Fig. 4g); SOM, _P_ > 0.2 comparing suppression at 40%, 60% and 80% response to that at 100% (Fig. 4h)). PV activation scaled response magnitude (Fig. 4i; +PV: _R_max 64.1 ± 3.5% of control; n = 17 cells; P < 0.001) without affecting half-saturation contrast (Fig. 4j; +PV: _C_50 95.0 ± 10% of control; P = 0.65). SOM activation significantly reduced _R_max (Fig. 4i; +SOM: 73.6 ± 5.3% of control; n = 16 cells; P < 0.001), but unlike PV, also significantly increased _C_50 (Fig. 4j; +SOM: 141 ± 17.7% of control; P < 0.05), with no effect on the response gain (P = 0.23 comparing slopes at _C_50 before and after SOM activation; versus P < 0.01 comparing slopes before and after PV activation; see also Supplementary Fig. 11). Thus, PV but not SOM activation contributes directly to controlling the gain of target-cell responses.
Figure 4. Modulation of response gain by PV and SOM cells during targeted cell-attached recordings.
a, Example responses from a neuron stimulated with drifting gratings of increasing contrast, at the cell’s preferred orientation. b, Naka–Rushton curve describing the increase in responses with increasing contrast. c, d, Four example cells whose control response–contrast curves (black) were suppressed by PV (blue) or SOM (pink) activation. _C_50 values are marked (green circles). e, Relationship between suppression and response when a PV cell was activated (slope = 0.15). f, The mean suppression versus response slope was larger when PV cells were activated than when SOM cells were activated. g, h, Population data for the amount of PV- and SOM-mediated suppression observed at different response levels. i, j, Effects of PV and SOM activation on response–contrast function parameters _R_max and _C_50 (see b). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Data are shown as mean ± s.e.m.
Single-cell circuit maps of network connections in vivo
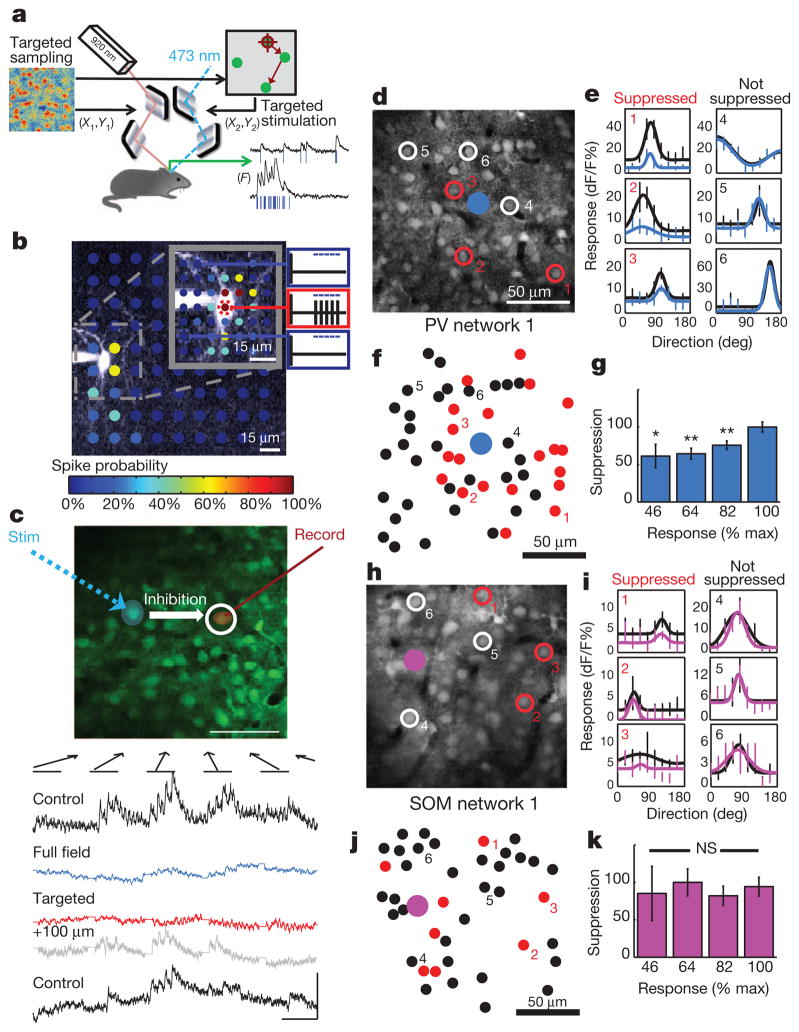
The functional roles of inhibitory neurons are manifested through the spatial distribution and functional targeting of subclasses of inhibition onto cortical cells in the local network. To define the output connections of single inhibitory neurons, we developed a system to focally stimulate an individual neuron while simultaneously imaging responses from large numbers of cells to assess their functional coupling (Fig. 5a). The ChR2-stimulating 473-nm beam was narrowed to a small effective radius and focused on sparsely distributed ChR2-positive cells (Fig. 5b and Supplementary Figs 12–16). Thus, we could optically activate chosen PV or SOM neurons in vivo while concurrently sampling population responses with targeted imaging (Fig. 5c and Supplementary Movie 4).
Figure 5. Dual-laser optical mapping of network connections to reveal maps of functional inhibition by single PV and SOM neurons.
a, Two laser systems are controlled independently and synchronized for concurrent high-speed imaging and targeted spatial stimulation of a ChR2-expressing inhibitory neuron. b, A SOM ChR2+ neuron in vivo was patched and recorded while systematically mapping target locations with a focused 473-nm laser; spikes were elicited only when stimulation was targeted on or very near to the soma. The colour bar indicates the spike probability at each location (0–100%). c, All-optical circuit mapping. Optical activation of one neuron (blue spot) during targeted recording of another neuron (red spot). Activity in the recorded neuron was measured with no stimulation (Control, black), full-field activation of PV neurons (blue), targeted activation of the PV neuron (red), and while aiming the stimulation beam 100 μm off-target (grey). Visual responses were suppressed by both full-field and single PV-cell activation. d, Mapping the influence of targeted PV activation on neighbouring cells revealed some cells that were significantly suppressed by PV activation (red, 1–3) which were clearly intermixed with other nearby cells that were not affected (white, 4–6). e, Tuning curves for cells 1–6 comparing cell responses during control (black) or targeted PV-cell activation (blue). f, Spatial distribution of all cells in the network that were either significantly suppressed (red) or not (black). g, Population data for the amount of PV- and SOM-mediated suppression observed at cells’ different response levels across all single cell networks. h–k, Same as d–g, in experiments in which focal stimulation was targeted to single SOM neurons. *P < 0.05; **P < 0.01; NS, not significant. Data are shown as mean ± s.e.m.
Mapping response modulation across a network while controlling a PV ChR2 cell or a SOM ChR2 cell (Fig. 5d, h), we found that visual responses of some neighbouring neurons were significantly suppressed while other cells were unaffected (Fig. 5e, i), resulting in maps of the functional suppression triggered during focal PV or SOM activation (Fig. 5f, j). Similar non-uniform, heterogeneous maps of functional PV and SOM cell connectivity were obtained from every imaged animal (Supplementary Fig. 17; PV, n = 4 networks; SOM, n = 5 networks). Focal PV activation resulted in the significant suppression of 43.1 ± 2.1% of neurons within the field of view, whereas focal SOM activation suppressed 16.2 ± 2.9% of neurons. Electrical stimulation of a single cell through whole-cell patch recording in vivo yielded a similar suppression map (Supplementary Fig. 18). Single PV or SOM neuron activation rarely triggered observable dis-inhibition (Supplementary Fig. 19). The nature of suppression was very similar to that observed with full-field activation: the amount of suppression by PV cells depended on the strength of the control response, whereas focal SOM stimulation resulted in more uniform suppression (Fig. 5h, m; see also Supplementary Fig. 20). Thus, the effects of SOM- and PV-mediated inhibition are distinct, whether they are evoked by populations or single SOM or PV neurons. Furthermore, these maps of affected neurons show remarkable diversity in the functional suppression exerted by specific PV or SOM neurons within their local neighbourhoods.
Functional connectivity of inhibitory networks
An important question is whether there is an underlying logic through which an inhibitory cell makes functional connections with its target cells. We tested whether, in vivo, functional suppression by single PV or SOM neurons can be predicted by spatial22,23 or functional28,29 relationships. The distance to potential targets did not predict whether a neuron was significantly (P < 0.05) suppressed by PV or SOM neurons within 100 μm (Supplementary Fig. 21a), and there was no significant relationship between suppression strength and distance for the population of either PV- or SOM-stimulated networks (Fig. 6a; PV, P = 0.80, R = −0.03, 4 networks; SOM, P = 0.1, R = −0.28, 5 networks). Furthermore, the spatial patterns of suppression by PV and SOM neurons were less clustered than a theoretical ordered distance model, and statistically similar to randomly targeted networks, although PV networks tended to be more spatially coherent than SOM networks (Supplementary Fig. 21b–d). Thus, individual PV and SOM neurons do not seem to functionally affect neurons along a distance gradient within their local network.
Figure 6. Spatial and functional analysis of targeting by single PV and SOM neurons.
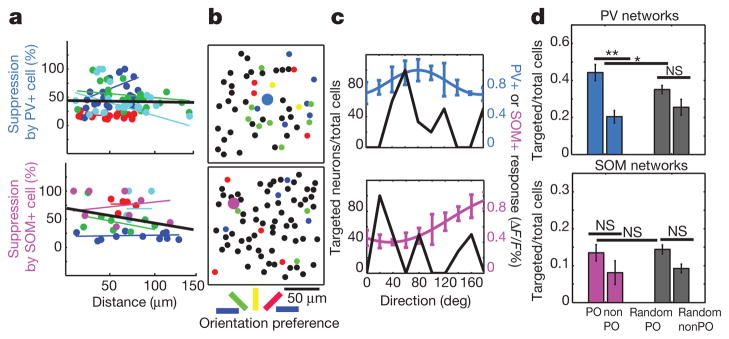
a, The amount of suppression in significantly suppressed cells is plotted against their distance from the stimulated PV cell (top) or SOM cell (bottom). Best fit coloured lines show individual networks; black line depicts pooled data. b, The orientation preferences of significantly suppressed cells are colour-coded for two example networks (top, PV (blue); bottom, SOM (pink)). c, Distribution of preferred orientations of target cells (black lines) for these example networks, superimposed with the orientation tuning curve of the stimulated PV (blue line, top) or SOM (pink line, bottom) neuron. d, Bar graphs show the mean proportion of neurons that matched the preferred orientation of stimulated PV neurons (blue bars) or SOM neurons (pink bars), and the orthogonal, non-preferred orientation. Grey bars show the same comparison when the preferred orientations were randomly re-sampled among the neurons in the fields of view (Random PO and Random nonPO). For PV but not SOM, the actual percentage of targets at the PO was greater than expected with random targeting, indicating that PV cells may preferentially suppress cells with similar functional response properties. *P < 0.05; **P < 0.01; NS, not significant. Data are shown as mean ± s.e.m.
We then asked whether there was any relationship between the preferred orientation of an inhibitory cell and the preferred orientations of its targets. Comparing the tuning curves of activated inhibitory neurons and the tuning of significantly suppressed cells (Fig. 6b, c), showed that PV cells targeted higher percentages of neurons that matched their own preferred orientations than the orthogonal or the expected percentage predicted by random targeting (Fig. 6d, top panel; preferred orientation (PO), 44.3 ± 7.6%; orthogonal, non-preferred orientation (nonPO), 20.4 ±4.8%; P < 0.05, treating each network as a single observation; n = 210 target cells from 4 PV networks; PO versus random PO, P < 0.05). However, for SOM cells and networks, the orientation distribution of suppressed cells was more uniform, with no significant difference between the percentage of targeted cells at the preferred orientation versus the orthogonal of the source SOM cells (Fig. 6d, bottom panel; PO, 13.5 ± 3.8%; nonPO, 8.1 ± 4.6%; P = 0.39, n = 238 target cells from 5 SOM networks). These results indicate that PV cells preferentially target other neurons that have similar preferred orientations, whereas SOM neurons seem to have a broader range of targets.
Discussion
Although a growing literature has started to examine the input and firing properties of specific inhibitory neuron classes19,20,30,31, little is understood about the functional nature of their output. By triggering inhibition and measuring its effects on connected cells in the functioning cortex, we have shown the computational impact of different forms of inhibition on sensory processing. PV neurons principally implement divisive normalization, whereas SOM neurons perform relatively uniform subtraction of responses in their targets, leading to complementary effects on neuronal responses: SOM neurons alter stimulus selectivity, whereas PV neurons preserve selectivity, and PV neurons modulate response gain, whereas SOM neurons shift response levels, leaving response gain unaffected. These effects are mediated by complementary local circuits: PV neurons preferentially target iso-oriented neurons, whereas SOM neurons target cells with a wide range of orientation preferences.
The role of inhibition in shaping stimulus selectivity of visual cortex neurons has been difficult to resolve with previous methods. Intracellular recordings from V1 neurons have found closely matched tuning of excitatory and inhibitory synaptic conductances8,28,29,32, indicating that inhibition might not sharpen orientation selectivity, and intracellular blockade of inhibition indeed does not seem to affect orientation tuning33. However, broadly tuned or untuned inhibition can in principle sharpen neuronal responses; such inhibition has also been described in V1 neurons10,34,35, and pharmacological blockade of network inhibition broadens orientation selectivity5–7. We show that SOM-mediated inhibition targets cells with a range of preferred orientations, and its presence on dendrites probably serves to sharpen the wide orientation preference of dendritic excitatory inputs36. Consistent with SOM neurons having this role, genetic reduction of a subset of dendrite-targeting interneurons broadens orientation selectivity37.
Inhibition has long been proposed to regulate the gain of cortical responses, and we now show that PV cells are crucial for this function. Consistent with our findings, a recent study26 has demonstrated that activating PV-interneuron populations in mouse visual cortex has a divisive scaling effect on responses of target neurons. Rapid PV-mediated inhibition matched to excitation in time could shape response gain as well as selectivity, as shown in the auditory9,38, somatosensory39,40 and prefrontal cortex41. Finally, the regulation of response gain by PV cells makes them an attractive mechanism for the developmental regulation of inputs during experience-dependent plasticity of cortical circuits42.
The distinctive effects of PV and SOM inhibition may arise from their cellular regions of impact43, and possibly synaptic differences between the cell types44,45. The methods we have described provide a basis for ‘functional connectomics’ in active cortical circuits, and also reveal the complementary computational roles of specific inhibitory cell classes in vivo during sensory processing. In bridging the gap between cellular and network function, these methods should be applicable to many cortical areas and cell types, to elucidate their functional connectivity and embodied computational principles.
METHODS SUMMARY
Details of mice and viral constructs used, animal surgical preparations, in vitro slice characterization of ChR2 function, interneuron expression, in vivo two-photon guided cell-attached recording, development of high-speed targeted scanning of calcium responses, development of simultaneous two-photon imaging and optogenetic stimulation, the focal stimulation system, and details of data analysis are described in the Supplementary Methods.
Supplementary Material
Supplementary Information
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Acknowledgments
We thank J. Huang for providing the SOM–Cre mouse line; C. Le for performing animal care support and viral injections; S. Yan and Y. Deng for help in the development of optogenetics and imaging methods in vitro; S. El-Boustani for collecting data for in vivo deconvolution; J. Sharma, M. Goard and A. Banerjee for comments and discussions on the manuscript; L.-H. Tsai, K. Meletis and M. Carlen for early provision of viral constructs and PV–Cre viral injections; and James Schummers and Hiroki Sugihara for participating in early pilot experiments testing optogenetics stimulation in vivo. This work was supported by postdoctoral fellowships from the US National Institutes of Health (NIH) and the Simons Foundation (N.R.W.), an NIH predoctoral fellowship (C.A.R.) and grants from the NIH and the Simons Foundation (M.S.).
Footnotes
Author Contributions N.R.W. conceived experiments, designed and engineered circuit interface and analysis systems, carried out in vivo and in vitro experiments, and performed analyses. C.A.R. conceived experiments, performed surgeries and viral injections, carried out in vivo experiments, and performed analyses. F.L.W. carried out in vivo experiments, and performed analyses. M.S. conceived experiments and contributed to analysis of experiments. N.R.W., C.A.R. and M.S. wrote the paper.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Freund TF, Buzsáki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 2.Markram H, et al. Interneurons of the neocortical inhibitory system. Nature Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 3.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis DA. GABAergic local circuit neurons and prefrontal cortical dysfunction in schizophrenia. Brain Res Brain Res Rev. 2000;31:270–276. doi: 10.1016/s0165-0173(99)00042-9. [DOI] [PubMed] [Google Scholar]
- 5.Sillito AM. The contribution of inhibitory mechanisms to the receptive field properties of neurones in the striate cortex of the cat. J Physiol (Lond) 1975;250:305–329. doi: 10.1113/jphysiol.1975.sp011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crook JM, Kisvárday ZF, Eysel UT. Evidence for a contribution of lateral inhibition to orientation tuning and direction selectivity in cat visual cortex: reversible inactivation of functionally characterized sites combined with neuroanatomical tracing techniques. Eur J Neurosci. 1998;10:2056–2075. doi: 10.1046/j.1460-9568.1998.00218.x. [DOI] [PubMed] [Google Scholar]
- 7.Tsumoto T, Eckart W, Creutzfeldt OD. Modification of orientation sensitivity of cat visual cortex neurons by removal of GABA-mediated inhibition. Exp Brain Res. 1979;34:351–363. doi: 10.1007/BF00235678. [DOI] [PubMed] [Google Scholar]
- 8.Ferster D, Miller KD. Neural mechanisms of orientation selectivity in the visual cortex. Annu Rev Neurosci. 2000;23:441–471. doi: 10.1146/annurev.neuro.23.1.441. [DOI] [PubMed] [Google Scholar]
- 9.Wehr M, Zador AM. Balanced inhibition underlies tuning and sharpens spike timing in auditory cortex. Nature. 2003;426:442–446. doi: 10.1038/nature02116. [DOI] [PubMed] [Google Scholar]
- 10.Monier C, Chavane F, Baudot P, Graham LJ, Frégnac Y. Orientation and direction selectivity of synaptic inputs in visual cortical neurons: a diversity of combinations produces spike tuning. Neuron. 2003;37:663–680. doi: 10.1016/s0896-6273(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 11.Carandini M, Heeger DJ. Summation and division by neurons in primate visual cortex. Science. 1994;264:1333–1336. doi: 10.1126/science.8191289. [DOI] [PubMed] [Google Scholar]
- 12.Somers DC, Nelson SB, Sur M. An emergent model of orientation selectivity in cat visual cortical simple cells. J Neurosci. 1995;15:5448–5465. doi: 10.1523/JNEUROSCI.15-08-05448.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds JH, Heeger DJ. The normalization model of attention. Neuron. 2009;61:168–185. doi: 10.1016/j.neuron.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohshiro T, Angelaki DE, DeAngelis GC. A normalization model of multisensory integration. Nature Neurosci. 2011;14:775–782. doi: 10.1038/nn.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louie K, Glimcher PW. Separating value from choice: delay discounting activity in the lateral intraparietal area. J Neurosci. 2010;30:5498–5507. doi: 10.1523/JNEUROSCI.5742-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schummers J, Yu H, Sur M. Tuned responses of astrocytes and their influence on hemodynamic signals in the visual cortex. Science. 2008;320:1638–1643. doi: 10.1126/science.1156120. [DOI] [PubMed] [Google Scholar]
- 19.Runyan CA, et al. Response features of parvalbumin-expressing interneurons suggest precise roles for subtypes of inhibition in visual cortex. Neuron. 2010;67:847–857. doi: 10.1016/j.neuron.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bock DD, et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshimura Y, Callaway EM. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nature Neurosci. 2005;8:1552–1559. doi: 10.1038/nn1565. [DOI] [PubMed] [Google Scholar]
- 22.Fino E, Yuste R. Dense inhibitory connectivity in neocortex. Neuron. 2011;69:1188–1203. doi: 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31:13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko H, et al. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473:87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lillis KP, Eng A, White JA, Mertz J. Two-photon imaging of spatially extended neuronal network dynamics with high temporal resolution. J Neurosci Methods. 2008;172:178–184. doi: 10.1016/j.jneumeth.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atallah BV, Bruns W, Carandini M, Scanziani M. Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron. 2012;73:159–170. doi: 10.1016/j.neuron.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katzner S, Busse L, Carandini M. GABAA inhibition controls response gain in visual cortex. J Neurosci. 2011;31:5931–5941. doi: 10.1523/JNEUROSCI.5753-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson JS, Carandini M, Ferster D. Orientation tuning of input conductance, excitation, and inhibition in cat primary visual cortex. J Neurophysiol. 2000;84:909–926. doi: 10.1152/jn.2000.84.2.909. [DOI] [PubMed] [Google Scholar]
- 29.Mariño J, et al. Invariant computations in local cortical networks with balanced excitation and inhibition. Nature Neurosci. 2005;8:194–201. doi: 10.1038/nn1391. [DOI] [PubMed] [Google Scholar]
- 30.Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofer SB, et al. Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nature Neurosci. 2011;14:1045–1052. doi: 10.1038/nn.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan AY, Brown BD, Scholl B, Mohanty D, Priebe NJ. Orientation selectivity of synaptic input to neurons in mouse and cat primary visual cortex. J Neurosci. 2011;31:12339–12350. doi: 10.1523/JNEUROSCI.2039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson S, Toth L, Sheth B, Sur M. Orientation selectivity of cortical neurons during intracellular blockade of inhibition. Science. 1994;265:774–777. doi: 10.1126/science.8047882. [DOI] [PubMed] [Google Scholar]
- 34.Xing D, Ringach DL, Hawken MJ, Shapley RM. Untuned suppression makes a major contribution to the enhancement of orientation selectivity in macaque v1. J Neurosci. 2011;31:15972–15982. doi: 10.1523/JNEUROSCI.2245-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu BH, et al. Broad inhibition sharpens orientation selectivity by expanding input dynamic range in mouse simple cells. Neuron. 2011;71:542–554. doi: 10.1016/j.neuron.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia H, Rochefort NL, Chen X, Konnerth A. Dendritic organization of sensory input to cortical neurons in vivo. Nature. 2010;464:1307–1312. doi: 10.1038/nature08947. [DOI] [PubMed] [Google Scholar]
- 37.Mao R, et al. Influence of a subtype of inhibitory interneuron on stimulus-specific responses in visual cortex. Cereb Cortex. 2011;22:493–508. doi: 10.1093/cercor/bhr057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan AY, Zhang LI, Merzenich MM, Schreiner CE. Tone-evoked excitatory and inhibitory synaptic conductances of primary auditory cortex neurons. J Neurophysiol. 2004;92:630–643. doi: 10.1152/jn.01020.2003. [DOI] [PubMed] [Google Scholar]
- 39.Okun M, Lampl I. Instantaneous correlation of excitation and inhibition during ongoing and sensory-evoked activities. Nature Neurosci. 2008;11:535–537. doi: 10.1038/nn.2105. [DOI] [PubMed] [Google Scholar]
- 40.Gentet LJ, Avermann M, Matyas F, Staiger JF, Petersen CC. Membrane potential dynamics of GABAergic neurons in the barrel cortex of behaving mice. Neuron. 2010;65:422–435. doi: 10.1016/j.neuron.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Haider B, Duque A, Hasenstaub AR, McCormick DA. Neocortical network activity in vivo is generated through a dynamic balance of excitation and inhibition. J Neurosci. 2006;26:4535–4545. doi: 10.1523/JNEUROSCI.5297-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yazaki-Sugiyama Y, Kang S, Câteau H, Fukai T, Hensch TK. Bidirectional plasticity in fast-spiking GABA circuits by visual experience. Nature. 2009;462:218–221. doi: 10.1038/nature08485. [DOI] [PubMed] [Google Scholar]
- 43.Koch C, Poggio T, Torre V. Nonlinear interactions in a dendritic tree: localization, timing, and role in information processing. Proc Natl Acad Sci USA. 1983;80:2799–2802. doi: 10.1073/pnas.80.9.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulik A, et al. Compartment-dependent colocalization of Kir3.2-containing K+ channels and GABAB receptors in hippocampal pyramidal cells. J Neurosci. 2006;26:4289–4297. doi: 10.1523/JNEUROSCI.4178-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nusser Z, Sieghart W, Benke D, Fritschy JM, Somogyi P. Differential synaptic localization of two major gamma-aminobutyric acid type A receptor alpha subunits on hippocampal pyramidal cells. Proc Natl Acad Sci USA. 1996;93:11939–11944. doi: 10.1073/pnas.93.21.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4